Abstract
Helicobacter pylori (H. pylori) is a spiral-shaped Gram-negative bacterium that causes the most common chronic infection in the human stomach. Approximately 1%-3% of infected individuals develop gastric cancer. However, the mechanisms by which H. pylori induces gastric cancer are not completely understood. The available evidence indicates a strong link between the virulence factor of H. pylori, cytotoxin-associated gene A (CagA), and gastric cancer. To further characterize H. pylori virulence, we established three cell lines by infecting the gastric cancer cell lines SGC-7901 and AGS with cagA+ H. pylori and transfecting SGC-7901 with a vector carrying the full-length cagA gene. We detected 135 differently expressed proteins from the three cell lines using proteome technology, and 10 differential proteins common to the three cell lines were selected and identified by LC-MS/MS as well as verified by western blot: β-actin, L-lactate dehydrogenase (LDH), dihydrolipoamide dehydrogenase (DLD), pre-mRNA-processing factor 19 homolog (PRPF19), ATP synthase, calmodulin (CaM), p64 CLCP, Ran-specific GTPase-activating protein (RanGAP), P43 and calreticulin. Detection of the expression of these proteins and genes encoding these proteins in human gastric cancer tissues by real-time PCR (RT-qPCR) and western blot revealed that the expression of β-ACTIN, LDH, DLD, PRPF19 and CaM genes were up-regulated and RanGAP was down-regulated in gastric cancer tissues and/or metastatic lymph nodes compared to peri-cancerous tissues. High gene expression was observed for H. pylori infection in gastric cancer tissues. Furthermore, the LDH, DLD and CaM genes were demethylated at the promoter -2325, -1885 and -276 sites, respectively, and the RanGAP gene was highly methylated at the promoter -570 and -170 sites in H. pylori-infected and cagA-overexpressing cells. These results provide new insights into the molecular pathogenesis and treatment targets for gastric cancer with H. pylori infection.
Introduction
Helicobacter pylori (H. pylori) causes the most common chronic stomach infection in humans worldwide. Approximately half of the world’s population is infected with H. pylori, and the majority of colonized individuals develop asymptomatic gastritis. Among infected individuals, approximately 10%-20% of individuals develop peptic ulcer diseases and 1%-3% develop gastric cancer [1,2]. In 1994, H. pylori was classified as a type I carcinogen for gastric cancer by the World Health Organization’s International Agency for Research on Cancer.
Gastric cancer is one of the most common types of cancers, and more than 70% of new cases and deaths occur in developing countries [3]. Although the global incidence rate has been declining for several decades, gastric cancer remains prevalent in most developing countries, including Japan, Korea and China [4–6]. In 2012, the Chinese Cancer Registry Annual Report indicated that gastric cancer morbidity and mortality are second and third among all malignant tumors, respectively.
The majority of H. pylori strains carry the cag pathogenicity island (cagPAI), which contains 27 to 31 genes that encode a bacterial type 4 secretion system (T4SS) [7]. The cytotoxin-associated gene A gene (cagA), which is located at the 3′ end of cagPAI, encodes the only known bacterial oncoprotein, CagA, which is translocated into host cells by T4SS after bacterial attachment to the stomach. Once inside the host cells, CagA is tyrosine phosphorylated by members of the Abl and Src kinase families and interacts with numerous intracellular effectors, leading to the activation of downstream signaling molecules [8]. In clinical studies, cagA-positive strains have been consistently linked to more severe gastric inflammation and ulcers, and a small fraction of individuals develop gastric cancer [9]. However, the mechanisms underlying the association of CagA with cancer have not been elucidated.
Proteomics has emerged as a promising technological platform for the rational identification of biomarkers and novel therapeutic targets for diseases and the determination of the underlying mechanisms of carcinogenesis [10]. However, most of the important proteins detected by proteomics in vitro have not been confirmed in vivo in clinical samples.
DNA methylation and demethylation plays an important role in the development and progression of cancers by blocking the binding of transcription factors to DNA and is occurs nearly exclusively in gene promoter CpG islands [11–13]. Among organs, the stomach exhibits the highest frequency of abnormal CpG island methylation, possibly H. pylori-mediated [14–16]. Although studies of DNA methylation are increasing, the gene methylation states induced by H. pylori are not yet clear.
In this work, we aimed to identify specific proteins related to H. pylori infection using comparative proteomics and characterize the gene expression and CpG island methylation of these proteins in gastric cancer tissues and cells.
Materials and Methods
Human tissues
Human tissues were obtained from surgery specimens from 30 gastric cancer patients and matched adjacent cancer tissues and metastatic lymph nodes at Guiyang Medical Hospital, Guiyang China, between January 2009 and June 2010. The diagnoses were confirmed by two pathologists. Among the patients, 23 were male, and 7 were female. The patients ranged in age from 38 to 77 years. Twenty-two patients had intestinal-type adenocarcinoma, and 8 had diffuse-type adenocarcinoma. The study protocol was approved by the Ethics Committee of Guiyang Medical Hospital, and all subjects provided written informed consent.
Cell culture
The human gastric carcinoma cell line AGS (ATCC CRL-1739TM) and SGC-7901 cells were purchased directly from American Type Culture Collection (ATCC) and Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China), respectively, and passaged for less than 3 months in our laboratory after receipt. Cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin (Gibco, Grand Island, NY, USA) at 37°C in a humidified incubator with 5% CO2.
H. pylori culture
H. pylori strain NCTC11637 (ATCC 43504, a gift from the Chinese Center of Helicobacter pylori strain Management and Preservation, Beijing, China), which is cagA positive, was grown in selective medium on a Columbia agar plate containing 10% fetal calf serum and H. pylori Selective Supplement (Oxoid Ltd, England) at 37°C under microaerobic conditions.
Cell infection with H. pylori
AGS and SGC-7901 cells (5×105) were seeded in 6-well plates for 24 h and infected with H. pylori for 6 h and 12 h at a multiplicities of infection (MOIs) of 1:100, 1:500 and 1:1000, respectively. Cells were infected for 12 h at a MOI of 1:1000 with H. pylori boiled for 15 min as controls.
