Abstract
The reproductive homeobox X-linked, Rhox, genes encode transcription factors that are selectively expressed in reproductive tissues. While there are 33 Rhox genes in mice, only Rhox and Rhox8 are expressed in Sertoli cells, suggesting that they may regulate the expression of somatic-cell gene products crucial for germ cell development. We previously characterized Rhox5-null mice, which are subfertile, exhibiting excessive germ cell apoptosis and compromised sperm motility. To assess the role of Rhox8 in Sertoli cells, we used a tissue-specific RNAi approach to knockdown RHOX8 in vivo, in which the Rhox5 promoter was used to drive Rhox8-siRNA transgene expression in the postnatal Sertoli cells. Western and immunohistochemical analysis confirmed Sertoli-specific knockdown of RHOX8. However, other Sertoli markers, Gata1 and Rhox5, maintained normal expression patterns, suggesting that the knockdown was specific. Interestingly, male RHOX8-knockdown animals showed significantly reduced spermatogenic output, increased germ cell apoptosis, and compromised sperm motility, leading to impaired fertility. Importantly, our results revealed that while some RHOX5-dependent factors were also misregulated in Sertoli cells of RHOX8-knockdown animals, the majority were not, and novel putative RHOX8-regulated genes were identified. This suggests that while reduction in levels of RHOX5 and RHOX8 in Sertoli cells elicits similar phenotypes, these genes are not entirely redundant. Taken together, our study underscores the importance of Rhox genes in male fertility and suggests that Sertoli cell-specific expression of Rhox5 and Rhox8 is critical for complete male fertility.
Keywords: gene regulation, rodents (rats, mice, guinea pigs, voles), sertoli cells, spermatogenesis, testis
INTRODUCTION
Homeobox genes encode transcription factors, which govern diverse embryonic developmental processes, including body-axis formation, organogenesis, and limb development [1, 2]. However, our knowledge of their postnatal roles governing cellular proliferation, differentiation, and survival in both normal tissues and cancers is still growing [3–5]. Rodent genomes contain ∼200 homeobox genes, 25% of which are known to be expressed in the testis [3, 5]. However, a clear function for the majority has yet to be identified, as their ablation often results in malformation of an essential organ, leading to pre- or perinatal lethality, precluding the examination of their role in fertility [3]. The formation of the male urogenital tract is regulated by some Hox subclass genes [6–9]. Non-Hox homeobox subfamily members, Emx2, Lhx9, Arx, and Pax2, also have a major role in dictating male reproductive tract development as well as the establishment of Leydig cells and primordial germ cells populations [2, 8, 10]. In contrast, little is known about the role of homeobox genes in reproductive events, including spermatogenesis. While more than 40 homeobox genes are expressed in postnatal and adult testes, only a few have published links to defects in male reproduction. For example, ablation of the germ cell homeobox gene Sprm1 results in subfertility of unknown origin [11]; disruption of Vax1 results in male subfertility, but the contribution of testis-expressed VAX1 is less clear, as the HPG axis is disrupted in these knockouts [12]; and loss of ESX1, which arose from the same precursor as the Rhox genes [3, 5, 13], is highly correlated with azoospermia [14], but the etiology is unknown.
The reproductive homeobox X-linked (Rhox) genes are good candidates to regulate both male and female reproductive tissue development and physiology, as they are selectively expressed in the gonads, epididymis, and placenta [3]. In the testis, the expression, regulation, and function of the founding member of the cluster, Rhox5, is the most complete. Rhox5 is androgen regulated and localized to Sertoli cells, and its ablation results in male subfertility due to increased germ cell apoptosis [13]. We recently identified Ins2 as a direct target of RHOX5, and its misregulation, in addition to other metabolism-supporting factors, is thought to underlie the phenotype of Rhox5-null males [15]. Analyses of the spatial and temporal mRNA expression patterns of the other Rhox genes have been reported in postnatal [13] and embryonic [16] testis. The majority of the cluster is expressed in germ cells at all time points examined. Interestingly, Rhox5, which is exclusively expressed in the Sertoli cells after birth, is expressed only in germ cells in the developing gonad [16]. In this report, we show that RHOX8 is exclusively expressed in Sertoli cells of the embryonic and postnatal testes. As RHOX5 and RHOX8 are the lone somatic-expressed RHOX factors present in the adult, we investigate the potential for redundancy with RHOX5 through the characterization of RHOX8 knockdown transgenic mice.
MATERIALS AND METHODS
Plasmids
To overexpress RHOX8 in cultured cells, Rhox8 coding sequence of various lengths (encompassing four distinct in-frame ATG start codons, indicated in Supplemental Fig. S1; Supplemental Data are available online at www.biolreprod.org) were amplified from testis cDNA by RT-PCR and cloned into HaloTag pHT2 (Promega) that expresses its insert under control of the CMV promoter. For Rhox8-siRNA knockdown, oligos containing the verified Rhox8 siRNA sequence (Supplemental Fig. S2) were annealed, cloned, and verified using previously described methods [17].
Animal Care and Breeding
All animal handling was done according to National Institutes of Health guidelines and in compliance with the Southern Illinois University Carbondale Institutional Animal Care and Use Committee. All animals were maintained under a 12L:12D schedule and fed NIH-31 mouse chow (Purina Labdiet 5008). The Rhox8-siRNA knockdown transgenic mice were made by freeing the transgene from the pMAN vector backbone [17] by digestion with NcoI and XhoI. Introduction of the transgene into the C57BL/6 strain resulted in 10 independent founder mice lines that harbored the transgene as assessed by PCR analysis using tail DNA as a template. Three of these lines failed to produce sufficient progeny for characterization, and two were found not to exhibit significant RHOX8 knockdown. However, five lines exhibited RHOX8 knockdown and equivalent male subfertility described in this report. As expected, female mice harboring the Rhox8-siRNA transgene (under the control of the androgen-dependent Rhox5 promoter) exhibited normal fertility. All mice were humanely sacrificed by carbon dioxide asphyxiation, followed by cervical dislocation, and both testes were removed. One testis was homogenized in 500 μl (prior to age 21 days) to 1 ml (after Postnatal Day [PND] 21) of Trizol (Invitrogen) for RNA isolation according to the manufacturer's recommendations, and the other was fixed in Bouins or 4% paraformaldehyde (PAF) for 12–16 h and then processed and embedded in paraffin for immunohistochemistry, apoptosis, proliferation, and morphometric analyses. Testosterone was extracted and measured in duplicate according to the manufacturer's instructions by ELISA (ADI-900-065; Enzo Life Sciences) as we reported previously [13]. Briefly, testes were homogenized and extracted three times with diethylether. Extracted samples were evaporated and dissolved in 250 μl Assay Buffer 3 supplied with the kit. Following pilot analysis of testosterone concentration, samples were diluted to ensure that unknown values would fall within the range detectable using the supplied standards. Final testosterone concentrations were determined after correction for dilution factor and mass of input testis tissue. The extraction efficiency was 90% as assessed by parallel extractions spiked with known quantities of hormone.
Quantitative Real-Time RT-PCR Analysis
The quantity and quality of total RNA was determined by spectrometry and denaturing agarose gel electrophoresis, respectively. The cDNA was synthesized from total RNA (2 μg) using iScript Select cDNA synthesis Kit (BioRad). Real-time quantitative RT-PCR (qPCR) analysis of mRNA expression was performed using a MyiQ Single-Color Real-Time PCR Detection System (BioRad) with iQ SYBR Green supermix (BioRad) as the detector according to the manufacturer's recommendations. Primers for the Rhox genes, testis markers, and normalization genes have been previously reported [15, 18–20]. Primers were designed to amplify cDNAs of around 200 bp, all of which spanned at least one exon/intron junction, and exhibited similar amplification efficiency (97 ± 3%) as assessed by amplification of cDNA dilution series. PCR cycle parameters were 95°C for 15 sec and 60°C for 1 min for 40 cycles. The threshold line was set in the linear region of the plots above the baseline noise, and threshold cycle (CT) values were determined as the cycle number at which the threshold line crossed the amplification curve. PCR without template or template substituted with total RNA were used as negative controls to verify experimental results. After amplification, the specificity of the PCR was determined by both melt-curve analysis and gel electrophoresis to verify that only a single product of the correct size was present. Data were normalized against Rpl19 and are shown as the average fold increase ± SEM.
Western Blot Analysis
Total protein from whole cell lysates were separated on SDS-PAGE gels and transferred to nitrocellulose membranes (EMD Millipore). Membranes were blocked and incubated overnight at 4°C with primary antibodies. Bound antibody was visualized with IRDye 700 or 800 conjugated affinity-purified secondary antibodies (Rockland Immunochemicals) and quantitated using the Odyssey infrared imaging system (LI-COR). For slot blots, double protein amounts were added and passed through wells directly onto nitrocellulose and processed as describe above.
