Abstract
Histone deacetylase 2 (HDAC2) is a key transcriptional coregulator that is suspected to play a role during oogenesis. It is known that RNA transcription in the cat germinal vesicle (GV) stops during folliculogenesis at the late antral follicle stage and is unrelated to histone deacetylation or chromatin condensation. The objective of the present study was to determine if and how HDAC2 participates in transcription regulation in the cat GV. Spatiotemporal HDAC2 protein expression was examined by immunostaining oocytes from primary to large antral follicles. HDAC2 was detected in the majority of GVs within oocytes from early, small, and large antral follicles. At early and small antral stages, HDAC2 was found primarily in the GV's nucleoplasm. There then was a significant shift in HDAC2 localization into the nucleolus, mostly in oocytes from large antral follicles. Assessments revealed that transcription was active in oocytes that contained nucleoplasm-localized HDAC2, whereas nucleolar-bound HDAC2 was associated with loss of both global transcription and ribosomal RNA presence at all antral stages. When oocytes were exposed to the HDAC inhibitor valproic acid, results indicated that HDAC regulated transcriptional activity in the nucleoplasm, but not in the nucleolus. Collective results suggest that nucleolar translocation of HDAC2 is associated with transcriptional silencing in the GV, thereby likely contributing to an oocyte's acquisition of competence.
Keywords: domestic cat, gamete biology, germinal vesicle, oocyte, transcription
INTRODUCTION
Immature oocytes are arrested at the first meiotic prophase and contain a nucleus called the germinal vesicle (GV). As folliculogenesis progresses from the primordial to the antral stage, the oocyte and its GV continue to grow in size and acquire the competence to undergo meiosis and support early embryo development after fertilization [1]. There has been increasing attention directed at immature oocytes because of their potential role as a fertility reservoir, particularly for oncological patients, biomedical models, livestock, and endangered species [2–6]. One of many critical unknowns is how the immature oocyte attains biological competence. A thorough understanding of the mechanisms regulating such a phenomenon will lay the foundation for formulating new fertility preservation approaches and taking advantage of the thousands of intraovarian oocytes that never contribute to sexual reproduction.
During oocyte growth, the GV undergoes chromatin remodeling and transcriptional regulation both locally at specific loci and globally throughout the genome [7, 8]. At the advanced stage of folliculogenesis, GV chromatin in many mammalian species (i.e., mouse, pig, cow, sheep, monkey, and human) undergoes large-scale rearrangement and forms a condensed ring around the nucleolus (also called the nucleolar-like body) [9–14]. This subnuclear domain mainly serves as the location for rRNA synthesis and ribosome biogenesis [15, 16]. Formation of this surrounded-nucleolus (SN) configuration, along with transcriptional repression of mRNA and rRNA, occurs in parallel with the acquisition of full oocyte competence [7, 12, 14, 17–21]. It is well established from studying somatic cells that chromatin condensation renders DNA inaccessible to transcriptional machinery, thereby leading to gene repression [22]. However, some evidence in growing oocytes indicates that global transcriptional repression is not strictly controlled by development of the SN configuration. For example, formation of an SN configuration in human and rabbit oocytes can be detected in early antral follicles, with transcription continuing until it stops at the large antral stage [12, 20, 23]. Disrupting chromatin remodeling in the mouse oocyte does not interfere with global transcriptional silencing [24]. In the domestic cat and goat, transcription ceases even though an SN-like configuration is never achieved [25, 26]. Collectively, these observations suggest that additional mechanisms are involved in the regulation of global transcriptional silencing. To explore further, we took advantage of a biological system where chromatin condensation and transcriptional arrest are uncoupled. The cat model serves such a purpose in that the chromatin remains uncondensed in a fully grown oocyte despite transcriptional silencing [25].
Histone deacetylases (HDACs) are the key transcriptional coregulators expressed in the oocyte. In the somatic cells of all species studied to date (from yeast to human), these proteins are well characterized for the ability to deacetylate core histones that, in turn, promote a closed chromatin conformation and reduce transcriptional potential [27, 28]. It now is clear that histone deacetylation serves as one key modification required for ensuring proper chromosome condensation and segregation during meiosis in the maturing oocyte [29–31]. However, at least for the growing mouse oocyte, histones are hyperacetylated, despite the presence of HDACs [32–36]. Therefore, the question remains—do HDACs participate in transcriptional control during oogenesis, and if so, how? Within the HDAC family, HDAC2 has been implicated as the major player in oocyte development for the mouse [33], cow [37], and nonhuman primate [38]. HDAC2 belongs to the class I HDACs that generally are detected in the nucleus and form the core deacetylation complex to mediate chromatin condensation and gene repression [39]. In the mouse, HDAC2 has been associated with transcriptional control in the immature oocyte, and deleting the gene encoding this protein renders the mouse subfertile [32]. Double deletion of Hdac1 and Hdac2 genes leads to global reduction in transcription and developmental arrest in the mouse oocyte [32]. However, beyond the latter model, there is minimal information on the role and function of HDAC2. Most studies are limited to some gene expression profiling during oocyte development, with no emphasis on the detailed developmental stage of the source follicle [38, 40, 41].
For these reasons, we designed a series of experiments with an overall aim to determine if and how HDAC2 participates in transcriptional regulation of the mammalian oocyte, but in a precise fashion during the course of specific stages of folliculogenesis. Using the cat model (because of minimal chromatin reorganization compared with other species), our specific objectives were to elucidate 1) the temporal and spatial expression pattern of HDAC2 in oocytes from defined follicular stages and 2) the role of HDAC2 in regulating global transcriptional activity.
MATERIALS AND METHODS
Collection of Follicles/Oocytes
Ovaries from adult, domestic cats were recovered after routine ovariohysterectomy at local veterinary clinics and transported in PBS supplemented with 100 IU/ml penicillin and 100 μg/ml streptomycin (Sigma-Aldrich) at 4°C to the laboratory within 6 h of excision. Follicles from primary through antral stages were collected by repeatedly slicing the ovaries with a scalpel blade in HEPES-buffered minimum essential medium (H-MEM; Gibco Laboratories) supplemented with 1 mM pyruvate, 2 mM l-glutamine, 100 IU/ml penicillin, 100 μg/ml streptomycin, and 4 mg/ml bovine serum albumin (BSA; Sigma-Aldrich). Follicles were then classified on the basis of diameter, numbers of surrounding granulosa cells, and presence of basal lamina, as previously described [42, 43]. As a general guideline, primary follicles (∼50–60 μm) contained a single layer of cuboidal granulosa cells; secondary follicles (60–90 μm) and preantral follicles (90–400 μm) contained more than one layer of granulosa cells; and antral follicles contained a fluid-filled antrum and were categorized into three size groups (≤0.5 mm, early antral; 0.6–1.0 mm, small antral; and ≥1.0 mm, large antral). Additionally, the developmental stage of each source follicle was double-checked by measuring oocyte and GV diameters, according to criteria characterized previously in our laboratory [25]. Specifically, the reference oocyte and GV diameters (in micrometers, mean ± SD) for each follicular stage were, respectively: primary follicle, 47.5 ± 3.2 and 16.9 ± 0.9; secondary follicle, 63.2 ± 2.7 and 17.0 ± 1.1; preantral follicle, 73.6 ± 4.2 and 24.8 ± 2.5; early antral follicle, 93.9 ± 3.5 and 31.0 ± 2.2; small antral follicle, 104.0 ± 3.6 and 36.5 ± 3.8; and large antral follicle, 113.7 ± 5.2 and 40.3 ± 3.0 [25]. Cumulus-oocyte complexes from antral follicles were then released and classified according to standard quality criteria [44], and only grade 1 (uniformly dark cytoplasm, five or more compact layers of cumulus cells) and grade 2 (same as grade 1, but with fewer than five cell layers) oocytes were selected. Oocytes from antral follicles were denuded of cumulus cells by exposure to 0.2% hyaluronidase (Sigma-Aldrich) for 15 min at 38°C, followed by vortexing and rinsing with H-MEM. Oocytes from primary, secondary, and preantral follicles did not require hyaluronidase treatment because of a paucity of surrounding cells [25].
