Abstract
Sex-reversal cases in humans and genetic models in mice have revealed that the fate of the bipotential gonad hinges upon the balance between pro-testis SOX9 and pro-ovary beta-catenin pathways. Our central query was: if SOX9 and beta-catenin define the gonad's identity, then what do the gonads become when both factors are absent? To answer this question, we developed mouse models that lack either Sox9, beta-catenin, or both in the somatic cells of the fetal gonads and examined the morphological outcomes and transcriptome profiles. In the absence of Sox9 and beta-catenin, both XX and XY gonads progressively lean toward the testis fate, indicating that expression of certain pro-testis genes requires the repression of the beta-catenin pathway, rather than a direct activation by SOX9. We also observed that XY double knockout gonads were more masculinized than their XX counterpart. To identify the genes responsible for the initial events of masculinization and to determine how the genetic context (XX vs. XY) affects this process, we compared the transcriptomes of Sox9/beta-catenin mutant gonads and found that early molecular changes underlying the XY-specific masculinization involve the expression of Sry and 21 SRY direct target genes, such as Sox8 and Cyp26b1. These results imply that when both Sox9 and beta-catenin are absent, Sry is capable of activating other pro-testis genes and drive testis differentiation. Our findings not only provide insight into the mechanism of sex determination, but also identify candidate genes that are potentially involved in disorders of sex development.
Keywords: beta-catenin, sex determination, Sox9, Sry, testis
INTRODUCTION
In mammals, both ovary and testis derive from a common embryonic structure called the gonadal primordium or bipotential gonads. The developmental path of bipotential gonads toward an ovary or a testis is orchestrated by activation or repression of signaling pathways in the supporting cell lineage (for a review, see [1]). In male or XY individual, SRY, the testis-determining gene on the Y-chromosome, induces differentiation of the supporting cell lineage into the testis-specific Sertoli cells and consequent transformation of the bipotential gonad into a testis [2–4]. During its short period of expression in the mouse fetal testis, from 10.5 to 12.5 days postcoitum (dpc) [5, 6], SRY induces the expression of Sox9 [7], which promotes testis organogenesis through the activation of pro-testis genes and the repression of pro-ovarian genes. Loss of Sox9 in XY gonads leads to complete testis-to-ovary sex reversal [8–10], whereas a gain of Sox9 expression in XX gonads results in complete ovary-to-testis sex reversal [11, 12]. Therefore, SOX9, acting as a downstream effector of SRY, is necessary and sufficient to drive testis differentiation.
In the absence of the Y chromosome (XX genetic background) or Sry gene (XY genetic background with Sry mutations), the supporting cell linages of the bipotential gonad commit to the ovary-specific granulosa cell fate (for a review, see [13]). In XX mouse embryos, global loss of Wnt4 [14, 15], Rspo1 [16, 17], or targeted loss of their downstream effector β-catenin in the somatic cells of the fetal gonad [18, 19] results in a partial ovary-to-testis sex-reversal characterized by the presence of a testis-specific coelomic vessel and steroidogenic cells. Close to birth in Wnt4−/− and Rspo1−/− ovaries, supporting cells start to differentiate into testis-specific Sertoli cells that express Sox9 [16, 20]. On the other hand, ectopic activation of β-catenin in the somatic cells of XY fetal gonads leads to repression of Sox9 expression and consequent testis-to-ovary sex reversal [21]. These studies implicate an antagonism between the pro-testis SOX9 and pro-ovary RSPO1/WNT4/β-catenin pathways in shaping the fate of the bipotential gonad. The characterization of global double knockout (DKO) models such as pro-ovary Wnt4/pro-testis Fgf9 DKO [22] and pro-ovary Rspo1/pro-testis Sox9 DKO [23] suggests that this antagonism operates in a sex chromosome-specific manner and that fate determination of the testis may involve factors other than Sox9.
The main goal of this study is to identify the early molecular changes that determine the gonadal identity when pro-testis Sox9 and pro-ovary β-catenin are inactivated in the somatic cells of bipotential gonads. We compared the outcome of sex determination in embryos lacking Sox9, β-catenin, or both, with a particular interest in the supporting cell lineage. To identify the candidate genes responsible for the fate determination in the absence of Sox9 and β-catenin, we characterized the transcriptomes of fetal gonads during early events of sex-reversal and made comparisons between XX and XY genetic backgrounds.
MATERIALS AND METHODS
Experimental Animals
All the embryos used in this study resulted from the crossing between Ctnnb1f/f; Sox9f/f females with Sf1-cre+/Tg; Ctnnb1+/f; Sox9+/f males. These Ctnnb1f/f; Sox9f/f females were generated by crossing B6.129-Ctnnb1tm2Kem/J (004152; Jackson Laboratory) to B6.129S7-Sox9tm1Crm/J (013106; Jackson Laboratory) mice, and the resulting Ctnnb1+/f; Sox9+/f mice were then intercrossed to generate double homozygotes. Males Sf1-cre+/Tg; Ctnnb1+/f; Sox9+/f were generated by crossing Ctnnb1f/f; Sox9f/f to Sf1-creTg/Tg transgenic mice [24]. The Cre recombinase expression under the control of steroidogenic factor 1 (Sf1) regulatory elements allowed the conditional KO of Ctnnb1 and Sox9 in the somatic cells of the gonads (Supplemental Fig. S1; Supplemental Data are available online at www.biolreprod.org). The Ctnnb1f/f; Sox9f/f females were mated overnight with Sf1-cre+/Tg; Ctnnb1+/f; Sox9+/f males, and checked for vaginal plug the next morning. Embryonic Day 0.5 corresponds to noon of the day of the vaginal plug detection. The genotype of the gonads used in this study is as follows: Sf1-cre+/Tg; Ctnnb1+/f; Sox9+/f as control, Sf1-cre+/Tg; Ctnnb1f/f; Sox9+/f as Ctnnb1 KO, Sf1-cre+/Tg; Ctnnb1+/f; Sox9f/f as Sox9 KO, and Sf1-cre+/Tg; Ctnnb1f/f; Sox9f/f as Ctnnb1/Sox9 DKO. All the animal procedures were approved by the National Institutes of Health Animals Care and Use Committee and were performed in accordance with an approved National Institute of Environmental Health Sciences animal study proposal.
Immunohistochemistry
Fetal gonads were collected and fixed overnight at 4°C in 4% paraformaldehyde. The 14.5 dpc gonads were immunostained as whole mounts, and 17.5 dpc gonads were cryosectioned to 8–10 μm sections before staining. Immunofluorescence analyses were performed as previously described [19]. The antibodies used in this study are listed in Supplemental Table S1.
Real-Time PCR Analyses
A single pair of gonads (14.5 or 17.5 dpc) was separated from the mesonephros and was used as one biological replicate (five biological replicates were included for each genotype). Total RNA was isolated using Picopure extraction kit (Arcturus). RNA quality and concentration were determined using the Nanodrop 2000c. Complementary DNA was synthetized using the Superscript II cDNA synthesis kit (Invitrogen); 250 ng of RNA was used for each biological replicate. Gene expression was analyzed by real-time PCR using Bio-Rad CFX96TM Real-Time PCR Detection system. Taqman probes (Bio-Rad) and primers used are listed in Supplemental Table S1. Relative expression was calculated as the percentage of the highest expression level recorded for each gene. All the values were expressed as mean ± SEM, and statistical analysis was performed using unpaired Student t-test (P < 0.05).
