SUMMARY
The mammalian eye-to-brain pathway includes more than twenty parallel circuits, each consisting of precise long-range connections between specific sets of retinal ganglion cells (RGCs) and target structures in the brain. The mechanisms that drive assembly of these parallel connections, and the functional implications of their specificity remain unresolved. Here we show that in absence of contactin 4 (CNTN4) or one of its binding partners, amyloid precursor protein (APP), a subset of direction selective retinal ganglion cells fail to target the nucleus of the optic tract (NOT) - the accessory optic system (AOS) target controlling horizontal image stabilization. Conversely, ectopic expression of CNTN4 biases RGCs to arborize in the NOT, and that process also requires APP. Our data reveal critical and novel roles for CNTN4/APP in promoting target-specific axon arborization and they highlight the importance of this process for functional development of a behaviorally-relevant parallel visual pathway.
INTRODUCTION
Perception, cognition and behavior all arise from highly precise patterns of connectivity between functionally specialized sets of neurons. Neural circuit precision emerges during development through a series of steps that collectively encompass a broad range of spatial scales; axonal growth cones must navigate long distances, often many millimeters, to link distantly located structures. At the other extreme, grow cones have to select where to form synapses on the dendrites of target neurons (Sanes and Yamagata, 2009; Lichtman and Denk, 2011). The ultimate goal of developmental neurobiology is to understand how specificity of circuit connections is established at all spatial scales and to determine how discrete alterations in specificity impact circuit function and behavior.
The cellular and molecular mechanisms underlying many steps of mammalian brain circuit assembly have been explored in detail. Prominent examples include growth cone navigation through intermediate choice points (Dickson, 2002), topographic mapping (Luo and Flanagan, 2007; Cang and Feldheim, 2013) and laminar specificity (Huberman et al., 2010; Robles and Baier, 2012; Baier, 2013). Among the lesser-understood steps involved in circuit assembly, however, is axon-target matching. As growth cones navigate through the brain, they encounter many target nuclei; they must recognize which of those targets to innervate and which to avoid. While data on axon-target matching in the mammalian brain are starting to emerge (e.g., Osterhout et al., 2011; 2014; Schmidt et al., 2014) our understanding of this crucial wiring step remains limited compared to the other aspects of circuit wiring.
The vertebrate eye-to-brain pathway is a longstanding model for addressing how CNS axons achieve connection specificity. The diversity of retinal ganglion cell (RGC) subtypes and their associated patterns of connections with distinct combinations of central targets (Robles et al., 2014; Dhande and Huberman, 2014) provide an ideal model to dissect the mechanisms of axon-target matching. Mammalian RGCs include ~20 different subtypes, each responding to a specific feature in the visual world and connecting to a stereotyped set of retinorecipient targets (Morin and Studholme, 2014). The distinct parallel pathways created by precise RGC axon-target matching are what enable different features in the visual world to drive the appropriate visual perception and behaviors (e.g., Güler et al., 2008; Sweeney et al., 2014), offering a unique opportunity to understand how axon-target matching relates to sensory circuit function and output.
The anatomical and functional specificity of a particular eye-to-brain circuit, the accessory optic system (AOS), make it particularly attractive for exploring axon-target matching in a behaviorally relevant context. The AOS evolved to control image stabilization and offset the visual “slip” that occurs when the head or eyes move at velocities too slow for the vestibular system to respond directly (reviewed in Simpson, 1984; Masseck and Hoffmann, 2009). The mammalian AOS consists of three target nuclei: the nucleus of the optic tract/dorsal terminal nucleus complex (NOT/DTN), the dorsal medial terminal nucleus (MTNd) and the ventral medial terminal nucleus (MTNv), which is also sometimes referred to as the lateral terminal nucleus (LTN) (Figure 1A–C) (Simpson et al., 1984; Pak et al., 1987; Yonehara et al., 2009; Dhande et al., 2013). These nuclei receive input from a functionally specialized collection of RGC subtypes: three subtypes of On-direction selective RGCs (On-DSGCs) and one subtype of On-Off DSGCs, all of which are tuned to slow speeds (Yonehara et al., 2009; Dhande et al., 2013). The axons that project to the NOT primarily target this region by terminal arbors or by collateral branching off an axon en route to the more distal DTN and/or superior colliculus (SC) whereas RGC inputs to the MTN are solely terminal arbors that arrive via the accessory optic tracts (Figure 1A–C) (Yonehara et al., 2009; Dhande et al., 2013). While progress has been made in identifying the RGC subtypes and targets that comprise the mammalian AOS (Yonehara et al., 2009; Kay et al., 2011; Dhande et al., 2013) the cellular mechanisms controlling development of this crucial visual circuit have only recently been investigated (Osterhout et al., 2014) and the molecular mechanisms are still completely unknown.
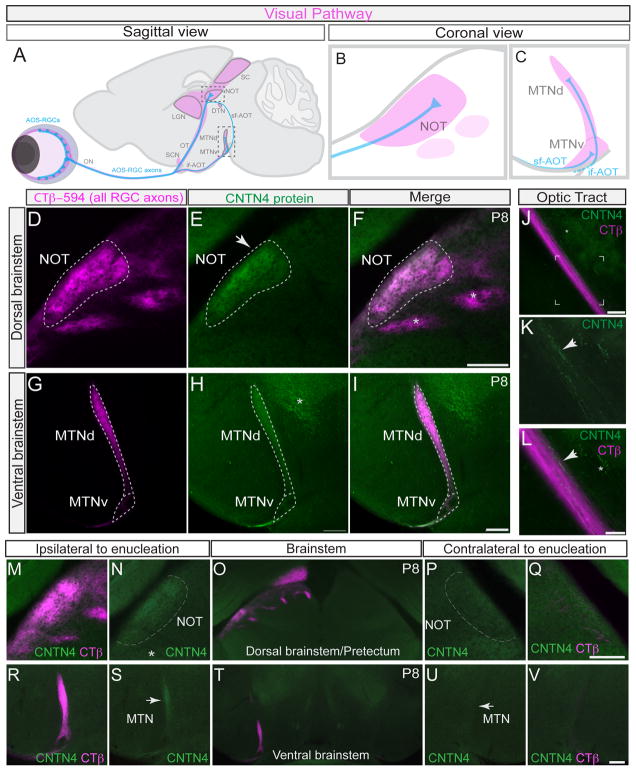
Figure 1. Contactin-4 is selectively expressed by axons of AOS-projecting RGCs.
(A–C) Mouse visual pathway. Blue: accessory optic system (AOS)-projecting RGCs and axons. Magenta: retinorecipient targets.
(A) Sagittal view, visual pathway; sf-AOT: superior fasciculus of the accessory optic tract. if-AOT: inferior fasciculus of the accessory optic tract; DTN: dorsal terminal nucleus; SCN: suprachiasmatic nucleus; LGN: lateral geniculate nucleus; SC: superior colliculus (B and C) coronal views of accessory optic system (AOS) targets, (B) the nucleus of the optic tract (NOT) and (C) the medial terminal nucleus, dorsal (MTNd) and ventral (MTNv) divisions.
(D–I) Contactin-4 (CNTN4) protein expression in AOS targets, the NOT and MTN.
(D) CTβ–594 (magenta) labeling of all RGC axons. (E) CNTN4 protein (green) is selectively expressed in the NOT. (F) Merge of D and E. Asterisks: retinorecipient targets that do not express CNTN4. Scale = 100μm
(G) CTβ–594 labeling all RGC axons. (H) CNTN4 protein expression in the medial terminal nucleus (MTN). Asterisk: CNTN4 expression outside of the visual system. (I) Merge of G and H. Scale = 100μm
(J–L) CNTN4 protein in axons in optic tract (OT). (J) CTβ–594/CNTN4 labeling. Scale in J = 200μm. (K) High magnification view framed region from (J). (L) Merged high magnification view of CTβ–594 and CNTN4. Scale in K, L = 100μm. Arrows in K and L: CNTN4+ axonal profiles. Asterisk: CNTN4 expression outside of the visual system.
(M–V) Effects of monocular enucleation on CNTN4 expression in the brain; Magenta: CTβ–594 labeling of RGC axons from intact eye; Green: CNTN4 protein expression.
(M–Q) CNTN4 expression in the NOT in the control (ipsilateral) hemisphere of the brain (M, N) and in the enucleated (contralateral) hemisphere of the brain (P, Q). Scale = 250μm. Asterisk: CNTN4 protein expression in other brain regions.
(R–V) CNTN4 expression in the MTN in the ipsilateral hemisphere (R, S) and contralateral hemisphere (U, V) of the brain. Scale = 250μm. Arrows: lateral border of the MTN.