HDAC2 Antibody and Western Blotting
A rabbit polyclonal HDAC2 antibody (H-54; Santa Cruz Biotechnology Inc.) was used to detect the HDAC2 protein. This antibody was raised against the C-terminal region of HDAC2 of human origin. The sequence in this region is highly conserved across species, including in the cat, yet is unique to HDAC2 and not shared with other HDAC members [45]. To further validate the specificity of this antibody in the cat, Western blot analysis was performed. Tissues from the ovary, oviduct, and uterus of two female adult cats were dissected and snap frozen. Proteins then were extracted with RIPA buffer (Sigma-Aldrich) supplemented with protease inhibitor mix (GE Healthcare) and 100 μM phenylmethylsulfonyl fluoride (Sigma-Aldrich). The extracts were mixed with SDS sample buffer (Boston Bioproducts) and resolved by SDS-PAGE with the use of 4%–15% Mini-PRO-TEAN TGX precast gels (Bio-Rad). Precision Plus Protein Dual Color Standards (Bio-Rad) were used as the molecular weight standard. Gels were transferred onto nitrocellulose membranes (Bio-Rad), blocked with 3% BSA (Sigma-Aldrich) in PBS, and then incubated at 4°C overnight with HDAC2 antibody at 1:300. Anti-rabbit immunoglobulin G (IgG) antibody conjugated with horseradish peroxidase (Santa Cruz Biotechnology) was used as the secondary antibody. Immunoreactivity was detected with Clarity Western ECL Substrate (Bio-Rad) and imaged with ChemiDoc XRS imaging system (Bio-Rad). The membranes then were stripped with Restore Western blot stripping buffer (Thermo Scientific) and reblotted with housekeeping control anti-GAPDH (Novus Biologicals) at a concentration of 1:500.
Immunofluorescent Staining and Imaging
To examine the subcellular distribution of HDAC2, immunofluorescent staining was performed using the HDAC2 antibody at 1:50 and a mouse monoclonal nucleolin antibody (a nucleolar marker that labeled the outer layer of the nucleoli [46, 47]) at 1:100 (Millipore). Oocytes from different ovaries and follicle stages were pooled and processed together for immunostaining. Oocytes first were fixed in 4% paraformaldehyde in PBS (USB Corporation) for 1 h at 38°C or for overnight at 4°C. After rinsing with wash solution (PBS with 2% fetal calf serum [FCS; Irvine Scientific] and 0.5% Triton X-100 [Sigma-Aldrich]) and blocking with saturation solution (PBS with 20% FCS and 1% Triton X-100) for 1 h at 38°C, oocytes were incubated with primary antibodies in wash solution overnight at 4°C. A negative control in which the primary antibodies were omitted was included in each trial. After three washes, oocytes were incubated with secondary antibodies (anti-rabbit IgG-fluorescein isothiocyanate [IgG-FITC] at 1:200 [Sigma-Aldrich] and/or anti-mouse IgG-Texas Red at 1:500 [Sigma-Aldrich]) for 1 h at 38°C, and then excess antibodies were washed away by three additional washes. To visualize DNA, oocytes were counterstained with 10 μg/ml Hoechst 33342 (Sigma-Aldrich) and/or 100 μg/ml propidium iodide (PI; Sigma-Aldrich) for 5–10 min before mounting with Vectashield mounting medium (Vector Laboratories). Hoechst only bound the DNA [48], whereas PI labeled all nucleic acids, including chromatin in the nucleus and rRNA in the nucleolus [49]. Resulting images were obtained using an Olympus BX41 epifluorescence microscope (Olympus Corporation) with SPOT advanced software 3.5.9 (Diagnostic Instruments Inc.). The excitation and detection wavelengths were 467–498 nm and 513–556 nm for FITC, respectively; 542–582 nm and 604–644 nm for Texas Red and PI; and 352–402 nm and 417–477 nm for Hoechst. Oocyte and GV diameters were measured with the SPOT advanced software, and the resulting values were used to confirm the developmental stages of the source follicles (as described above). Additionally, a portion of samples were also visualized with a Zeiss LSM 710 confocal microscope with Zen 2009 software (Zeiss) to secure higher-resolution images. The excitation and detection wavelengths were 488 nm and 492–544 nm for FITC, respectively, and 405 nm and 415–487 nm for Hoechst.
Transcription Assay
To determine global transcriptional activity in the GV, nascent RNAs were labeled and imaged with the Click-iT RNA imaging kit (Life Technologies) following the manufacturer's instructions. Denuded oocytes from antral follicles were incubated with 1 mg/ml 5-ethynyl uridine (5-EU) in MEM medium (supplemented as above; Sigma-Aldrich) for 1 h in a 38°C incubator with 5% CO2 before being fixed in 4% paraformaldehyde in PBS for 1 h at 38°C or overnight at 4°C. A negative control without 5-EU incorporation was included in each trial. To validate the transcription assay, a portion of oocytes also were incubated for 2 h with 0.2 μg/ml actinomycin D (Sigma-Aldrich), a drug known to inhibit RNA synthesis, before the addition of 5-EU. To detect 5-EU-labeled RNA, oocytes were rinsed with PBS containing 1 mg/ml polyvinylpyrrolidone (Sigma-Aldrich) and permeabilized with 0.5% Triton X-100 at 38°C for 30 min, followed by a 30-min incubation in the dark with reaction cocktails containing Alexa Fluor 594 azide. After reaction cocktail was washed off, oocytes were subjected to immunofluorescent staining of HDAC2 (as described above). Oocytes were visualized and measured with the Olympus BX41 epifluorescence microscope and SPOT software (as described above). Transcriptional activity was determined by quantifying fluorescence intensity in each GV with ImageJ software (National Institutes of Health). Integrated fluorescent density (IntDen) within the GV was measured, along with the mean fluorescent value (Mfv) of three additional selections around the GV for background readings. Fluorescence intensity (arbitrary units) was calculated using the following formula:
 |
The experimental background fluorescence intensity level was determined from the average fluorescence intensity measurement of non-5-EU-labeled, control GVs.
Valproic Acid Treatment
To examine the impact of adding an HDAC inhibitor, denuded oocytes in each replicate were randomly allocated and incubated in MEM medium (supplemented as above) in the presence or absence of valproic acid (VPA; Sigma-Aldrich). For oocytes exposed to the inhibitor, 0.5 mg/ml VPA was added to the oocyte cohort, and then both the treated (+VPA) and the untreated (–VPA) groups were incubated for 1 h (38°C, 5% CO2). Then, 5-EU was added immediately (without removing VPA in the treated group) to assess transcription (as described above). To validate that VPA effectively inhibited HDAC activity, some of the treated and the untreated oocytes were subjected to immunostaining with a rabbit polyclonal antibody against pan-acetylated Histone H3 (Millipore).
Experimental Design and Statistical Analysis
Oocytes selected from a pool of ovaries collected on a given day were considered as one replicate. For each of the three described experiments, oocytes were randomly allocated to different treatment groups, including for controls.
Experiment 1 examined HDAC2 protein expression in the cat. Western blotting was used to demonstrate the specificity of the HDAC2 antibody and to confirm expression of HDAC2 in tissues from the adult cat reproductive tract. Next, oocytes (a total of 212 from 33 cats) collected from a diversity of follicles (primary to large antral stages) were immunostained with anti-HDAC2 antibody across six replicates to examine the temporal and spatial expression pattern of HDAC2 protein in the GV.