Construction of the expression vector pcDNA3.1/cagA
DNA was extracted from H. pylori, and the full-length cagA sequence was synthesized by PCR and cloned into pMD18-T plasmids to construct pMD18-T/cagA. The cagA gene was identified by sequencing (GQ161098). pMD18-T/cagA was digested with the restriction enzymes PstI and BamHI, and cagA was ligated into pcDNA3.1/Zeo (-) (Invitrogen, USA) to construct the eukaryotic expression vector pcDNA3.1/cagA. The sequences of the primers are provided in Table 1.
Table 1. Primers used in PCR and quantitative RT- PCR.
| Gene | Primer sequence | Length (bp) | GenBank accession no. |
|---|---|---|---|
| cagAa | Sense: 5′-ACAATGACTAACGAAACCA-3′ | 3467 | GQ161098 |
| Antisense: 5′-TTTTGGTAT TCCTTAATCCT-3′ | |||
| cagAb | Sense: 5′-AATACACCAACGCCTCCAAG-3′ | 397 | GQ161098 |
| Antisense: 5′- AAAAGCTTCTGCAGAGCTGGGAGGTGTG-3′ | |||
| β-ACTIN | Sense: 5′-TGGAGAAAATCTGGCACCAC-3′ | 190 | BC 016045 |
| Antisense: 5′-GAGGCGTACAGGGATAGCAC-3′ | |||
| LDH | Sense: 5′-ACGTCAGCAAGAGGGAGAA-3′ | 194 | NM_005566.3 |
| Antisense: 5′-AACCGCTTCCAATAACACG-3′ | |||
| DLD | Sense: 5′-TTCCCATTTGCTGCTAACA-3′ | 214 | NM_000108.3 |
| Antisense: 5′-CTGATAAGGTCGGATGTGC-3′ | |||
| PRPF19 | Sense: 5′-GGACGGAGATTCTTCACTTTACAG-3′ | 183 | NM_014502.4 |
| Antisense: 5′-CAAACCCTAATTCTACCCCTCTACT-3′ | |||
| ATP synthase | Sense: 5′-GCTGCCACTCAACAACTT-3′ | 113 | NM_001001937.1 |
| Antisense: 5′-AGACCCGCATAGATAACAG-3′ | |||
| CaM | Sense: 5′-TGGAGACGGACAAGTCAACTAT-3′ | 243 | NM_006888.3 |
| Antisense: 5′-GACAGGACCACCAACCAATAC-3′ | |||
| p64 CLCP | Sense: 5′-CCTTTGCCACTTGCTCAT-3′ | 198 | NM_001288.4 |
| Antisense: 5′-CCTTTGCCACTTGCTCAT-3′ | |||
| RanGAP | Sense: 5′-AGGAGGAAGATGAGGAAGAGG-3′ | 180 | NM_002883.2 |
| Antisense: 5′-AAGCCAGGAAGGTGGAGAC-3′ | |||
| calreticulin | Sense: 5′-GCCGAGCCTGCCGTCTACTT-3′ | 238 | NM_004343.3 |
| Antisense: 5′-TGAACTGCACCACCAGCGTCT-3′ | |||
| HPRT | Sense: 5′-TGAGGATTTGGAAAGGGTGT-3′ | 118 | NM_000194.2 |
| Antisense: 5′-GAGCACACAGAGGGCTACAA-3′ |
a Primers used to amplify full-length cagA.
b Primers used to detect cagA expression in cells. LDH, L-lactate dehydrogenase. DLD, Dihydrolipoamide dehydrogenase. PRPF19, pre-mRNA processing factor 19 homolog. CaM, calmodulin. p64 CLCP, nuclear chloride ion channel protein. RanGAP, Ran-specific GTPase-activating protein. HPRT, Hypoxanthine-guanine phosphoribosyltransferase.
Cell transfection with pcDNA3.1/cagA
SGC-7901 cells were incubated for 12 h in 6-well plates to obtain 80% confluent cells. The cells were then transfected according to the manufacturer’s instructions. After 48 h, the cells were divided 1:10 into new 6-well plates and incubated for an additional 24 h and then maintained in selective zeocin-containing (250 μg/mL) standard medium for 2 weeks until clone formation. Cells transfected with empty vector pcDNA3.1/Zeo (-) were used as controls.
Protein extraction and western blot
Protein extracts were prepared by resuspending cell pellets in a RIPA buffer or homogenizing 200 mg tissues in a RIPA buffer. A total of 30–50 μg of protein extract was subjected to SDS-PAGE gel electrophoresis, transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA, USA) and blotted overnight with mouse monoclonal anti-CagA antibody (1:800, sc-28368), mouse monoclonal anti-phosphotyrosine antibody (PY99, 1:300, sc-7020), rabbit polyclonal anti-beta actin antibody (1:500, ab189073), rabbit polyclonal anti-LDH antibody (1:350, ab125683), mouse monoclonal anti-DLD (1:800, sc-376890), rabbit polyclonal anti-PRPF19 (1:400, ab27692), mouse monoclonal anti-ATP synthase antibody (1:300, ab54880), rabbit polyclonal anti-calmodulin anbody (1:250, ab208911), rabbit polyclonal anti-RanGAP antibody (1:200, ab92360), rabbit polyclonal anti-p64CLCP antibody(1:300, ab28722), rabbit polyclonal anti-calreticulin antibody (1:350, ab4) and mouse monoclonal anti-glyceraldehyde-3-phosphate -dehydrogenase antibody (GAPDH, 1:8000) from Santa Cruz (CA, USA), Abcam (Cambridge, UK) and CangChen (Shanghai, China), respectively, in 5% BSA in Tris-buffered saline and 0.01% Tween-20. Peroxidase-conjugated secondary antibodies (1:5000, SC-2371, Santa Cruz, CA, USA) were used and developed with the chemiluminescence reagent ECL Plus using hyperfilm (Amersham Biosciences, Buckinghamshire, UK). Quantification of the western blots was performed using Quantity One software. Each experiment was performed 3 times, and a representative result is shown.