Microarray Analysis
Three to four biological replicas (one single pair of 14.5 dpc gonads) were used for each genotype. RNA quality and concentration were determined using N Agilent 2100 BioAnalyzer (Agilent Technologies). Gene expression analysis was conducted using Affymetrix Mouse Genome 430 2.0 GeneChip arrays (Affymetrix). A total of 40 ng of total RNA was amplified as directed in the Ovation Pico RNA Amplification System protocol and labeled with biotin following the Encore Biotin Module. Biotin-amplified RNAs (4.6 μg) were fragmented and hybridized to each array for 18 h at 45°C in a rotating hybridization. Array slides were stained with streptavidin/phycoerythrin utilizing a double-antibody staining procedure and then washed for antibody amplification according to the GeneChip Hybridization, Wash and Stain Kit and user manual following protocol FS450-0004. Arrays were scanned in an Affymetrix Scanner 3000 and data (GEO accession no. GSE67463, http://www.ncbi.nlm.nih.gov/geo/) was obtained using the GeneChip Command Console Software (AGCC Version 1.1) using the MAS5 algorithm to generate .CHP files. Partek software was used for data analysis. Principal component analysis (PCA) was performed on all biological replicates plotted in a three-dimensional manner using the three principal components that captures most of the variance in the dataset. The hierarchical clustering was created using a one-way ANOVA comparing the robust multiarray average normalized log-2 intensities of 2215 probes that were significantly different between control XX and XY (fold change ≥ 2, P < 0.05). The gene lists comparing two genotypes were filtered with fold-change cut-off of 1.5 with P < 0.05. To identify the genes normally expressed in Sertoli cells, a published cell-type-specific microarray dataset performed on fetal gonads was used [25]. This dataset permitted the identification of genes that are more highly expressed in fetal Sertoli cells than in fetal granulosa cells (from 11.5 to 13.5 dpc, fold change ≥ 1.5, P < 0.05). In addition, genes that were more highly expressed in Leydig cells than Sertoli cells (fold change ≥ 1.5, P < 0.05) were removed from this list. This leads to the creation of a list of 796 genes representative of the Sertoli cell identity (see green circle in Fig. 6A). To compare our data with this gene list based on Jameson et al. microarray data [25], a Venn diagram was generated using the gene-symbol as the identifier because the probes identification from these two microarrays are different.
RESULTS
Phenotypes in the XX Gonads: Consequences of Inactivation of Both β-Catenin and Sox9 at 14.5 dpc
To determine what the bipotential gonad becomes in the absence of β-catenin (or Ctnnb1) and Sox9, we inactivated Sox9 and Ctnnb1 genes using the Sf1-Cre transgenic mouse [24]. Expression of the Sf1-Cre transgene in the somatic compartment of the gonadal ridge around 9.5/10.5 dpc [24] allowed the specific inactivation of Sox9 and Ctnnb1 genes in the somatic cells of the bipotential gonads (Supplemental Fig. S1). We first characterized the mutant gonads in the XX genetic context at 14.5 dpc, when dimorphic development of the gonads is apparent (Fig. 1, A–I). Consistent with previous observations by our group and others [18, 19], XX Ctnnb1 single KO gonads developed morphologically similar to the control ovaries (Fig. 1B), with the exception of ectopic testis-specific vasculature (Fig. 1E, arrow) and steroidogenic cells (see Supplemental Fig. S2). The supporting cells of XX Ctnnb1 KO gonads (Fig. 1H) had an ovarian identity with the presence of FOXL2 (granulosa cell marker) and absence of AMH (Sertoli cell marker). However, Foxl2 mRNA level was significantly reduced compared to the control ovaries (Fig. 1S). XX Ctnnb1/Sox9 DKO gonads were indistinguishable from XX Ctnnb1 KO gonads with the same vasculature and steroidogenic phenotypes (Fig. 1F and Supplemental Fig. S2). The supporting cells of XX DKO gonads maintained their FOXL2-positive ovarian identity (Fig. 1I) similar to XX Ctnnb1 KO gonads. These results indicate that Sox9 is not required for the appearance of testis characteristics observed in the XX Ctnnb1 KO gonads at 14.5 dpc.
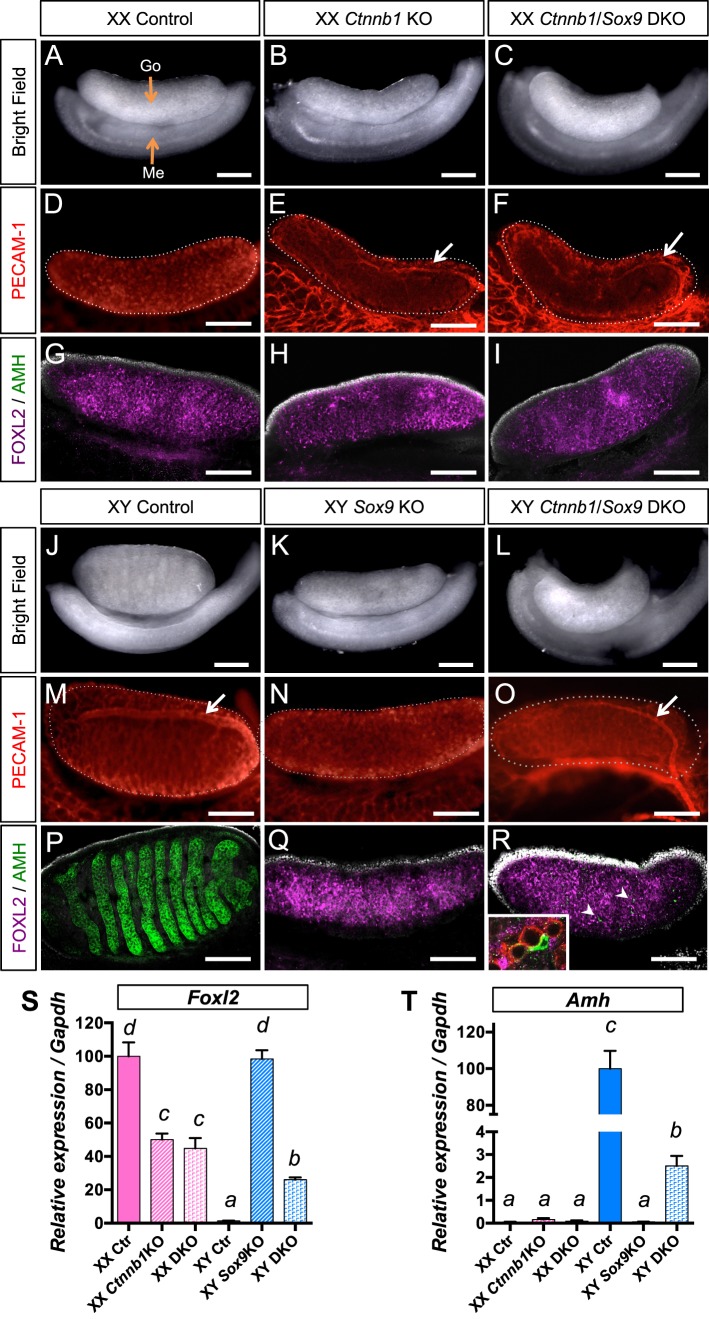
FIG. 1.
Consequences of inactivation of both β-catenin and Sox9 in the gonads at 14.5 dpc. The 14.5 dpc XX (A–I) and XY (J–R) gonads of various genotypes (control, Ctnnb1 KO, Sox9 KO, and Ctnnb1/Sox9 DKO) were analyzed by bright field microscopy (A–C, J–L), immunofluorescence for germ cell/vasculature marker PECAM-1 (D–F, M–O) and supporting cell markers FOXL2/AMH (G–I, P–R). White arrows show the testis-specific vessel at the surface of the gonad (E, F, M, O). Nuclei are labeled with 4′,6-diamidino-2-phenylindole ([DAPI] grey) in G–I and P–R. R) Arrowheads show the AMH+ cells, and the inset is a higher magnification of a part of the XY Ctnnb1/Sox9 DKO gonads that contain PECAM+ germ cells (red) surrounded by FOXL2+ granulosa cells (magenta) and one AMH+ Sertoli-like cell (green). Go, gonad; Me, mesonephros. The white dotted lines outline the gonads. Bars = 200 μm. S and T) represent quantitative PCR analysis of Foxl2 (S) and Amh (T) mRNA expression in 14.5 dpc gonads. Error bars represent SEM of five biological replicates (Student t-test, P < 0.05; a < b < c < d).