Experiment 2 evaluated the relationship between HDAC2 localization and transcriptional activity in GVs within oocytes recovered from one of the three antral stage follicles (early, small, and large). A total of 184 oocytes from 32 donors underwent transcriptional assay followed by HDAC2 antibody staining across five replicates. Transcriptional activity was determined by quantifying the intensity of 5-EU-labeled nascent RNAs. All oocytes from the same batch were immunostained at the same time, and all images were taken and processed with the same setting to allow comparison. Only oocytes demonstrating expression of HDAC2 were analyzed on the basis of one of two patterns of localization. The first focused on HDAC2 expression exclusively in the nucleoplasm, and the second on nucleolar localization with or without residual nucleoplasmic expression. As a control, 72 oocytes from 14 cats were incubated with or without actinomycin D before the assay of transcriptional activity across two replicates. Additionally, another 69 oocytes from 18 cats were tristained with HDAC2 antibody, PI, and Hoechst 33342 across four replicates to better elucidate the relationship between HDAC2 localization and rRNA expression.
Experiment 3 investigated the requirement of HDAC activity for transcriptional regulation and HDAC2 translocation. A total of 271 denuded oocytes from early, small, or large antral follicles from 41 cats were treated with or without VPA and then assayed for transcriptional activity, HDAC2 expression, and chromatin structure (Hoechst staining) across six replicates. Another 64 oocytes from 15 cats, with or without VPA treatment, were immunostained with pan-acetylated Histone H3 antibody to evaluate the effectiveness of the inhibitor. All oocytes from the same batch were immunostained at the same time, and all images were taken and processed with the same setting to allow comparison.
For HDAC2 localization analysis in experiments 1 and 3, data from all replicates were pooled to determine the proportion of each distribution pattern in each follicle category. For HDAC2 distribution patterns, comparisons between follicle categories were assessed by χ2 testing. Quantified transcriptional activity data in experiments 2 and 3 were evaluated by ANOVA followed by Tukey multiple test. Differences were considered significant at P < 0.05 (GraphPad Prism 6; GraphPad Software).
RESULTS
Experiment 1: Temporospatial Expression Pattern of HDAC2 in the GVs of Intraovarian, Immature Oocytes at Different Follicular Stages
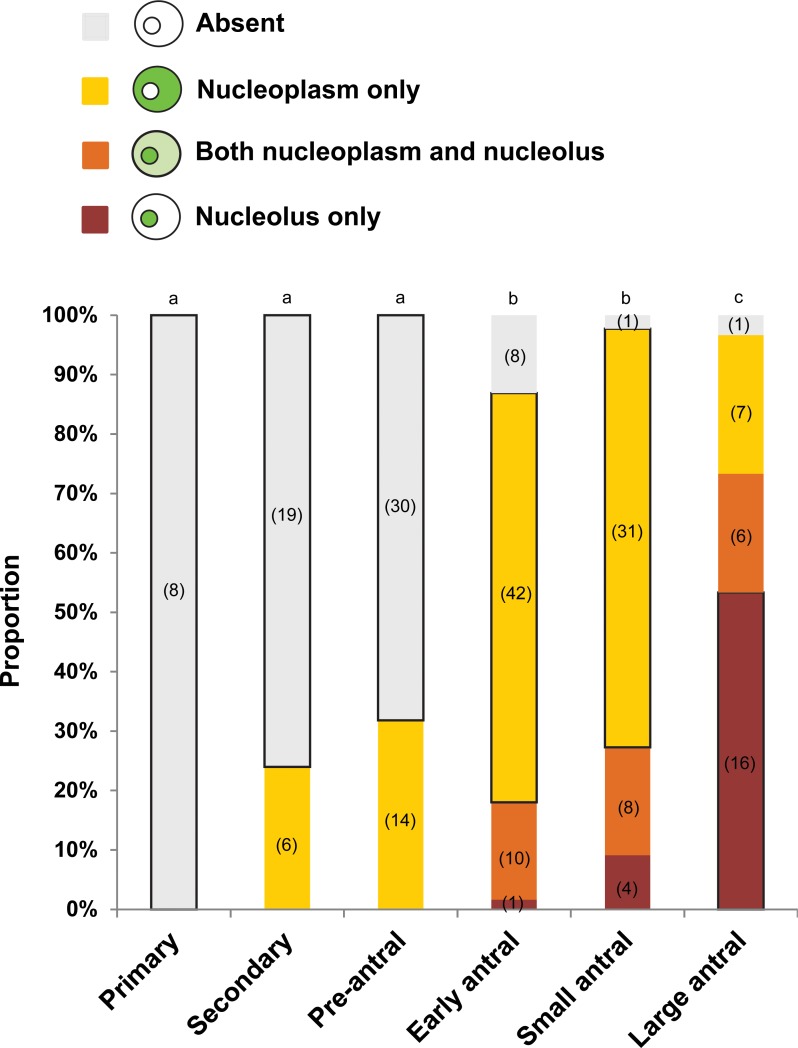
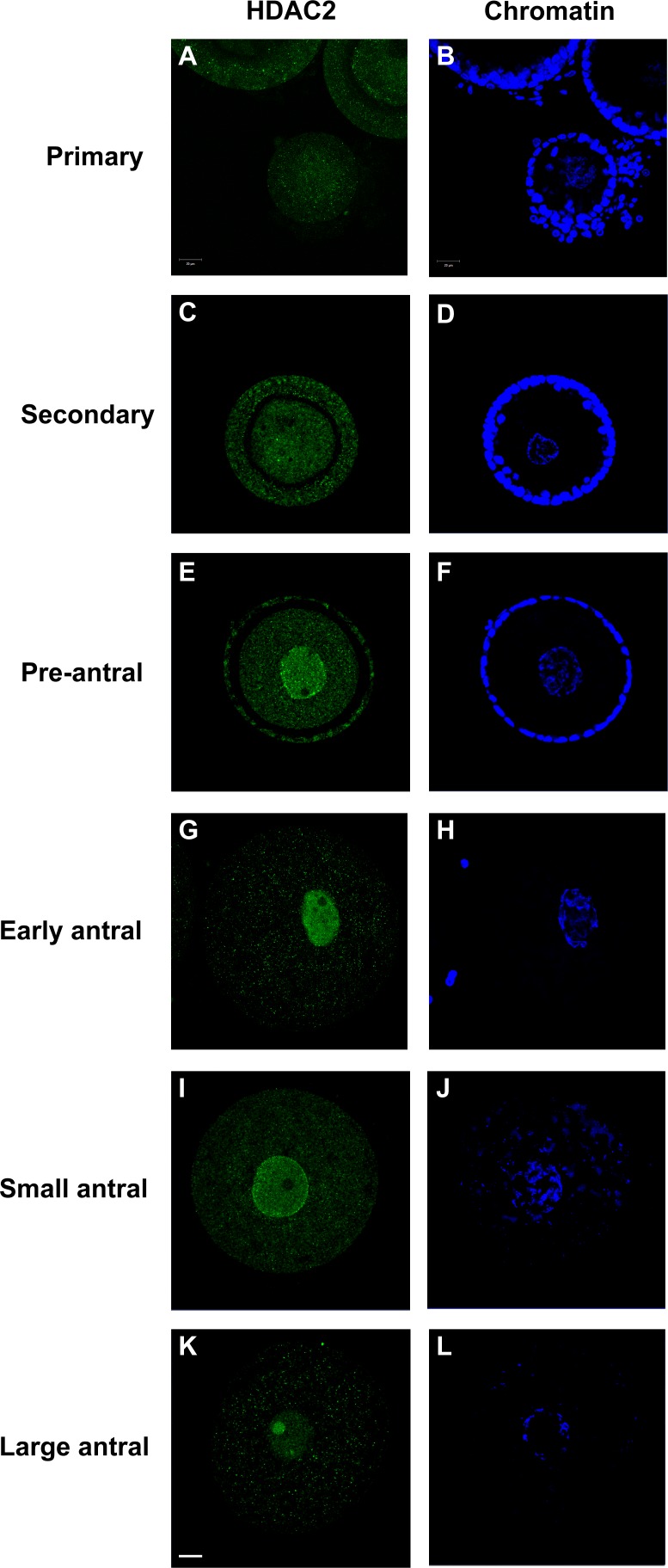
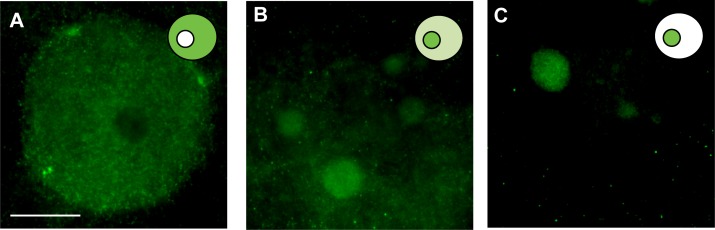
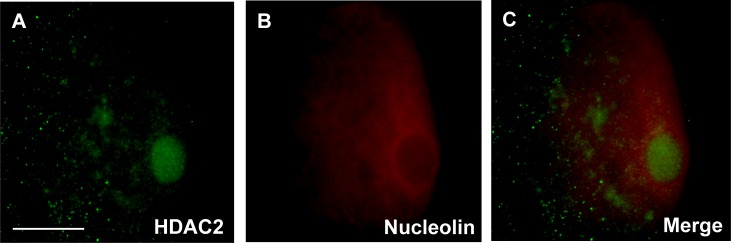
The HDAC2 antibody used in this study specifically recognized a single protein product corresponding to the predicted size (52 kDa) of cat HDAC2 in cat reproductive tract tissue (Supplemental Fig. S1; Supplemental Data are available online at www.biolreprod.org). Examining the temporospatial expression of HDAC2 using this antibody demonstrated that the protein was largely absent in the GVs of oocytes from primary, secondary, and preantral follicles (Fig. 1). No HDAC2 staining was detected in oocytes from primary follicles (Figs. 1 and 2, A and B). In oocytes from the secondary and preantral follicular stages, HDAC2 was only detected in 24% and 32% of the GVs, respectively (Figs. 1 and 2, C–F). As follicles progressed into antral stages, the proportion of GVs with positive HDAC2 staining increased (P < 0.05) up to 87% in early antral, 98% in small antral, and 97% in