Protein extraction and two-dimensional gel electrophoresis
Cell pellets were dissolved in cell lysis buffer overnight at 4°C, and protein was precipitated with three volumes of ice-cold acetone by incubating at 4°C for 2 h. The samples were then centrifuged at 20,000 rpm for 30 min, and the pellets were resuspended in cell lysis buffer and stored at 4°C overnight.
A total of 800 μg of protein was adjusted to a volume of 250 μL with rehydration solution, and isoelectric focusing (IEF) was performed using an Ettan IPGphor II Isoelectric Focusing system (Amersham Biosciences) according to the manufacturer’s instructions. The protocol for IEF was 300 V for1 h, 500 V for 2.5 h, 1,000 V for 2 h; 8,000 V for 8 h, 60 kVh (total).
After completing IEF, the IPG strips (Amersham Biosciences) were equilibrated in equilibration buffer for 15 min and placed on a 12% SDS-PAGE gel for two-dimensional electrophoresis at 30 mA/gel. The resulting SDS-PAGE gel was fixed in 20% TCA for 30 min and then stained with colloidal Coomassie G-250 [5].
The protein spots on the gel were scanned and analyzed automatically. Differentially expressed protein spots were confirmed with Imaging Master 2D 5.0 analytical software (Amersham Biosciences). Student’s t-test was performed for the quantitative analysis of the 2D gels. Differential expression of a specific protein was defined as a ≥2-fold change in spot optical density between the two matched sets in duplicates. The differential spots were then excised from the SDS-PAGE gels for further identification by LC-MS/MS.
RNA extraction and quantitative RT-PCR
Total RNA was extracted from 50 mg of tissue and treated with DNase I (RNase-free). The genes were amplified with SYBR Green (Applied Biosystems, Australia). Hypoxanthine-guanine phosphoribosyltransferase (HPRT) was used as a normalization control, and relative mRNA levels were calculated by a comparative Ct method using Step-one software (Applied Biosystems, Australia) [8]. Each sample was assayed in triplicate, and the results are expressed as the mean±SD. The primers used are listed in Table 1.
DNA extraction and methylation analysis of CpG islands
DNA was isolated from cells and modified with sodium bisulfite using an EZ DNA Methylation-Gold Kit™ (Zymo Research, CA, USA) according to the manufacturer’s instructions. Promoter CpG islands were predicted using Methyl Primer Express software and amplified from bisulfite-modified DNA by PCR using the following procedures: denaturing at 96°C for 10 min, followed by 40 cycles of denaturing at 94°C for 40 sec, annealing at 60°C for 40 sec, and elongation at 72°C for 40 sec, with a final extension at 72°C for 10 min. The amplified PCR products were cloned into pMD19-T vectors and sequenced. In addition, DNA isolated from tumor tissues was used to detect H. pylori 16S rRNA gene and cagA gene. The primer sequences are listed in Table 2.
Table 2. Primers used to amplify promoter CpG islands in methylation-PCR.
| Gene | Primer sequence (5′-3′) | Length (bp) |
|---|---|---|
| Hp16S rRNA | Sense: 5′-GCTAAGAGAT CAGCCTATGTC-3′ | 118 |
| Antisense: 5′- CCGTGTCTCAGTTCCAGTGT-3′ | ||
| LDH1 | Sense: 5′-CGGAATAAGGATATGATAGGT-3′ | 427 |
| Antisense: 5′-ATCCCTAACTATCTCCTAACTTT-3′ | ||
| LDH2 | Sense: 5′-GGGGTATTTATTAGGTTTGAAGTT-3′ | 532 |
| Antisense: 5′-CCTCCTAAAAA TTCACCCATC-3′ | ||
| DLD1 | Sense: 5′-TGTGGATATAGGAGGTGA ATTTT-3′ | 447 |
| Antisense: 5′-CAATCAAATCCCAAAAACAATA-3′ | ||
| DLD2 | Sense: 5′-TTTATATGGTTGTTGTAAGGATGAA-3′ | 473 |
| Antisense: 5′-CCTTAACCAAAAAACAATACACAC-3′ | ||
| RanGAP1 | Sense: 5′-TTAGTATAGTGGTATGGATGGTAGG-3′ | 314 |
| Antisense: 5′-TCTACTAACCCAACCCTACTCTATT-3′ | ||
| RanGAP2 | Sense: 5′-GGGATTGATAGGATATATGGGAT-3′ | 301 |
| Antisense: 5′-AATAAATCTAACACCAAAATAACCC-3′ | ||
| CaM1 | Sense: 5′-AAGAGGATTAATTTTTTTTAGGAGG-3′ | 250 |
| Antisense: 5′-CAACCTCACCCCACCTAAATA-3′ | ||
| CaM2 | Sense: 5′-GTTGAGGTGGGAGGGTTATTTA-3′ | 392 |
| Antisense: 5′-TCCCAACACCACTACCGAA-3′ | ||
| CaM3 | Sense: 5′-TTTTGGTAGTGGTGTTGGGA-3′ | 285 |
| Antisense: 5′-AACAAACAAAACAACTAAAAATCTAAA-3′ |
Hp, H. pylori; LDH, L-lactate dehydrogenase; DLD, Dihydrolipoamide dehydrogenase; RanGAP, Ran-specific GTPase-activating protein; CaM, calmodulin.
Statistical analysis
Results are expressed as the means ± SD. Statistical analyses were performed using SPSS 15.0 software. One-way analysis of variance (ANOVA) and Student’s t-test were used to analyze the data. P<0.05 (two-sided) was considered significant.