Immunohistochemistry and Immunofluorescence
Immunolocalization of RHOX8 was performed in cross sections (5 μm) of paraffin-embedded testis sections using rabbit polyclonal antibody at 1:2000 dilution (2223B; IMGENEX) and a Vectastain Elite ABC Kit (Vector Laboratories) as described previously [18]. Negative controls were performed in which the primary antibody was substituted with preimmune serum from the rabbit used by IMGENEX prior to injection of the peptide QEMKEREENAGIQDC corresponding to amino acids 30–44 of the amino domain of mouse RHOX8. Antigen retrieval using a boiling citrate buffer was performed as described previously [21]. Three recently generated anti-RHOX8 rabbit polyclonal antisera were also characterized: ARP36806, ARP36807, and ARP37849 (Aviva Systems Biology). All three of these antisera were generated against distinct RHOX8 peptides and produced similar results to the previously characterized IMGENEX RHOX8 antibody (Supplemental Fig. S3). Immunofluorescence was performed on similarly prepared sections, except that donkey serum was used as blocking agent to allow the use of AlexaFluor-conjugated AffiniPure F (ab′) secondary antibodies (711-546-152, 711-546-152, 711-516-152; Jackson Immuno Research). Primary antibodies for established testis marker proteins: SOX9 (ab5535; (Millipore; 1:1000 dilution), STRA8 (ab49602; Abcam; 1:3000 dilution), HSD3B1 (kindly provided by the late Anita Payne), and SOX8 (MABN268; Millipore; 1:250 dilution) were applied overnight at 4°C and were used for identification of germ, Leydig, and Sertoli cells, respectively. For STRA8 staining, PAF produced stronger staining than Bouins fixed sections. For SOX8 staining, sections were additionally blocked with mouse-on-mouse blocker (MOM kit; Vector Laboratories) for 3 h prior to addition of primary antibodies. Slides were viewed and photographed on a Leica fluorescent microscope using Leica Application Suite X (LAS X). When comparisons between wild-type (WT) and RHOX8-KD were made, identical advanced fluorescence software settings were used. Sertoli cell numbers within individual testis tubules, cut in circular cross section (i.e., obliquely cut tubules were omitted from the assessment), were determined as we previously reported for Rhox5-null and WT-knockdown mice [13, 22]. Sertoli cells were identified by a combination of RHOX8, SOX8, and SOX9 staining where appropriate. Residual RHOX8 and SOX9 expression in RHOX8-KD animals could be visually enhanced by adjusting contrast levels in LAS X preview images that allowed visualization of all Sertoli cells. Leydig cells were identified by HSD3B1 immunofluorescence and manually counted using within photomicrographs. Because Leydig cell clusters varied in number at different locations in the interstitium, we normalized the total Leydig cell counts to surface area examined as determined by LAS X following calibration using a stage micrometer.
Apoptosis Assay
Using tissue sections prepared in the same method as those for immunohistochemistry (IHC), TUNEL assays were performed to quantify the level of apoptosis in Rhox8-siRNA mice compared to WT controls. These assays were conducted using the ApopTag Fluorescin In Situ Apoptosis Detection Kit S7110 (Millipore). Tissue sections were processed according to the manufacturer's protocol and counterstained with DAPI diluted 1:1500 in 1× TBS for 5 min. Tubule cross sections containing at least one positively stained cell were considered positive tubules. The percentage of positive tubules and the number of positively fluorescing cells within each tubule were recorded and analyzed as described previously [13, 22].
Cell Culture
The MSC1 Sertoli cell line was kindly provided by Michael Griswold (Washington State University) and cultured in DMEM supplemented with 10% fetal bovine serum (Atlanta Biologicals) and 50 μg/ml penicillin and streptomycin. Transient transfection of Rhox8 expression constructs and/or Rhox8 siRNA was performed using the Attractene transfection reagent (Qiagen), and visualization of recombinant fusion proteins was performed using fluorescent dye substrates bound to the Halotag moiety.
Sperm Count
Sperm counts were performed as described previously with minor modifications [13, 22]. First, the caudal region of the epididymis was dissected from both WT (control) and Rhox8-KD animals and placed in 500-μl EmbyoMax M2 (Millipore) medium heated to 37°C. The caudal epididymis was spread open using a back-cutting method and then left undisturbed for 30 sec (giving sperm time to escape into liquid). Subsequently, 5 μl of sperm-containing liquid was diluted in 45 μl of unused EmbyoMax M2 media (1:10 dilution), and 10 μl of this dilution were placed in the center of a Cell-Vu disposable counting chamber and covered with a glass slip containing a counting grid. Finally, motile and nonmotile sperm were counted to determine sperm motility defects and differences in total sperm populations as well as severed sperm heads to eliminate speculation of processing differences between samples.
Statistical Analysis
All qPCR, transfection, cell counts, histological measurements, and subfertility data were subjected to one-way ANOVA, and differences between individual means were tested by a Tukey multiple-range test using Prism 5.0 (GraphPad). All qPCR data were corrected for differences in sample loading using the Rpl19 data as a covariate. Tests of significance were performed using the appropriate error terms according to the expectation of the mean squares for error. A P-value of 0.05 or less was considered significant. Data are presented as least-square means with SEM.
RESULTS
Rhox8 Is Expressed Exclusively in Sertoli Cells of the Testes
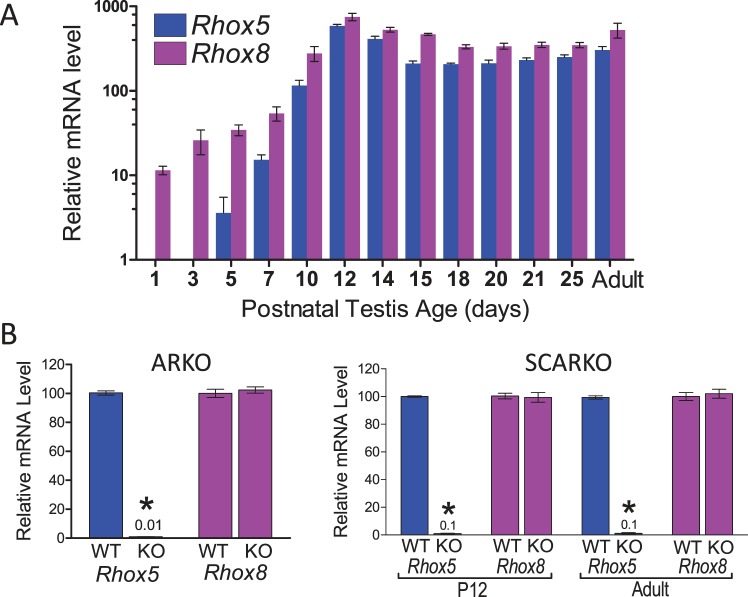
The Rhox8 gene is highly expressed in the testis, epididymis, and ovary and weakly in the placenta [13, 18, 19, 23]. We previously examined the temporal expression patterns of the Rhox cluster during the first wave of spermatogenesis by qPCR [13]. We found that Rhox5 and Rhox8 were induced by PND 7 and peaked at PND 12, when Sertoli cells make up the majority of the cells within testis tubules [13]. We extended the analysis to earlier time points and found that Rhox8 mRNA is present in the testes from birth (Fig. 1A). The regulatory regions of Rhox5 in the male reproductive tract and ovary have been well defined [3, 24, 25]. Two distinct promoters exist: the distal promoter, which drives granulosa-specific expression, and an androgen-dependent proximal promoter, responsible for testis and epididymal expression. We previously characterized the promoter region employed by granulosa cells to drive Rhox8 transcription [18] and were curious if the two-promoter arrangement of Rhox5 is also conserved for Rhox8. While Rhox5 expression begins to spike ∼PND 5 in conjunction with the induction of androgen receptor activity, the relatively high expression level of Rhox8 suggested that it is not androgen dependent. In support of this prediction, Rhox8 mRNA levels remain unchanged in mice with complete ablation of androgen receptors (ARKO) and (SCARKO) mice, which possess Sertoli-specific androgen receptor ablation (Fig. 1B).
FIG. 1.
Postnatal expression and regulation of Rhox5 and Rhox8. A) qPCR analysis of total cellular RNA from testes obtained from the indicated age (n = 5 per time point). Values were normalized against ribosomal L19 (Rpl19) mRNA and are expressed as fold above background (±SEM), which was arbitrarily given a value of 1. Oligo pairs specific for Rhox5 and Rhox8 exhibited 99% amplification efficiency. Equivalent results were obtained when cyclophilin mRNA was used as the internal control (data not shown). B) The dependence on androgen on Rhox gene induction was assessed by comparison of relative mRNA levels in testes from control, ARKO, and SCARKO mice (n = 3 per genotype and age). Rhox5 mRNA was significantly reduced (*P < 0.0001) with residual qPCR signal indicated above bar. However, Rhox8 expression level was not reduced on androgen receptor ablation.
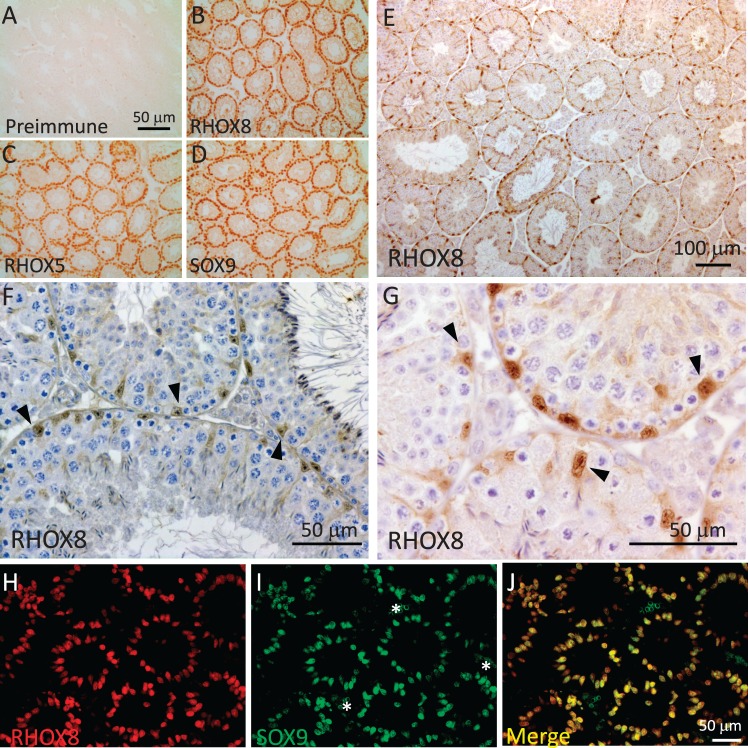
At peak expression on PND 12, RHOX5, RHOX8, and the hallmark Sertoli cell marker SOX9 [26] were found to be expressed in all Sertoli cells (Fig. 2, A–D, and Supplemental Fig. S4). Interestingly, our results showed that after PND 30, when RHOX5 is known to become confined to Sertoli cells of stages IV–VIII of the spermatogenic cycle [27], RHOX8 protein is found in all Sertoli cells regardless of spermatogenic stage (Fig. 2, E–G). This postnatal expression pattern matches that of SOX9 [26], and we confirmed colocalization of RHOX8 and SOX9 by sequential immunofluorescence staining in PND 12 testes (Fig. 2, H–J). In this experiment and subsequent analyses at PND 18 and PND 30 and of adult testes, very few Sertoli cells expressed predominantly SOX9 or RHOX8, with most exhibiting equal expression levels.