large antral follicles (Fig. 1). In the majority of oocytes from early (69%) and small (70%) antral follicles, HDAC2 appeared to be uniformly distributed in the GV nucleoplasm (Figs. 1, 2, G–J, and 3A). In 18% of early antral and 27% of small antral follicles, HDAC2 was localized primarily in a single globular structure within the GV, sometimes accompanied by expression in one or more other smaller structures (Figs. 1 and 3, B and C). In these oocytes, nucleoplasmic expression of HDAC2 was diminished (Fig. 3B) or completely absent (Fig. 3C). Control, double immunostaining of HDAC2 and nucleolin (nucleolar marker) confirmed that HDAC2 expression was inside the nucleolus of these oocytes (Fig. 4). A shift (P < 0.05) in the proportion of each HDAC2 distribution pattern was observed in oocytes from large antral follicles (Figs. 1 and 2, K and L). At this point in folliculogenesis, nucleolar HDAC2 expression was detected in 73% of the GVs, including both with (20%) or without (53%) residual nucleoplasmic expression (Fig. 1).
FIG. 1.
Proportion of diverse distribution patterns of HDAC2 in the cat GV within oocytes recovered from different stages of intraovarian follicles. Proportions with different letters across each follicle category differ (P < 0.05). For a given follicle category, the majority percentage is outlined with a black line. Numbers in parentheses indicate numbers of oocytes examined in each category at each stage.
FIG. 2.
Expression patterns of HDAC2 in the cat GV within oocytes from different stages of intraovarian follicles. Confocal images of immunostaining of HDAC2 (A, C, E, G, I, and K) from primary stage to large antral stage, with the respective chromatin counterstained with Hoechst (B, D, F, H, J, and L). Oocytes from early, small, and large antral follicles were stripped of surrounding cumulus cells before evaluation. Bar = 20 μm.
FIG. 3.
Representative HDAC2 expression patterns observed in the cat GV within oocytes from antral stage follicles. A) Nucleoplasmic expression. B) Distribution in the nucleolus and small globular structures with residual nucleoplasmic expression. C) Solely nucleolar expression. Bar = 20 μm.
FIG. 4.
Double staining of HDAC2 (A) and nucleolin (B), and the merged image of both (C) in a cat GV with nucleolar-localized HDAC2. Bar = 20 μm.
Experiment 2: Relationship Between HDAC2 Localization and Transcriptional Activity in the GVs of Oocytes from Different Antral Follicle Stages
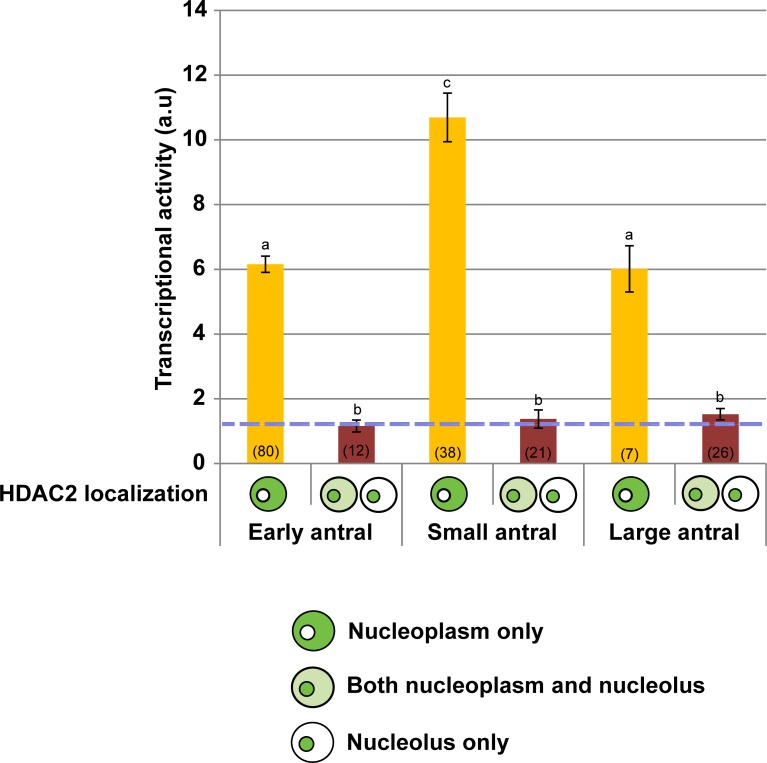
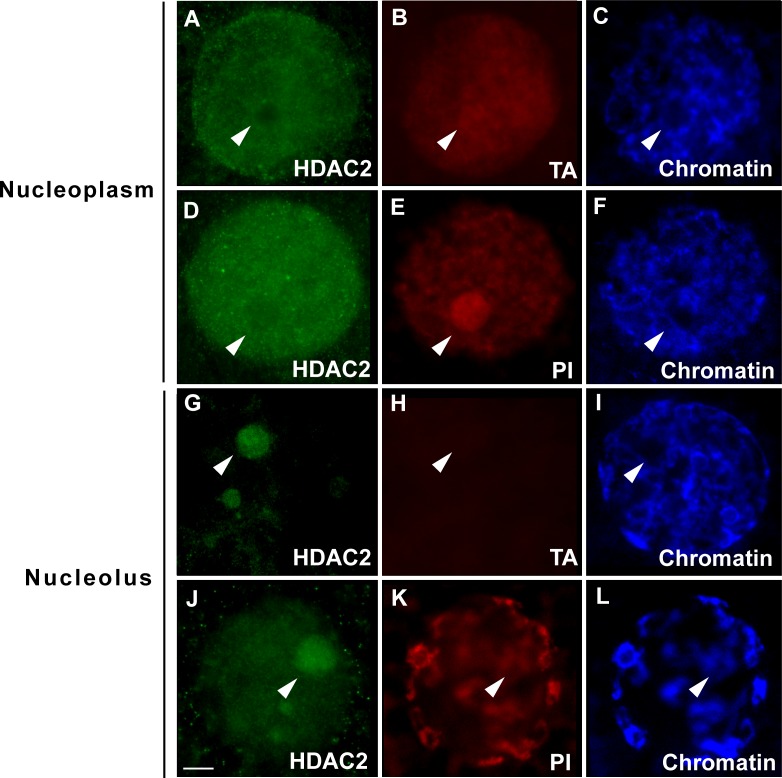
High transcriptional activity was detected in the GVs of oocytes from all three antral stage follicles when HDAC2 was present solely in the nucleoplasm (Figs. 5 and 6, A–C). Nascent RNAs were detected not only in the nucleoplasm, but also in the nucleolus of the transcriptionally active oocytes (Fig. 6B, arrowhead). Consistent with results from the transcriptional assays, strong rRNA expression (based on PI staining) was observed in the nucleolus when HDAC2 expression was excluded from the structure (Fig. 6, D–F, arrowheads). In contrast, oocytes that contained nucleolar-bound HDAC2 from all stages exhibited less (P < 0.05) or no transcriptional activity in both the nucleoplasm and nucleolus (Figs. 5 and 6, G–I, arrowheads). Similarly, when HDAC2 proteins were detected in the nucleolus, nucleolar PI staining was diminished (Fig. 6, J–L, arrowheads). In the control experiment that validated the transcription assay, actinomycin D treatment led to reduced signals in both nucleoplasm and the nucleolus (Supplemental Fig. S2).