Results
Introduction of CagA into gastric cancer cells
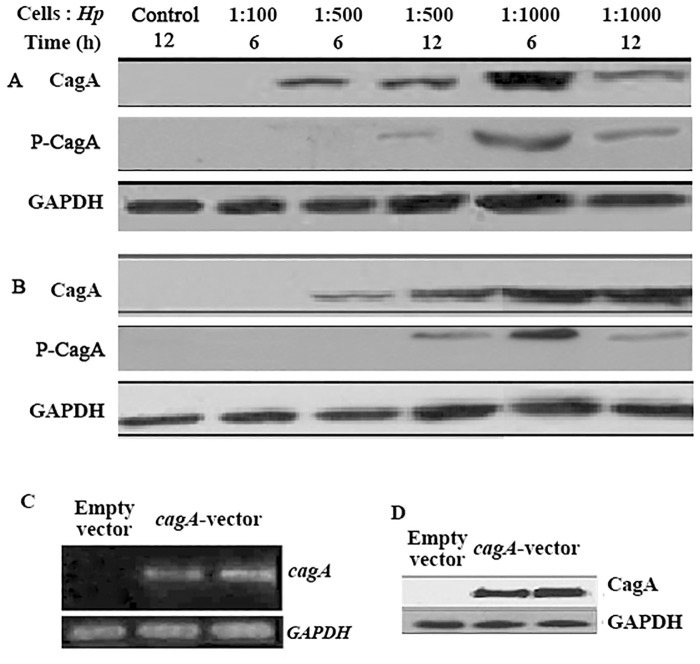
Because CagA of H. pylori is a critical virulence factor in the development and progression of gastric cancer, CagA was detected in gastric cancer cell lines by RT-PCR and western blot after infection of SGC-7901 and AGS cells with H. pylori and transfection of SGC-7901 cells with cagA-vector. The CagA protein began to appear at a ratio of cells to bacteria of 1:500 in cultured cells at 6 h, and the content was highest at a ratio of 1:1000 at 6 h (Fig 1A and 1B). However, phosphorylated CagA was observed in cells at a ratio of 1:500 after culturing for 12 h, and the highest content was observed in cells at a ratio of 1:1000 after 6 h of culture. CagA mRNA and protein were also observed in stably cagA-overexpressing SGC-7901 cells (Fig 1C and 1D). These data suggest that CagA was successfully introduced into the three cell lines and phosphorylated.
Fig 1. Introduction of CagA into gastric cancer cells.
(A and B) Western blot analysis of CagA and phosphorylated CagA in H. pylori-infected SGC-7901(A) and AGS (B) cells. The cells infected with the indicated ratio of cells to H. pylori for the indicated time were collected and lysed, and the proteins were separated by SDS-PAGE. Cells infected with H. pylori boiled for 15 min at a MOI of 1:1000 were used as a control. (C and D) Detection of CagA mRNA and protein in cagA-overexpressing SGC-7901 cells by RT-PCR (C) and western blot (D). GAPDH served as the loading control. The data are representative of three independent experiments. Hp, H. pylori; P-CagA, phosphorylated CagA; GAPDH, Glyceraldehyde-3-phosphate- dehydrogenase.
Identification of differential proteins in gastric cancer cells
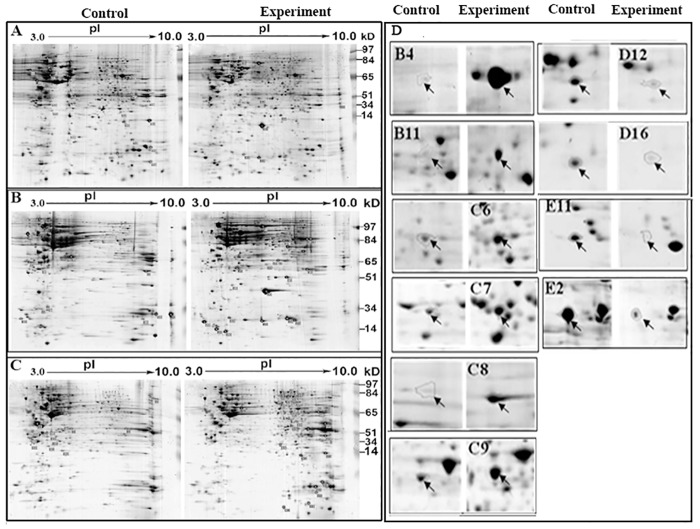
After cells were infected with H.pylori for 6 h at a MOI of 1:1000 or transfected with cagA-vector, a proteomic technique was used to create six two-dimensional electrophoresis (2-DE) maps from the three cell lines and their respective controls (Fig 2), and 135 differential spots were detected, of which 73 were up-regulated and 62 were down-regulated. Ten differential spots common to all three cell lines were identified by LC- MS/MS and verified by western blot, including 6 up-regulated proteins: β-actin, LDH, DLD, PRPF19, ATP synthase and calmodulin (CaM), and 4 down-regulated proteins such as RanGAP, p64 CLCP, P43 and calreticulin (Table 3 and Fig 3A).
Fig 2. Representative 2-DE maps and magnified image of differential spots in three cell lines.
SGC-7901 and AGS cells infected with H. pylori for 6 h at a MOI of 1:1000 (cell to H. pylori) and SGC-7901 cells transfected with pcDNA3.1/cagA for 48 h were collected and lysed, and the protein concentrations were determined using Bradford colorimetry. A total of 800 μg of protein was loaded for two-dimensional electrophoresis. Cells infected with boiled H. pylori or transfected with empty vector served as controls for the infected or transfected cells, respectively. (A) SGC-7901 cells infected with H. pylori. (B) AGS cells infected with H. pylori. (C) SGC-7901 cells transfected with the cagA-vector. (D) Magnified image of 10 differential spots. B4, B11, C6, C7, C8 and C9 spots were up-regulated, whereas D12, D16, E2 and E11 spots were down-regulated. These spots are identified in Table 3.