FIG. 2.
Rhox8 encodes a Sertoli-specific homeobox factor. Serial testis sections from PND 12 mice were incubated with either preimmune serum (1:250 dilution; A) or RHOX8 polyclonal antiserum (1:2500 dilution; B) from IMGENEX rabbit #2223B immunized with a RHOX8 amino domain peptide. DAB visualization reveals that RHOX8 protein is localized to Sertoli cells as compared to the established Sertoli cell markers RHOX5 and SOX9 (C and D). In adult testes (E–G), RHOX8 staining is present in the nuclei of all Sertoli cells (arrowheads). H–J) Immunofluorescence analysis confirms colocalization of RHOX8 and SOX9 in all Sertoli cells at PND 12. * indicates that nonspecific autofluorescence in erythrocytes was observed in the green channel.
Sertoli-Specific Knockdown of RHOX8 (RHOX8-KD) In Vivo
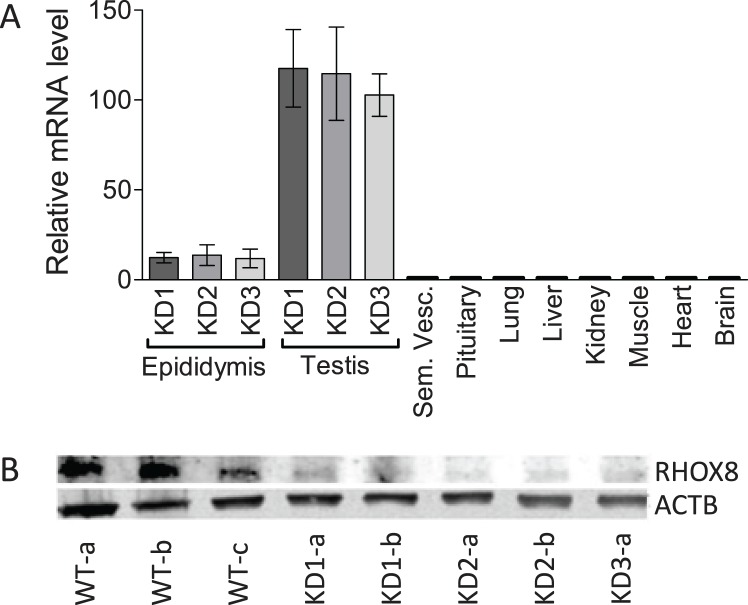
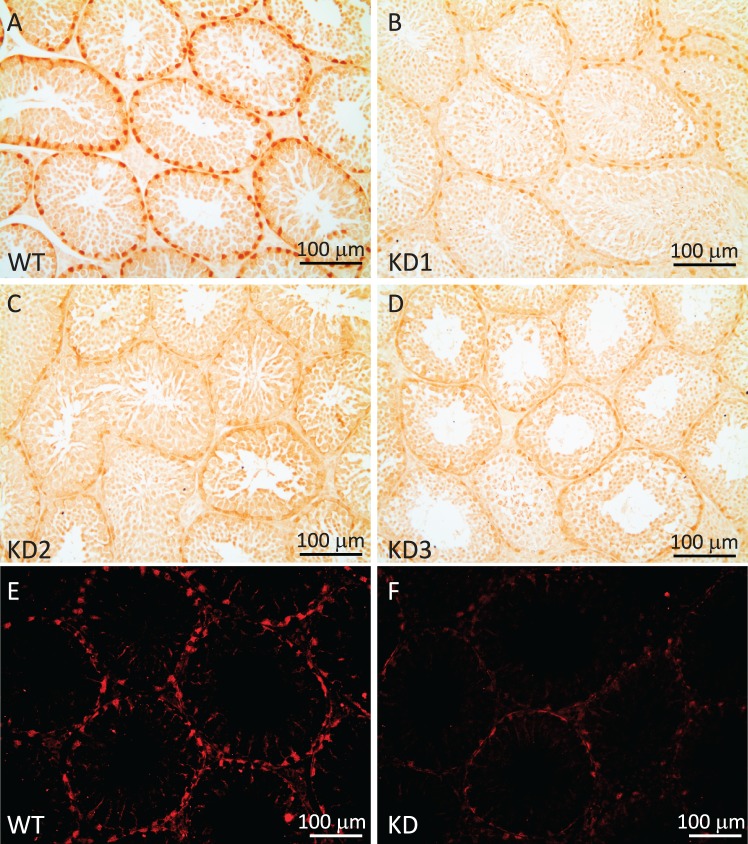
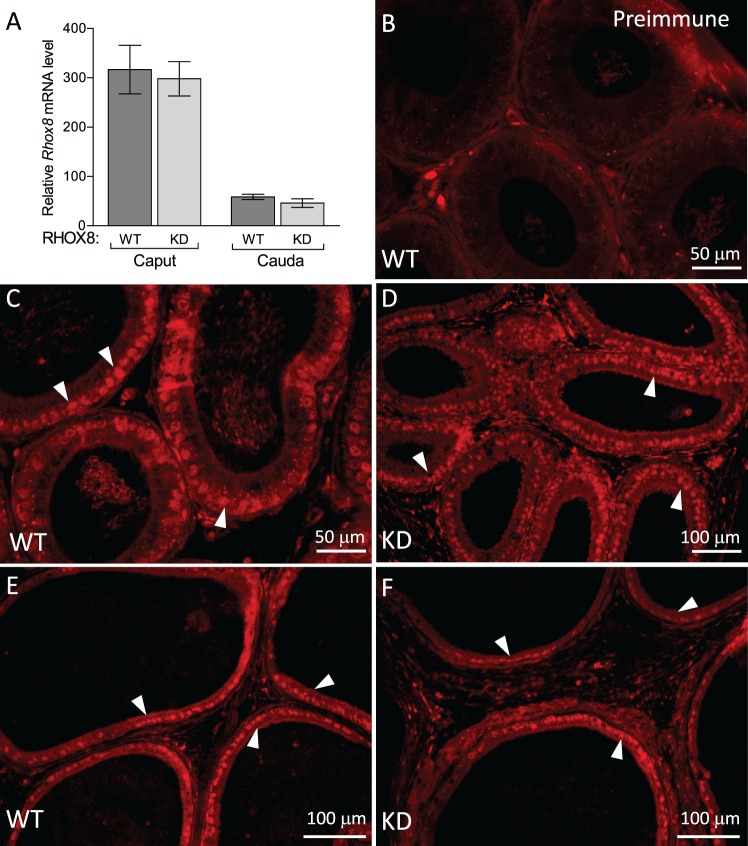
To elucidate RHOX8's function in Sertoli cells, we used an in vivo tissue-specific RNA interference knockdown approach that we previously employed to ablate WT1 in Sertoli cells [17, 22]. This approach mimics the pathway by which naturally occurring micro-RNAs are made and uses a portion of Rhox5's androgen-dependent proximal promoter to drive the expression of a hairpin RNA containing the siRNA sequence in Sertoli cells in the testes and principle caput cells of the epididymis (Fig. 3A; [17]). While Rhox8 was not found to be androgen dependent, this promoter fragment lacks the stage-specific elements of the Rhox5 Pp and thus would drive knockdown of Rhox8 in all Sertoli cells. Ten founder lines (four male, six female) harboring the Rhox8-siRNA transgene were generated. Two male founders failed to produce any litters when mated with WT partners and were lost, suggesting that the activated transgene was detrimental to male fertility (these mice had 64% smaller testes at 1 yr of age than the other founders and f1 progeny from female founders). Three lines failed to exhibit detectable transgene expression or consistent subfertility in pilot assays and were discontinued (data not shown). Because the transgene is driven by a Sertoli-specific promoter, female founders should harbor but not express the transgene. RHOX8-KD females exhibited normal fertility and thus were capable of generating litters for characterization of RHOX8 knockdown in male progeny. Five independent lines contributed to our initial analyses; however, the majority of the data comes the three lines that proved easier to maintain in the colony (KD1, KD2, and KD3). Because the transgene was integrated randomly, we expected to see differences in the level of knockdown depending on what enhancer/silencing elements were present near the integration site. However, the remaining lines exhibited indistinguishable differences in Rhox8-siRNA transgene expression (Fig. 3A) and RHOX8 knockdown (Fig. 3B). RHOX8 protein levels were visibly reduced as early as PND 7 and predominant at PND 12 but were variable by animal and testis tubule until PND 18, where consistent and persistent knockdown was achieved. Aggregate data from Western blot and protein slot-blot quantification indicated that more than 87% RHOX8 protein was depleted in knockdown animals, further confirming the efficacy of our in vivo siRNA approach. The reduction of RHOX8 was visually apparent in Sertoli cells as determined by IHC; Fig. 4, A–D) and immunofluorescence (Fig. 4, E and F) analyses. In contrast to testes, while the siRNA transgene was detectable in epididymis (Fig. 3A), no apparent reduction in Rhox8 mRNA or RHOX8 protein was observed in knockdown animals (Fig. 5). RHOX8 protein is localized to the nuclei of luminal epithelium in both the caput (Fig. 5, C and D) and the caudal (Fig. 5, E and F) regions of the epididymis.
FIG. 3.
