Abstract
The main nonmedullary form of thyroid cancer is papillary thyroid carcinoma (PTC) that accounts for 80–90% of all thyroid malignancies. Only 3–10% of PTC patients have a positive family history of PTC yet the familiality is one of the highest of all cancers as measured by case control studies. A handful of genes have been implicated accounting for a small fraction of this genetic predisposition. It was therefore of considerable interest that a mutation in the HABP2 gene was recently implicated in familial PTC. The present work was undertaken to examine the extent of HABP2 variant involvement in PTC. The HABP2 G534E variant (rs7080536) was genotyped in blood DNA from 179 PTC families (one affected individual per family), 1160 sporadic PTC cases and 1395 controls. RNA expression of HABP2 was tested by qPCR in RNA extracted from tumor and normal thyroid tissue from individuals that are homozygous wild-type or heterozygous for the variant. The variant was found to be present in 6.1% familial cases, 8.0% sporadic cases (2 individuals were homozygous for the variant) and 8.7% controls. The variant did not segregate with PTC in one large and 6 smaller families in which it occurred. In keeping with data from the literature and databases the expression of HABP2 was highest in the liver, much lower in 3 other tested tissues (breast, kidney, brain) but not found in thyroid. Given these results showing lack of any involvement we suggest that the putative role of variant HABP2 in PTC should be carefully scrutinized.
Introduction
Thyroid cancer represents 3.8% of all new cancer cases in the United States. It is estimated that 62,450 new cases will be diagnosed in 2015 in the United States (www.seer.cancer.gov).
PTC has been shown to be one of the most heritable cancers of all [1–5]. It is therefore notable that only few gene variants have been suggested, let alone proven to predispose to PTC. In early reports linkage analyses pointed to several loci for genetic predisposition to PTC but causal variants have not been found [6]. More recently several studies based on the candidate gene approach have found variants associated with predisposition to PTC but the variants appear to be rare being found in just 1 family [7], present in a few families [8] or present in one family and a small number of “sporadic” cases [9,10]. Other variants emanate from genome-wide association studies (GWAS) but account for just some 10% of the estimated genetic predisposition [11]. In conclusion, the predisposition from all the above variants accounts for just a minority of all PTC.
Recently Gara et al. [12] reported a germline HABP2 mutation causing familial nonmedullary thyroid cancer. This missense mutation (c.1601G>A; p.G534E; rs7080536) was found in a family affected with PTC and was shown to segregate with PTC in that family. The variant was present in 4.7% of 423 patients in the TCGA database and was reported to occur in 0.7% when combining controls from 1000 Genomes Project (phase 3; www.1000genomes.org) and HapMap3 (release 3; http://hapmap.ncbi.nlm.nih.gov/), suggesting a relatively high population attributable fraction. The present study was undertaken to elaborate further on the role of the HABP2 variant in PTC risk.
Materials and Methods
Patients and controls
The studies were approved by the Institutional Review Board at The Ohio State University Medical Center. All subjects gave written informed consent before participation.
Individuals with thyroid cancer from The Ohio State University (OSU) collection were histologically confirmed as having traditional PTC or follicular variant PTC. The majority of the patients had no family history and were designated as “sporadic”. We defined “familial” broadly as the occurrence of two or more first- or second-degree relatives with PTC; there were in total 179 families meeting this criterion. Of these families 103 have 3 or more first or second degree relatives (Group 1) while 76 have only 2 individuals with PTC (Group 2). The majority of our families are of Caucasian origin. Information on sporadic cases and controls used in this study can be found in Tomsic et al. 2015 [10] and S1 Table.
DNA and RNA extraction
Genomic DNA was extracted from blood following standard phenol-chloroform extraction procedures. RNA was extracted from white blood cells using TRIzol reagent. RNA quality was checked using an Agilent 2100 Bioanalyzer.