Table 3. Identification of differential proteins in three cell lines by LC-MS/MS.
| SpotNo. | Accession No. | Protein | M(dalton) | pI | Sequencecoverage (%) | Score | Fold change | Function |
|---|---|---|---|---|---|---|---|---|
| B4 | gi|4501885 | beta actin | 41710.7 | 5.29 | 8 | 30 | ↑106.355 | Skeleton rearrangement |
| B11 | gi|13786847 | L-Lactate Dehydrogenase | 36485.1 | 5.72 | 20 | 90 | ↑3.3682 | Energy metabolism |
| C6 | gi|91199540 | dihydrolipoamide dehydrogenase | 54144 | 7.95 | 9 | 60 | ↑2.59701 | Energy metabolism |
| C7 | gi|7657381 | pre-mRNA processing factor 19 homolog | 55147.4 | 6.14 | 15 | 70 | ↑2.73432 | pre-mRNA splicing |
| C8 | gi|4757810 | ATP synthase | 59714.6 | 9.16 | 24 | 218 | ↑5.45285 | Energy metabolism |
| C9 | gi|825635 | Calmodulin | 17153 | 4.06 | 21 | 40 | ↑3.55856 | Signal transduction |
| D12 | gi|895845 | p64 CLCP | 23592.1 | 5.12 | 65 | 328 | ↓6.11339 | Ion transport |
| D16 | gi|542991 | Ran-specific GTPase-activating protein | 23439.7 | 5.21 | 12 | 30 | ↓4.55474 | Signal transduction |
| E11 | gi|833999 | P43 | 49503.1 | 7.69 | 31 | 200 | ↓7.66244 | anti-angiogenesis |
| E2 | gi|4757900 | calreticulin | 48112.8 | 4.29 | 55 | 898 | ↓11.9006 | calcium homeostasis |
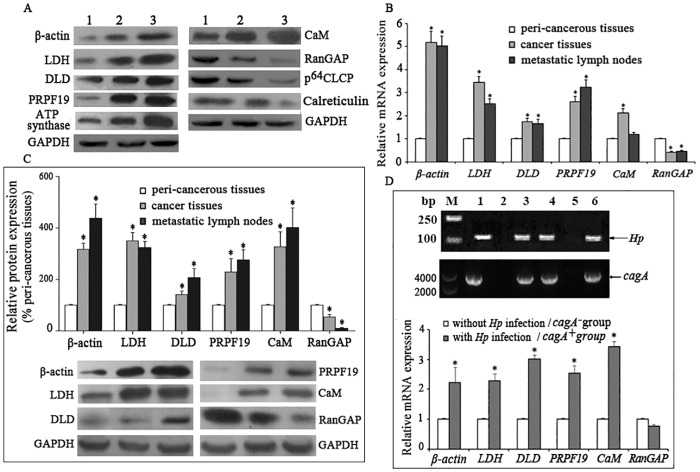
Fig 3. H. pylori infection promotes the genes and proteins expression in gastric cancer tissues.
(A) Western blot analysis of the indicated proteins in the control cells (1), H.pylori-infected SGC-7901 cells (2) and cagA-overexpressed SGC-7901 cells (3). GAPDH served as the loading control. The data are representative of three independent experiments. (B) Quantitative RT-PCR analysis of the indicated genes in 30 gastric cancer tissues. Values are represented as average Ct fold compared to peri-cancerous tissues, where peri-cancerous tissues were set to 1. (C) Western blot analysis of the indicated proteins in 30 gastric cancer tissues. 200 mg tissues were homogenized and total proteins were collected. A total of 50 μg protein extracts were subjected to SDS-PAGE gel electrophoresis. GAPDH served as the loading control. (D) Detection of the H. pylori 16S rRNA gene and cagA gene in gastric cancer tissues by PCR (Upper). M represents the DNA molecular weight marker. Lane 1 is the positive control. Lane 2 is the negative control. Lanes 3, 4 and 6 are positive samples. Lanes 5 are negative samples. Quantitative RT-PCR analysis of the indicated genes in gastric cancer tissues with and without H. pylori infection (Lower). Values are presented as the average Ct fold compared to the group without H. pylori infection, which was set to 1. The figure presents the average of 30 samples. Data are presented as the means ± SD. Error bars represent standard deviations. LDH, L-lactate dehydrogenase. DLD, Dihydrolipoamide dehydrogenase. PRPF19, pre-mRNA processing factor 19 homolog. RanGAP, Ran-specific GTPase-activating protein. CaM, calmodulin. p64 CLCP, nuclear chloride ion channel protein. *, P<0.05 compared to peri-cancerous tissue (B and C) and tissues without H. pylori infection (D).
H. pylori infection promotes the expression of differential proteins in gastric cancer tissues
Because H. pylori selectively colonizes the human stomach to activate a set of pathological processes, the gene expression of 10 differential proteins in 30 human gastric cancer samples was evaluated by quantitative RT-PCR. Of the 10 genes, the expression of β-ACTIN, LDH, DLD, PRP19 and CaM in gastric cancers and/or metastatic lymph nodes were up-regulated compared to peri-cancerous tissues, whereas the expression of RanGAP was down-regulated. Similarily, the 6 proteins were also abnormally expressed in the same tissues (Fig 3B and 3C). No significant differences in the expression of the other genes were observed.
H. pylori colonization in gastric cancer tissues was measured by detecting the 16S rRNA gene and cagA gene of H. pylori by PCR. H. pylori was present in 20 of the 30 samples (positive rate 67%), and all of H. pylori strains are cagA positive. The mRNA levels of β-ACTIN, LDH, DLD, PRP19 and CaM were higher in H. pylori-positive tissues than in the negative samples (Fig 3D).