Here we explored the molecular mechanisms of axon-target matching in the assembly of the mammalian AOS. Using genetic labeling of AOS-projecting RGCs, knockout mouse analyses and single cell over-expression experiments, we show that the IgG superfamily member contactin 4 (CNTN4/BIG-2) is necessary for AOS-projecting RGC axons to innervate the AOS target required for horizontal image stabilization (the NOT) and is sufficient to bias RGCs to selectively arborize in that target - a process that is contingent on expression of amyloid precursor protein (APP). Our findings reveal that axon-target matching in the mammalian brain involves target-specific axonal arborization that is ultimately, crucial for the function of brain circuits linked to specific behaviors.
RESULTS
Contactin-4 is expressed by RGCs that target AOS nuclei
To explore the molecular signals controlling development of parallel eye-to-brain circuits, we screened the expression patterns of IgG superfamily proteins in retinorecipient targets and identified Contactin-4 (CNTN4) as a candidate. CNTN4 belongs to a small family of axon-associated IgG cell adhesion molecules within the IgG superfamily that has six Ig domains, four fibronectin type-III domains and is GPI-anchored to the plasma membrane (Yoshihara et al., 1995; Kaneko-Goto et al., 2008; reviewed in Shimoda and Watanabe, 2009). Labeling of retinorecipient targets by intravitreal injections of cholera toxin beta (CTβ-594) followed by staining of tissue sections with an antibody specific for CNTN4 (Kaneko-Goto et al., 2008; Figure S1E and S1H) revealed it was selectively expressed in AOS targets: it was present at high levels in the developing NOT (Figures 1D–F) and at lower levels in the MTNd and MTNv (Figures 1G–I). By contrast, there was little or no CNTN4 protein expression in other retinorecipient nuclei (Figures S1A–G), even those situated adjacent to NOT (Figure 1F, asterisks).
CNTN4 protein expression was absent from RGC somas, but was clearly expressed by a small subset of RGC axons within the optic tract (Figures 1J–L), which is consistent with previous findings that CNTN4 expression is axonally-localized (Yoshihara et al., 1995). To determine if CNTN4 expression in AOS targets arises from RGC axons we removed one eye at postnatal day 0 (P0) mice and allowed a period of 7 days for RGC axons from that eye to degenerate. We then labeled the RGC axons from the intact eye with CTβ-594 and compared the patterns of CNTN4 expression in AOS targets on the two sides of the brain at P8. CNTN4 protein was present in AOS targets contralateral to the intact eye (Figures 1M, 1N, 1R, 1S) but was absent in AOS targets contralateral to the enucleated eye (Figures 1P, 1Q, 1U, 1V). Thus, we conclude that CNTN4 expression in AOS targets arises from RGC axons.
Developmental expression of CNTN4 protein in the nucleus of the optic tract
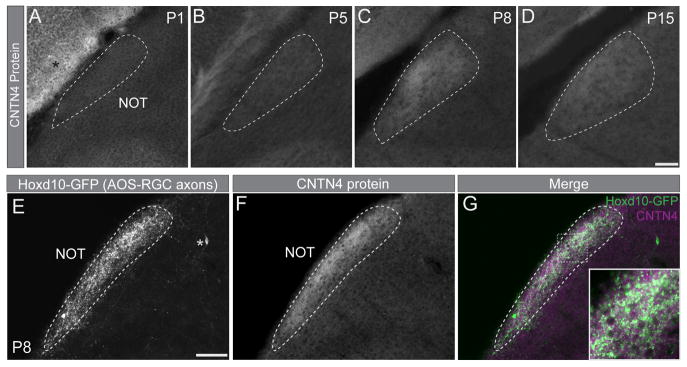
Next we analyzed the timing of CNTN4 expression in AOS targets during the developmental time frame when RGC axons arrive to these targets. CNTN4 protein expression was weak in the NOT from P1–P5 (Figures 2A, and 2B) but increased by P8 (Figure 2C), the stage when most AOS-projecting RGC axons innervate this target (Osterhout et al., 2014). After P8, CNTN4 protein expression in the NOT diminished (Figure 2D). We also analyzed CNTN4 expression in the NOT of P8 Hoxd10-GFP transgenic mice in which all four subtypes of AOS-projecting RGCs (3 On-DSGC subtypes and one On-Off DSGC subtype) selectively express GFP (Dhande et al., 2013). The Hoxd10-GFP+ axonal profiles overlapped with CNTN4 protein in the NOT (Figures 2E–G). In addition, the specificity of CNTN4 expression in the entire visual pathway matched the projection pattern of Hoxd10-GFP RGC axons (Figure S1). Taken together, these data indicate that CNTN4 is expressed by the axons of AOS-projecting RGCs during the developmental phase when they grow into their targets in the brain.
Figure 2. CNTN4 protein expression is developmentally regulated and correlates with Hoxd10-RGC innervation of the NOT.
(A–D) CNTN4 protein in the NOT at P1 (A), P5 (B), P8 (C), and P15 (D). Scale = 200μm. Asterisk: CNTN4 expression in overlying cortex.
(E–G) Hoxd10-GFP RGC axons (AOS-axons; E) and CNTN4 protein expression (F) in the NOT at P8. Asterisk: GFP-expressing cell body outside the NOT.
(G) Merge; inset high magnification of boxed area. Scale = 250μm.
Reduced innervation of the NOT by AOS-DSGC axons in CNTN4 mutant mice
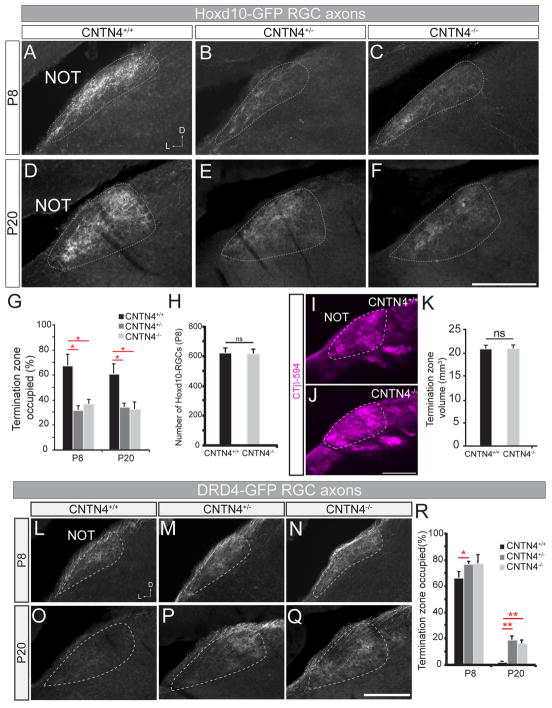
Does CNTN4 play a functional role in generating axonal connectivity between AOS-projecting RGCs and their targets? To test this, we crossed CNTN4 null mutant mice (CNTN4−/−; Kaneko-Goto et al., 2008) to Hoxd10-GFP reporter mice and analyzed the projection patterns of the GFP-expressing axons at P8 and P20. In wildtype (CNTN4+/+), Hoxd10-GFP mice the GFP+ axons densely filled the entire NOT at both P8 and P20 (Figures 3A and 3D). By contrast, there was a reduction in the density of NOT-projecting Hoxd10-GFP RGC axons in CNTN4+/−, Hoxd10-GFP mice (Figures 3B and 3E), and in CNTN4−/−, Hoxd10-GFP mice (Figures 3C and 3F). At P8 and P20, a small number of Hoxd10-GFP RGC axons still terminated along the lateral edge of the NOT (e.g., Figure 3F) but there was a clear and consistent defect in overall innervation density within this target. This defect was confirmed by comparison of the percentage of NOT area occupied by Hoxd10-GFP RGC axons in CNTN4 mutant mice versus their wildtype littermates at both P8 and P20 (Figure 3G; p<0.05; n= 4–6 mice per genotype, per age; see Experimental Procedures). By contrast, there were no observable changes in the pattern of projections from Hoxd10-GFP RGC axons to the MTNd or MTNv (Figures S2A–C). In addition, bulk whole-eye labeling of RGC axons showed that the timeframe of NOT innervation was unchanged in CNTN4 mutants (Figure S2D–G). Dual-color whole-eye labeling also showed that the pattern of binocular RGC targeting to other retinorecipient targets also appeared normal (Figure S2H–K).
Figure 3. Loss of CNTN4 results in a decrease of innervation by Hoxd10-GFP RGC axons to the NOT.
(A–F) Hoxd10-GFP RGC axons in the NOT at P8 (A–C) and at P20 (D–F), in wildtype (A, D), CNTN4+/− (B, E), and CNTN4−/− mice (C, F). Scale = 250μm; D = dorsal; L = lateral.
(G) Fraction of the termination zone (in NOT) occupied by Hoxd10-GFP RGC axons in wildtype (black), CNTN4+/− (dark grey) and CNTN4−/− (light grey) mice (±SEM). *= p<0.05 (n=4–6 mice per genotype, per age).
(H) Total number of Hoxd10-RGCs in the retinas of wildtype and CNTN4−/− mice at P8 (±SEM). n= 5–6 mice per genotype.