Genotyping
SNaPshot assay (Life Technologies) was used on genomic DNA from blood of familial PTC patients for genotyping HABP2 G534E variant (SNP rs7080536) as previously described [9] (He at al. 8q24 2009). The primers for PCR were as follows: forward 5’-TATGCCTCTGTTTCCCTTAG-3’ and reverse 5’- TGAGGTCCAGAAGACAGTAC-3’.
PCR assays were performed according to a standard protocol using AmpliTaq Gold DNA polymerase (Life Technologies) as follows: 10 min at 94°C; followed by 32 cycles of 15 s at 94°C, 15 s at 58°C, and 1 min at 72°C; followed by a final extension of 5 min at 72°C.
Screening for the SNP 7080536 in Ohio PTC patients and controls was carried out by deCODE Genetics in Reykjavik, Iceland, applying the Centaurus (Nanogen) single-track assay [13,10]. To confirm the A allele at rs7080536 as identified from single-track assay, we used SNaPshot assay (as described above) on unamplified DNA from roughly 200 samples extracted from blood and the results were 100% concordant.
Quantitative PCR (qPCR) measurement of gene expression
One μg of RNA was reverse transcribed using the High Capacity Reverse Transcription kit (Applied Biosystems). The expression of HABP2 was measured using PrimeTime® qPCR Assays (Hs.PT.58.592181, Hs.PT.58.39116864,and Hs.PT.58.38507028; Integrated DNA Technologies) and the expression of GAPDH as internal control was measured using Human GAPD (GAPDH) (Life Technologies). The real time PCR reaction was run in the ABI Prism 7900HT Sequence Detection System (Applied Biosystems). Fast TaqMan assay reaction mix was used and the conditions were as follows: 95°C for 5 min followed by 40 cycles at 95°C, 5 s and 60°C, 5 s. At the end of the assay an aliquot of the reaction was loaded on 1.5% agarose gel.
Results
Frequency of the G534E variant of HABP2 in patients and controls
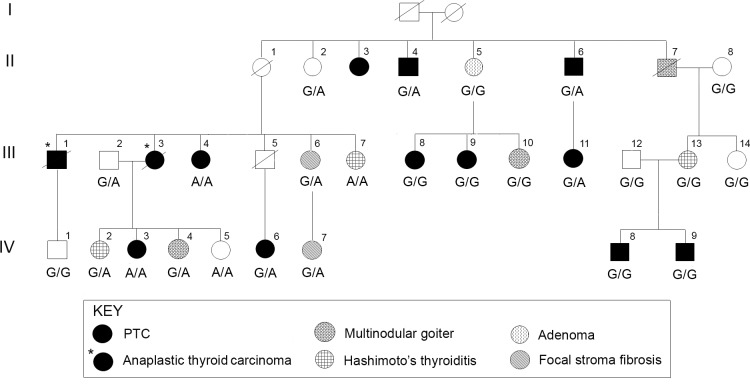
Using SNaPshot analysis on one affected individual from each of the 179 available families we found 11 heterozygous variant carriers (6.1%; Table 1) corresponding to a minor allele frequency of 3.1% (Table 1). Six of the eleven variant carriers are found among the 101 families with 3 or more affected individuals (Group 1) while five are found in the remaining 75 families where only 2 individuals are affected (Group 2). We genotyped the available family members of the 8 families that had blood DNA available for at least two individuals; six belonging to Group 1 and two belonging to Group 2. One of these families (Family 1) is a large family in which after genotyping the 25 available samples the variant clearly disclosed no cosegregation with the disease (simplified pedigree shown in Fig 1). Moreover, as many as four individuals were homozygous for the variant (two with PTC–III,4 and IV,3; one with Hashimoto’s thyroiditis–III,7; and one unaffected–IV,5). There are several PTC patients in Family 1 that are not carriers of the risk allele (A) (III,8 and 9 and IV,8 and 9). Interestingly, a causative genomic alteration in this family has been described previously [7]. The mutation is a single nucleotide change located in a long-range enhancer element on chromosome 4q.