RHOX8 knockdown in Rhox8-siRNA (RHOX8-KD) mice. A) Quantitative RT-PCR analysis of total cellular tissue RNA, obtained from three independent transgenic lines (n = 4 per tissue), confirms that the Rhox8-siRNA transgene is expressed only in the testis and epididymis and that there is no significant differences in relative transgene expression between lines. B) Western blot analysis of total testis lysate (20 μg) from PND 25 animals using RHOX8 or ACTB loading control antibodies.
FIG. 4.
In vivo knockdown of RHOX8 in Rhox8-siRNA transgenic mice. A–D) Immunohistochemical analysis of testes from PND 25 animals confirmed RHOX8 knockdown in three independent transgenic mice lines. The WT (A) and knockdown (B–D) testes shown were collected, processed, and embedded in the same block. Thus, single tissue sections containing all testis samples were exposed to the same 1° antibody, 2° antibody, and DAB reagent, ensuring that reduced protein levels were not from differences in development. E and F) RHOX8 knockdown was confirmed by immunofluorescence at PND 30.
FIG. 5.
RHOX8 is expressed in caput and caudal regions of the adult epididymis. A) Rhox8 is highly expressed in the caput and caudal epididymis as assessed by qPCR. B–F) Epididymal sections from adult mice were incubated with either preimmune serum (1:250 dilution; B) or RHOX8 polyclonal antiserum (1:600 dilution; C and D). RHOX8 protein is localized to the basally located nuclei (arrowheads) of luminal epithelium of the caput (C and D) and caudal (E and F) epididymis. Taken together, these results indicate that Rhox8 expression is not disturbed in RHOX8-KD mice.
Subfertility of Male RHOX8-KD Mice
RHOX8-KD male mice exhibited several reproductive defects. Because we observed similar protein knockdown and subfertility in all of the independent RHOX8-KD lines in pilot studies, henceforth we will present aggregate data from three to five lines per experiment. Copulation rates, as assessed by the observation of vaginal plugs, were the same between RHOX8-KD males and controls, indicating that mating behavior was not altered (data not shown). However, in 5-day timed mating experiments, only one pair in six was able to produce a litter (Table 1). When pairs were monitored for 8 wk and 6 mo, RHOX8-KD male mice produced half as many litters and yielded significantly smaller litters than control males, most of which were nontransgene harboring littermates (Table 1). The subfertility of RHOX8-KD male mice was similar to that of Rhox5-null mice (Table 1; [13]).
TABLE 1.
Reproductive fitness of Rhox8- and Rhox5-deficient mice.
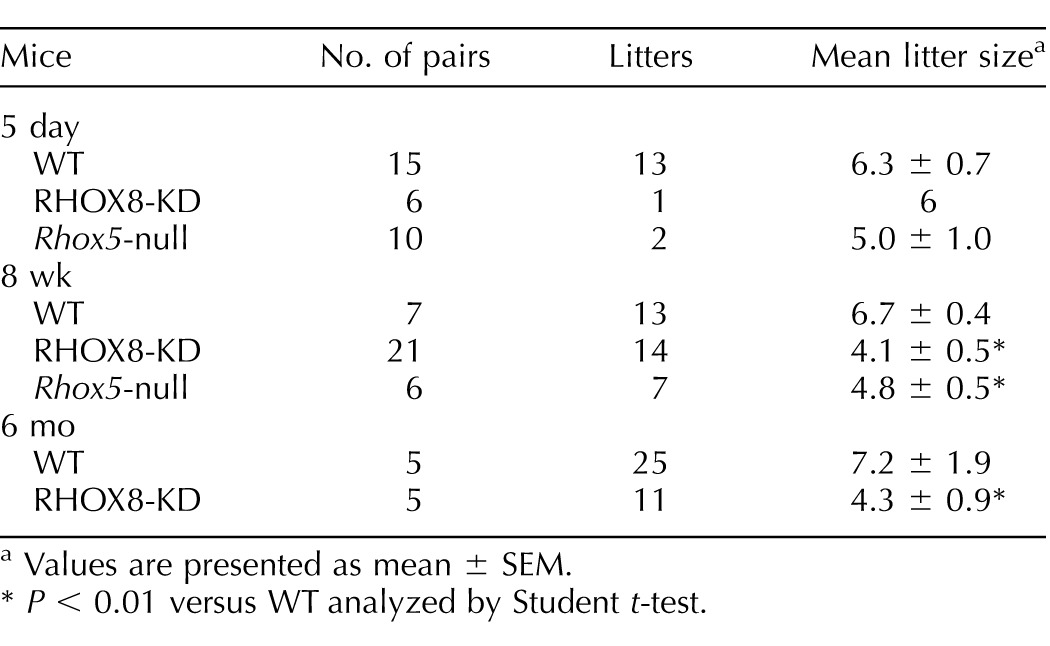
Values are presented as mean ± SEM.
P < 0.01 versus WT analyzed by Student t-test.
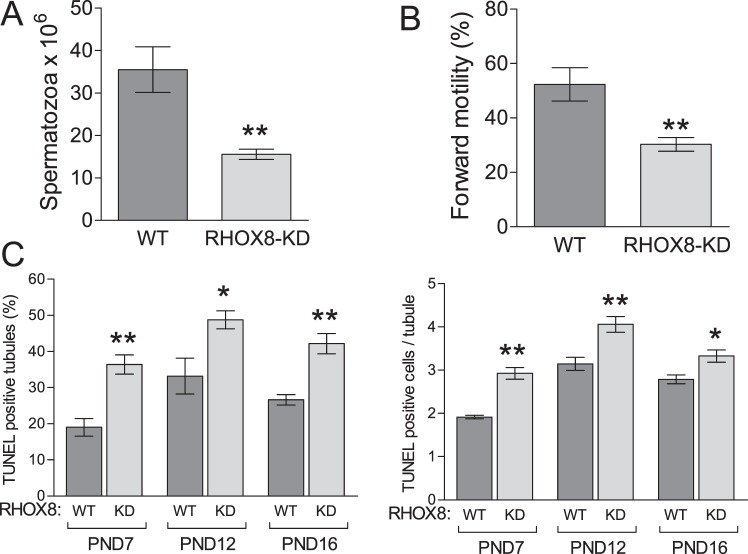
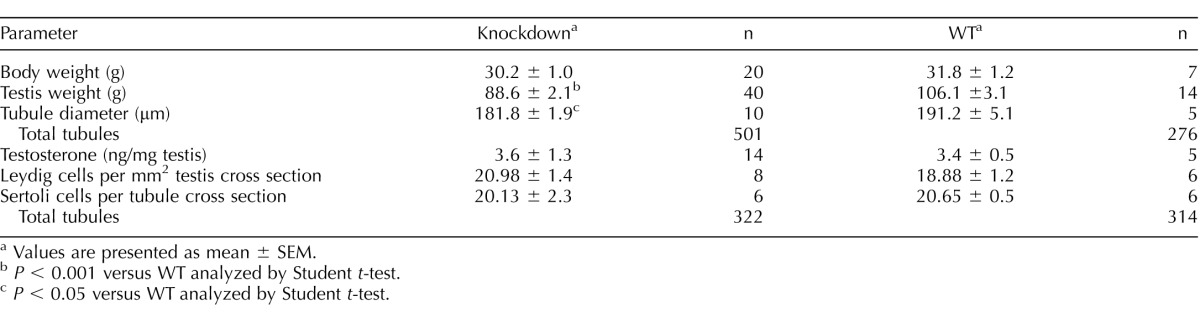
Subfertility of RHOX8-KD mice could be explained in part by their significantly reduced caudal sperm counts, assessed to be 47% of the normal WT spermatogenic output (Fig. 6A). In addition, these sperm exhibited a mean drop in forward motility to 57% of control animals (Fig. 6B). Because RHOX8 expression was apparently unaffected in RHOX8-KD epididymis, the observed motility defects are most likely to arise from improper germ cell development in the testes. In our previous studies, the low spermatogenic output of Rhox5-null and WT1-knockdown animals was attributed to poor survival and increased germ cell apoptosis [13, 15, 22]. Therefore, we counted TUNEL-positive cells in testis cross sections from RHOX8-KD mice at several postnatal ages. TUNEL-positive cells were most likely germ cells based on their luminal localization within testis tubules (Supplemental Fig. S5) as well as our finding that Leydig and Sertoli cell numbers were not significantly reduced in RHOX8-KD testes (Table 2). Leydig cell function was also unaffected, as no differences in intratesticular androgen levels were detected between WT and RHOX8-KD mice (Table 2). Interestingly, at PND 7, 12, and 16, a significantly larger (∼2-fold) proportion of spermatogenic tubules exhibited evidence of programmed cell death. In addition, positive tubules possessed a higher number of TUNEL-positive cells per TUNEL-positive tubule (Fig. 6C). While apoptotic cells were commonly found in testes of mice older than PND 16, the proportion of positive tubules and apoptotic cell counts were variable, and significant differences between WT and RHOX8-KD mice were not observed (data not shown). However, net disruptions in germ cell production over 1 yr in RHOX8-KD mice results in significantly smaller testes and a slightly smaller average testis tubule diameter (Table 2).
FIG. 6.
Analysis of testis defects in RHOX8-KD mice. Caudal sperm counts (A) and motility (B) were determined in control (n = 16) and RHOX8-KD (n = 45) mice. C) RHOX8-KD mice exhibited an increased proportion of TUNEL-positive testis tubules and apoptotic cells per TUNEL-positive tubule compared to control animals. At least six animals and 800 tubule cross sections were counted per genotype for each time point. *P < 0.01, **P < 0.001.
TABLE 2.
Physical and histological differences between Rhox8-deficient and control mice.
Values are presented as mean ± SEM.
P < 0.001 versus WT analyzed by Student t-test.
P < 0.05 versus WT analyzed by Student t-test.