(I–J) RGC axons in NOT of wildtype (I) and CNTN4−/− mice (J) labeled by intraocular CTβ-594. Scale = 200μm.
(K) Total volume of the NOT (mm3 ±SEM) in wildtype (black) and CNTN4−/− mice (grey) (n = 3 mice per genotype).
(L–Q) DRD4-GFP RGC axons in the NOT at P8 (L–N) and at P20 (O–P), in wildtype (L, O), CNTN4+/− (M, P) and CNTN4−/− mice (N,Q). Scale = 250μm.
(R) Fraction of the termination zone (in NOT) with DRD4-GFP RGC axons in wildtype (black), CNTN4+/− (dark grey) and CNTN4−/− (light grey) mutants (±SEM).
*= p<0.05 **=p<0.01, (n=5–6 mice per genotype, per age).
One possible explanation for the axonal projection phenotype we observed in the NOT is that there was a reduction in number of Hoxd10-GFP RGCs present in CNTN4 mutants. However, when we quantified the total number of Hoxd10-GFP RGCs in wildtype CNTN4+/+ mice and in CNTN4−/− mice at P8, we found no significant differences between these groups (Figure 3H). Additionally, the dendritic stratification patterns of Hoxd10-GFP RGCs appeared normal (Figures S2L–S). We also studied the electrophysiological properties of Hoxd10-RGCs in retinal whole mounts from wildtype and CNTN4 mutant mice (Figure S3). In both wildtype and CNTN4 mutant retinas, Hoxd10-GFP RGCs included the three expected On-DSGCs subtypes and one On-Off DSGCs subtype (Figure S3A–B), all of which were tuned to the correct axes of motion and displayed no significant differences in magnitude of direction tuning compared to control Hoxd10-GFP RGCs (Figure S3E–F). Together, our data indicate that loss in CNTN4 expression results in perturbed projections to the NOT by AOS-projecting RGCs and that the altered patterns of NOT innervation are not the consequence of changes in Hoxd10-GFP RGC number, subtype identity or retinal wiring.
Interestingly, although Hoxd10-GFP RGCs comprise the majority of AOS-projecting RGCs (Dhande et al., 2013), whole eye labeling showed that the total volume of projections to this target was normal in CNTN4−/− mice (Figures 3I–K; n=3 mice per genotype), suggesting non-AOS RGCs fill in the target zone. Indeed, when we explored the axon targeting patterns of another category of RGCs: the On-Off DSGCs labeled in DRD4-GFP transgenic mice (Huberman et al., 2009), we found they had abnormal NOT projections in CNTN4 mutant mice (Figure 3L–R). In mature, wildtype DRD4-GFP mice, the GFP+ RGCs project to the NOT only transiently during development and by P20 they have retracted from the NOT (Figure 3L, 3O, 3R) (Huberman et al., 2009; Kay et al., 2011; Osterhout et al., 2014). By exploring multiple ages of CNTN4−/−, DRD4-GFP mice, we discovered that CNTN4 mutations cause an abnormal maintenance of DRD4-GFP RGC projections to the NOT (Figure 3M, 3N, 3P, 3Q). Targeting of DRD4-GFP RGC axons to the dLGN and SC by contrast, appeared normal (Figure S4A–D), indicating the impact of CNTN4 mutations on these RGCs was AOS target-specific.
To determine whether other RGCs subtypes also alter their axon projection patterns in CNTN4 mutant mice, we crossed CNTN4 mutants to Cdh3-GFP mice. These mice selectively express GFP in a subset of intrinsically photosensitive RGCs (ipRGCs) that project to non-image-forming targets important for pupil reflex and circadian-related behaviors (Osterhout et al., 2011; 2014). The projection patterns of Cdh3-GFP RGC axons were unaltered in CNTN4 mutants (Figure S4E–L) and they avoided the NOT (Figure S4K–L) just as they typically would in wildtype mice (Osterhout et al., 2011; 2014). Together, the analyses of Hoxd10-, DRD4- and Cdh3-GFP mice carrying CNTN4 mutations indicate that the developmental influence of CNTN4 on RGC axon-target matching is limited to the axons that interface with AOS targets during development.
Axonal arborization as a key step in CNTN4-mediated axon-target matching
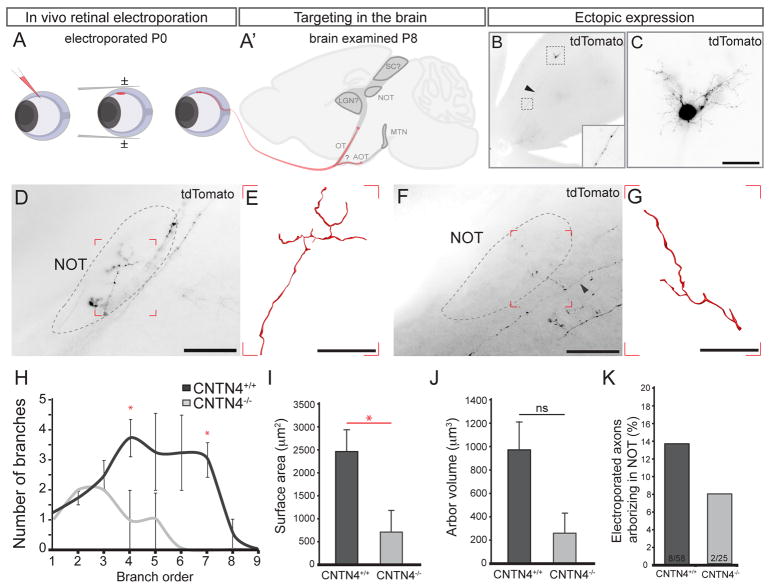
To gain a better understanding of the mechanisms by which CNTN4 mediates targeting of RGC axons to the NOT, we tested whether the phenotypes present in CNTN4 mutants reflected a decrease in the number of axons targeting the NOT or decreased arbor complexity of correctly targeted axons. Either theory could result in an apparent reduction of Hoxd10-GFP RGC axons in the NOT. We used sparse in vivo RGC electroporation (Dhande et al., 2011) to ectopically express tdTomato in individual RGCs of wildtype and CNTN4 mutant mice at P0 and then examined their axons in the brain one week later, on P8 (Figure 4A–A′). Figure panels 4B and 4C show an example of an electroporated RGC expressing tdTomato throughout its cell body, dendritic arbor, and axon. By sparse-electroporating large numbers of wildtype mice, we succeeded in labeling 4 individual NOT-projecting RGC axons in separate animals. Labeling of individual NOT-projecting RGC axons in CNTN4−/− mice proved even more challenging. We did, however, successfully label two individual NOT-innervating RGC axons in CNTN4−/− mice (e.g., Figure 4F, G). Reconstruction and quantification of the morphological features of these axons (branch number, arbor area, etc.) revealed that wildtype NOT-targeted RGC axons were significantly more complex than CNTN4 mutant axons (compare Figure 4E, and 4G; quantified in 4H). The surface area and volume of CNTN4−/− arbors were also reduced compared to wildtype (Figure 4I–J). Notably, the fraction of electroporated RGCs that projected to the NOT was also greatly reduced in CNTN4 as compared to wildtype mice (Figure 4K). These data suggest that the phenotype of reduced Hoxd10-GFP RGC input to the NOT (Figure 3A–F) likely arises from two sources: i) a reduction in the number of AOS-projecting RGCs targeting the NOT and ii) reduced branching and overall complexity of the arbors that do manage to innervate this target.
Figure 4. CNTN4 is required for accurate targeting and efficient arborization in the NOT.
(A, A′) In vivo electroporation. (A) Plasmid DNA is injected into the eye on P0, receives square waves pulses. (A′) P8 brains are examined for labeled RGC axons.
(B) Example of pCMV-tdTomato electroporated RGC. Arrowhead and inset: RGC axon expressing tdTomato. (C) High magnification of the RGC shown in (B) the RGC soma, dendrites and axon (arrowhead) express high levels of tdTomato. Scale = 125μm.
(D–G) Example of NOT-projecting tdTomato+ axons in wildtype (D, E) and CNTN4−/− mice (F, G); arrowhead: parent axon. (E) Wildtype, NOT-projecting RGC axon reconstruction from boxed region in D. (G) CNTN4−/−, NOT-projecting axon reconstruction from boxed region in F. Scale in D, F= 250μm., Scale in E, G = 50μm.
(H) Quantification of the average number (±SEM) of branches from branch order 1–8 for wildtype and CNTN4−/−, NOT-projecting axon (n=4 axons/mice wildtype, n=2 axons/mice CNTN4−/−).
(I) Quantification of the average arbor surface area for wildtype and CNTN4−/− mice (±SEM), p=0.040.
(J) Quantification of the average arbor volume for wildtype and CNTN4−/− mice (±SEM), p=0.062.