Phenotypes in the XY Gonads: Consequences of Inactivation of Both β-Catenin and Sox9 at 14.5 dpc
Next we examined the effects of loss of both Ctnnb1 and Sox9 in the male or XY genetic context, and we compared these gonads to control and XY Sox9 KO gonads at 14.5 dpc (Fig. 1, J–R). Consistent with the findings by others [10, 26], XY Sox9 single KO gonads underwent complete testis-to-ovary sex reversal, characterized by its ovarian shape and size (Fig. 1K), presence of AMH-negative/FOXL2-positive granulosa cells (Fig 1Q), absence of the testis-specific vessel (Fig. 1N) and steroidogenic cells (Supplemental Fig. S2). In the XY Ctnnb1/Sox9 DKO gonads, the supporting cells were mostly FOXL2-positive, resembling the XY Sox9 KO gonads (Fig. 1, Q and R). However, XY DKO gonads developed a testis-specific coelomic vessel (Fig. 1O), steroidogenic cells (Supplemental Fig. S2), and a few AMH-positive Sertoli cells (Fig. 1R, inset). Real-time PCR analyzes confirmed the expression of Amh in XY DKO gonads and its absence in XY Sox9 KO gonads (Fig. 1T). Moreover, Foxl2 expression in the XY Sox9 KO gonads was sex-reversed to the level of control XX gonads (Fig. 1S) whereas Foxl2 expression in the XY DKO gonad was significantly reduced (Fig. 1S). The restoration of some testicular characteristics by additional inactivation of Ctnnb1 in the Sox9 KO XY gonads indicates that Ctnnb1 is partially responsible for the appearance of ovarian phenotypes in the absence of Sox9. In addition, masculinization phenotypes in the XX and XY DKO further support that certain testicular characteristics such as formation of the coelomic vessels do not require Sox9.
Y Chromosome-Specific Effects of Loss of β-Catenin and Sox9 on Gonadal Differentiation
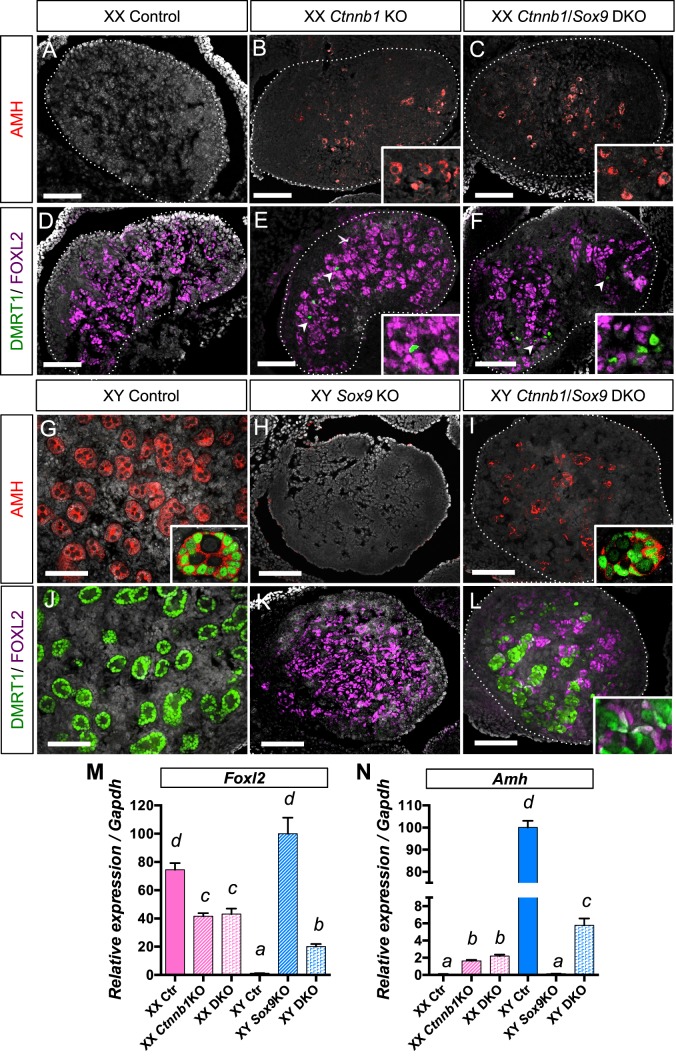
When comparing the XX and XY Ctnnb1/Sox9 DKO phenotypes at 14.5 dpc (Fig. 1, C, F, and I vs. L, O, and R), we noticed several subtle differences: a few AMH-positive cells were detected in XY DKO gonads (Fig. 1R) but not in XX DKO gonads (Fig. 1I). Moreover, Foxl2 expression was significantly reduced in XY DKO gonads compared to XX DKO gonads (Fig. 1S). These results imply that loss of both Ctnnb1 and Sox9 has a sex chromosome-specific effect (XX/XY) on the supporting cells, similar to Wnt4/Fgf9 DKO [22] and Rspo1/Sox9 DKO [23]. To test this hypothesis, we allowed the gonads of various genotypes to develop to 17.5 dpc and investigated whether the differences between XX and XY Ctnnb1/Sox9 DKO gonads become more pronounced (Fig. 2). At 17.5 dpc, the XX DKO gonads expressed the Sertoli-cell markers AMH and DMRT1 in a few cells, similarly to XX Ctnnb1 KO (Fig. 2, B, C, E, F, and N). In contrast, the XY DKO gonads contained many more AMH+/DMRT1+ Sertoli cells that clustered and formed cordlike structures in the center of the gonads (Fig. 2, I and L). FOXL2 and DMRT1 cells in the XY DKO gonads were predominantly mutually exclusive with the exception of a few double-positive cells (inset in Fig. 2L). This observation correlated with a lower expression of the granulosa cell-specific marker Foxl2 (Fig. 2M) and a higher expression of the Sertoli cell-specific marker Amh (Fig. 2N) in the XY DKO gonads compared to the XX DKO gonads. The impact of loss of both β-catenin and Sox9 on supporting cell fate determination is clearly dependent upon the genetic context (XX vs. XY) of the gonadal environment.
FIG. 2.
Consequences of inactivation of both β-catenin and Sox9 in the gonads at 17.5 dpc. The supporting cell identity of 17.5 dpc XX (A–F) and XY (G–L) gonads of various genotypes (control, Ctnnb1 KO, Sox9 KO, and Ctnnb1/Sox9 DKO) was analyzed by immunofluorescence for Sertoli cell markers AMH (red) and DMRT1 (green) and the granulosa cell marker FOXL2 (magenta). Nuclei are labeled with 4′,6-diamidino-2-phenylindole (grey). Arrowheads in E and F show the DMRT1+ cells. Insets in G and I show testis-cord structures formed by AMH+ (red, cytoplasmic)/DMRT1+ (green, nuclear) Sertoli cells. Bars = 100 μm. M and N) Quantitative PCR analysis of Foxl2 (M) and Amh (N) mRNA expression in 17.5 dpc gonads. Error bars represent SEM of five biological replicates (Student t-test, P < 0.05; a < b < c < d).
Sex-Specific Masculinization of Gonadal Transcriptome in the Absence of β-Catenin and Sox9
To obtain a global picture of the early molecular changes that lead to the different phenotypes between Ctnnb1/Sox9 DKO XX and XY fetal gonads, we compared the transcriptomes of control, Ctnnb1 KO, Sox9 KO, and Ctnnb1/Sox9 DKO gonads of both sexes at 14.5 dpc, the onset of the sex-specific differences. Gene expression analysis was conducted using Affymetrix Mouse Genome 430 2.0 GeneChip arrays with three to four biological replicas for each genotype. The raw microarray data are available in GEO (accession no. GSE67463, http://www.ncbi.nlm.nih.gov/geo/). A full dataset containing the normalized log-2 intensity of all probes for each genotype and a graphic view of their expression are provided in Supplemental Dataset S1. Data for XX Sox9 KO and XY Ctnnb1 KO gonads are shown in Supplemental Dataset S1 but are not included in the following analysis because they do not differ from the control XX and XY, respectively.