Table 1. Genotyping results of variant HABP2 G534E (rs7080536) in Ohio cohorts.
| Genotype | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| GG | GA | AA | Minor Allele Frequency (%) | p-value* | |||||
| Sample | Individuals | Count | Proportion (%) | Count | Proportion (%) | Count | Proportion (%) | ||
| Familial PTC | 179 | 168 | 93.9 | 11 | 6.1 | 0 | 0.0 | 3.1 | 0.32 |
| Group 1 (3+) | 103 | 97 | 94.2 | 6 | 5.8 | 0 | 0.0 | 2.9 | 0.46 |
| Group 2 (2) | 76 | 71 | 93.4 | 5 | 6.6 | 0 | 0.0 | 3.3 | 0.67 |
| Sporadic PTC | 1160 | 1067 | 92.0 | 91 | 7.8 | 2 | 0.17 | 4.1 | 0.24 |
| Controls | 1395 | 1274 | 91.3 | 121 | 8.7 | 0 | 0.0 | 4.3 | |
* Fisher's Exact test p-value, comparing genotype distributions with Controls
Fig 1. Genotyping of HABP2 G534E variant in Family 1.
The pedigree shows genotypes of the HABP2 variant (c.1601G>A; p.G534E). Circle, female; square, male. Please note that in this family the basis of the predisposition to PTC has been determined. It is a single nucleotide variant in a long-range enhancer on chromosome 4q (He et al., 2013) [7].
The genotyping results for families 1 to 6 (Fig 1 and S1 Fig) clearly show no segregation between PTC and the G534E variant. There is at least one affected individual in each family that does not carry the HABP2 variant (marked with asterisk). The results in Family 7 and Family 8 are compatible with cosegregation but this can obviously be just a matter of chance.
Genotyping was further performed on blood DNA from 1160 sporadic PTC cases and 1395 controls (Table 1). The G534E variant is present in a total of 93 patients (8.0%), with 2 patients being homozygous for the variant (0.17%) leaving 91 heterozygous patients (7.8%). Among the controls there are 121 individuals (8.7%) heterozygous for the variant but no homozygous variant carriers. We calculated Fisher’s exact test p-value comparing genotype distributions of different groups of cases with controls and we found no statistically significant difference in distribution (Table 1). The minor allele (A) frequency is 4.1% in cases and 4.3% in controls showing no statistically significant difference between the two groups (Fisher’s exact test p-value = 0.676).
RNA expression of HABP2
Gara et al. [12] reported a higher mRNA expression of HABP2 in the tumor tissue as compared with adjacent normal tissue. This finding is different from the data in Human Gene Atlas [14] and Illumina Body Map (Array Express accession: E-MTAB-513; http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-513/) according to which HABP2 mRNA is expressed primarily in liver and not expressed in normal thyroid. We studied RNA expression of the gene in fresh frozen paired tumor and normal thyroid tissue from heterozygotes for the variant (n = 3) as well as from wild-type homozygotes (n = 5), all representing sporadic PTC. HABP2 expression was not detected in any of the samples (the Ct value was undetermined for HABP2 while in the range of 20–22 for GAPDH endogenous control; S2 Fig). We also performed qPCR on cDNAs derived from human breast, kidney, liver, and brain tissue and expression was seen in liver with traces of expression in breast (S3 Fig). When using two different qPCR assays expression of the gene is still the highest in liver but expression also is detectable in brain and kidney (S4 Fig). Reactions were loaded on agarose gel to visualize qPCR products and results can be seen in Fig 2. On the gel the HABP2 amplicon can be seen in liver but not in thyroid (tumor or adjacent normal tissue) that either carry or don’t carry the variant.
Fig 2. The HABP2 gene is mainly expressed in liver and not expressed in thyroid tumors or adjacent normal thyroid tissue from patients carrying or not carrying the HABP2 G534E variant.