(K) The percentage of electroporated axons arborizing in the NOT after electroporation in wildtype (8/58) and CNTN4−/− (2/25) mice.
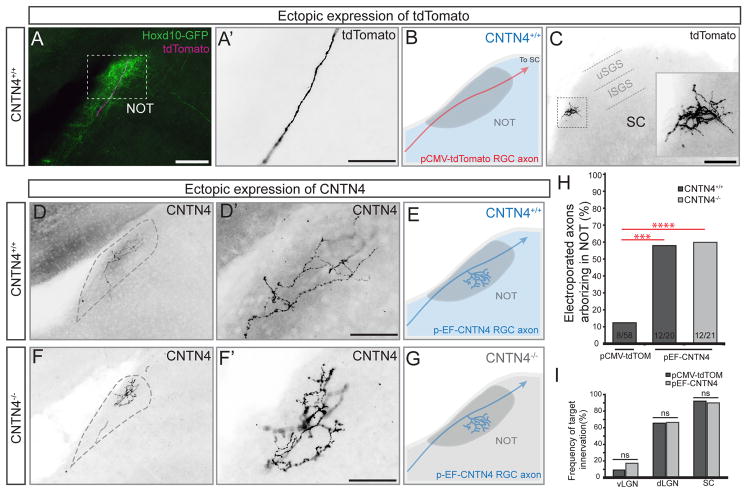
Ectopic expression of CNTN4 biases RGCs to branch in an AOS target
To further explore the role of CNTN4 in RGC axon-target matching, we again utilized sparse in vivo RGC electroporation to ectopically expressed either control, tdTomato plasmid or a plasmid encoding full-length CNTN4 in RGCs that normally bypass the NOT. We accomplished this by electroporating one or the other plasmid into the retina of Hoxd10-GFP mice at P0 and examined the targeting of non-Hoxd10-GFP RGC axons in the brain a week later, on P8. Of the 58 control/tdTomato+ RGC axons we examined, 8 of them (~14%) innervated the NOT; the remaining 50/58 tdTomato+ axons traveled through or over the NOT to arborize in more distal retinorecipient targets such as the SC (Figures 5A–C and 5H). By contrast, ectopic expression of CNTN4 in individual RGC axons strongly biased them to arborize in the NOT. 60% of CNTN4-electroporated RGCs (12/20) targeted and the elaborated axonal arbors in the NOT (Figures 5D–E) – a nearly five-fold increase over what was observed for control RGCs electroporated with tdTomato (Figure 5H; p=0.0002). We note that 10/12 of the CNTN4+ RGC axons that elaborated arbors in the NOT also projected to the SC. Thus, CNTN4 expression in individual RGC axons biases them to form axonal arbors in the NOT but does not appear to otherwise alter their growth or patterning of retinorecipient targeting.
Figure 5. CNTN4 expression in RGC axons is sufficient to bias arborization in the NOT.
(A–F) Electroporation of RGCs with control/pCMV-tdTomato plasmid (A–C) or CNTN4 plasmid (D–G).
(A) Td-tomato+ axon (magenta) projecting through the NOT in a Hoxd10-GFP mouse (green). Scale = 250μm.
(A′) High magnification of tdTomato+ RGC axons from the boxed region in (A). Scale = 125μm.
(B) Schematic of typical outcome of td-Tomato expression (red) in RGCs of wildtype mice (wildtype denoted by blue color). (C) Axon terminal in the SC; this the same axon as shown in (A, A′). Inset: high magnification of boxed region in (C), Scale = 250μm. lSGS: lower stratum griseum superficialis. uSGS: upper stratum griseum superficialis.
(D–E) Example RGC axon electroporated with pEF-CNTN4 plasmid in wildtype background. (D′) High magnification of axon in (D); (E) Schematic of typical outcome of electroporation of CNTN4 in RGCs in a wildtype background.
(F–G) Example RGC axon electroporated with pEF-CNTN4 in a CNTN4−/− mouse. (F′) High magnification of axon in (F). (G) Schematic of typical outcome of electroporating CNTN4 into RGCs of CNTN4−/− mice. Scales in D′ and F′ = 125μm.
(H) Percentage of electroporated RGC axons arborizing in the NOT after electroporation of tdTomato or CNTN4. Statistical significance determined by Fisher analysis; ** = p<0.01; the fraction of electroporated RGCs for each experiment given at the bottom of the bars.
(I) Percentage of electroporated axons arborizing in other major retinorecipient targets after electroporation of tdTomato or CNTN4. Statistical significance calculated using Fisher analysis.
The results of our expression analyses described above indicate that normally CNTN4 is expressed by a subset of AOS-projecting RGC axons but not by cells in retinorecipient targets. In theory, however, CNTN4 expressed by other RGC axons could bias RGCs to elaborate arbors in the NOT. To test this, we electroporated CNTN4 into individual RGCs in CNTN4−/− mice and examined their resulting pattern of axon targeting in the brain. Remarkably, even in the CNTN4−/− background, ectopic expression of CNTN4 in an individual RGC axon strongly biased that axon to arborize in the NOT. Approximately 55% (12/21 axons) of the CNTN4-electroporated RGCs arborized in the NOT in CNTN4−/− mutants (Figures 5F–H), which was not significantly different from the impact of ectopically expressing CNTN4 in mice of wildtype background (Figure 5H).
We also addressed the possibility that CNTN4 non-specifically promotes axon arborization independent of target region, by comparing the frequency of innervation within other major visual targets by axons electroporated with tdTomato or CNTN4 plasmid. The frequency of arborization in the ventral and dorsal lateral terminal nuclei (vLGN and dLGN, respectively) or the SC was indistinguishable between control and CNTN4-electroporated groups (Figure 5I). Thus, the expression of CNTN4 in an individual RGC axon biases that axon to target and establish arbors specifically in the NOT.
Amyloid precursor protein (APP) is necessary for CNTN4-mediated AOS-targeting
What are the possible binding partners required for CNTN4’s influence on axon targeting to the NOT? Previously the Flanagan lab used biochemical and in vitro growth cone assays to discover the binding partners of β-Amyloid Precursor Protein (APP) and found that CNTN4 is one of the major binding partners in chick neurons, and that CNTN4 can impact RGC growth cone guidance (Osterfield et al., 2008). Intrigued by those findings, we sought to determine whether in mice, CNTN4 and APP interact and contribute to targeting of RGCs to the AOS. We first co- immunoprecipitated APP from P8 whole brain lysate and immunoblotted for CNTN4 protein. We were able to pull down CNTN4 protein with APP in lysates from wildtype mice but not from lysates in CNTN4 mutant mice (Figure 6A and B). While these experiments do not rule out other binding partners for APP or CNTN4, they establish a direct relationship between the two. We next analyzed the developing visual pathway for APP protein by immunohistochemistry and found it is expressed by the vast majority of cells within the developing ganglion cell layer of the retina, including by Hoxd10-RGCs (Figures 6C, and S5I–K). APP protein is also expressed within most retinorecipient targets, including the NOT, where it overlaps with the expression of CNTN4 (Figures 6D–F). Eye removal abolished APP staining in the NOT (Figure 6G) indicating that, like CNTN4, APP is expressed by RGC axons that target the NOT and not by cells within the NOT.
Figure 6. APP is a CNTN4 binding partner and is required for normal RGC targeting of the NOT.
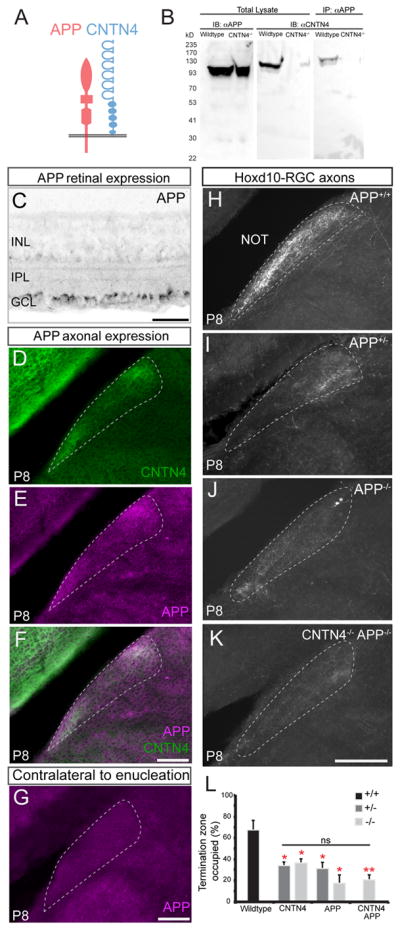
(A) Schematic of proposed interaction between CNTN4 and APP (Osterfield et al., 2008)
(B) Co-immunoprecipitation: APP protein was immunoprecipitated from wildtype or from CNTN4−/− brain tissue and ran on a western blot that was blotted for CNTN4 and APP antibodies.