FIG. 5.
Association between HDAC2 subcellular localization and transcriptional activity (arbitrary units) in the cat GV within oocytes collected from early, small, or large antral stage follicles. Blue dashed line represents the average background determined from non-5-EU-labeled control oocytes. Diagrams represent HDAC2 localization only in the nucleoplasm, in both nucleoplasm and nucleolus, or only in the nucleolus. Values are mean ± SEM. Values with different letters differ (P < 0.05). Numbers in parentheses indicate the number of oocytes examined in each group.
FIG. 6.
Association between HDAC2 subcellular localization (A–F, solely within the nucleoplasm; G–L, detected in the nucleolus) and transcriptional activity and rRNA expression in the cat GV within oocytes collected from antral stage follicles. HDAC2 stained with anti-HDAC2 antibody (green), nascent RNA labeled with 5-EU to determine transcriptional activity (TA; red), PI labeling for both chromatin and rRNA (red), and Hoechst staining only for chromatin (blue). Arrowheads indicate locations of the nucleoli. Bar = 10 μm.
Experiment 3: Involvement of HDAC Activity in HDAC2 Localization-Dependent Transcriptional Regulation
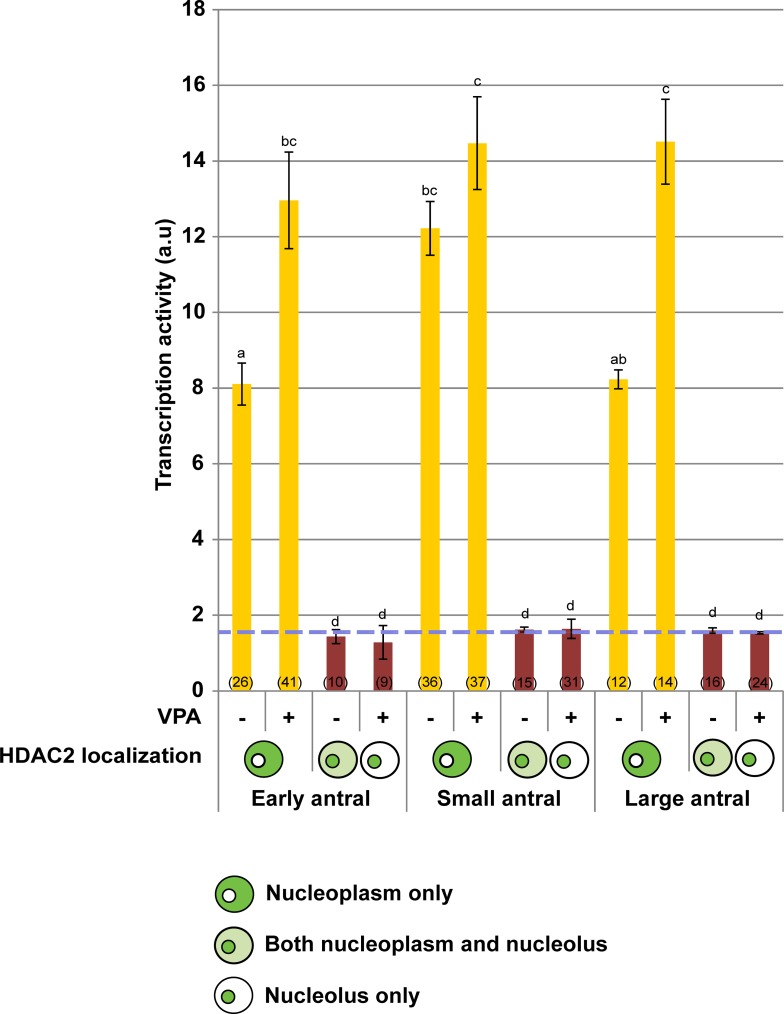
Supplementation with VPA inhibitor effectively increased overall histone acetylation (Supplemental Fig. S3). When HDAC2 was exclusively expressed in the nucleoplasm, transcriptional activity was the highest in oocytes from small antral follicles and was unaffected (P > 0.05) in the presence of VPA (Fig. 7). Blocking HDAC activity at the early and large antral stages allowed transcriptional activity to increase (P < 0.05) to levels that were similar to what was observed at the small antral stage (Fig. 7) without changing chromatin conformation (Supplemental Fig. S4A). There were no differences (P > 0.05) between the VPA-treated and control groups when nucleolar-bound HDAC2 was present, regardless of follicular stage (Fig. 7). Analysis also revealed that the proportion of GVs expressing HDAC2 in the nucleoplasm, nucleolus, or both from any given antral stage was not altered by the treatment (P > 0.05; Supplemental Fig. S4B).
FIG. 7.
Influence of presence (+) or absence (−) of HDAC inhibitor VPA on transcriptional activity (arbitrary units) in the cat GV within oocytes exhibiting different HDAC2 localizations from early, small, or large antral follicles. Blue dashed line represents the average background determined from non-5-EU-labeled control oocytes. Diagrams represent HDAC2 localization only in the nucleoplasm, in both nucleoplasm and nucleolus, or only in the nucleolus. Values are mean ± SEM. Values with different letters differ (P < 0.05). Numbers in parentheses indicate the number of oocytes examined in each group.
DISCUSSION
Using the domestic cat model, we demonstrated that HDAC2, a key transcriptional coregulator, was translocated from the nucleoplasm to the nucleolus of the oocyte's GV toward the late stages of follicle development. Secondly, nucleolar localization of HDAC2 was associated with transcriptional silencing, the latter normally considered a prerequisite to acquire full oocyte competence [17, 50]. This is the first report of nucleolar translocation of HDAC2 for any species. We propose that, as the end of folliculogenesis approaches, HDAC2 is removed from the nucleoplasm to prevent this protein from further regulating transcriptional activity in the GV, thereby facilitating the oocyte to achieve biocompetence. The use of a nontypical model, the cat, allowed us to uncouple the potential confounding influence of chromatin reorganization from the regulation of gene silencing while studying the functional role of HDAC2.
The expression of this protein began in the majority of cat oocytes when follicles grew to an early antral stage. HDAC2 transcripts have been reported to increase in bovine oocytes recovered from preantral and small antral follicles compared with their secondary stage counterparts [37]. In the mouse, HDAC2 protein expression increases from primary to the secondary/preantral stage and then appears sustained through the remainder of the growth phase [32]. Despite a slight discordance at onset, it appears that HDAC2 mRNA and/or protein expressions are most prominent at the antral follicular stage, at least in the three species examined to date.