Germ Cell Defects in Mice with Reduced Levels of RHOX8 in Sertoli Cells
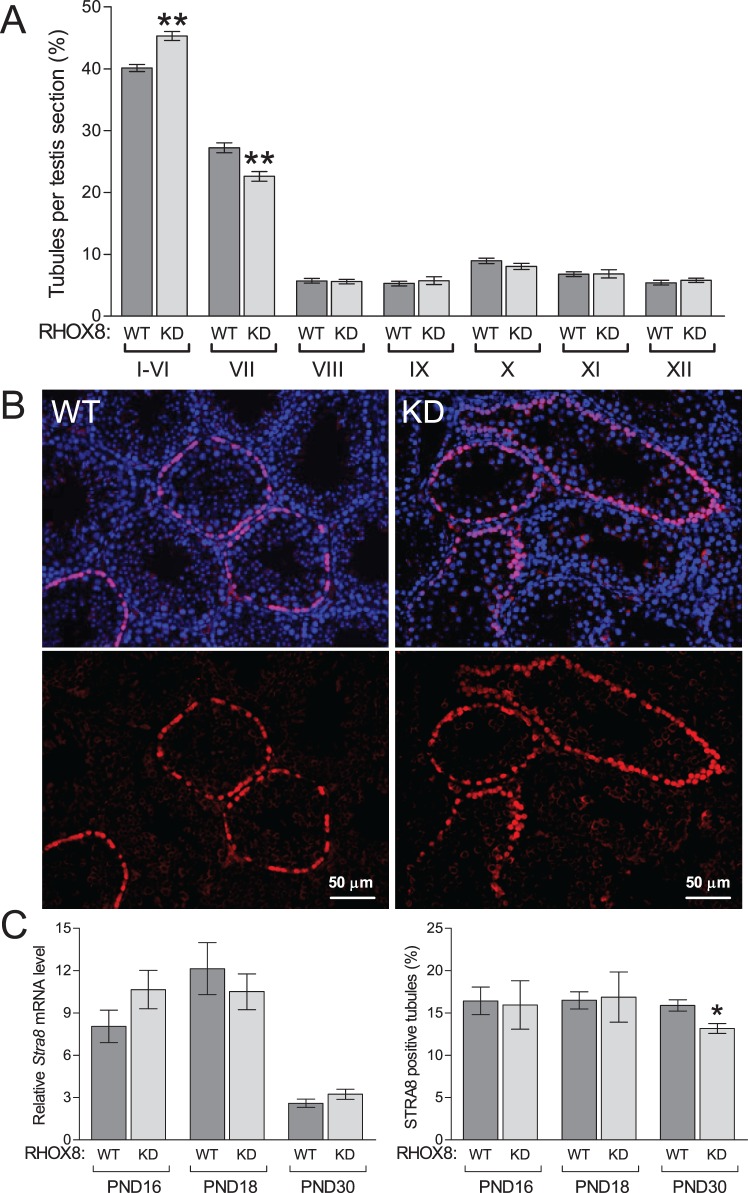
The spermatogenic cycle of the seminiferous epithelium can be divided into 12 stages that distinguish the steps of spermatid development. To assess whether germ cell development was compromised in RHOX8-KD animals, we quantified the distribution of testis tubule stages as described previously [13, 28, 29]. RHOX8-KD mice exhibited a significantly increased proportion of tubules in stages I–VI compared to control testes (Fig. 7A). Concomitantly, fewer tubules were observed at stage VII. At stage VIII and afterward, there were no significant differences in tubule distribution between RHOX8-KD and control animals, suggesting that once spermiation was reached, germ cell development can proceed normally.
FIG. 7.
Germ cell defects in RHOX8-KD mice. A) Comparison of the relative proportions of the 12 distinct stages of the spermatogenic cycle in control and RHOX8-KD animals. Only round cross sections of a single, clearly defined stage were counted. For each genotype, eight animals were assessed, with the number of qualifying tubules ranging from 189 to 248 per animal. B) STRA8 immunolocalization of preleptotene spermatocytes in PND 30 testes of WT and RHO8-KD mice. C) While no apparent changes in Stra8 mRNA were found (n = at least 6 per genotype and time point), a reduced percentage of STRA8 positive testis tubules was observed at PND 30 (n = 12) compared to PND 16 (n = 5) and PND 18 (n = 6). *P < 0.01, **P < 0.001.
In late stage VI and early stage VII, B spermatogonia divide to form preleptotene spermatocytes [29]. These preleptotene spermatocytes can be visualized by STRA8 immunostaining as rings near the basement membrane at PND 20 and beyond [30]. We assessed STRA8 expression at PND 16, 18, and 30 (Fig. 7B and data not shown) in WT and RHOX8-KD mice. When present, we found no obvious differences in preleptotene spermatocyte staining intensity between genotypes at any time point, and Stra8 mRNA levels between RHOX8-KD mice and controls were not significantly different (Fig. 7B). However, the prevalence of STRA8-positive tubules exhibiting the characteristic staining of the preleptotene spermatocyte layer was significantly lower in PND 30 RHOX8-KD mice, suggesting that a partial block in spermatogonial to spermatocyte transition may be present.
Identification of Somatic Genes Exhibiting Altered Expression in RHOX8-KD Testes
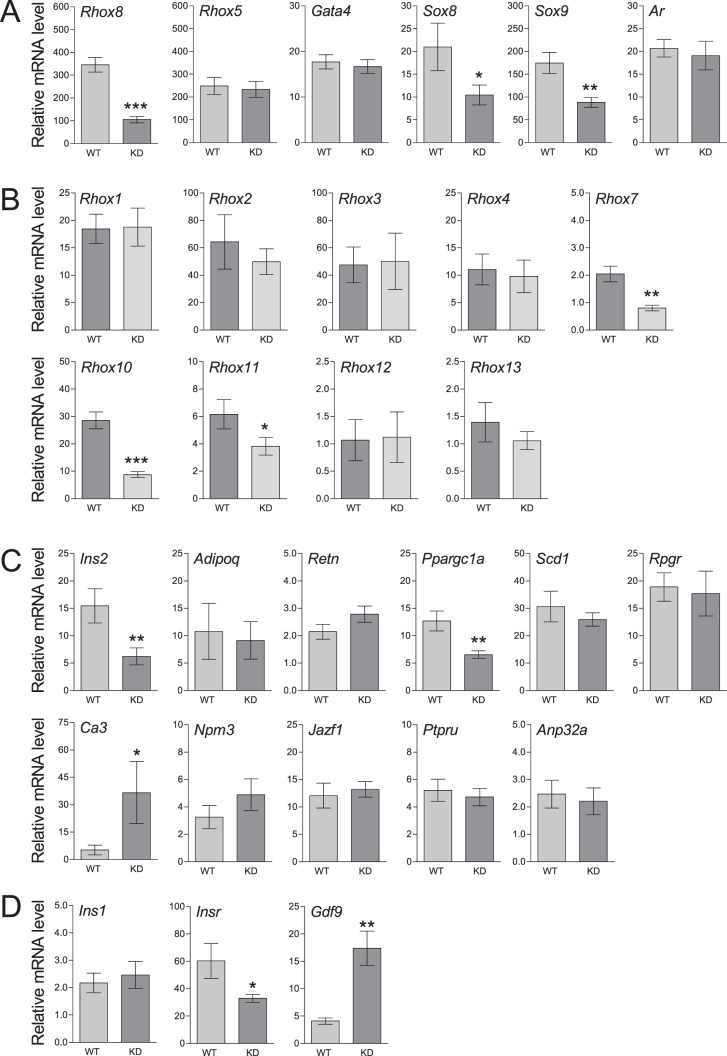
To begin to identify putative RHOX8-regulated genes as well as determine whether the actions of the Rhox8-siRNA were specific, we examined a panel of established Sertoli cell markers using qPCR at PND 18 (Fig. 8A). PND 18 was chosen, as it represents the first postnatal time point where maximal RHOX8 knockdown is evenly distributed. Rhox8 mRNA levels were significantly reduced in RHOX8-KD mice, but transcription of neither Rhox5 nor Gata4 was significantly altered, suggesting that the presence of the Rhox8-siRNA did not globally impact Sertoli cell transcription. Two genes key to Sertoli development and function, Sox8 and Sox9, were significantly reduced in RHOX8-KD testes. As further evidence that the androgen signaling is not directly impaired, Rhox5 and Ar mRNA levels were unchanged in RHOX8-KD testes (Fig. 8A), and AR protein levels were not obviously different in either peritubular myoid or Sertoli cells as determined by IHC (data not shown). At PND 30, Gata1 were unaltered, but similar downregulation was observed for Rhox8, Sox8, and Sox9 as was observed at PND 18 (Supplemental Fig. S6).
FIG. 8.
Analysis of differentially expressed genes in PND 18 testes. Relative expression of the indicated genes in total cellular RNA from WT (n = 6–10) and RHOX8-KD (n = 6–12) mice was determined by qPCR as described in Fig. 1. A) Somatic cell-specific genes. B) Germ cell-specific Rhox family genes. C) Candidate RHOX5-regulated genes previously identified by microarray analysis. D) Additional genes of potential relevance to germ cell-Sertoli cell communication. *P < 0.05, **P < 0.01, ***P < 0.001.