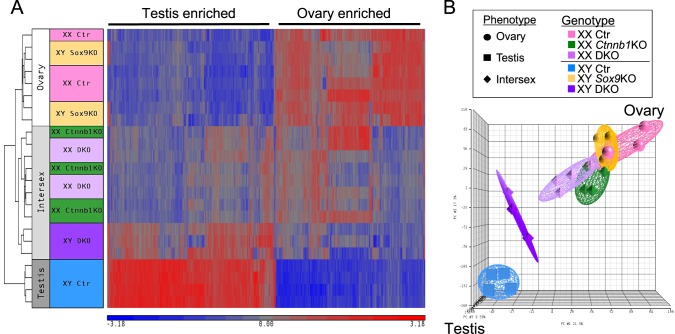
A heat map based on the expression of sexually dimorphic genes (Fig. 3A) revealed three clusters that were correlated to the morphological phenotypes (Fig. 1): ovary, intersex (defined as gonads containing features of both testis and ovary), and testis. The ovary group included XX control and XY Sox9 KO gonads, and the testis group contained only XY control gonads. The intersex group consisted of XX Ctnnb1 KO, XX DKO, and XY DKO gonads. Within the intersex group, transcriptomes of XX Ctnnb1 KO and XX DKO clustered together while XY DKO belonged to its own distinct group. PCA provided a simplified visualization of the dataset distribution (Fig. 3B). Transcriptomes of control XX gonads (in pink) and control XY gonads (in blue) clustered in opposed positions as expected. Both XX Ctnnb1 KO (in green) and XX DKO (in lilac) clusters were located close to XX control cluster, with a slight shift toward the XY control cluster. This slight shift toward a testis profile was due to the upregulation of testis specific genes such as Hsd17b3, Cbln1, Nrg1, or Ptgds, and the downregulation of ovarian genes such as Fst, Wnt4, or Bmp2 (Supplemental Fig. S3 and Supplemental Datasets S2 and S3). XX Ctnnb1 KO and XX DKO transcriptomes were not significantly different, consistent with the identical morphological phenotypes observed at 14.5 and 17.5 dpc (Figs. 1 and 2). These results further support that Sox9 is not responsible for the testis characteristics that appear in XX Ctnnb1 KO gonads, including the appearance of Sertoli-like cells.
FIG. 3.
Transcriptome analyses of 14.5 dpc control, Ctnnb1 single KO, Sox9 single KO, and Ctnnb1/Sox9 DKO XX and XY gonads. A) The heat map of the sexually dimorphic genes at 14.5 dpc (fold change ≥ 2 and P < 0.05) consists of three clusters that follow the gonadal phenotypes: ovary (XX control and XY Sox9 KO), intersex (XX Ctnnb1 KO, XX Ctnnb1/Sox9 DKO, and XY Ctnnb1/Sox9 DKO), and testis (XY control). Red indicates high expression and blue for low expression. B) Principal component analysis (PCA) shows that XX Ctnnb1 KO and XX Ctnnb1/Sox9 DKO transcriptomes show a shift toward a testis profile, whereas XY Ctnnb1/Sox9 DKO transcriptome is even more masculinized.
PCA also illustrated that while XY Sox9 KO (in yellow) transcriptome was almost identical to that of the XX control (Supplemental Dataset S4), XY DKO gonads (in purple) clearly shifted toward a testis profile. Compared to XY Sox9 KO gonads, XY DKO gonads had 1783 genes differentially expressed, with higher expression of testis-enriched genes such as Cyp11a1, Cbln1, or Ptgds and lower expression of ovary-enriched genes such as Fst, Wnt4, or Irx3 (Supplemental Dataset S5 and Supplemental Fig. S3). However, XY DKO remained significantly different from the XY control, with 1745 genes differentially expressed (Supplemental Dataset S6).
Identification of Candidate Genes Underlying the Y Chromosome-Specific Masculinization of β-Catenin/Sox9 DKO gonads
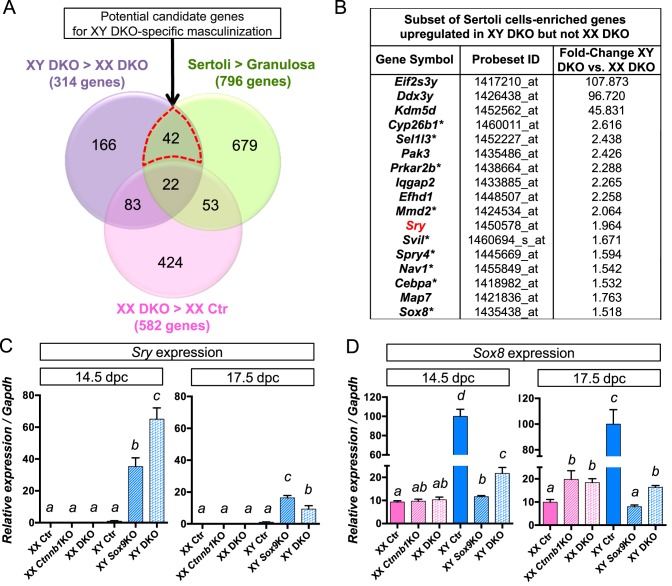
While only little morphological differences were observed between XX and XY Ctnnb1/Sox9 DKO gonads at 14.5 dpc (Fig. 1), XY DKO transcriptome was clearly more masculinized than XX DKO gonads (Fig. 3; Supplemental Dataset S7). To determine what genes are potentially involved in these sex-specific differences, we performed extensive transcriptome comparisons. We first identified the genes expressed higher in XY DKO than in XX DKO (314 genes in the purple circle, Fig. 4A; fold-change ≥ 1.5, P < 0.05). Next, in order to focus on the molecular changes that occur in the supporting cells, we took advantage of a cell-type specific microarray performed in fetal gonads [25] to create a list of 796 genes representatives of fetal Sertoli cell identity (green circle, Fig. 4A; see Materials and Methods). Comparison of our data with this gene list led to the identification of 64 genes (42 + 22 in Fig. 4A) specific for Sertoli cells. Interestingly, out of these 64 Sertoli-specific genes that are more highly expressed in XY DKO gonads than their XX counterpart, 21 genes are potential direct targets of SRY according to SRY chromatin immunoprecipitation (ChIP) sequencing data [27, 28], such as Sox8, Cyp26b1, Dhh, or Cbln4 (Supplemental Dataset S8 and Supplemental Fig. S3). We then hypothesized that the genes responsible for the more pronounced masculinization in XY DKO gonads must be upregulated only in XY DKO and not in XX DKO (pink circle in Fig. 4A). This analysis led to the identification of 42 Sertoli-cell genes that are specifically upregulated in XY DKO (the red dotted area in Fig. 4A and Supplemental Dataset S8). Several Y-linked genes were present at the top of the list, such as Eif2s3y, Ddx3y, or Kdm5d (Fig. 4B), but these genes are not expected to play a role in the early differentiation of Sertoli cells [25, 29].
FIG. 4.
Identification of the potential mechanisms underlying the differences between XX and XY Ctnnb1/Sox9 DKO gonads. A) Venn diagram comparing three groups of genes: 1) purple circle: genes upregulated in XY Ctnnb1/Sox9 DKO versus XX Ctnnb1/Sox9 DKO gonads, 2) green circle: genes enriched in fetal Sertoli cells versus granulosa cells from a published dataset [25], and 3) pink circle: genes upregulated in XX Ctnnb1/Sox9 DKO versus XX control gonads. The red dotted area represents the 42 genes upregulated in XY but not XX Ctnnb1/Sox9 DKO gonads. A subset of genes from this 42 genes list is shown in B; asterisk (*) represents putative SRY target genes. C and D) Quantitative PCR analysis for Sry and Sox8 at 14.5 and 17.5 dpc. Error bars represent SEM of five biological replicates (Student t-test, P < 0.05; a < b < c < d).
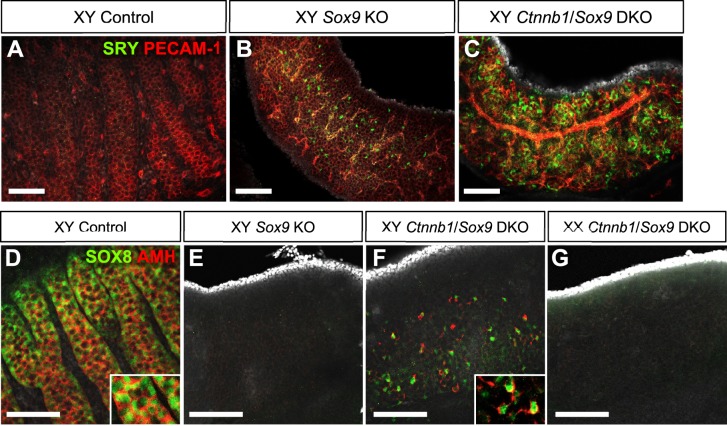
Among the genes that were only upregulated in XY Ctnnb1/Sox9 DKO at 14.5 dpc, Sry and Sox8 were the most promising candidates for the sex-specific difference (Fig. 4B and Supplemental Dataset S8) due to the fact that these two genes are known to be involved in acquisition of Sertoli cell identity [2–4, 9, 30]. In the control XY gonads, Sry expression was not detected at 14.5 and 17.5 dpc as described previously [5, 31]. However in the XY DKO gonads, Sry expression was maintained at 14.5 dpc, and still detected at low levels at 17.5 dpc (Figs. 4C and 5, A–C and Supplemental Dataset S6). Expression of Sry mRNA and protein was also detected in XY Sox9 KO gonads similar to other observations [9, 10], albeit at a level much lower than in the XY DKO gonads (Figs. 4C and 5B). Sox8, a close homologue of Sox9 and target of SRY, was also upregulated in XY DKO gonads at 14.5 dpc (Figs. 4D and 5, D–G). In 14.5 dpc XY DKO gonads, most of the SOX8-positive cells start to express AMH, a potential direct target gene of SOX8 [32] (Fig. 5F).