HDAC2 is known to be predominantly expressed and functional in the nucleoplasm [39]. In the mouse, HDAC2 appears to be exclusively localized in the GV nucleoplasm, even in the fully grown oocyte [32]. This was different than what we observed in the cat, where a nucleoplasm-to-nucleolus transition of HDAC2 localization occurred toward the end of folliculogenesis. By the large antral stage, HDAC2 had completely translocated into the nucleolus in the majority of oocytes. Occasionally HDAC2 staining also was found in what probably were small nucleoli, similar to what has been described in human antral oocytes [12]. It has been suggested that only a subset of HDACs may be localized within the nucleolus to mediate rRNA transcription [51]. The present study demonstrated the first direct evidence of a complete nucleolar translocation of a class I HDAC in a follicular stage-dependent fashion. More importantly, this transition may represent an avenue for regulating HDAC function.
It was particularly interesting that the timing of the nucleolar translocation coincided with transcriptional silencing in the growing cat oocyte [25]. This prompted speculation that these two events may have been correlated. We already knew that gene transcription in the cat oocyte begins at the primary follicle stage [25] and before HDAC2 is expressed. Therefore, this protein was not involved in initiating global transcription, but rather probably playing some role in maintaining or regulating gene expression in oocytes from antral follicles. Indeed, we then discovered that oocytes were transcriptionally active when HDAC2 was expressed in the nucleoplasm, but were largely inactive when HDAC2 was concentrated in the nucleolus. This subnuclear compartment is critical for rRNA and ribosome biosynthesis [15, 16], as well as protein sequestration as a regulatory mechanism for functionally inactivating specific proteins [51–53]. We suspected two possible mechanisms explaining the impact of HDAC2 nucleolar translocation on transcriptional activity in the GV. First, HDAC2 may have facilitated protein modifications and/or rRNA synthesis in the nucleolus that, in turn, indirectly reduced global transcriptional activity in the nucleoplasm. This rationalization is supported by previous evidence suggesting that HDACs are involved in regulating rRNA transcription by controlling deacetylation of histones and upstream-binding factor in NIH 3T3 cells [51]. Alternatively, HDAC2 may have controlled transcriptional activity exclusively in the nucleoplasm and, upon translocation, caused functional suppression via nucleolar sequestration. In the latter case, HDAC2 could be considered a positive regulator of global transcription in the nucleoplasm. Consistent with this idea, it has been suggested that HDACs indirectly up-regulate transcription in the mouse oocyte by altering expression of a histone demethylase [32]. However, it remains unclear if this same mechanism could exist in the cat, especially because the latter species has different methylation patterns from the rodent [34, 52].
To distinguish these two possible mechanisms, we exposed cat oocytes to the class I HDAC inhibitor VPA, which is capable of inhibiting HDAC2 activity while eliminating potential compensatory influences of other class I HDAC members [32, 53, 54]. Results demonstrated that VPA treatment of oocytes containing nucleolar-bound HDAC2 exerted no significant change on RNA synthesis, which supported the concept that HDAC2 was inactive within the nucleolus. By contrast, VPA increased transcriptional activity in the oocytes that exhibited nucleoplasmic HDAC2 expression in the early and large antral stages, thereby suggesting that HDAC activity was required to at least partially repress gene transcription. It also was noteworthy that VPA treatment resulted in transcriptional activity for oocytes from all three antral stages comparable with that of untreated ones from the small antral stage. We speculated that perhaps as transcriptional activity increased during the course of oogenesis, a ceiling level was reached in the oocytes from small antral follicles, thus explaining the inability to provoke more activity from that cohort. Lastly, it was important that general chromatin conformation and HDAC2 distribution across antral stages were indeed unaffected by VPA treatment, reaffirming that these factors did not contribute to the change in transcriptional activity.
Our observation that supplementation with VPA increased (rather than decreased) transcriptional activity appeared to be contradictory to the original prediction that HDAC2 was a positive transcriptional regulator in the nucleoplasm. One possible explanation is that cat HDAC2 may have been involved in modulating transcriptional activity at specific loci essential for regulating gene expression. Its removal from the nucleoplasm may have disrupted the equilibrium among multiple regulatory elements to indirectly accelerate cessation of transcription. Alternatively, as VPA inhibits HDAC activity by blocking substrate access without interfering with other protein interactions [54], it was plausible that HDAC2 influenced global transcription through uncharacterized protein-protein interaction rather than deacetylation [39]. A third alternative was that the stimulatory influence of VPA on transcriptional activity in the present study may have represented a combinational inhibitional outcome, given that VPA has been known to inhibit all class I HDACs [53, 54]. However, the roles of these other proteins (i.e., HDAC1 and HDAC3) are currently unknown during oogenesis in the cat, largely because of a lack of effective antibodies. Nonetheless, it was unlikely that an HDAC-dependent global chromatin remodeling was involved, because chromatin structure was unperturbed after VPA treatment. Overall, although the use of inhibitor provided clues to understanding the role of HDAC2 in the cat GV, it also unveiled the complexity of the network regulating transcriptional activity.
It can be concluded that nucleolar translocation of HDAC2 represented a novel mechanism that was associated with the regulation of transcriptional silencing in the domestic cat. Although the detailed molecular mechanism requires more investigation, there appears to be some cross-species conservation, because decreased HDAC2 mRNA expression in late-stage cow oocytes also has been linked recently to competence acquisition [37]. Thus, reducing functional HDAC2, via decreased synthesis or nucleolar sequestration, may be vital for transcriptional arrest in preparing an oocyte for becoming biologically competent. Because previous studies have linked nucleolar localization with protein degradation [55, 56], it is plausible that HDAC2 may have undergone a similar process before GV breakdown. A critical next step is validating the relationship between HDAC2 nucleolar translocation and global transcription, perhaps by finding a means to manipulate its localization. This could include determining the signals that induce HDAC2 translocation and mapping the nucleolar localization sequences (thus far, no known nucleolar localization sequences have been detected in the cat HDAC2 sequence). The capacity to manipulate protein functionality along with a clear understanding of the molecular regulatory events occurring within the GV would have practical benefits, such as evaluating the status and competence of the GV for in vitro studies and improving the ability to stimulate and retain competence during oocyte preservation [57–61]. Moreover, the HDAC2 shuttling mechanism could serve as an attractive contraceptive target for disrupting competence acquisition of the oocyte. Preventing the HDAC2 transport could specifically inhibit the localization-dependent effect within the GV without influencing other cellular functions of the protein, thereby minimizing side effects.
ACKNOWLEDGMENT
We thank Dr. Brent Whitaker (Animal Rescue Inc.) and Dr. Keiko Antoku (Paw Prints Animal Hospital) for providing domestic cat ovaries, and the Imaging Core at the University of Maryland (College Park) for access to the confocal microscope.
Footnotes
Supported by the National Center for Research Resources (R01 RR026064), a component of the National Institutes of Health (NIH), and currently by the Office of Research Infrastructure Programs/Office of the Director (R01 OD010948).