Altered Rhox Cluster Expression in RHOX8-KD Mice
We previously reported that ablation of Rhox5 in the epididymis results in misregulation of the majority of the Rhox cluster, including Rhox8, in the caput region where RHOX5 is expressed [19]. However, our lab and others have been unable to identify significant changes in Rhox gene expression in the testes of Rhox5-null animals. We used qPCR to determine the relative expression of the Rhox cluster at PND 18 (Fig. 8B). Rhox5 and Rhox8 are the only members of the cluster expressed postnatally in somatic cells; all others are restricted to germ cells [16; unpublished data]. Rhox6 and Rhox9 are difficult to reproducibly detect in postnatal testes and were omitted from the analysis. Rhox12 was the lowest expressed and was used as the baseline for relative expression of the rest of the cluster. None of the alpha subcluster genes (Rhox1-Rhox4) or Rhox13 exhibited significantly altered expression in RHOX8-KD mice. For this analysis, pan oligos for Rhox2, Rhox3, and Rhox4, which would not distinguish between the seven copies of those recently duplicated genes, were used; thus, potential RHOX8-regulation of a specific paralog cannot be ruled out. However, current evidence suggests that these gene copies may be coregulated in the gonads [31]. The predicted spermatocyte-specific gene Rhox7 [13] was lowly expressed but significantly reduced 2-fold in RHOX8-KD testes. Rhox11 was significantly reduced but by less than 2-fold. It should be noted that PND 18 represents the initiation point of Rhox11 transcription [13; unpublished data], so this reduction could reflect a delay in normal expression and not necessarily direct regulation by RHOX8. The most substantial downregulation was observed for Rhox10. Rhox10 transcripts can be detected in all germ cells, but mRNA and protein levels are highest in spermatogonia and spermatocytes through the zygotene phase [32]. Future studies will reveal whether the reduced expression of these Rhox genes is due to loss of a direct activating factor induced by RHOX8 or a shift in temporal expression patterns indicative of a developmental delay or perhaps an indication that the germ cells expressing these factors are the ones that are undergoing apoptosis in response to reduced RHOX8.
Analysis of RHOX5-Regulated Genes in RHOX8-KD Mice
We recently reported the characterization of RHOX5-regulated genes in PND 12 testes [15]. In that study, PND 12 was chosen to maximize the opportunity to identify Sertoli-specific genes, as Sertoli transcripts are known to exhibit peak expression and represent ∼40% of testis cells at that stage of development. Unfortunately, in our pilot analyses, we did not observe consistent knockdown of putative RHOX8-regulated genes until PND 18. We are not certain if this was due to residual RHOX8 protein existing in Sertoli cells prior to Rhox8-siRNA transgene expression or whether it represents the need for accumulation of a critical mass of knockdown necessary to see changes in gene expression within Sertoli cells of total testis extracts. The majority of the characterized RHOX5-regulated genes from our prior study involved metabolism as either secreted hormones, transcriptional regulators, or enzymes and transporters. We used qPCR to profile the 11 genes exhibiting the largest differential expression in Rhox5-null testes (Fig. 8C). Only three of these genes, Ins2, Ppargc1a, and Ca3, exhibited significantly different expression in RHOX8-KD testes, suggesting that RHOX5 and RHOX8 are likely to govern the expression of largely distinct subsets of Sertoli-specific genes. Of these targets, direct regulation by RHOX5 in Sertoli cells has been demonstrated for Ins2 only by using a combination of in vitro ChIP, EMSA, and promoter deletion analyses [15]. The observed 2.5-fold downregulation of Ins2 in RHOX8-KD testes suggests that RHOX8 may bind to the RHOX5-binding element within the Ins2 promoter. However, continued expression of RHOX5 in RHOX8-KD mice likely prevents the severe crash in Ins2 expression observed in Rhox5-null mice. Two genes, Ins1 and Insr, related to insulin signaling that were not part of the original microarray but were logical to examine were found to be unaffected and downregulated 2-fold, respectively (Fig. 8D). A panel screen of gonad development factors employed for another study serendipitously identified Gdf9 as a putative RHOX8 downstream gene that was upregulated 3-fold in RHOX8-KD testes (Fig. 8D).
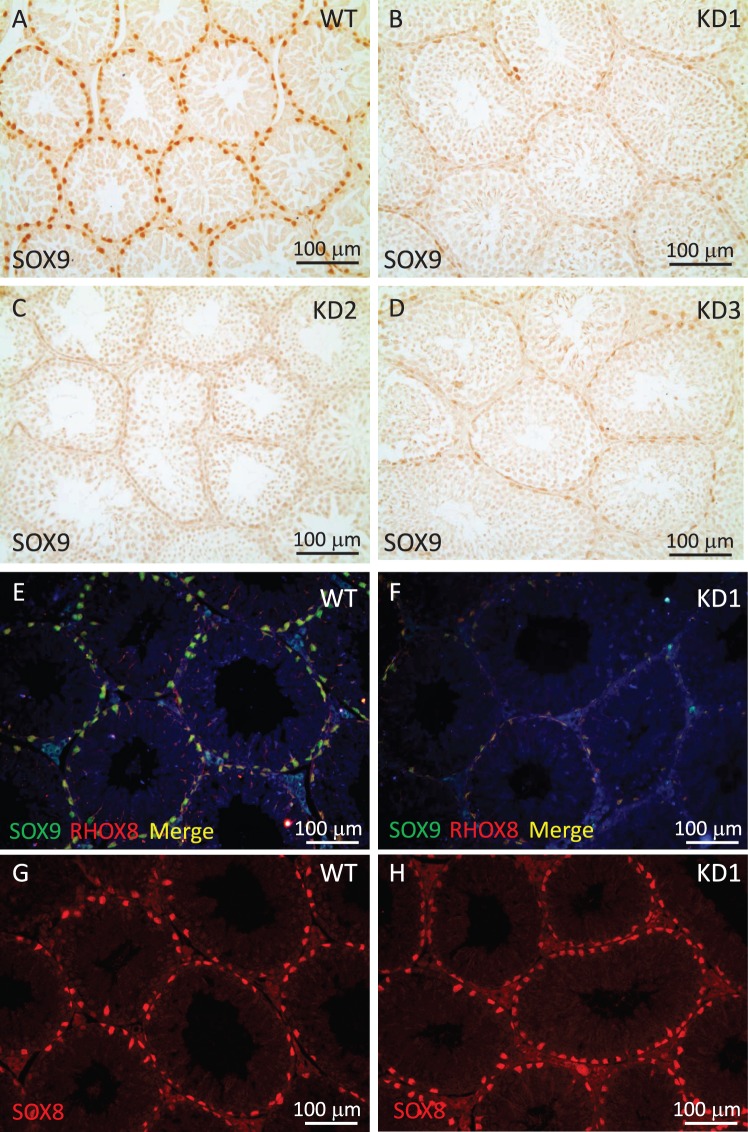
Knockdown of RHOX8 Leads to Downregulation of Sox9
While assessing the specificity of RHOX8 knockdown using qPCR analysis of established Sertoli marker genes, we observed that Sox8 and Sox9 transcription were reproducibly diminished (Fig. 8A). As there were no global changes in Sertoli-specific gene expression (Rhox5, Gata1, and Gata4 are unchanged), this observation indicated that Sox8 and Sox9 may be a potential RHOX8-regulated genes. We assessed SOX9 levels in RHOX8-KD testis sections and found that SOX9 protein was highly reduced in all Sertoli cells (Fig. 9, A–D). RHOX8 and SOX9 were colocalized to all Sertoli cells (Fig. 9E), and while not sufficient evidence for direct regulation, it was interesting to note that residual expression of RHOX8 and SOX9 in knockdown testes (Fig. 9F and Supplemental Fig. S7) seemed matched (i.e., very few cells expressed only RHOX8 or SOX9 in RHOX8-KD testes). In contrast, while Sox8 mRNA was reduced ∼2-fold in RHOX8-KD testes at PND 18 and 30, no apparent differences in SOX8 protein were observed (Fig. 9, G and H) at PND 30. This difference may result from the relatively greater downregulation 10-fold or more in Sox9 mRNA at PND 30 (Supplemental Fig. S6).
FIG. 9.
Sox9 misregulation in RHOX8-KD testes. A–D) Immunohistochemical analysis of SOX9 protein in control (A) and three independent RHOX8-KD (B–D) transgenic mouse lines was performed as described in Fig. 4. Knockdown of RHOX8 and SOX9 was confirmed by immunofluorescence (E and F; individual channels are shown in Supplemental Fig. S7), but despite a reduction in Sox8 mRNA, no changes in SOX8 protein were observed in RHOX8-KD mice (G and H).
DISCUSSION
Homeobox genes encode transcription factors that have well-established roles in embryonic development. We recently discovered the Rhox genes, a new family of homeobox genes [13] that are selectively expressed in the developing embryo, postnatal and adult gonads, and accessory tissues associated with mammalian reproduction [3]. The largest and best-characterized Rhox cluster is found in mice [5]. However, all mammals examined to date possess a set of Rhox genes that, while they may vary in number by species, appear relevant to reproduction and are located in the syntenic region of the X chromosome. All but three of the 33 mouse Rhox genes are highly expressed in the male reproductive tract, and many are androgen regulated. When we first characterized the Rhox genes [13], cellular fractionation of the adult testis suggested that half of the cluster was expressed in Sertoli cells. However, subsequent analyses have revealed that most Rhox genes are specific to germ cells and are differentially regulated in spermatogonia and spermatocytes [16, 32–34; unpublished data]. Only Rhox5 and Rhox8 are expressed postnatally in Sertoli “nurse” cells, suggesting that they alone regulate the expression of somatic cell gene products crucial for germ cell development. Indeed, we previously reported that targeted deletion of Rhox5 results in male subfertility due to increased germ cell apoptosis and abnormal sperm motility [13].
Results of the present study indicate that RHOX8 is also necessary for complete fertility in male mice. Knockdown of RHOX8 expression in the testes, using a Sertoli-specific Rhox5-promoter-driven Rhox8-siRNA, results in increased germ cell apoptosis as well as a potential developmental delay in the seminiferous epithelium that ultimately combine to elicit decreased spermatogenic output. The diminished sperm counts and motility defects in RHOX8-KD mice are similar to those observed in Rhox5-null animals [13] and Sertoli-specific WT1-knockdown animals previously generated using the Rhox5 promoter-driven RNAi system [17, 22]. However, several lines of evidence suggest that it is unlikely that RHOX5 and RHOX8 are completely redundant in postnatal testes. First, while RHOX8 may regulate Ins2, a recently identified RHOX5 target in Sertoli cells, it does so weakly [15]. The key residues within the homeodomain that confer DNA binding specificity vary between RHOX5 and RHOX8, suggesting that they have evolved to bind distinct promoters [13, 15]. In support of this, most of the RHOX5-regulated genes identified to date were not found to be misregulated in Sertoli cells of RHOX8-KD mice. Conversely, Rhox10 and Sox9 do not exhibit altered expression in Rhox5-null testes. Second, Rhox8 possesses a long trinucleotide repeat region that encodes a glutamate repeat domain that is unique to RHOX8 that may confer unique function. Third, RHOX8 is highly expressed in Sertoli cells of all seminiferous tubules, whereas RHOX5 is restricted to Sertoli cells of stages IV–VIII of the spermatogenic cycle [27], consistent with its well-established regulation by androgen receptor signaling [3, 35–38]. Finally, unlike Rhox5, Rhox8 is not androgen responsive, as its expression was not diminished in testes lacking androgen receptors.