FIG. 5.
Immunostaining for SRY and SOX8 in control and different KO gonads at 14.5 dpc. A–C) Whole mount immunofluorescence for SRY and PECAM-1 showed the presence of SRY in XY Sox9 KO (B) and XY Ctnnb1/Sox9 DKO (C) gonads. D–G) Whole mount immunofluorescence for SOX8 and AMH at 14.5 dpc showed the presence of SOX8+ cells in XY Ctnnb1/Sox9 DKO (F), and most of these cells coexpressed AMH (inset). SOX8 was absent in 14.5 dpc XY Sox9 KO (E) and XX Ctnnb1/Sox9 DKO (G) gonads. Nuclei are labeled with 4′,6-diamidino-2-phenylindole (grey). Bars = 100 μm.
In addition to Sox8, the list of 42 Sertoli-specific genes that were upregulated only in XY DKO gonads contained nine other potential SRY target genes, such as Cyp26b1 [27] (Fig. 4B). Cyp26b1 is involved in the male germ cells fate by repressing germ cells entry into meiosis during fetal life [33]. Interestingly, this expression of Cyp26b1 in XY DKO was correlated with a reduced expression of meiosis markers compared to XX DKO (Supplemental Fig. S3 and Supplemental Dataset S1). Finally, this list also contained Map7, a gene whose function in the testis is unknown but its mutation (Map7mshi) leads to reduced testis size and sterility [34]. Most of the other genes in this list are of unknown functions in testis differentiation.
Our analysis of the early events of the sex-chromosome specific differences in the masculinization of Ctnnb1/Sox9 DKO gonads demonstrate that XY DKO gonads develop much more pronounced testicular characteristics than the XX DKO gonads, potentially due to the expression of Sry and the activation of several of its target genes recently identified by ChiP-sequencing [27], including Sox8 and Cyp26b1.
Identification of Candidate Genes Commonly Involved in the Masculinization of Ctnnb1/Sox9 DKO XX and XY Gonads
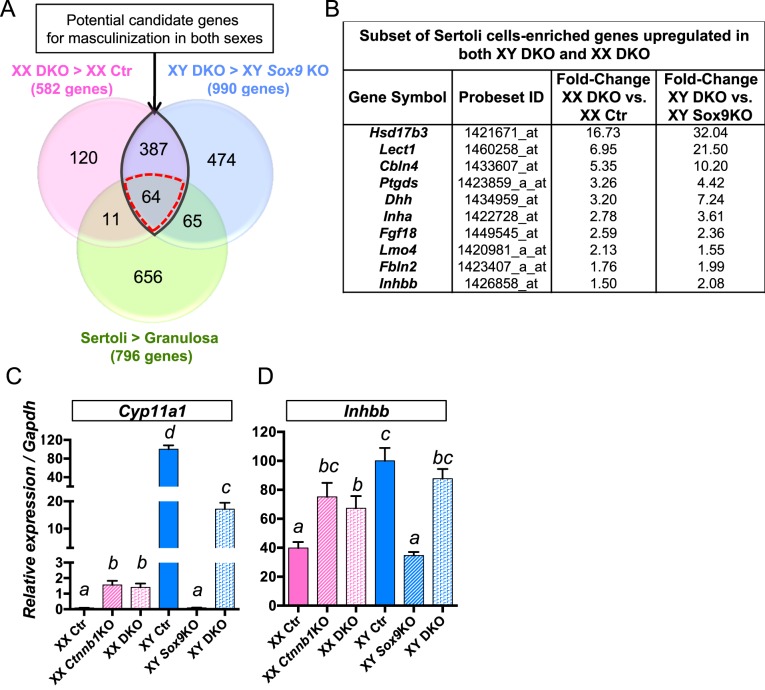
Despite the sex chromosome-specific differences in phenotypes and transcriptomes between XX and XY DKO gonads, gonads of these two genotypes share similar features such as the appearance of a testis-specific coelomic vessel and steroidogenic cells. We suspected that some pro-testis genes commonly responsible for these phenotypes were activated in Ctnnb1/Sox9 DKO gonads regardless of their genetic contexts (XX vs. XY). To identify these common pro-testis genes, we first identified the genes upregulated in XX DKO by comparing its transcriptome to XX control transcriptome (582 genes with a fold-change ≥ 1.5, P < 0.05; pink circle in Fig. 6A and Supplemental Dataset S3). Second, we identified the genes upregulated in XY DKO in comparison to XY Sox9 KO gonads (990 genes with a fold-change ≥ 1.5, P < 0.05; blue circle in Fig. 6A and Supplemental Dataset S5). We cross-referenced these two lists of genes and identified 451 (387 + 64; Fig. 6A, purple shaded area with black outline) genes that commonly associate with the masculinization of both XX and XY DKO gonads. Finally, we narrowed down our search to the genes known to be expressed in Sertoli cells by comparing our data with the list of genes representative of fetal Sertoli cell identity based on Jameson et al. microarray data [25] (Fig. 6A, green circle, see Materials and Methods). This comparison removed 387 genes that were not Sertoli cell specific, such as genes involved in steroidogenesis, including Cyp11a1, Hsd17b, or Cyp17a1 (Fig. 6C and Supplemental Dataset S9). This series of comparisons led to the identification of 64 Sertoli cell-specific genes that were potential components of the common pathway responsible for the masculinization of both XX and XY Ctnnb1/Sox9 DKO gonads (Fig. 6 and Supplemental Dataset S9). Desert hedgehog or Dhh, a gene expressed in Sertoli cells and involved in specification of the Leydig cells lineage [35], was upregulated in both XX and XY DKO gonads (Fig. 6B). In addition, several genes involved in the acquisition of Sertoli cell fate were upregulated in both XX and XY DKO gonads. These genes included Ptgds, a paracrine signaling factor involved in the recruitment of somatic cells into the Sertoli fate [36, 37], and the transcription cofactor Lmo4, a novel regulator of early testis differentiation [38] (Fig. 6B and Supplemental Dataset S9). Inhibin beta B or Inhbb, a gene involved in the testis-specific vasculature [39], was also a part of this candidate list (Fig. 6D). This list also contains many genes whose function during gonads differentiation remains undetermined (Supplemental Dataset S9).
FIG. 6.
Identification of candidate genes commonly involved in the masculinization of both XX and XY Ctnnb1/Sox9 DKO gonads. A) Venn diagram comparing three groups of genes: 1) pink circle: genes upregulated in XX Ctnnb1/Sox9 DKO versus XX control gonads, 2) blue circle: genes upregulated in XY Ctnnb1/Sox9 DKO versus XY Sox9 KO gonads, and 3) green circle: genes enriched in fetal Sertoli cells versus granulosa cells from a published dataset [25]. A subset of these Sertoli-specific genes is shown in B. Quantitative PCR analyses of Cyp11a1 (C) and Inhbb (D), two genes that were upregulated in both XX and XY Ctnnb1/Sox9 DKO gonads in non-Sertoli cells (387 genes in the purple area) and Sertoli cells (64 genes in the red-dotted area), respectively. Error bars represent SEM of five biological replicates (Student t-test, P < 0.05; a < b < c < d).
DISCUSSION
The fate determination of the fetal gonads hinges upon the balance between pro-testis SOX9 and pro-ovary β-catenin pathways. By analyzing the gonadal identity of Sox9 and β-catenin DKO embryos, we discovered that in the absence of Sox9 and β-catenin in the somatic cells that 1) both XX and XY gonads progressively lean toward a male fate, with appearance of testis-specific vasculature, steroidogenic cells, and Sertoli cells and 2) XY gonads become more masculinized than their XX counterparts. This Y-chromosome specific masculinization is associated with the expression of Sertoli-specific genes such as Sry and several putative SRY target genes.