REFERENCES
- Zuccotti M, Merico V, Cecconi S, Redi CA, Garagna S. What does it take to make a developmentally competent mammalian egg? Hum Reprod Update. 2011;17:525–540. doi: 10.1093/humupd/dmr009. [DOI] [PubMed] [Google Scholar]
- Trudgen K, Ayensu-Coker L. Fertility preservation and reproductive health in the pediatric, adolescent, and young adult female cancer patient. Curr Opin Obstet Gynecol. 2014;26:372–380. doi: 10.1097/GCO.0000000000000107. [DOI] [PubMed] [Google Scholar]
- Berwanger AL, Finet A, El Hachem H, le Parco S, Hesters L, Grynberg M. New trends in female fertility preservation: in vitro maturation of oocytes. Future Oncol. 2012;8:1567–1573. doi: 10.2217/fon.12.144. [DOI] [PubMed] [Google Scholar]
- Comizzoli P, Songsasen N, Wildt DE. Protecting and extending fertility for females of wild and endangered mammals. Cancer Treat Res. 2010;156:87–100. doi: 10.1007/978-1-4419-6518-9_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto S. Application of in vitro maturation to assisted reproductive technology. J Reprod Dev. 2009;55:1–10. doi: 10.1262/jrd.20127. [DOI] [PubMed] [Google Scholar]
- Wang X, Catt S, Pangestu M, Temple-Smith P. Successful in vitro culture of pre-antral follicles derived from vitrified murine ovarian tissue: oocyte maturation, fertilization, and live births. Reproduction. 2011;141:183–191. doi: 10.1530/REP-10-0383. [DOI] [PubMed] [Google Scholar]
- Lodde V, Modina S, Galbusera C, Franciosi F, Luciano AM. Large-scale chromatin remodeling in germinal vesicle bovine oocytes: interplay with gap junction functionality and developmental competence. Mol Reprod Dev. 2007;74:740–749. doi: 10.1002/mrd.20639. [DOI] [PubMed] [Google Scholar]
- De La Fuente R. Chromatin modifications in the germinal vesicle (GV) of mammalian oocytes. Dev Biol. 2006;292:1–12. doi: 10.1016/j.ydbio.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Russo V, Martelli A, Berardinelli P, Di Giacinto O, Bernabo N, Fantasia D, Mattioli M, Barboni B. Modifications in chromatin morphology and organization during sheep oogenesis. Microsc Res Tech. 2007;70:733–744. doi: 10.1002/jemt.20462. [DOI] [PubMed] [Google Scholar]
- Liu Y, Sui HS, Wang HL, Yuan JH, Luo MJ, Xia P, Tan JH. Germinal vesicle chromatin configurations of bovine oocytes. Microsc Res Tech. 2006;69:799–807. doi: 10.1002/jemt.20349. [DOI] [PubMed] [Google Scholar]
- Sun XS, Liu Y, Yue KZ, Ma SF, Tan JH. Changes in germinal vesicle (GV) chromatin configurations during growth and maturation of porcine oocytes. Mol Reprod Dev. 2004;69:228–234. doi: 10.1002/mrd.20123. [DOI] [PubMed] [Google Scholar]
- Miyara F, Migne C, Dumont-Hassan M, Le Meur A, Cohen-Bacrie P, Aubriot FX, Glissant A, Nathan C, Douard S, Stanovici A, Debey P. Chromatin configuration and transcriptional control in human and mouse oocytes. Mol Reprod Dev. 2003;64:458–470. doi: 10.1002/mrd.10233. [DOI] [PubMed] [Google Scholar]
- Zuccotti M, Piccinelli A. Giorgi Rossi P, Garagna S, Redi CA. Chromatin organization during mouse oocyte growth. Mol Reprod Dev. 1995;41:479–485. doi: 10.1002/mrd.1080410410. [DOI] [PubMed] [Google Scholar]
- Schramm RD, Tennier MT, Boatman DE, Bavister BD. Chromatin configurations and meiotic competence of oocytes are related to follicular diameter in nonstimulated rhesus monkeys. Biol Reprod. 1993;48:349–356. doi: 10.1095/biolreprod48.2.349. [DOI] [PubMed] [Google Scholar]
- Cisterna B, Biggiogera M. Ribosome biogenesis: from structure to dynamics. Int Rev Cell Mol Biol. 2010;284:67–111. doi: 10.1016/S1937-6448(10)84002-X. [DOI] [PubMed] [Google Scholar]
- Moss T, Langlois F, Gagnon-Kugler T, Stefanovsky V. A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis. Cell Mol Life Sci. 2007;64:29–49. doi: 10.1007/s00018-006-6278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodde V, Modina S, Maddox-Hyttel P, Franciosi F, Lauria A, Luciano AM. Oocyte morphology and transcriptional silencing in relation to chromatin remodeling during the final phases of bovine oocyte growth. Mol Reprod Dev. 2008;75:915–924. doi: 10.1002/mrd.20824. [DOI] [PubMed] [Google Scholar]
- Wu D, Cheung QC, Wen L, Li J. A growth-maturation system that enhances the meiotic and developmental competence of porcine oocytes isolated from small follicles. Biol Reprod. 2006;75:547–554. doi: 10.1095/biolreprod.106.051300. [DOI] [PubMed] [Google Scholar]
- Zuccotti M, Ponce RH, Boiani M, Guizzardi S, Govoni P, Scandroglio R, Garagna S, Redi CA. The analysis of chromatin organisation allows selection of mouse antral oocytes competent for development to blastocyst. Zygote. 2002;10:73–78. doi: 10.1017/s0967199402002101. [DOI] [PubMed] [Google Scholar]
- Combelles CM, Cekleniak NA, Racowsky C, Albertini DF. Assessment of nuclear and cytoplasmic maturation in in-vitro matured human oocytes. Hum Reprod. 2002;17:1006–1016. doi: 10.1093/humrep/17.4.1006. [DOI] [PubMed] [Google Scholar]
- Ledda S, Bogliolo L, Leoni G, Naitana S. Follicular size affects the meiotic competence of in vitro matured prepubertal and adult oocytes in sheep. Reprod Nutr Dev. 1999;39:503–508. doi: 10.1051/rnd:19990409. [DOI] [PubMed] [Google Scholar]
- Swygert SG, Peterson CL. Chromatin dynamics: interplay between remodeling enzymes and histone modifications. Biochim Biophys Acta. 2014;1839:728–736. doi: 10.1016/j.bbagrm.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HL, Sui HS, Liu Y, Miao DQ, Lu JH, Liang B, Tan JH. Dynamic changes of germinal vesicle chromatin configuration and transcriptional activity during maturation of rabbit follicles. Fertil Steril. 2009;91:1589–1594. doi: 10.1016/j.fertnstert.2008.10.071. [DOI] [PubMed] [Google Scholar]
- De La Fuente R, Viveiros MM, Burns KH, Adashi EY, Matzuk MM, Eppig JJ. Major chromatin remodeling in the germinal vesicle (GV) of mammalian oocytes is dispensable for global transcriptional silencing but required for centromeric heterochromatin function. Dev Biol. 2004;275:447–458. doi: 10.1016/j.ydbio.2004.08.028. [DOI] [PubMed] [Google Scholar]
- Comizzoli P, Pukazhenthi BS, Wildt DE. The competence of germinal vesicle oocytes is unrelated to nuclear chromatin configuration and strictly depends on cytoplasmic quantity and quality in the cat model. Hum Reprod. 2011;26:2165–2177. doi: 10.1093/humrep/der176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui HS, Liu Y, Miao DQ, Yuan JH, Qiao TW, Luo MJ, Tan JH. Configurations of germinal vesicle (GV) chromatin in the goat differ from those of other species. Mol Reprod Dev. 2005;71:227–236. doi: 10.1002/mrd.20251. [DOI] [PubMed] [Google Scholar]
- Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol. 2014;6:a018713. doi: 10.1101/cshperspect.a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallinari P, Di Marco S, Jones P, Pallaoro M, Steinkuhler C. HDACs, histone deacetylation and gene transcription: from molecular biology to cancer therapeutics. Cell Res. 2007;17:195–211. doi: 10.1038/sj.cr.7310149. [DOI] [PubMed] [Google Scholar]
- Yang F, Baumann C, Viveiros MM, De La Fuente R. Histone hyperacetylation during meiosis interferes with large-scale chromatin remodeling, axial chromatid condensation and sister chromatid separation in the mammalian oocyte. Int J Dev Biol. 2012;56:889–899. doi: 10.1387/ijdb.120246rd. [DOI] [PubMed] [Google Scholar]
- Wang Q, Yin S, Ai JS, Liang CG, Hou Y, Chen DY, Schatten H, Sun QY. Histone deacetylation is required for orderly meiosis. Cell Cycle. 2006;5:766–774. doi: 10.4161/cc.5.7.2627. [DOI] [PubMed] [Google Scholar]