(C) Expression of APP protein in RGC somas within the ganglion cell layer (GCL). IPL: inner plexiform layer. INL: inner nuclear layer.
Scale = 100μm.
(D) CNTN4, and APP (E) protein expression and (F) their merge, in the P8 NOT. Scale = 250μm.
(G) Lack of APP protein expression in the NOT on P8 after binocular enucleation on P0. Scale = 250μm.
(H–K) Hoxd10-RGC axons in the NOT of P8 wildtype APP+/+ (H), APP+/− (I), APP−/− (J), and APP−/−, CNTN4−/− double-knockout mice (K). Scale = 250μm.
(L) Percentage of the NOT occupied by Hoxd10-RGC axons in wildtype mice, CNTN4 and APP mutant mice (±SEM); Statistical comparison to wildtype mice by students one-tailed t-test. * = p<0.05; ** = p<0.01; ns: no significant difference (n = 3–6 mice per group).
To examine the role of APP in RGC axon-target matching in the AOS, we generated APP−/−, Hoxd10-GFP mice and analyzed the projection patterns of the GFP+ RGCs in the brain. The density and extent of Hoxd10- RGC axon targeting in the NOT was significantly reduced in both APP+/−, Hoxd10-GFP and APP−/−, Hoxd10-GFP mice, compared to their wildtype littermate controls (Figures 6H–J, 6L; n=4–5 mice per genotype). Indeed, the fraction of the termination zone occupied by Hoxd10-GFP RGC axons was qualitatively and quantitatively similar between CNTN4 and APP heterozygous and homozygous null mice (Figure 6L). Similar to CNTN4 mutants, Hoxd10-GFP RGC inputs to the MTN and whole-eye innervation and binocular segregation of RGC projections to the visual thalamus and SC appeared normal in APP mutant mice (Figure S5A–C, 5E–G). Also, the number of Hoxd10-RGCs was comparable between APP mutant and wildtype retinas (Figure S5D). Thus, altering APP expression does not generally perturb RGC targeting or survival but it does have a significant impact on axon-target matching to the AOS in a manner resembling CNTN4.
To address whether APP and CNTN4 mediate axon-target matching of AOS-projecting RGCs through convergent or independent molecular pathways, we generated double-mutant, transgenic CNTN4−/−, APP−/−, Hoxd10-GFP mice. Our prediction was that if CNTN4 and APP reside in independent pathways then removal of both of them would cause an additive reduction to NOT targeting beyond that observed in APP or CNTN4 single mutants alone. Instead, however, we observed a similar reduction in Hoxd10-GFP RGC innervation of the NOT in CNTN4−/−, APP−/− double mutant mice as we observed in CNTN4-only or APP-only mutant mice (Figure 6K and 6L).
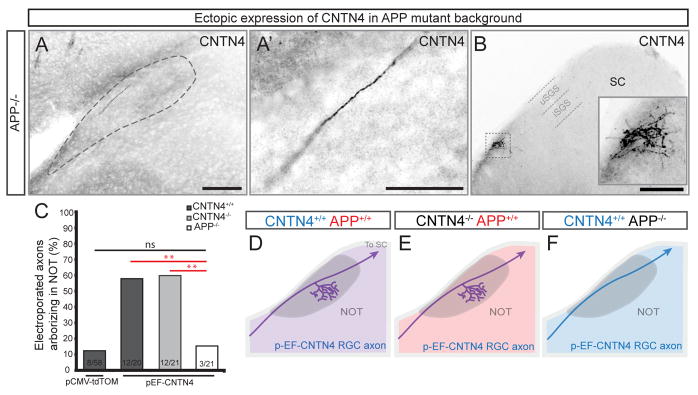
To further address the role of APP in CNTN4-mediated RGC targeting, next we used an in vivo genetic approach. We ectopically expressed CNTN4 in individual RGC axons by electroporation in APP mutants and examined their patterns of targeting in the brain. As described above, regardless of whether this manipulation is carried out in wildtype mice or in CNTN4−/− mice, the CNTN4-electroporated RGCs were biased to arborize in the NOT. By contrast, ectopic expression of CNTN4 in individual RGC axons failed to produce this targeting bias in mice lacking APP (Figures 7A, 7C): only 3/21 CNTN4-electroporated RGCs arborized in the NOT of APP mutants, the same low frequency observed when RGCs were electroporated with tdTomato plasmids (Figure 7C). Importantly, the vast majority of CNTN4-electroporated RGC axons still managed to form terminal arbors in the SC of APP−/− mice (Figure 7B) indicating that the loss of APP does not generally disrupt an RGC’s ability to grow the full distance of the subcortical visual pathway or to elaborate axonal arbors within visual targets. Collectively, these results suggest that APP is required for normal targeting of Hoxd10-GFP RGCs to the NOT, and that CNTN4 and APP, both of which are expressed by NOT-projecting RGC axons, physically interact with each other to influence axon-target matching.
Figure 7. CNTN4-mediated axon arborization in the NOT requires APP.
(A) Individual CNTN4-electroporated RGC axon in the NOT of an APP−/− mouse.
(A′) High magnification of axon in (A); Scale in A = 250μm; Scale in A′ = 125μm.
(B) Terminal arborization of the same CNTN4-electroporated RGC axon in the SC; Scale = 500μm.
(C) Probability of selective arborization in the NOT in wildtype, CNTN4 or APP mutants. Statistical significance determined by Fisher analysis; ** = p<0.01.
(D–F) Schematics depicting typical outcomes of each electroporation experiment.
purple: intact CNTN4 and APP expression; red: APP expression only; blue; CNTN4 expression only
Functional defects in AOS circuits and their behavioral outputs in CNTN4 mutant mice
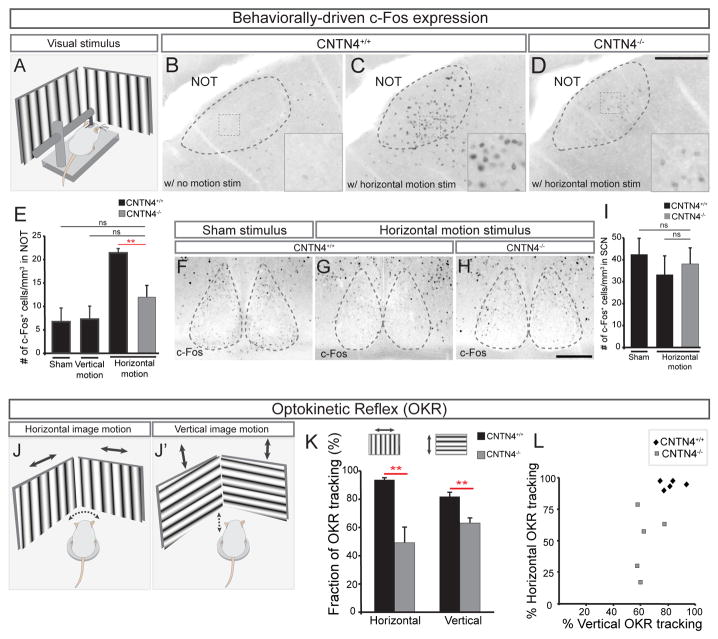
What are the functional consequences of altering Hoxd10-RGC input to the NOT? The NOT is responsible for generating horizontal compensatory eye movements in response to slow speed motion (Simpson, 1984). To determine whether there are defects in the activity of NOT neurons in CNTN4−/− mice, we presented dark-adapted head-fixed wildtype mice and CNTN4−/− mice with horizontal or vertical stimuli optimal for driving slip-compensating eye movements (Figure 8A) (Dhande et al., 2013). Mice were exposed to 2 hours of stimulation with either a sham stimulus (gray screen), a horizontal motion stimulus, or a vertical motion stimulus, and then were perfused and their brains processed for c-Fos immunoreactivity, which is an indirect readout of neural activation (Omori et al., 2005; Yonehara et al., 2009). In wildtype mice, large numbers of intensely labeled c-Fos+ cells were observed in the NOT after horizontal motion stimulus as compared to a sham stimulus (Figure 8B and 8C) However, in CNTN4−/− mice shown an identical horizontal motion stimulus, the number of c-Fos+ cells was significantly reduced and the few c-Fos+ cells that were activated appeared dimmer (Figures 8D, E). Notably, the number of c-Fos+ cells in the NOT in response to vertical stimulation was similar to the number of c-Fos+ cells elicited by sham stimulus (Figure 8E) and c-Fos activation in a different retinorecipient target which responds to overall ambient luminance but not to motion, the suprachiasmatic nucleus (SCN), revealed no significant differences between wildtype or CNTN4−/− mice using either sham or horizontal motion stimuli (Figure 8F–I). These data indicate that loss of CNTN4 causes a significant defect in the activation of NOT neurons in response to stimuli that normally drive this retinofugal pathway.
Figure 8. Loss of CNTN4 perturbs function and behavioral output of AOS circuitry.