β-Catenin in the XX Gonad Promotes Ovarian Fate by Suppressing Pro-Testis Genes Other than Sox9
Based on the observation that WNT/β-catenin pathway represses the key effector of testis differentiation Sox9 in gonads and other tissues [21, 40, 41], it was therefore hypothesized that when β-catenin is absent in the ovary, Sox9 is upregulated and responsible for the partial ovary-to-testis sex-reversal. However, we found that loss of Sox9 does not rescue any aspects of the masculinization phenotypes in β-catenin KO XX gonads. Similarly, loss of Sox9 did not restore the partial ovary-to-testis sex reversal of XX Rspo1 KO gonads [23]. Lavery et al. [23] characterized the postnatal phenotypes of Rspo1/Sox9 DKO gonads and found that newborn Rspo1 KO and Rspo1/Sox9 DKO ovaries developed phenotypes similar to our observations at 17.5 dpc, characterized by the presence of a few Sertoli-like cells. The analyses of Rspo1/Sox9 DKO ovaries mainly focused on postnatal stages, and the early molecular changes that contribute to their masculinization were not known. Regardless of the presence or absence of a functional Sox9 gene, both Ctnnb1 KO and Rspo1 KO ovaries underwent a shift toward the testis fate. These observations imply that pro-testis genes, other than Sox9, induce the appearance of Sertoli-like cells and the other testis characteristics observed in the absence β-catenin or Rspo1. We identified a list of candidate genes associated with the fetal appearance of testis features in the Ctnnb1 KO ovary independent of Sox9, including Inhbb, a Sertoli-specific gene responsible for the formation of the coelomic vessel [39]. We speculate that in the wild-type fetal ovary, Inhbb expression and formation of testis-specific coelomic vessel are absent as a result of repression by the β-catenin pathway, rather than a lack of direct activation by SOX9. Other Sertoli cell-enriched genes induced in fetal Ctnnb1/Sox9 DKO ovaries, including Ptgds [36], Lmo4 [38], Dhh [42], Cbln4 [28], and later, close to birth, Sox8 [9] and Amh [32], might be responsible for the differentiation of supporting cells into Sertoli-like cells. Interestingly, Ptgds, Amh, and Cbln4 are known to be direct target genes of SOX9 [28, 43, 44]; however, our results indicate that these genes can be induced in a Sox9-independent manner. We conclude that in the supporting cells of the ovary, β-catenin suppresses the testis fate by inhibiting not only Sox9, but also other Sox9-independent pro-testis genes (Fig. 7). It remains unclear whether β-catenin acts directly to repress these pro-testis genes, or if other pro-ovarian genes such as Foxl2 are involved in this process. The antagonism between testis and ovary pathways is clearly more complex than previously thought.
FIG. 7.
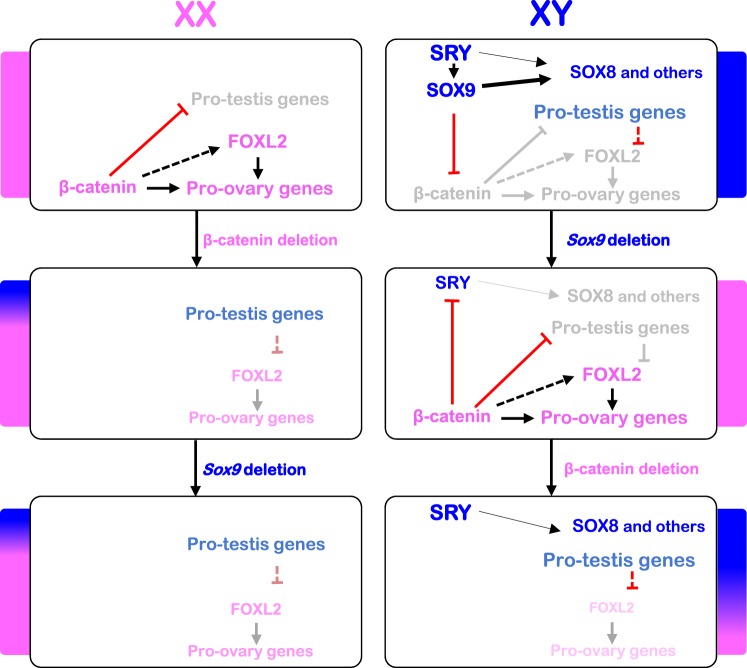
Model for the molecular pathways controlling supporting cell fate in the gonads. In the XX gonad, β-catenin is involved in expression of pro-ovary genes, including Foxl2, and blocks Sertoli cell fate through repression of some pro-testis genes. In the absence of β-catenin, some pro-testis genes such as Inhbb, Dhh, and Ptgds are activated, and expression of pro-ovarian genes is reduced, leading to a partial ovary-to-testis sex-reversal. Additional deletion of Sox9 results to an identical phenotype, indicating that Sox9 is not required for the activation of the pro-testis genes in the absence of β-catenin. In the XY gonad, Sox9 expression is induced by SRY to drive Sertoli cell differentiation. SOX9 activates the expression of pro-testis genes directly and indirectly through repression of β-catenin. SRY may also be involved in the expression of pro-testis genes other than Sox9, such as Sox8. When Sox9 is deleted in the supporting cell lineages, β-catenin becomes activated, subsequently leading to repression of the pro-testis genes, the expression of pro-ovary genes, and the acquisition of granulosa cell identity. In the absence of both Sox9 and β-catenin, SRY activates pro-testis genes other than Sox9 (such as Sox8) to drive testis differentiation.
Testis Differentiation in XY Embryos Requires Sox9 in the Supporting Cell Lineage in Order to Repress the Pro-Ovary β-Catenin Pathway
Absence of β-catenin partially reverses the complete testis-to-ovary sex-reversal phenotype observed in XY Sox9 KO gonads. In XY gonads lacking both Sox9 and β-catenin, some supporting cells transdifferentiate from FOXL2-positive granulosa cells to a DMRT1-positive Sertoli cell identity, resulting in the formation of ovotestes. A similar restoration of testis differentiation was observed by other groups in Wnt4/Fgf9 DKO [22] and Rspo1/Sox9 DKO models [23]. Our findings along with these observations suggest that during normal testis differentiation, SOX9 promotes testis differentiation directly, via the transcriptional activation of pro-testis genes, and indirectly, via repression of the β-catenin pathway (Fig. 7). The repression of β-catenin by SOX9 is necessary for not only preventing the expression pro-ovary genes downstream of β-catenin, but also promoting the expression of pro-testis genes that are negatively regulated by β-catenin.
A Potential Involvement of SRY in the Y Chromosome-Specific Masculinization of the Gonads in the Absence of SOX9 and β-Catenin
We observed a Y-chromosome dependent appearance of Sertoli cells in Ctnnb1/Sox9 DKO gonads. Among the Y-linked genes, Sry is an obvious candidate because of its role in directing supporting cell progenitors toward the male fate [45]. SRY ChIP analyses in normal fetal testes have revealed that Sox9 is not the only target of SRY [7, 27, 28, 46]. Interestingly, we found that among the Sertoli-specific genes upregulated in XY DKO gonads, 21 genes were recently identified as potential SRY target genes [27]. We hypothesize that SRY is able to activate these genes and therefore drive the Y-chromosome dependent masculinization in the absence of SOX9 and β-catenin. However, Sry expression alone cannot explain the activation of these genes and consequent appearance of Sertoli cells in XY Ctnnb1/Sox9 DKO gonads. Indeed, Sry is also expressed in XY Sox9 KO gonads, in which a complete testis-to-ovary sex reversal still occurs [9, 10]. In vitro studies have shown that SRY requires the cooperation of cofactors to activate its target genes and fulfill its testis-inducing functions [7, 27]. SRY acts in synergy with SF1 to activate many target genes, including Sox8 and Mmd2 [27]. Interestingly, β-catenin is able to prevent SF1 binding to the SRY target promoter [7, 40]. In XY control and XY Ctnnb1/Sox9 DKO, the conditions are met to allow the expression of SRY target genes: SRY is expressed and SF1 cannot be repressed by β-catenin. On the other hand in XY Sox9 KO, SRY is expressed, but the presence of β-catenin may prevent SF1 binding, consequently blocking the expression of SRY target genes. We propose that SRY, in association with SF1 activates pro-testis genes other than Sox9 to initiate the appearance of Sertoli cells in the XY Ctnnb1/Sox9 DKO gonads (Fig. 7).