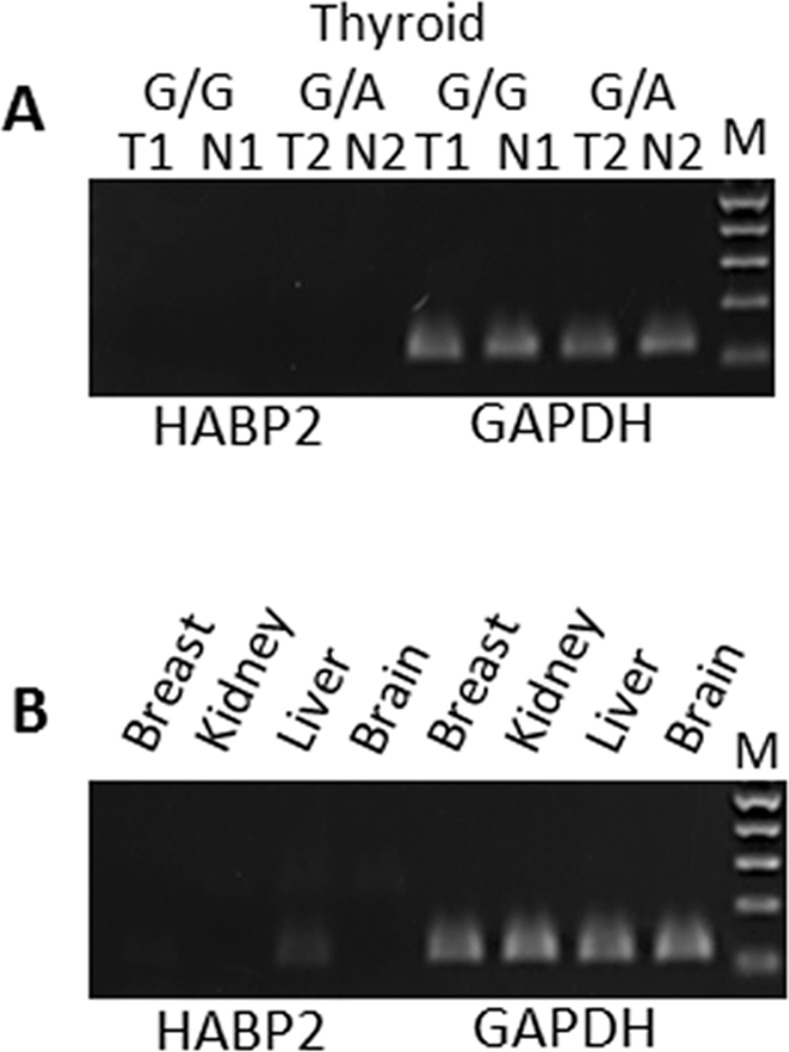
(A) Detection of the HABP2 gene expression in thyroid tumor (T) or adjacent normal (N) sample. Patient 1 was heterozygous for the variant (G/A), patient 2 was homozygous wild-type (G/G). (B) qPCR in normal tissue. HABP2 showing faint but clear expression in liver. GAPDH was used as internal control. Details for qPCR reactions are described in Materials and Methods.
Discussion
In our study we find no evidence supporting a role for the HABP2 G534E variant (SNP rs7080536) in PTC. There is no difference in the frequency of variant carriers among familial PTC cases (grouped by the number of affected family members), sporadic PTC cases, and controls (Table 1). We found no cosegregation of the variant in six out of eight families tested. It is noticeable that in two medium size families (Family 7 and Family 8, S1 Fig) the variant is segregating with PTC and this finding is compatible with cosegregation as well as with the result of chance alone. The fact that the cosegregation occurs in only two out of eight tested families speaks in favor of our interpretation of the data, that is, that the variant is not associated with PTC.