(A) Schematic of visual stimulation. Head-fixed mouse is placed in a chamber surrounded by video monitors on all 4 sides.
(B–E) Analysis of c-Fos expression in NOT cells to assess the level of activity induced by a visual stimulus in wildtype (B, C) or CNTN4−/− mice (D). Insets in (B–D) high magnification views of boxed regions. Scale = 250μm. (E) Number of c-Fos+ cells in the NOT per mm3. ±SEM. *** = p<0.001 (n = 5–6 mice per group; age = P22–P23).
(F–I) c-Fos activation in the suprachiasmatic nucleus (SCN) of wildtype (F, G) or CNTN4−/− (H) mice. Scale = 100μm. (H) Number of c-Fos+ cells per mm3 of the SCN (±SEM); ns: no significant difference.
(J–L) Schematic if the optokinetic reflex (OKR) behavioral analysis; head movements in response to horizontal (J) or vertical drifting stimuli (J′) measured (see text and Experimental Procedures).
(K) The average percentage of trials tracked by CNTN4+/+ or CNTN4−/− mice in response to horizontal or vertical motion (n=5 mice per genotype).
(L) Plot of percentage of trials tracked in vertical versus horizontal motion for each animal.
What are the behavioral consequences of reduced afferent input to the NOT from AOS-RGCs in CNTN4 mutants? To address this, we measured optokinetic reflex (OKR) behavior where mice display compensatory head movements in response to bar-grating stimuli drifting slowly in either the horizontal or vertical axis (Figures 8J, J′, K; Prusky et al., 2008; Wang et al., 2009; Tschetter et al., 2011). We quantified the percentage of 15 second horizontal or vertical motion trials tracked by P25-P30 wildtype and CNTN4−/− (Figures 8J–L). Wildtype mice tracked ~95% of the horizontal-stimulus trials and tracked ~80% of the vertical-stimulus trials (Figure 8K; n=5 mice) whereas CNTN4−/− mice tracked only ~50% of horizontal trials and ~60% of vertical trials, both of which represent significant reductions in OKR behavior compared to controls (Figure 8K, L; n=5 mice; p<0.01). These differences are highly unlikely to be caused by defects in retinal wiring as the number and physiology of RGCs that drive this system was normal in CNTN4 mutants at equivalent ages (Figures S3 and S5G). These indicate that the reduction in Hoxd10-GFP RGC projections to the NOT and the diminished c-Fos activation of NOT neurons are both also associated with a defect in AOS circuit performance.
The defects in vertical OKR tracking in CNTN4−/− mice may seem somewhat surprising given that Hoxd10-GFP RGC input to the MTN appears normal in these mice (Figures S2B and S2C), it is noteworthy that NOT neurons are known to project to and modulate activity of a subset of neurons in the MTN (Simpson, 1984). Indeed, in wildtype mice, there were a greater number of c-Fos+ cells in the MTN after horizontal full-field motion as compared to a sham stimulus, and this effect was lost in CNTN4−/− mice (Figure S6A, 6C–E). Importantly, there were no defects in the pupillary light reflex in CNTN4−/− mice, even when tested at several irradiance levels (Figure S6F). Together, these data indicate that loss of CNTN4 and the associated defects in axon-target matching to the NOT selectively perturb AOS circuit function and its behavior-generating capacity.
DISCUSSION
Here we report a molecular mechanism controlling a specific and functionally essential aspect of parallel pathway assembly: axon-target matching. We identified CNTN4 as a factor required for a subset of RGCs to connect to AOS brain targets for image stabilization. Then, through loss-of-function and gain-of-function experiments in wildtype, CNTN4 and APP mutant mice, we determined that i) CNTN4 expressed by RGC axons is required for collateral branching and target-specific arborization of those RGCs in the NOT, ii) this process requires APP, and iii) functional and behavioral consequences result from disruption of axon-target specificity in this pathway.
Axon-target matching by target-specific arborization
By exploring the effects of ectopically expressing CNTN4 in individual RGCs, we identified key aspects of the axon-target matching process in the developing optic pathway: target-specific axon arborization. In mice >90% of RGCs project to the most distal retinorecipient target, the SC, and the vast majority of projections to other retinorecipient targets represent locally-arborized collateral branches of those same SC-projecting axons (Hofbauer and Drager, 1985; Huberman et al., 2009; Dhande et al., 2011). When we electroporated RGCs with CNTN4, we found that the axons of those RGCs tended to establish branched arbors specifically in the NOT. We found that their axons still traveled to the SC where they formed arbors and that there was no increased propensity to establish arbors in other major retinorecipient targets – indicating a role in specificity as opposed to non-specific growth. Our results suggest several possible mechanisms by which CNTN4 exerts its effects on AOS wiring. CNTN4 may promote NOT targeting which in turn initiates arbor formation, or CNTN4 may promote axon arborization in the NOT in a manner that reflects ‘targeting’ as the final outcome. In the absence of in vivo time-lapse imaging, these two possibilities are difficult to separate. Nonetheless, given that CNTN4 mutations both reduced the frequency NOT targeting and reduced arbor complexity it is clear that these processes are linked. Taken with previous findings, these results indicate that axon-target matching reflects molecular programs that can bias axons to i) locally steer toward groups of targets (e.g., Ringstedt et al., 1998), ii) remain within those targets once they arrive (Sun et al., submitted; Osterhout et al., 2011) and, iii) promote target-specific arborization (the present data). Recent work in Drosophila also underscores the extent to which axon-target matching is a multi-step process that reflects a series of axon-navigation and arbor maturation events (Joo et al., 2013).
What factors control RGC targeting to the MTN? An accompanying paper describes a key role for semaphorin-plexin reverse signaling in the development of retino-MTN circuitry (Sun et al., in press). It is possible that semaphorin-plexin signaling may override the requirement for CNTN4 in the MTN-projection pathway. Also, it is notable that DSGC inputs to the MTN comprise a distinct axonal trajectory from the other AOS-projecting RGCs and may require unique molecular cues (e.g., Figure 1A; Pak et al., 1987; Yonehara et al., 2009; Dhande et al., 2013; Dhande and Huberman, 2014). It is also possible that CNTN4 is important for arborization in the MTN but the compact, “tract-like” architecture of the MTN nuclei make it challenging to detect abnormal arborization patterns within them. Nevertheless, we note that ectopic expression of CNTN4 in individual RGC axons did not bias RGC axons to terminate in the MTN, suggesting that other pre-target and within-target molecular signals govern development of that pathway; an accompanying paper from Kolodkin and co-workers supports that model (e.g., Sun et al., in press).
APP is required for CNTN4-mediated RGC axon targeting
Our data reveal a novel role for APP in axon-target matching. Removal of APP caused an overall reduction in NOT innervation by Hoxd10-GFP RGCs. Removal of APP was also sufficient to occlude CNTN4-mediated biasing of RGC axon arborization in the NOT. Since we find that APP is expressed by most RGCs, it seems unlikely that APP directly imparts target-specificity. Rather, because both APP and CNTN4 are expressed by RGC axons, we favor a model in which APP acts as co-receptor with CNTN4 to allow CNTN4 to impart its role in target specificity. At this time, the identity of the putative target-derived ligand is unknown but there is a general precedent for this model; Kaltschmidt and co-workers recently showed that a closely related contactin, CNTN5, operates with a co-receptor to influence axon connectivity in the developing vertebrate spinal cord (Ashrafi et al., 2014). It is also interesting that both in our study and that of Ashrafi et al., (2014) axonal targeting defects were observed in heterozygous and homozygous CNTN mutant mice, which points to the importance of maintaining correct CNTN levels for CNS wiring.
The fact that whole-eye labeling of all RGCs in APP mutant mice did not reveal any overt changes in retinofugal targeting may be due the redundancy between APP and its related family members APLP1 and APLP2, which are also expressed by RGCs (Reinhard et al., 2005; Walsh et al., 2007). In any case, the results presented here that CNTN4-mediated targeting requires APP, add to growing evidence obtained in vitro (Osterfield et al., 2008) and in vivo (Nikolaev et al., 2009; Olsen et al., 2014) that APP is an important molecular determinant of RGC axon targeting.