Because of their differences in regulation and expression patterns in Sertoli cells, it is tempting to speculate that RHOX8 may synergize with RHOX5 to coordinate the expression of unique downstream targets that govern the androgen-independent and androgen-dependent arms of spermatogenesis, respectively. In support of this notion, our previous characterization revealed that germ cell defects in Rhox5-null mice resulted in fewer stage VII round and elongated spermatids in one of the androgen critical phases of spermatogenesis [13]. While a complete investigation was not pursued, preliminary analyses (∼150 tubules from four mice per genotype) found that putative reductions in round and elongated spermatid numbers were highly variable and not statistically significant in RHOX8-KD mice (data not shown). Additionally, while there was an apparent disruption in stage VI–VII transition, spermatogenesis appeared to proceed normally after that point in RHOX8-KD testes. The precise cause of accumulation at stage VI and reduced appearance of preleptotene spermatocytes (as assessed by STRA8 staining) cannot be determined until an established factor with such a role is identified as a putative RHOX8-regulated target for characterization. However, Rhox10 was downregulated in RHOX8-KD testes. The function of RHOX10 is unknown, but its expression pattern throughout the spermatogenic cycle has been determined [32]. We have not examined which cell types Rhox10 is downregulated in RHOX8-KD testes, but it is most highly expressed in the nuclei of spermatogonia of all stages followed by preleptotene to zygotene spermatocytes of stages IX and XI when RHOX5 is not present but RHOX8 is abundant. Thus, RHOX10 is positioned as a factor to contribute to B spermatogonia transition to preleptotene spermatocytes. However, its relevance in this process remains to be determined.
In addition to potential alteration of germ cell development, low spermatogenic output in RHOX8-KD mice may result from reduced survivability of germ cells. Downregulation of Ins2 and Sox9 and upregulation of Gdf9, all of which were misregulated in RHOX8-KD testes, may be responsible for increased germ cell death. In our recent report, Ins2 was found to be a direct target of RHOX5 [15]. Disruptions in insulin signaling are known to elicit cell death in a variety of tissues, but Ins2 became of great interest when it was discovered that the Akita mouse, a diabetes model organism with a naturally occurring point mutation in Ins2, exhibited similar defects in male germ cell survival as Rhox5-null mice [13, 39, 40]. Sox8 and Sox9 are members of the group E sox transcription factors. Conditional ablation of Sox8 [41] or Sox9 [42] results in male sterility at 5–6 mo, which in double mutants accelerates to PND 18, suggesting compensation between these genes [42]. SOX9 was reduced in RHOX8-KD mice, and Sox8 mRNA was downregulated but not SOX8 protein, suggesting that while loss of RHOX8 may result in downregulation of SOX9 eliciting germ cell apoptosis, residual SOX9 and/or continued expression of SOX8 preserve subfertility. Finally, the serendipitous discovery that Gdf9 may be downstream of RHOX8 is intriguing, as, in addition to its well-established role in female gonad development and fertility, it has been shown to be a germ cell regulator of Sertoli function [43]. In rat testis cultures, recombinant GDF9 disrupted the integration of junctional proteins responsible for forming Sertoli tight junctions. We do not know if the blood-testis barrier is disrupted in RHOX8-KD mice, but its integrity is essential for normal spermatogenesis and fertility [44].
Efforts to clarify the role of homeobox genes in the male reproductive tract using conventional knockout approaches have been challenging, as targeted disruption often causes embryonic lethality, thereby precluding analysis of their role in postnatal and adult sexual development and fertility [8, 38]. Knockout mice for many homeobox genes do not exhibit any obvious defects in fertility, most likely due to functional redundancy. Recently, the expression of the Rhox cluster during embryonic gonad development was examined by quantitative real-time RT-PCR (qPCR) and in situ hybridization [16]. Daggag et al. [16] found that 8 of the 13 basic Rhox genes were expressed in a dynamic and sexually dimorphic manner in embryonic germ cells. While Rhox5 expression in the adult is restricted to Sertoli cells, their qPCR fractionation study showed that like Ddx4 [45] and the other members of the Rhox cluster, Rhox5 is expressed only in Oct4-GFP-positive germ cells purified from the embryonic gonad. Knockdown of RHOX8 in postnatal Sertoli cells results in similar male subfertility as our Rhox5-null mice [1]. However, unlike all other RHOX factors, Rhox8 is expressed in the somatic cells of the embryonic gonad and remains expressed in all Sertoli cells from birth to adult. Because our current Rhox8-siRNA mice do not activate their inhibitory transgene until PND 7–10, when androgen signaling is established, it is quite likely that we have missed a significant portion of RHOX8's potential functions (i.e., if RHOX8 ultimately regulates Sox8/Sox9 in the embryonic gonad, its loss may have widespread effects on testis development). Thus, while our present results provide a key addition to our long-term goal of understanding the relative contributions of each RHOX factor to promote fertility, a definitive answer will require analysis of conditional RHOX8-KD models that may be employed at earlier developmental stages.
ACKNOWLEDGMENT
The authors would like to thank Dr. Miles Wilkinson (University of California, San Diego), Dr. Karel De Gendt (Catholic University of Leuven, Belgium), and Dr. Guido Verhoeven (Catholic University of Leuven, Belgium) for supplying the testis samples from androgen receptor-deficient mice. We thank Prema Narayan (Southern Illinois University) for assistance with Leydig cell counting and helpful comments during the editing of the manuscript. The authors would like to thank Yao-Fu Chang (University of Texas Health Science Center at San Antonio) for preparation of the pMAN vector. We would also like to thank Lisa Stein (IMGENEX) and Sherleen Cepeda (Aviva Systems Biology) for supplying the RHOX8 antisera used in this study.
Footnotes
Supported by National Institutes of Health/NICHD HD055268 to J.A.M., SIU ORDA Faculty Seed Grant (to J.A.M.), and the SIU Research Rookies program.
These authors contributed equally to this work.
REFERENCES
- Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- Svingen T, Koopman P. Involvement of homeobox genes in mammalian sexual development. Sex Dev. 2007;1:12–23. doi: 10.1159/000096235. [DOI] [PubMed] [Google Scholar]
- MacLean JA, II, Wilkinson MF. The Rhox genes. Reproduction. 2010;140:195–213. doi: 10.1530/REP-10-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MK, Wayne CM, Wilkinson MF. Pem homeobox gene regulatory sequences that direct androgen-dependent developmentally regulated gene expression in different subregions of the epididymis. J Biol Chem. 2002;277:48771–48778. doi: 10.1074/jbc.M209417200. [DOI] [PubMed] [Google Scholar]
- MacLean JA., II The role of Rhox homeobox factors in tumorigenesis. Front Biosci. 2013;18:474–492. doi: 10.2741/4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph A, Yao H, Hinton BT. Development and morphogenesis of the Wolffian/epididymal duct, more twists and turns. Dev Biol. 2009;325:6–14. doi: 10.1016/j.ydbio.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey S, Wilkinson MF. Homeobox genes and male reproductive development. J Assist Reprod Genet. 1996;13:182–192. doi: 10.1007/BF02072542. [DOI] [PubMed] [Google Scholar]
- Rao M, Wilkinson MF. Homeobox genes and the male reproductive tract. In: Robaire B, Hinton BT, editors. The Epididymis. New York: Kluwer Academic/Plenum Publishers;; 2002. pp. 269–283. In. (eds.) [Google Scholar]
- Snyder EM, Small CL, Bomgardner D, Xu B, Evanoff R, Griswold MD, Hinton BT. Gene expression in the efferent ducts, epididymis, and vas deferens during embryonic development of the mouse. Dev Dyn. 2010;239:2479–2491. doi: 10.1002/dvdy.22378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyabayashi K, Katoh-Fukui Y, Ogawa H, Baba T, Shima Y, Sugiyama N, Kitamura K, Morohashi K. Aristaless related homeobox gene, Arx, is implicated in mouse fetal Leydig cell differentiation possibly through expressing in the progenitor cells. PLoS One. 2013;8:e68050. doi: 10.1371/journal.pone.0068050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearse RV, II, Drolet DW, Kalla KA, Hooshmand F, Bermingham JR, Jr, Rosenfeld MG. Reduced fertility in mice deficient for the POU protein sperm-1. Proc Natl Acad Sci U S A. 1997;94:7555–7560. doi: 10.1073/pnas.94.14.7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann HM, Tamrazian A, Xie H, Perez-Millan MI, Kauffman AS, Mellon PL. Heterozygous deletion of ventral anterior homeobox (Vax1) causes subfertility in mice. Endocrinology. 2014;155:4043–4053. doi: 10.1210/en.2014-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean JA, II, Chen MA, Wayne CM, Bruce SR, Rao M, Meistrich ML, Macleod C, Wilkinson MF. Rhox: a new homeobox gene cluster. Cell. 2005;120:369–382. doi: 10.1016/j.cell.2004.12.022. [DOI] [PubMed] [Google Scholar]