When genotyping our cases (n = 1160) and controls (n = 1395) (mostly Caucasian; for clinical and demographic information see S1 Table) we found the minor allele frequency (MAF) to be 4.1% and 4.3% respectively. When examining the MAF of the HABP2 G534E variant in a database of controls (http://evs.gs.washington.edu/EVS/; Table 2) we found it to be 4.0% in a European American population (n = 4300) while it was remarkably different at 0.66% in an African American population (n = 2203). In another database of controls, the 1000 Genomes Project Phase 3 (www.1000genomes.org; Table 3), MAF for the total population is 0.81% but we found pronounced differences in MAF among 5 populations with the highest MAF of 2.7% reported for the individuals of European ancestry, while the frequency is 0% in East Asian Ancestry.
Table 2. Genotyping results for SNP rs7080536 from NHLBI GO Exome Sequencing Project (ESP; http://evs.gs.washington.edu/EVS/).
| Genotype | ||||||||
|---|---|---|---|---|---|---|---|---|
| GG | GA | AA | Minor Allele Frequency (%) | |||||
| Sample | Individuals | Count | Proportion (%) | Count | Proportion (%) | Count | Proportion (%) | |
| European American | 4300 | 3971 | 92.3 | 324 | 7.6 | 5 | 0.1 | 4.0 |
| African American | 2203 | 2174 | 98.7 | 29 | 1.3 | 0 | 0.0 | 0.66 |
Table 3. Race distribution in 1000 Genome final release Phase 3 (total of 2504 individuals) and genotyping results for SNP rs7080536.
| Race | Individuals | Genotype (GA or AA) | Minor Allele Frequency (%) | ||
|---|---|---|---|---|---|
| Count | Proportion (%) | Count | Proportion (%) | ||
| East Asian Ancestry (EAS) | 504 | 20.1 | 0 | 0.0 | 0 |
| South Asian Ancestry (SAS) | 489 | 19.5 | 4 | 0.8 | 0.4 |
| African Ancestry (AFR) | 661 | 26.4 | 0 | 0.0 | 0 |
| European Ancestry (EUR)** | 503 | 20.1 | 26* | 5.2 | 2.7 |
| American Ancestry (AMR)** | 347 | 13.9 | 10 | 2.9 | 1.4 |
*25 samples are GA, 1 sample is AA
** EUR and AMR can be grouped as White
As reported by Gara et al. [12] the SNP rs7080536 was found in 4.7% of 423 thyroid cancer patients that had the sequence of the variant nucleotide position reported in the TCGA. Thyroid cancer patients in TCGA were selected not to be familial. When analyzing the race distribution of the 507 available thyroid cancer patients we note that the majority of individuals (65.7%) can be categorized as white (Table 4). The 404 individuals with UCSC mutation calling info (TCGA database—Nov 24th 2015) we find 21 heterozygous carriers of the variant, 19 of them being of European Ancestry and for two of them race is not specified. This confirms that most commonly carriers of the A allele are of European origin for PTC cases and controls (MAF in controls is 2.7% (Table 3), 4.0%(Table 2) or 4.3% (controls in Table 1)).
Table 4. Race distribution of 507 Thyroid Carcinoma Samples (THCA) from The Cancer Genome Atlas (09/21/15) (http://cancergenome.nih.gov/).
| Individuals | ||
|---|---|---|
| Race | Count | Proportion (%) |
| Not Specified | 94 | 18.5 |
| American Indian or Alaska Native | 1 | 0.2 |
| Asian | 52 | 10.3 |
| Black or African American | 27 | 5.3 |
| White | 333 | 65.7 |
Based on the results of our qPCR assays and gene expression data published in different databases, we conclude that the HABP2 gene is not expressed in healthy thyroid or tumor thyroid tissue from sporadic PTC cases. Our genetic findings and gene expression findings allow us to say that the HABP2 G534E variant is not a player in the predisposition to PTC.