Circuit alterations in CNTN4 mutants cause altered function and behavioral output of the AOS
One of the central goals of developmental neuroscience is to parse how specific developmental wiring steps influence circuit function and behavior. We found that loss of CNTN4 causes a dramatic decrease in the number and intensity of c-Fos+ cells in AOS targets – an indirect readout of neural activity (Omori et al., 2005). In addition, CNTN4 mutant mice displayed defects in their ability to track drifting gratings, as compared to wildtype controls. We interpret these defects as a reduced efficiency of the stimuli to drive NOT neurons that generate OKR behavior (Yonehara et al., 2009; Dhande et al., 2013; Tschetter et al., 2013). The defect in horizontal tracking is especially intriguing because even though Hoxd10-GFP RGC inputs to the NOT were significantly reduced in CNTN4 mutants, whole-eye labeling revealed that RGC axons still fill this entire target. This suggests that RGCs that target the NOT in CNTN4 mutants, such as DRD4-RGCs are less efficient in driving NOT neurons – Likely because they are not optimally tuned for full-field slow moving horizontal stimuli. A non-mutually-exclusive idea is the RGC axons that abnormally target the NOT are unable to establish normal functional connections with NOT target cells. This could be due to a mismatch in cell-cell recognition factors, delays in developmental timing, or other key steps of synaptic circuit assembly. Regardless, our data support the idea that axon-target matching is a crucial step leading to normal activation patterns of target neurons and the overall circuits in which they participate.
Disease and broader implications
Finally, although the present study was limited to analysis of optic pathway eye-to-brain connections, it is worth noting that CNTN4 is expressed in multiple vertebrate CNS regions (Yoshihara et al., 1995) and has been shown to be important for circuit assembly in other contexts, including olfactory sensory neuron axonal convergence in the olfactory bulb (Kaneko-Goto et al., 2008), and dendritic fasciculation and laminar targeting in the chick retina (Yamagata and Sanes, 2012). CNTN4 mutations are also associated with multiple neurological disorders, including 3p-deletion syndrome, bipolar disorder and autism spectrum disorder (ASD; Fernandez et al., 2004; Roohi et al., 2009; Kerner et al., 2011; Zuko et al., 2013). In addition, CNTN4 maps to a chromosome 3p26, which harbors a genetic link to Alzheimer’s Disease (Blacker et al., 2003). Understanding the full range of ways in which CNTN4 and APP control neural circuit assembly may eventually inform better understanding of the specific defects that underlie these diseases (Zuko et al., 2011).
Experimental procedures
Animals
Homeobox D10-EGFP (Hoxd10-GFP) mice were obtained from MMRRC (Dhande et al., 2013), Contactin-4 deficient mice (CNTN4−/−) were generated as previously described (Kaneko-Goto et al, 2008), Beta-amyloid precursor protein knockout mice (APPtm1dbo) were obtained from Jax. All experimental comparisons were littermate controls. Postnatal day 0 is day of birth. All procedures carried out in accordance with institutionally approved protocols at UCSD.
Tissue processing
See: Osterhout et al., 2014. Briefly, animals were overdosed and perfused with saline followed by 4% paraformaldehyde (PFA). Brains were post-fixed overnight (retinas were post-fixed for 2–4 hours). After sectioning on a freezing microtome or cryostat (brains, 45μm thickness; retinas, 30μm thickness), tissue was incubated with blocking solution for 2 hours [10% goat serum, and 0.25% Triton-X by volume in PBS] then incubated in primary antibodies overnight at 4 degrees Celsius. After a series of washes (3 × 20 minutes each in 1XPBS), secondary antibody staining was performed for 1.5 hours at room temperature by diluting secondary antibodies in blocking solution as above. Primary antibodies: rabbit anti-GFP (1:1000; Invitrogen), guinea pig anti-GFP (1:1000; Synaptic Systems), guinea pig anti-CNTN4 (1:1000–1500; Kaneko-Goto et al., 2008), rabbit anti-APP (1:1000; Calbiochem) guinea pig anti-VAChT (1:1000; Millipore), rabbit anti-c-Fos (1:10,000; Calbiochem)
Monocular enucleation
Postnatal mice (P0/1) were anesthetized on ice, eyelid was parted and eye removed, taking care to limit bleeding from the ophthalmic artery. Tissue was harvested one week later, at P8.
Quantification of target area occupied by Hoxd10-GFP RGC axons and total target volume
See Osterhout et al., 2014; Retinorecipient nuclei labeled by intraocular CTβ-594 and target boundaries determined by landmark shape. Target volume was measured by adding values from every section through the target (using the area tool in ImageJ) and multiplying that value by the thickness of the individual sections (45μm).
Retinal ganglion cell electroporation
See: Dhande et al., 2011. Mice (P0/1) were anesthetized on ice, the eyelid opened, and DNA plasmid (~0.5–1μl; pEF-CNTN4 or pCA-tdTomato) injected by picospritzer. Six square wave pulses (25V, 50 msec each, 1s gap x3 pulses of each polarity) were applied with forcep electrodes. To quantify the number of electroporated axons that arborized in the NOT, Hoxd10-GFP brains were sectioned and stained for GFP and CNTN4. Axons with secondary branch confined to the boundaries of the NOT were considered ‘arborizing’ in the target. Statistical significance was determined using Fisher analysis (n=20–58 axons per plasmid/genetic background condition; 10–20 mice per electroporation group).
Single axon reconstruction
7 days post-electroporation, brains were sectioned at 200μm, imaged with Zeiss 710 confocal microscope and axons reconstructed and analyzed with Neurolucida.
Co-immunoprecipitation
Dynabeads Co-immunoprecipitation kit (Life technologies) and protocol were used to co-immunoprecipitate APP in P8 whole brain lysate from wildtype and CNTN4−/− mice. Lysate was spun at 840 Gs × 2 min. Antibody coupled Dynabeads, were incubated with lysate for 45 min. Western blot was analyzed using guinea pig anti-CNTN4 (1:2000; Kaneko-Goto et al., 2008) and rabbit anti-APP (1:1000; Calbiochem) antibodies.
c-Fos
See: Dhande et al., (2013). P20 mice were anesthetized and fitted with a small metal head post adaptor using dental cement; recovery: 2–3 days. Mice were dark adapted for 24 hours. Head-fixed mice were presented with sham stimulus, horizontal-drifting or vertical-drifting, bars (0.16 cycles/degree, 1.25 degrees/second) for 2 hours (Yonehara et al., 2009; Dhande et al., 2013). Immediately after stimulation brains were collected and processed for c-Fos. Number of c-Fos+ cells was measured from throughout the NOT in both groups (n=5–6 mice per genotype for horizontal stimulus and n=3 for vertical and sham stimulus).
Analysis of optokinetic reflex behavior
See: Wang et al. (2009). Wildtype and mutant mice (P25–P30) were placed on an elevated platform surrounded by 4 computer monitors. Each trial consisted of horizontal or vertical drifting-bar stimulus at 0.16 cycles/degree and 12 deg/sec. Each trial lasted 15 seconds; if the head of the mouse moved in concert with the gratings, that trial was scored as “tracked”. Each mouse was presented with 10–12 trials per day, for 3–4 consecutive days, the same time of day. Responses were averaged to generate a mean % of trials tracked; n = 5 mice per genotype.
Analysis of pupil reflex
See Sweeney et al. (2014). Mice were dark-adapted for 1 hour. Infrared video of the right eye was recorded just prior to and at the finish of a 30s trial of blue light stimulation directed at the left eye (Güler et al., 2008). The percent change in pupil area measured in ImageJ; n = 5 mice per genotype.
Electrophysiology
See: Dhande et al., 2013. Briefly, retinas were harvested and dissected in gassed (95% O2 and 5% CO2) Ames medium under infrared illumination, and cut along the dorsal-ventral axis (Wei et al., 2010; Stafford et al., 2014). Only ventral pieces were used. A piece of retina was placed in a chamber and superfused with gassed Ames medium heated to 33–35 C. GFP+ ganglion cells were visualized at by attenuated mercury light passed through a GFP dichroic mirror, and then targeted for recording under IR illumination. Cells were recorded with borosilicate glass pipettes (4–6 MΩ) filled with intracellular solution containing (in mM): 120 K-methanesulphonate 10 HEPES, 5 NaCl, 0.1 EGTA, 2 ATP-Mg2+, and 0.3 GTP-Na, titrated to pH 7.3.
Light stimuli were generated using Psychophysics Toolbox and MATLAB. Stimuli were projected onto the retina using a Dell video projector custom fitted with a UV LED (398nm), attenuated by a 1.0 ND filter, and focused to the level of rod and cone outer segments. The wavelength of the light stimulus is equally efficient at stimulating mouse M- and S-cones (Borghuis et al., 2013) and stable S-cone mediated responses can be recorded from GFP+ cells in the ventral mouse retina that have been targeted using epifluorescence (Stafford et al., 2014). Stimuli were presented over the receptive field center as a contrast pulse or as drifting square-wave gratings modulated against a mean luminance. The spatial (500 μm/cycle) and temporal frequency (1 Hz) of the gratings were near the peak sensitivity of Hoxd10-GFP RGCs (Dhande et al., 2013).
Directional preference was determined by drifting gratings in 12 directions for 4 seconds with an inter-stimulus interval of 10 seconds. The number of spikes obtained during a presentation of the gratings in a given direction was considered the response for that direction. Responses were normalized by the total number of spikes in all directions, and the preferred direction was the angle of the vector sum of the normalized response. The DSI was calculated as: DSI = (preferred − null)/(preferred+null), where preferred is the response in the stimulus direction closest to the preferred direction, and null is the response to the stimulus 180° opposite. The tuning width was determined by fitting the cell’s response as a function of stimulus direction with the von Mises distribution; the width was defined as the full width at half height of the von Mises fit (Elstrott et al., 2008).