H. pylori induces aberrant DNA methylation in gastric cancer cells
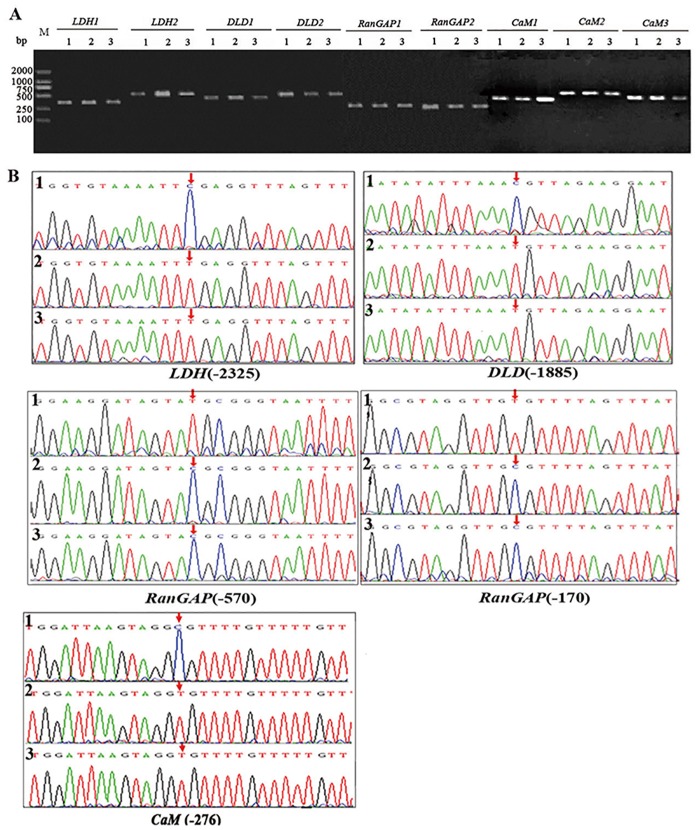
In three cell lines, PCR products of the CpG islands of the above genes were successfully acquired following bisulfite treatment and sequenced. In these genes, LDH, DLD and CaM genes were demethylated at the promoter -2325, -1885 and -276 sites, respectively, and the RanGAP gene was highly methylated at the promoter -570 and -170 sites (Fig 4).
Fig 4. H. pylori induces aberrant DNA methylation in cells.
(A) Electrophoretic analysis of the promoter CpG islands of the indicated genes by bisulfite modification-PCR. (B) Methylation sites of the CpG islands at the indicated gene promoters. LDH, L-lactate dehydrogenase. DLD, Dihydrolipoamide dehydrogenase. RanGAP, Ran-specific GTPase-activating protein. CaM, Calmodulin. M, Molecular weight marker. 1, untreated SGC-7901 cells; 2, H. pylori-infected SGC-7901 cells; 3, cagA-overexpressing SGC-7901 cells. The arrows indicate abnormal methylation sites.
Discussion
Accumulative evidence indicates that H. pylori infection is an important factor for H. pylori–associated gastric diseases and that CagA is a promoting factor for gastric cancer [17,18]. We constructed three experimental cell lines, including two gastric cancer cell lines infected with H. pylori (cagA+) and a gastric cancer cell line overexpressing cagA, and determined that CagA began to appear in cells 6 h after infection and for up to 12 h. Subsequently, phosphorylated CagA began to appear, consistent with the injection and subsequent phosphorylation of CagA into gastric epithelial cells by the T4SS of H. pylori after infection of the human gastric mucosa. Phosphorylated CagA activates downstream signaling pathways and plays a pathological role. We also observed CagA in cells stably transfected with the cagA-vector. These data confirm the successful construction of the three experimental cell lines.
Proteomics has been used to study the relationship between H. pylori infection and gastric diseases, and many differentially expressed proteins have been identified [19–22]. However, the association of these proteins with CagA and their expression in human gastric cancer tissues remain unclear. We obtained a total of 135 differential spots from the three cell lines, of which 73 were up-regulated and 62 were down-regulated. Ten differential spots were common to all three cell lines, including 6 up-regulated proteins (β-actin, LDH, DLD, PRP19, ATP synthase, and CaM) and 4 down-regulated proteins (p64 CLCP, RanGAP, P43 and calreticulin), and the 10 proteins’ expression were verified by western blot in these cell lines. These proteins are involved in energy metabolism, skeleton rearrangement, pre-mRNA processing, signal transduction, and other proteins closely associated with the development and progression of many human cancers [23,24]. We quantitatively detected the expression of the genes encoding these 10 proteins in human gastric cancer tissues, which revealed that β-ACTIN, LDH, DLD, PRP19 and CaM were consistently highly expressed and RanGAP was poorly expressed in both cancer tissues and/or metastatic lymph nodes compared to peri-cancerous tissue. The aberrant expression of these proteins was also verified by western blot in gastric cancer and metastatic lymph nodes. Next, we found that cagA+ H. pylori colonized in 20 of 30 gastric cancer tissues and promoted or inhibited the expression of these genes in vivo.
LDH and DLD are closely correlated with energy metabolism. Disorder of energy metabolism is considered an important factor for the development and progression of cancer. In contrast to normal cells, cancer cells exhibit increased dependence on the glycolytic pathway with sufficient oxygen, termed aerobic glycolysis. A large quantity of pyruvic acid, a final product of the pathway, is converted to lactic acid by LDH, promoting the glycolytic pathway and energy metabolism imbalance [25]. Lactic acid can also decrease the pH of the cell microenvironment, thus increasing neovascular response to angiogenesis factors to promote tumor cell metastasis [26,27]. Consistent with our results, LDH was highly expressed in cancer tissues in 61.8% of patients with gastric cancer; patients with LDH overexpression had shorter survival compared to patients with low expression [28]. Kim et al. [29] observed that DLD expression increases with tumor progression, thus providing more energy for tumor cell growth. Therefore, high expression of LDH and DLD is a crucial factor for the development and progression of gastric cancer, and H. pylori infection promotes the expression of these two genes in gastric cancer tissues.