- Bonaparte E, Moretti M, Colpi GM, Nerva F, Contalbi G, Vaccalluzzo L, Tabano S, Grati FR, Gazzano G, Sirchia SM, Simoni G, Gallina A, et al. ESX1 gene expression as a robust marker of residual spermatogenesis in azoospermic men. Hum Reprod. 2010;25:1398–1403. doi: 10.1093/humrep/deq074. [DOI] [PubMed] [Google Scholar]
- MacLean JA, II, Hu Z, Welborn JP, Song HW, Rao MK, Wayne CM, Wilkinson MF. The RHOX homeodomain proteins regulate the expression of insulin and other metabolic regulators in the testis. J Biol Chem. 2013;288:34809–34825. doi: 10.1074/jbc.M113.486340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daggag H, Svingen T, Western PS, van den Bergen JA, McClive PJ, Harley VR, Koopman P, Sinclair AH. The rhox homeobox gene family shows sexually dimorphic and dynamic expression during mouse embryonic gonad development. Biol Reprod. 2008;79:468–474. doi: 10.1095/biolreprod.107.067348. [DOI] [PubMed] [Google Scholar]
- Rao MK, Wilkinson MF. Tissue-specific and cell type-specific RNA interference in vivo. Nat Protoc. 2006;1:1494–1501. doi: 10.1038/nprot.2006.260. [DOI] [PubMed] [Google Scholar]
- Brown RM, Davis MG, Hayashi K, MacLean JA. Regulated expression of Rhox8 in the mouse ovary: evidence for the role of progesterone and RHOX5 in granulosa cells. Biol Reprod. 2013;88:126. doi: 10.1095/biolreprod.112.103267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean JA, II, Hayashi K, Turner TT, Wilkinson MF. The Rhox5 homeobox gene regulates the region-specific expression of its paralogs in the rodent epididymis. Biol Reprod. 2012;86:189. doi: 10.1095/biolreprod.112.099184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon NA, Handyside AH, Joyce IM. Expression of Sox8, Sf1, Gata4, Wt1, Dax1, and Fog2 in the mouse ovarian follicle: implications for the regulation of Amh expression. Mol Reprod Dev. 2005;70:271–277. doi: 10.1002/mrd.20208. [DOI] [PubMed] [Google Scholar]
- Reardon SN, King ML, MacLean JA, II, Mann JL, DeMayo FJ, Lydon JP, Hayashi K. CDH1 is essential for endometrial differentiation, gland development, and adult function in the mouse uterus. Biol Reprod. 2012;86:141. doi: 10.1095/biolreprod.112.098871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MK, Pham J, Imam JS, MacLean JA, Murali D, Furuta Y, Sinha-Hikim AP, Wilkinson MF. Tissue-specific RNAi reveals that WT1 expression in nurse cells controls germ cell survival and spermatogenesis. Genes Dev. 2006;20:147–152. doi: 10.1101/gad1367806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang YL, Li H, Chen WH, Tzeng YS, Lai YL, Hsieh-Li HM. A novel PEPP homeobox gene, TOX, is highly glutamic acid rich and specifically expressed in murine testis and ovary. Biol Reprod. 2004;70:828–836. doi: 10.1095/biolreprod.103.021048. [DOI] [PubMed] [Google Scholar]
- Bhardwaj A, Song HW, Beildeck M, Kerkhofs S, Castoro R, Shanker S, De Gendt K, Suzuki K, Claessens F, Issa JP, Orgebin-Crist MC, Wilkinson MF. DNA demethylation-dependent AR recruitment and GATA factors drive Rhox5 homeobox gene transcription in the epididymis. Mol Endocrinol. 2012;26:538–549. doi: 10.1210/me.2011-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean JA, II, Rao MK, Doyle KM, Richards JS, Wilkinson MF. Regulation of the Rhox5 homeobox gene in primary granulosa cells: preovulatory expression and dependence on SP1/SP3 and GABP. Biol Reprod. 2005;73:1126–1134. doi: 10.1095/biolreprod.105.042747. [DOI] [PubMed] [Google Scholar]
- Barrionuevo F, Scherer G. SOX. E genes: SOX9 and SOX8 in mammalian testis development. Int J Biochem Cell Biol. 2010;42:433–436. doi: 10.1016/j.biocel.2009.07.015. [DOI] [PubMed] [Google Scholar]
- Rao MK, Wayne CM, Meistrich ML, Wilkinson MF. Pem homeobox gene promoter sequences that direct transcription in a Sertoli cell-specific, stage-specific, and androgen-dependent manner in the testis in vivo. Mol Endocrinol. 2003;17:223–233. doi: 10.1210/me.2002-0232. [DOI] [PubMed] [Google Scholar]
- Ahmed EA, de Rooij DG. Staging of mouse seminiferous tubule cross-sections. Methods Mol Biol. 2009;558:263–277. doi: 10.1007/978-1-60761-103-5_16. [DOI] [PubMed] [Google Scholar]
- Russell LD. Histological and Histopathological Evaluation of the Testis Clearwater, FL: Cache River Press; 1990. [Google Scholar]
- Zhou Q, Nie R, Li Y, Friel P, Mitchell D, Hess RA, Small C, Griswold MD. Expression of stimulated by retinoic acid gene 8 (Stra8) in spermatogenic cells induced by retinoic acid: an in vivo study in vitamin A-sufficient postnatal murine testes. Biol Reprod. 2008;79:35–42. doi: 10.1095/biolreprod.107.066795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean JA, II, Lorenzetti D, Hu Z, Salerno WJ, Miller J, Wilkinson MF. Rhox homeobox gene cluster: recent duplication of three family members. Genesis. 2006;44:122–129. doi: 10.1002/gene.20193. [DOI] [PubMed] [Google Scholar]
- Song HW, Dann CT, McCarrey JR, Meistrich ML, Cornwall GA, Wilkinson MF. Dynamic expression pattern and subcellular localization of the Rhox10 homeobox transcription factor during early germ cell development. Reproduction. 2012;143:611–624. doi: 10.1530/REP-11-0479. [DOI] [PubMed] [Google Scholar]
- Geyer CB, Eddy EM. Identification and characterization of Rhox13, a novel X-linked mouse homeobox gene. Gene. 2008;423:194–200. doi: 10.1016/j.gene.2008.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song HW, Anderson RA, Bayne RA, Gromoll J, Shimasaki S, Chang RJ, Parast MM, Laurent LC, de Rooij DG, Hsieh TC, Wilkinson MF. The RHOX homeobox gene cluster is selectively expressed in human oocytes and male germ cells. Hum Reprod. 2013;28:1635–1646. doi: 10.1093/humrep/det043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gendt K, Swinnen JV, Saunders PT, Schoonjans L, Dewerchin M, Devos A, Tan K, Atanassova N, Claessens F, Lecureuil C, Heyns W, Carmeliet P, et al. A Sertoli cell-selective knockout of the androgen receptor causes spermatogenic arrest in meiosis. Proc Natl Acad Sci U S A. 2004;101:1327–1332. doi: 10.1073/pnas.0308114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdcraft RW, Braun RE. Androgen receptor function is required in Sertoli cells for the terminal differentiation of haploid spermatids. Development. 2004;131:459–467. doi: 10.1242/dev.00957. [DOI] [PubMed] [Google Scholar]
- Hu Z, Dandekar D, O'Shaughnessy PJ, De Gendt K, Verhoeven G, Wilkinson MF. Androgen-induced Rhox homeobox genes modulate the expression of AR-regulated genes. Mol Endocrinol. 2010;24:60–75. doi: 10.1210/me.2009-0303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean JA, II, , Wilkinson MF. Gene regulation in spermatogenesis. Curr Top Dev Biol. 2005;71:131–197. doi: 10.1016/S0070-2153(05)71005-X. [DOI] [PubMed] [Google Scholar]
- Izumi T, Yokota-Hashimoto H, Zhao S, Wang J, Halban PA, Takeuchi T. Dominant negative pathogenesis by mutant proinsulin in the Akita diabetic mouse. Diabetes. 2003;52:409–416. doi: 10.2337/diabetes.52.2.409. [DOI] [PubMed] [Google Scholar]
- Kim ST, Moley KH. Paternal effect on embryo quality in diabetic mice is related to poor sperm quality and associated with decreased glucose transporter expression. Reproduction. 2008;136:313–322. doi: 10.1530/REP-08-0167. [DOI] [PubMed] [Google Scholar]
- O'Bryan MK, Takada S, Kennedy CL, Scott G, Harada S, Ray MK, Dai Q, Wilhelm D, de Kretser DM, Eddy EM, Koopman P, Mishina Y. Sox8 is a critical regulator of adult Sertoli cell function and male fertility. Dev Biol. 2008;316:359–370. doi: 10.1016/j.ydbio.2008.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrionuevo F, Georg I, Scherthan H, Lecureuil C, Guillou F, Wegner M, Scherer G. Testis cord differentiation after the sex determination stage is independent of Sox9 but fails in the combined absence of Sox9 and Sox8. Dev Biol. 2009;327:301–312. doi: 10.1016/j.ydbio.2008.12.011. [DOI] [PubMed] [Google Scholar]
- Nicholls PK, Harrison CA, Gilchrist RB, Farnworth PG, Stanton PG. Growth differentiation factor 9 is a germ cell regulator of Sertoli cell function. Endocrinology. 2009;150:2481–2490. doi: 10.1210/en.2008-1048. [DOI] [PubMed] [Google Scholar]
- Morrow CM, Mruk D, Cheng CY, Hess RA. Claudin and occludin expression and function in the seminiferous epithelium. Philos Trans R Soc Lond B Biol Sci. 2010;365:1679–1696. doi: 10.1098/rstb.2010.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara Y, Komiya T, Kawabata H, Sato M, Fujimoto H, Furusawa M, Noce T. Isolation of a DEAD-family protein gene that encodes a murine homolog of Drosophila vasa and its specific expression in germ cell lineage. Proc Natl Acad Sci U S A. 1994;91:12258–12262. doi: 10.1073/pnas.91.25.12258. [DOI] [PMC free article] [PubMed] [Google Scholar]