- Akiyama T, Nagata M, Aoki F. Inadequate histone deacetylation during oocyte meiosis causes aneuploidy and embryo death in mice. Proc Natl Acad Sci U S A. 2006;103:7339–7344. doi: 10.1073/pnas.0510946103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P, Pan H, Montgomery RL, Olson EN, Schultz RM. Compensatory functions of histone deacetylase 1 (HDAC1) and HDAC2 regulate transcription and apoptosis during mouse oocyte development. Proc Natl Acad Sci U S A. 2012;109:E481–489. doi: 10.1073/pnas.1118403109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P, Schultz RM. Histone deacetylase 1 (HDAC1) regulates histone acetylation, development, and gene expression in preimplantation mouse embryos. Dev Biol. 2008;319:110–120. doi: 10.1016/j.ydbio.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama S, Liu H, Kaneko N, Ooga M, Nagata M, Aoki F. Alterations in epigenetic modifications during oocyte growth in mice. Reproduction. 2007;133:85–94. doi: 10.1530/REP-06-0025. [DOI] [PubMed] [Google Scholar]
- Akiyama T, Kim JM, Nagata M, Aoki F. Regulation of histone acetylation during meiotic maturation in mouse oocytes. Mol Reprod Dev. 2004;69:222–227. doi: 10.1002/mrd.20121. [DOI] [PubMed] [Google Scholar]
- Zuccotti M, Bellone M, Longo F, Redi CA, Garagna S. Fully-mature antral mouse oocytes are transcriptionally silent but their heterochromatin maintains a transcriptional permissive histone acetylation profile. J Assist Reprod Genet. 2011;28:1193–1196. doi: 10.1007/s10815-011-9562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessa IR, Nishimura RC, Franco MM, Dode MA. Transcription profile of candidate genes for the acquisition of competence during oocyte growth in cattle. Reprod Domest Anim. 2013;48:781–789. doi: 10.1111/rda.12162. [DOI] [PubMed] [Google Scholar]
- Zheng P, Patel B, McMenamin M, Paprocki AM, Schramm RD, Nagl NG, Jr., , Wilsker D, Wang X, Moran E, Latham KE. Expression of genes encoding chromatin regulatory factors in developing rhesus monkey oocytes and preimplantation stage embryos: possible roles in genome activation. Biol Reprod. 2004;70:1419–1427. doi: 10.1095/biolreprod.103.023796. [DOI] [PubMed] [Google Scholar]
- Joshi P, Greco TM, Guise AJ, Luo Y, Yu F, Nesvizhskii AI, Cristea IM. The functional interactome landscape of the human histone deacetylase family. Mol Syst Biol. 2013;9:672. doi: 10.1038/msb.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw S, Robert C, Massicotte L, Sirard MA. Quantification of histone acetyltransferase and histone deacetylase transcripts during early bovine embryo development. Biol Reprod. 2003;68:383–389. doi: 10.1095/biolreprod.102.005991. [DOI] [PubMed] [Google Scholar]
- Segev H, Memili E, First NL. Expression patterns of histone deacetylases in bovine oocytes and early embryos, and the effect of their inhibition on embryo development. Zygote. 2001;9:123–133. doi: 10.1017/s0967199401001137. [DOI] [PubMed] [Google Scholar]
- Bristol-Gould S, Woodruff TK. Folliculogenesis in the domestic cat (Felis catus) Theriogenology. 2006;66:5–13. doi: 10.1016/j.theriogenology.2006.03.019. [DOI] [PubMed] [Google Scholar]
- Jewgenow K. Role of media, protein and energy supplements on maintenance of morphology and DNA-synthesis of small preantral domestic cat follicles during short-term culture. Theriogenology. 1998;49:1567–1577. doi: 10.1016/s0093-691x(98)00102-2. [DOI] [PubMed] [Google Scholar]
- Wood TC, Wildt DE. Effect of the quality of the cumulus-oocyte complex in the domestic cat on the ability of oocytes to mature, fertilize and develop into blastocysts in vitro. J Reprod Fertil. 1997;110:355–360. doi: 10.1530/jrf.0.1100355. [DOI] [PubMed] [Google Scholar]
- Khochbin S, Wolffe AP. The origin and utility of histone deacetylases. FEBS Lett. 1997;419:157–160. doi: 10.1016/s0014-5793(97)01423-3. [DOI] [PubMed] [Google Scholar]
- Maddox-Hyttel P, Bjerregaard B, Laurincik J. Meiosis and embryo technology: renaissance of the nucleolus. Reprod Fertil Dev. 2005;17:3–14. doi: 10.1071/rd04108. [DOI] [PubMed] [Google Scholar]
- Ma N, Matsunaga S, Takata H, Ono-Maniwa R, Uchiyama S, Fukui K. Nucleolin functions in nucleolus formation and chromosome congression. J Cell Sci. 2007;120:2091–2105. doi: 10.1242/jcs.008771. [DOI] [PubMed] [Google Scholar]
- Otto F, Tsou KC. A comparative study of DAPI, DIPI, and Hoechst 33258 and 33342 as chromosomal DNA stains. Stain Technol. 1985;60:7–11. doi: 10.3109/10520298509113885. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Fujikura K, Higashiyama T, Takata K. DNA staining for fluorescence and laser confocal microscopy. J Histochem Cytochem. 1997;45:49–53. doi: 10.1177/002215549704500107. [DOI] [PubMed] [Google Scholar]
- Liu H, Aoki F. Transcriptional activity associated with meiotic competence in fully grown mouse GV oocytes. Zygote. 2002;10:327–332. doi: 10.1017/s0967199402004069. [DOI] [PubMed] [Google Scholar]
- Hirschler-Laszkiewicz I, Cavanaugh A, Hu Q, Catania J, Avantaggiati ML, Rothblum LI. The role of acetylation in rDNA transcription. Nucleic Acids Res. 2001;29:4114–4124. doi: 10.1093/nar/29.20.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips TC, Wildt DE, Comizzoli P. Increase in histone methylation in the cat germinal vesicle related to acquisition of meiotic and developmental competence Reprod Domest Anim 2012. 47 (suppl 6): 210 214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlicher M. Valproic acid: an old drug newly discovered as inhibitor of histone deacetylases Ann Hematol 2004. 83 (suppl 1): S91 S92 [DOI] [PubMed] [Google Scholar]
- Gottlicher M, Minucci S, Zhu P, Kramer OH, Schimpf A, Giavara S, Sleeman JP, Lo Coco F, Nervi C, Pelicci PG, Heinzel T. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001;20:6969–6978. doi: 10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao T, Shi H, Guan Y, Huang D, Chen Y, Lane DP, Chen J, Peng J. Def defines a conserved nucleolar pathway that leads p53 to proteasome-independent degradation. Cell Res. 2013;23:620–634. doi: 10.1038/cr.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Wu M. Identification of a novel nucleolar localization signal and a degradation signal in Survivin-deltaEx3: a potential link between nucleolus and protein degradation. Oncogene. 2005;24:2723–2734. doi: 10.1038/sj.onc.1208097. [DOI] [PubMed] [Google Scholar]
- Wang TY, Li Q, Li Q, Li H, Zhu J, Cui W, Jiao GZ, Tan JH. Non-frozen preservation protocols for mature mouse oocytes dramatically extend their developmental competence by reducing oxidative stress. Mol Hum Reprod. 2014;20:318–329. doi: 10.1093/molehr/gat088. [DOI] [PubMed] [Google Scholar]
- Niu HR, Zi XD, Xiao X, Xiong XR, Zhong JC, Li J, Wang L, Wang Y. Developmental competence of frozen-thawed yak (Bos grunniens) oocytes followed by in vitro maturation and fertilization. Cryobiology. 2014;68:152–154. doi: 10.1016/j.cryobiol.2014.01.012. [DOI] [PubMed] [Google Scholar]
- Luvoni GC. Cryosurvival of ex situ and in situ feline oocytes Reprod Domest Anim 2012. 47 (suppl 6): 266 268 [DOI] [PubMed] [Google Scholar]
- Gosden R, Nagano M. Preservation of fertility in nature and ART. Reproduction. 2002;123:3–11. doi: 10.1530/rep.0.1230003. [DOI] [PubMed] [Google Scholar]
- Wang L, Fu X, Zeng Y, Zhu S. Epinephrine promotes development potential of vitrified mouse oocytes. Pak J Biol Sci. 2014;17:254–259. doi: 10.3923/pjbs.2014.254.259. [DOI] [PubMed] [Google Scholar]