Beta-actin, a key component of the cytoskeleton, maintains the structure, motion and division of cells under normal condition. β-ACTIN is abnormally expressed in many diseases [30]. The PRPF19 protein is believed to function in pre-mRNA splicing. Ubiquitination of PRPF19 could lead to DNA damage, and abnormal DNA repair is an important cause of tumorigenesis [31]. Calmodulin is a calcium-binding protein that regulates many signaling pathways and thus participates in cell proliferation, mitosis and gene transcription. Calmodulin promotes cell proliferation in liver carcinomas in combination with PI3K and the transition from G1 to S phase in combination with cyclin E [32]. An inhibitor of calmodulin arrests cells in G1 phase to inhibit cell division [33]. Consistent with these results, we determined that H. pylori may participate in cancer development of gastric tissues by inducing the high expression of these three proteins.
In addition, we observed down-regulation of the RanGAP gene in cancer tissues with H. pylori infection. RanGAP is a GTPase-activating protein. After hydrolysis of GTP to GDP by a GTPase, active RanGTP becomes inactive RanGDP, leading to the blockade of cell signaling to inhibit cancer progression [34]. Similarly, decreased RanGAP activity induced by ubiquitination promotes cancer development [35].
DNA methylation can induce abnormal gene expression. H. pylori infection increases the methylation levels of some genes and results in carcinogenesis of the gastric mucosa [36,37]. DNA methylation usually appears in the CpG islands of gene promoters. We detected the methylation status of the CpG islands of these genes in the constructed cell lines and determined that the LDH, DLD and CaM genes were demethylated at the promoter -2325, -1885 and -276 sites, respectively, and the RanGAP gene was highly methylated at the promoter -570 and -170 sites, consistent with the high expression of the LDH, DLD and CaM genes and the low expression of the RanGAP gene in gastric cancer tissues with cagA+ H. pylori infection.
In conclusion, these results suggest that H. pylori infection, via CagA translocation, may induce the aberrant methylation of these genes to lead to dysfunctional gene expression in gastric cancer tissues and cells. Our results provide a new understanding of the molecular pathogenesis of gastric cancer with H. pylori infection. Future studies will explore the effects of these altered methylation patterns on the pathogenesis of gastric cancer in clinical samples.
Acknowledgments
We thank the Chinese Center of Helicobacter pylori strain Management and Preservation for providing H. pylori NCTC11637 and Dr Yang Wenxiu and Wu Jiahong for technical assistance.
Data Availability
All relevant data are within the paper.
Funding Statement
The work was supported by National Natural Science Foundation of China (81260303, 31560326), Guizhou province foundation (QianJiaoHe [2014]06) and Key Project of Science and Technology of Guizhou Province (QianKeHe SY[2011]3067, QianKeHe J[2012]2039). In the original version, the work was supported by National Natural Science Foundation of China (81260303, 31560326) and Key Project of Science and Technology of Guizhou Province (QianKeHe SY(2011) 3067).
References
- 1.Peek RM Jr, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2: 28–37. [DOI] [PubMed] [Google Scholar]
- 2.Peek RM Jr, Crabtree JE. Helicobacter infection and gastric neoplasia. J Pathol. 2006;208: 233–248. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61: 69–90. 10.3322/caac.20107 [DOI] [PubMed] [Google Scholar]
- 4.Bertuccio P, Chatenoud L, Levi F, Praud D, Ferlay J, Negri E, et al. Recent patterns in gastric cancer: a global overview. Int J Cancer. 2009;125: 666–673. 10.1002/ijc.24290 [DOI] [PubMed] [Google Scholar]
- 5.Venerito M, Selgrad M, Malfertheiner P. Helicobacter pylori: gastric cancer and extragastric malignancies—clinical aspects. Helicobacter. 2013;18 (Suppl 1): 39–43. 10.1111/hel.12078 [DOI] [PubMed] [Google Scholar]
- 6.Bauer B, Meyer TF. The human gastric pathogen Helicobacter pylori and its association with gastric cancer and ulcer disease. Ulcers. 2011;2011: 1–23. [Google Scholar]
- 7.Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397: 176–180. [DOI] [PubMed] [Google Scholar]
- 8.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- 9.Wu AH, Crabtree JE, Bernstein L, Hawtin P, Cockburn M, Tseng CC, et al. Role of Helicobacter pylori CagA+ strains and risk of adenocarcinoma of the stomach and esophagus. Int J Cancer. 2003;103: 815–821. [DOI] [PubMed] [Google Scholar]
- 10.Li JJ, Qi RZ, Ng GK, Xie D. Proteomics in gastric cancer research: benefits and challenges. Proteomics Clin Appl. 2009;3: 185–196. 10.1002/prca.200800151 [DOI] [PubMed] [Google Scholar]
- 11.Gigek CO, Chen ES, Calcagno DQ, Wisnieski F, Burbano RR, Smith MA. Epigenetic mechanisms in gastric cancer. Epigenomics. 2012;4: 279–294. 10.2217/epi.12.22 [DOI] [PubMed] [Google Scholar]
- 12.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28: 1057–1068. 10.1038/nbt.1685 [DOI] [PubMed] [Google Scholar]
- 13.Kang GH. CpG island hypermethylation in gastric carcinoma and its premalignant lesions. Korean J Pathol. 2012;46: 1–9. 10.4132/KoreanJPathol.2012.46.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baylin SB, Jones PA. A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer. 2011;11: 726–734. 10.1038/nrc3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding SZ, Goldberg JB, Hatakeyama M. Helicobacter pylori infection, oncogenic pathways and epigenetic mechanisms in gastric carcinogenesis. Future Oncol. 2010;6: 851–862. 10.2217/fon.10.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ando T, Yoshida T, Enomoto S, Asada K, Tatematsu M, Ichinose M, et al. DNA methylation of microRNA genes in gastric mucosae of gastric cancer patients: its possible involvement in the formation of epigenetic field defect. Int J Cancer. 2009;124: 2367–2374. 10.1002/ijc.24219 [DOI] [PubMed] [Google Scholar]