A certain degree of redundancy appears to exist between SOX transcription factors. For instance, SRY and SOX9 are able to activate common target genes such as Sox9, Sox8, or Cyp26b1 [7, 27]. This redundancy is probably due to the conserved DNA-binding high-mobility group box among members of the SOX family. Lavery et al. [23] showed that Sox8 and Sox10 were expressed in Rspo1KO Sox9KO mutants at Postnatal Day 12 and 0, respectively. In our XY Ctnnb1/Sox9 DKO model, in addition to Sry, Sox8 is the only Sox gene expressed during the initial events of masculinization. While Sox8 inactivation does not affect testis determination [47], analyses of Sox8/Sox9 double mutants showed that SOX8 is able to compensate for the loss of Sox9 after sex determination [9, 30, 48]. Moreover, SOX8 and SOX9 activate common target genes such as Amh and Cbln4 [32, 48]. Therefore, we propose that SRY and its target gene SOX8 partially compensate for the absence of SOX9 in XY Ctnnb1/Sox9 DKO supporting cells and promote Sertoli cell differentiation (Fig. 7).
Altogether, our findings shed light onto the complex antagonisms between pro-ovary and pro-testis pathways. Testis differentiation, which was thought to be the result of a direct action of SOX9, requires both SOX9-dependent and -independent morphogenetic changes along with the repression of the β-catenin pathway. Moreover, the Y-chromosome dependent masculinization in the absence of SOX9 and β-catenin indicates that SRY, along with its downstream target genes, is capable of guiding Sertoli cell differentiation in a SOX9-independent manner. In humans, the majority of the congenital disorders of sex development are of unknown genetic origin [49]. The microarray analyses we provide in this mouse sex-reversal study are useful tools to identify new genes potentially associated with disorders of sex development. Many of the genes associated with testis differentiation in our mouse models, such as Lmo4, Cbln4, or Nrg1, and several potential SRY targets genes have an unknown function in the fetal gonads. It would be interesting to determine the role of these genes in testis differentiation and investigate if these genes are associated with disorders of sex development in humans.
ACKNOWLEDGMENT
We thank Keith Parker (UT Southwestern Medical Center) for the Sf1-Cre mice, Dagmar Wilhelm (Monash University) for FOXL2 and SRY antibodies, David Zarkower (University of Minnesota) for DMRT1 antibody, and M. Wegner (Universität Erlangen) for SOX8 antibody. We thank the NIEHS Molecular Genomics Core and Comparative Medicine Branch for their technical support, Karina Rodriguez for the initial establishment of the mice lines, and the Yao laboratory members for their critical comments.
Footnotes
REFERENCES
- Svingen T, Koopman P. Building the mammalian testis: origins, differentiation, and assembly of the component cell populations. Genes Dev. 2013;27:2409–2426. doi: 10.1101/gad.228080.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Munsterberg A, Vivian N, Goodfellow P, Lovellbadge RA. Gene-mapping to the sex-determining region of the mouse Y-chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990;346:245–250. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. Male development of chromosomally female mice transgenic for Sry. Nature. 1991;351:117–121. doi: 10.1038/351117a0. [DOI] [PubMed] [Google Scholar]
- Hacker A, Capel B, Goodfellow P, Lovell-Badge R. Expression of Sry, the mouse sex determining gene. Development. 1995;121:1603–1614. doi: 10.1242/dev.121.6.1603. [DOI] [PubMed] [Google Scholar]
- Hiramatsu R, Matoba S, Kanai-Azuma M, Tsunekawa N, Katoh-Fukui Y, Kurohmaru M, Morohashi K, Wilhelm D, Koopman P, Kanai Y. A critical time window of Sry action in gonadal sex determination in mice. Development. 2009;136:129–138. doi: 10.1242/dev.029587. [DOI] [PubMed] [Google Scholar]
- Sekido R, Lovell-Badge R. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature. 2008;453:930–934. doi: 10.1038/nature06944. [DOI] [PubMed] [Google Scholar]
- Foster JW, Dominguez-Steglich MA, Guioli S, Kwok C, Weller PA, Stevanovic M, Weissenbach J, Mansour S, Young ID, Goodfellow PN, Brook JD, Schafer AJ. Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature. 1994;372:525–530. doi: 10.1038/372525a0. [DOI] [PubMed] [Google Scholar]
- Chaboissier MC, Kobayashi A, Vidal VI, Lutzkendorf S, van de Kant HJ, Wegner M, de Rooij DG, Behringer RR, Schedl A. Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development. 2004;131:1891–1901. doi: 10.1242/dev.01087. [DOI] [PubMed] [Google Scholar]
- Barrionuevo F, Bagheri-Fam S, Klattig J, Kist R, Taketo MM, Englert C, Scherer G. Homozygous inactivation of Sox9 causes complete XY sex reversal in mice. Biol Reprod. 2006;74:195–201. doi: 10.1095/biolreprod.105.045930. [DOI] [PubMed] [Google Scholar]
- Huang B, Wang S, Ning Y, Lamb AN, Bartley J. Autosomal XX sex reversal caused by duplication of SOX9. Am J Med Genet. 1999;87:349–353. doi: 10.1002/(sici)1096-8628(19991203)87:4<349::aid-ajmg13>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Vidal VP, Chaboissier MC, de Rooij DG, Schedl A. Sox9 induces testis development in XX transgenic mice. Nat Genet. 2001;28:216–217. doi: 10.1038/90046. [DOI] [PubMed] [Google Scholar]
- Nicol B, Yao HH. Building an ovary: insights into establishment of somatic cell lineages in the mouse. Sex Dev. 2014;8:243–251. doi: 10.1159/000358072. [DOI] [PubMed] [Google Scholar]
- Vainio S, Heikkila M, Kispert A, Chin N, McMahon AP. Female development in mammals is regulated by Wnt-4 signalling. Nature. 1999;397:405–409. doi: 10.1038/17068. [DOI] [PubMed] [Google Scholar]
- Jeays-Ward K, Hoyle C, Brennan J, Dandonneau M, Alldus G, Capel B, Swain A. Endothelial and steroidogenic cell migration are regulated by WNT4 in the developing mammalian gonad. Development. 2003;130:3663–3670. doi: 10.1242/dev.00591. [DOI] [PubMed] [Google Scholar]
- Chassot AA, Ranc F, Gregoire EP, Roepers-Gajadien HL, Taketo MM, Camerino G, de Rooij DG, Schedl A, Chaboissier MC. Activation of beta-catenin signaling by Rspo1 controls differentiation of the mammalian ovary. Hum Mol Genet. 2008;17:1264–1277. doi: 10.1093/hmg/ddn016. [DOI] [PubMed] [Google Scholar]
- Tomizuka K, Horikoshi K, Kitada R, Sugawara Y, Iba Y, Kojima A, Yoshitome A, Yamawaki K, Amagai M, Inoue A, Oshima T, Kakitani M. R-spondin1 plays an essential role in ovarian development through positively regulating Wnt-4 signaling. Hum Mol Genet. 2008;17:1278–1291. doi: 10.1093/hmg/ddn036. [DOI] [PubMed] [Google Scholar]
- Manuylov NL, Smagulova FO, Leach L, Tevosian SG. Ovarian development in mice requires the GATA4-FOG2 transcription complex. Development. 2008;135:3731–3743. doi: 10.1242/dev.024653. [DOI] [PubMed] [Google Scholar]