Supporting Information
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
This work was supported by National Cancer Institute Grants P01CA124570 and P30CA16058. The OSUCCC Genomics Shared Resource performed SNP genotyping. Genotyping of rs7080536 in Ohio samples was kindly performed by Dr. Julius Gudmundsson at deCODE Inc, Reykjavik, Iceland. This work was supported in part by an allocation of computing time from the Ohio Supercomputer Center. The Body Map data were kindly provided by the Gene Expression Applications research group at Illumina. Pedigrees shown were kindly drawn by Megan Knapke. The results published here are in part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by National Cancer Institute (US) grants P01CA124570 and P30CA16058. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Risch N. The genetic epidemiology of cancer: Interpreting family and twin studies and their implications for molecular genetic approaches. Cancer Epidemiol Biomarkers Prev. 2001;10:733–741. [PubMed] [Google Scholar]
- 2.Czene K, Lichtenstein P,Hemminki K. Environmental and heritable causes of cancer among 9.6 million individuals in the Swedish family-cancer database. Int J Cancer. 2002;99: 260–266. [DOI] [PubMed] [Google Scholar]
- 3.Amundadottir LT, Thorvaldsson S, Gudbjartsson DF, Sulem P, Kristjansson K, Aranson S, et al. Cancer as a complex phenotype: Pattern of cancer distribution within and beyond the nuclear family. PLoS Med. 2004;1: 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albright F, Teerlink C, Werner T, Cannon-Albright L. Significant evidence for a heritable contribution to cancer predisposition: a review of cancer familiality by site. BMC Cancer. 2012;12: 138–144. 10.1186/1471-2407-12-138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fallah M, Sundquist K, Hemminki K. Risk of thyroid cancer in relatives of patients with medullary thyroid carcinoma by age at diagnosis. Endocr Relat Cancer. 2013;20: 717–724. 10.1530/ERC-13-0021 [DOI] [PubMed] [Google Scholar]
- 6.Bonora E, Tallini G, Romeo G. Genetic predisposition to familial nonmedullary thyroid cancer: An update of molecular findings and state-of-the-art studies. J Oncol. 2010;385: 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He H, Li W, Wu D, Nagy R, Liyanarachchi S, Akagi K, et al. Ultra-rare mutation in long-range enhancer predisposes to thyroid carcinoma with high penetrance. PLoS One. 2013;8: e61920 10.1371/journal.pone.0061920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He H, Bronisz A, Liyanarachchi S, Nagy R, Li W, Huang Y, et al. SRGAP1 is a candidate gene for papillary thyroid carcinoma susceptibility. J Clin Endocrin Metab. 2013: c.2012–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He H, Nagy R, Liyanarachchi S, Jiao H, Li W, Suster S, Kere J, de la Chapelle A. A susceptibility locus for papillary thyroid carcinoma on chromosome 8q24. Cancer Res. 2009;69: 625–631. 10.1158/0008-5472.CAN-08-1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomsic J, He H, Akagi K, Liyanarachchi S, Pan Q, Bertani B, et al. A germline mutation in SRRM2, a splicing factor gene, is implicated in papillary thyroid carcinoma predisposition. Sci Rep. 2015;5: 10566 10.1038/srep10566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liyanarachchi S, Wojcicka A, Li W, Czertwertynska M, Stachlewska E, Nagy R, et al. Cumulative risk impact of five genetic variants associated with papillary thyroid carcinoma. Thyroid. 2013;23: 1532–1540. 10.1089/thy.2013.0102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gara SK, Jia L, Merino MJ, Agarwal SK, Zhang L, Cam M, et al. Germline HABP2 Mutation Causing Familial Nonmedullary Thyroid Cancer. N Engl J Med. 2015;373: 448–455. 10.1056/NEJMoa1502449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kutyavin IV, Milesi D, Belousov Y, Podyminogin M, Vorobiev A, Gorn V, et al. A novel endonuclease IV post-PCR genotyping system. Nucleic Acids Res. 2006;34:e128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101: 6062–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.