- 17.Yan HB, Wang XF, Zhang Q, Tang ZQ, Jiang YH, Fan HZ, et al. Reduced expression of the chromatin remodeling gene ARID1A enhances gastric cancer cell migration and invasion via downregulation of E-cadherin transcription. Carcinogenesis. 2014;35: 867–876. 10.1093/carcin/bgt398 [DOI] [PubMed] [Google Scholar]
- 18.Targosz A, Brzozowski T, Pierzchalski P, Szczyrk U, Ptak-Belowska A, Konturek SJ, et al. Helicobacter pylori promotes apoptosis, activates cyclooxygenase (COX)-2 and inhibits heat shock protein HSP70 in gastric cancer epithelial cells. Inflamm Res. 2012;61: 955–966. 10.1007/s00011-012-0487-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franco AT, Friedman DB, Nagy TA, Romero-Gallo J, Krishna U, Kendall A, et al. Delineation of a carcinogenic Helicobacter pylori proteome. Mol Cell Proteomics. 2009;8: 1947–1958. 10.1074/mcp.M900139-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holland C, Schmid M, Zimny-Arndt U, Rohloff J, Stein R, Jungblut PR, et al. Quantitative phosphoproteomics reveals link between Helicobacter pylori infection and RNA splicing modulation in host cells. Proteomics. 2011;11: 2798–2811. 10.1002/pmic.201000793 [DOI] [PubMed] [Google Scholar]
- 21.Momynaliev KT, Kashin SV, Chelysheva VV, Selezneva OV, Demina IA, Serebryakova MV, et al. Functional divergence of Helicobacter pylori related to early gastric cancer. J Proteome Res. 2010;9: 254–267. 10.1021/pr900586w [DOI] [PubMed] [Google Scholar]
- 22.Wu MS, Chow LP, Lin JT, Chiou SH. Proteomic identification of biomarkers related to Helicobacter pylori-associated gastroduodenal disease: challenges and opportunities. J Gastroenterol Hepatol. 2008;23: 1657–1661. 10.1111/j.1440-1746.2008.05659.x [DOI] [PubMed] [Google Scholar]
- 23.Cho SO, Lim JW, Jun JH, Kim KH, Kim H. Helicobacter pylori in a Korean isolate expressed proteins differentially in human gastric epithelial cells. Dig Dis Sci. 2010;55: 1550–1564. 10.1007/s10620-009-0908-z [DOI] [PubMed] [Google Scholar]
- 24.Obeid M, Tesniere A, Ghiringhelli F, Fimia GM, Apetoh L, Perfettini JL, et al. Calreticulin exposure dictates the immunogenicity of cancer cell death. Nat Med. 2007;13: 54–61. [DOI] [PubMed] [Google Scholar]
- 25.Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330: 1340–1344. 10.1126/science.1193494 [DOI] [PubMed] [Google Scholar]
- 26.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452: 181–186. 10.1038/nature06667 [DOI] [PubMed] [Google Scholar]
- 27.Hunt TK, Aslam RS, Beckert S, Wagner S, Ghani QP, Hussain MZ, et al. Aerobically derived lactate stimulates revascularization and tissue repair via redox mechanisms. Antioxid Redox Signal. 2007;9: 1115–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolev Y, Uetake H, Takagi Y, Sugihara K. Lactate dehydrogenase-5 (LDH-5) expression in human gastric cancer: association with hypoxia-inducible factor (HIF-1α) pathway, angiogenic factors production and poor prognosis. Ann Surg Oncol. 2008;15: 2336–2344. 10.1245/s10434-008-9955-5 [DOI] [PubMed] [Google Scholar]
- 29.Kim H. Activity of human dihydrolipoamide dehydrogenase is largely reduced by mutation at isoleucine-51 to alanine. J Biochem Mol Biol. 2006;39: 223–227. [DOI] [PubMed] [Google Scholar]
- 30.He JQ, Sandford AJ, Wang IM, Stepaniants S, Knight DA, Kicic A, et al. Selection of housekeeping genes for real-time PCR in atopic human bronchial epithelial cells. Eur Respir J. 2008;32: 755–762. 10.1183/09031936.00129107 [DOI] [PubMed] [Google Scholar]
- 31.Lu X, Legerski RJ. The Prp19/Pso4 core complex undergoes ubiquitylation and structural alterations in response to DNA damage. Biochem Biophys Res Commun. 2007;354: 968–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao L, Zhao Q, Lu R, Fu Z, Zhu Z, Jia J, et al. Effects of tyroserleutide on gene expression of calmodulin and PI3K in hepatocellular carcinoma. J Cell Biochem. 2008;103: 471–478. [DOI] [PubMed] [Google Scholar]
- 33.Choi J, Chiang A, Taulier N, Gros R, Pirani A, Husain M. A calmodulin-binding site on cyclin E mediates Ca2+-sensitive G1/s transitions in vascular smooth muscle cells. Circ Res. 2006;98: 1273–1281. [DOI] [PubMed] [Google Scholar]
- 34.Zhao Q, Brkljacic J, Meier I. Two distinct interacting classes of nuclear envelope-associated coiled-coil proteins are required for the tissue-specific nuclear envelope targeting of Arabidopsis RanGAP. Plant Cell. 2008;20: 1639–1651. 10.1105/tpc.108.059220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, Sarge KD. Mel-18 interacts with RanGAP1 and inhibits its sumoylation. Biochem Biophys Res Commun. 2008;375: 252–255. 10.1016/j.bbrc.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shin CM, Kim N, Jung Y, Park JH, Kang GH, Kim JS, et al. Role of Helicobacter pylori infection in aberrant DNA methylation along multistep gastric carcinogenesis. Cancer Sci. 2010;101: 1337–1346. 10.1111/j.1349-7006.2010.01535.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niwa T, Tsukamoto T, Toyoda T, Mori A, Tanaka H, Maekita T, et al. Inflammatory processes triggered by Helicobacter pylori infection cause aberrant DNA methylation in gastric epithelial cells. Cancer Res. 2010;70: 1430–1440. 10.1158/0008-5472.CAN-09-2755 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.