- Liu CF, Bingham N, Parker K, Yao HH. Sex-specific roles of beta-catenin in mouse gonadal development. Hum Mol Genet. 2009;18:405–417. doi: 10.1093/hmg/ddn362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maatouk DM, Mork L, Chassot AA, Chaboissier MC, Capel B. Disruption of mitotic arrest precedes precocious differentiation and transdifferentiation of pregranulosa cells in the perinatal Wnt4 mutant ovary. Dev Biol. 2013;383:295–306. doi: 10.1016/j.ydbio.2013.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maatouk DM, DiNapoli L, Alvers A, Parker KL, Taketo MM, Capel B. Stabilization of beta-catenin in XY gonads causes male-to-female sex-reversal. Hum Mol Genet. 2008;17:2949–2955. doi: 10.1093/hmg/ddn193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson SA, Lin Y-T, Capel B. Testis development requires the repression of Wnt4 by Fgf signaling. Dev Biol. 2012;370:24–32. doi: 10.1016/j.ydbio.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavery R, Chassot AA, Pauper E, Gregoire EP, Klopfenstein M, de Rooij DG, Mark M, Schedl A, Ghyselinck NB, Chaboissier MC. Testicular differentiation occurs in absence of R-spondin1 and Sox9 in mouse sex reversals. PLoS Genet. 2012;8:e1003170. doi: 10.1371/journal.pgen.1003170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham NC, Verma-Kurvari S, Parada LF, Parker KL. Development of a steroidogenic factor 1/Cre transgenic mouse line. Genesis. 2006;44:419–424. doi: 10.1002/dvg.20231. [DOI] [PubMed] [Google Scholar]
- Jameson SA, Natarajan A, Cool J, DeFalco T, Maatouk DM, Mork L, Munger SC, Capel B. Temporal transcriptional profiling of somatic and germ cells reveals biased lineage priming of sexual fate in the fetal mouse gonad. PLoS Genet. 2012;8:e1002575. doi: 10.1371/journal.pgen.1002575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavery R, Lardenois A, Ranc-Jianmotamedi F, Pauper E, Gregoire EP, Vigier C, Moreilhon C, Primig M, Chaboissier MCXY. Sox9 embryonic loss-of-function mouse mutants show complete sex reversal and produce partially fertile XY oocytes. Dev Biol. 2011;354:111–122. doi: 10.1016/j.ydbio.2011.03.029. [DOI] [PubMed] [Google Scholar]
- Li Y, Zheng M, Lau YF. The sex-determining factors SRY and SOX9 regulate similar target genes and promote testis cord formation during testicular differentiation. Cell Rep. 2014;8:723–733. doi: 10.1016/j.celrep.2014.06.055. [DOI] [PubMed] [Google Scholar]
- Bradford ST, Hiramatsu R, Maddugoda MP, Bernard P, Chaboissier MC, Sinclair A, Schedl A, Harley V, Kanai Y, Koopman P, Wilhelm D. The cerebellin 4 precursor gene is a direct target of SRY and SOX9 in mice. Biol Reprod. 2009;80:1178–1188. doi: 10.1095/biolreprod.108.071480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Riel JM, Stoytcheva Z, Ward MA. Two Y genes can replace the entire Y chromosome for assisted reproduction in the mouse. Science. 2014;343:69–72. doi: 10.1126/science.1242544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrionuevo F, Georg I, Scherthan H, Lecureuil C, Guillou F, Wegner M, Scherer G. Testis cord differentiation after the sex determination stage is independent of Sox9 but fails in the combined absence of Sox9 and Sox8. Dev Biol. 2009;327:301–312. doi: 10.1016/j.ydbio.2008.12.011. [DOI] [PubMed] [Google Scholar]
- Bullejos M, Koopman P. Delayed Sry and Sox9 expression in developing mouse gonads underlies B6-Y(DOM) sex reversal. Dev Biol. 2005;278:473–481. doi: 10.1016/j.ydbio.2004.11.030. [DOI] [PubMed] [Google Scholar]
- Schepers G, Wilson M, Wilhelm D, Koopman P. SOX8 is expressed during testis differentiation in mice and synergizes with SF1 to activate the Amh promoter in vitro. J Biol Chem. 2003;278:28101–28108. doi: 10.1074/jbc.M304067200. [DOI] [PubMed] [Google Scholar]
- Bowles J, Knight D, Smith C, Wilhelm D, Richman J, Mamiya S, Yashiro K, Chawengsaksophak K, Wilson MJ, Rossant J, Hamada H, Koopman P. Retinoid signaling determines germ cell fate in mice. Science. 2006;312:596–600. doi: 10.1126/science.1125691. [DOI] [PubMed] [Google Scholar]
- Magnan DR, Spacek DV, Ye N, Lu YC, King TR. The male sterility and histoincompatibility (mshi) mutation in mice is a natural variant of microtubule-associated protein 7 (Mtap7) Mol Genet Metab. 2009;97:155–162. doi: 10.1016/j.ymgme.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao HHC, Whoriskey W, Capel B. Desert Hedgehog/Patched 1 signaling specifies fetal Leydig cell fate in testis organogenesis. Genes Dev. 2002;16:1433–1440. doi: 10.1101/gad.981202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm D, Martinson F, Bradford S, Wilson MJ, Combes AN, Beverdam A, Bowles J, Mizusaki H, Koopman P. Sertoli cell differentiation is induced both cell-autonomously and through prostaglandin signaling during mammalian sex determination. Dev Biol. 2005;287:111–124. doi: 10.1016/j.ydbio.2005.08.039. [DOI] [PubMed] [Google Scholar]
- Moniot B, Declosmenil F, Barrionuevo F, Scherer G, Aritake K, Malki S, Marzi L, Cohen-Solal A, Georg I, Klattig J, Englert C, Kim Y, et al. The PGD2 pathway, independently of FGF9, amplifies SOX9 activity in Sertoli cells during male sexual differentiation. Development. 2009;136:1813–1821. doi: 10.1242/dev.032631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger SC, Natarajan A, Looger LL, Ohler U, Capel B. Fine time course expression analysis identifies cascades of activation and repression and maps a putative regulator of mammalian sex determination. PLoS Genet. 2013;9:e1003630. doi: 10.1371/journal.pgen.1003630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao HH, Aardema J, Holthusen K. Sexually dimorphic regulation of inhibin beta B in establishing gonadal vasculature in mice. Biol Reprod. 2006;74:978–983. doi: 10.1095/biolreprod.105.050286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Ryan J, Sim H, Czech DP, Sinclair AH, Koopman P, Harley VR. Wnt signaling in ovarian development inhibits Sf1 activation of Sox9 via the Tesco enhancer. Endocrinology. 2012;153:901–912. doi: 10.1210/en.2011-1347. [DOI] [PubMed] [Google Scholar]
- Kormish JD, Sinner D, Zorn AM. Interactions between SOX factors and Wnt/beta-catenin signaling in development and disease. Dev Dyn. 2010;239:56–68. doi: 10.1002/dvdy.22046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierucci-Alves F, Clark AM, Russell LD. A developmental study of the Desert hedgehog-null mouse testis. Biol Reprod. 2001;65:1392–1402. doi: 10.1095/biolreprod65.5.1392. [DOI] [PubMed] [Google Scholar]
- Wilhelm D, Hiramatsu R, Mizusaki H, Widjaja L, Combes AN, Kanai Y, Koopman P. SOX9 regulates prostaglandin D synthase gene transcription in vivo to ensure testis development. J Biol Chem. 2007;282:10553–10560. doi: 10.1074/jbc.M609578200. [DOI] [PubMed] [Google Scholar]
- De Santa Barbara P, Bonneaud N, Boizet B, Desclozeaux M, Moniot B, Sudbeck P, Scherer G, Poulat F, Berta P. Direct interaction of SRY-related protein SOX9 and steroidogenic factor 1 regulates transcription of the human anti-Mullerian hormone gene. Mol Cell Biol. 1998;18:6653–6665. doi: 10.1128/mcb.18.11.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekido R, Lovell-Badge R. Genetic control of testis development. Sex Dev. 2013;7:21–32. doi: 10.1159/000342221. [DOI] [PubMed] [Google Scholar]
- Bhandari RK, Haque MM, Skinner MK. Global genome analysis of the downstream binding targets of testis determining factor SRY and SOX9. PLoS One. 2012;7:e43380. doi: 10.1371/journal.pone.0043380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sock E, Schmidt K, Hermanns-Borgmeyer I, Bosl MR, Wegner M. Idiopathic weight reduction in mice deficient in the high-mobility-group transcription factor Sox8. Mol Cell Biol. 2001;21:6951–6959. doi: 10.1128/MCB.21.20.6951-6959.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmel F, Lardenois A, Georg I, Barrionuevo F, Demougin P, Jegou B, Scherer G, Primig M. Genome-wide identification of Sox8-, and Sox9-dependent genes during early post-natal testis development in the mouse. Andrology. 2013;1:281–292. doi: 10.1111/j.2047-2927.2012.00049.x. [DOI] [PubMed] [Google Scholar]
- Ohnesorg T, Vilain E, Sinclair AH. The genetics of disorders of sex development in humans. Sex Dev. 2014;8:262–272. doi: 10.1159/000357956. [DOI] [PubMed] [Google Scholar]