Supplementary Material
HIGHLIGHTS.
CNTN4 is expressed by axons of image-stabilizing DSGCs
Loss of CNTN4 or APP alters RGC projections to a specific AOS target
CNTN4 is sufficient to bias RGC axonal arborization in the NOT
Loss of CNTN4 alters AOS-driven behaviors for image stabilization
Acknowledgments
We thank Dr. Onkar Dhande, Dr. Lu Sun and Dr. Alex Kolodkin for helpful comments and suggestions on an earlier version of this manuscript, Jared Sewell and Dr. Jeff Long for technical assistance with biochemistry experiments and Pritha P. Multani for histology help. J.A.O. and A.D.H conceived the experiments, J.A.O. and P.L.N. performed histology. J.A.O imaged and analyzed all the data. B.K.S recorded wildtype and mutant RGC receptive fields and analyzed the data. J.A.O., B.K.S, Y.Y. and A.D.H wrote the paper. Supported by NIH RO1 EY022157 (A.D.H.), the E. Matilda Ziegler Foundation (A.D.H), a Pew Scholar Award (A.D.H.) and National Science Foundation Graduate Research Fellowship under grant no. DGE-1144086 (J.A.O.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashrafi S, Betley JN, Comer JD, Brenner-Morton S, Bar V, Shimoda Y, Watanabe K, Peles E, Jessell TM, Kaltschmidt JA. Neuronal Ig/Caspr recognition promotes the formation of axoaxonic synapses in mouse spinal cord. Neuron. 2014;81:120–129. doi: 10.1016/j.neuron.2013.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier H. Synaptic laminae in the visual system: molecular mechanisms forming layers of perception. Annu Rev Cell Dev Biol. 2013;29:385–416. doi: 10.1146/annurev-cellbio-101011-155748. [DOI] [PubMed] [Google Scholar]
- Blacker D, Bertram L, Saunders AJ, Moscarillo TJ, Albert MS, Wiener H, Perry RT, Collins JS, Harrell LE, Go RCP, et al. Results of a high-resolution genome screen of 437 Alzheimer’s disease families. Hum Mol Genet. 2003;12:23–32. doi: 10.1093/hmg/ddg007. [DOI] [PubMed] [Google Scholar]
- Borghuis BG, Marvin JS, Looger LL, Demb JB. Two-Photon Imaging of Nonlinear Glutamate Release Dynamics at Bipolar Cell Synapses in the Mouse Retina. J Neurosci. 2013;33:10972–10985. doi: 10.1523/JNEUROSCI.1241-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cang J, Feldheim DA. Developmental mechanisms of topographic map formation and alignment. Annu Rev Neurosci. 2013;36:51–77. doi: 10.1146/annurev-neuro-062012-170341. [DOI] [PubMed] [Google Scholar]
- Dhande OS, Hua EW, Guh E, Yeh J, Bhatt S, Zhang Y, Ruthazer ES, Feller MB, Crair MC. Development of single retinofugal axon arbors in normal and β2 knock-out mice. J Neurosci. 2011;31:3384–99. doi: 10.1523/JNEUROSCI.4899-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhande OS, Estevez ME, Quattrochi LE, El-Danaf RN, Nguyen PL, Berson DM, Huberman AD. Genetic dissection of retinal inputs to brainstem nuclei controlling image stabilization. J Neurosci. 2013;33:17797–17813. doi: 10.1523/JNEUROSCI.2778-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhande OS, Huberman AD. Retinal ganglion cell maps in the brain: implications for visual processing. Curr Opin Neurobiol. 2014;24:133–142. doi: 10.1016/j.conb.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson BJ. Molecular mechanisms of axon guidance. Science. 2002;298:1959–1964. doi: 10.1126/science.1072165. [DOI] [PubMed] [Google Scholar]
- Elstrott J, Anishchenko A, Greschner M, Sher A, Litke AM, Chichilnisky EJ, Feller MB. Direction selectivity in the retina is established independent of visual experience and cholinergic retinal waves. Neuron. 2008;58:499–506. doi: 10.1016/j.neuron.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez T, Morgan T, Davis N, Klin A, Morris A, Farhi A, Lifton RP, State MW. Disruption of Contactin 4 (CNTN4) results in developmental delay and other features of 3p deletion syndrome. Am J Hum Genet. 2008;82:1385. doi: 10.1016/j.ajhg.2008.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güler AD, Ecker JL, Lall GS, Haq S, Altimus CM, Liao HW, Barnard AR, Cahill H, Badea TC, Zhao H, et al. Melanopsin cells are the principal conduits for rod-cone input to non-image-forming vision. Nature. 2008;453:102–105. doi: 10.1038/nature06829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofbauer A, Drager U. Depth segregation of retinal ganglion cells projecting to the mouse superior colliculus. J Comp Neurol. 1985;234:465–74. doi: 10.1002/cne.902340405. [DOI] [PubMed] [Google Scholar]
- Huberman AD, Wei W, Elstrott J, Stafford BK, Feller MB, Barres BA. Genetic identification of an On-Off direction-selective retinal ganglion cell subtype reveals a layer-specific subcortical map of posterior motion. Neuron. 2009;62:327–334. doi: 10.1016/j.neuron.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman AD, Clandinin TR, Baier H. Molecular and cellular mechanisms of lamina-specific axon targeting. Cold Spring Harb Perspect Biol. 2010;2:a001743. doi: 10.1101/cshperspect.a001743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo WJ, Sweeney LB, Liang L, Luo L. Linking Cell Fate, Trajectory Choice, and Target Selection: Genetic Analysis of Sema-2b in Olfactory Axon Targeting. Neuron. 2013;78:673–686. doi: 10.1016/j.neuron.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko-Goto T, Yoshihara SI, Miyazaki H, Yoshihara Y. BIG-2 mediates olfactory axon convergence to target glomeruli. Neuron. 2008;57:834–846. doi: 10.1016/j.neuron.2008.01.023. [DOI] [PubMed] [Google Scholar]
- Kay JN, De la Huerta I, Kim IJ, Zhang Y, Yamagata M, Chu MW, Meister M, Sanes JR. Retinal ganglion cells with distinct directional preferences differ in molecular identity, structure, and central projections. J Neurosci. 2011;31:7753–7762. doi: 10.1523/JNEUROSCI.0907-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerner B, Lambert CG, Muthén BO. Genome-wide association study in bipolar patients stratified by co-morbidity. PLoS ONE. 2011;6:e28477. doi: 10.1371/journal.pone.0028477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtman JW, Denk W. The big and the small: challenges of imaging the brain’s circuits. Science. 2011;334:618–623. doi: 10.1126/science.1209168. [DOI] [PubMed] [Google Scholar]
- Luo L, Flanagan JG. Development of continuous and discrete neural maps. Neuron. 2007;56:284–300. doi: 10.1016/j.neuron.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Masseck OA, Hoffmann KP. Comparative neurobiology of the optokinetic reflex. Ann N Y Acad Sci. 2009;1164:430–439. doi: 10.1111/j.1749-6632.2009.03854.x. [DOI] [PubMed] [Google Scholar]
- Morin LP, Studholme KM. Retinofugal projections in the mouse. J Comp Neurol. 2014;522:3733–3753. doi: 10.1002/cne.23635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev A, McLaughlin T, O’Leary DDM, Tessier-Lavigne M. APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature. 2009;457:981–989. doi: 10.1038/nature07767. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Olsen O, Kallop DY, McLaughlin T, Huntwork-Rodriguez S, Wu Z, Duggan CD, Simon DJ, Lu Y, Easley-Neal C, Takeda K, et al. Genetic Analysis Reveals that Amyloid Precursor Protein and Death Receptor 6 Function in the Same Pathway to Control Axonal Pruning Independent of -Secretase. Journal of Neuroscience. 2014;34:6438–6447. doi: 10.1523/JNEUROSCI.3522-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori T, Kawashima H, Kizuka T, Ohiwa N, Tateoka M, Soya H. Increased c-fos gene expression in alpha motoneurons in rat loaded hindlimb muscles with inclined locomotion. Neuroscience Letters. 2005;389:25–29. doi: 10.1016/j.neulet.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Osterfield M, Egelund R, Young LM, Flanagan JG. Interaction of amyloid precursor protein with contactins and NgCAM in the retinotectal system. Development. 2008;135:1189–1199. doi: 10.1242/dev.007401. [DOI] [PubMed] [Google Scholar]
- Osterhout JA, Josten N, Yamada J, Pan F, Wu S, Nguyen PL, Panagiotakos G, Inoue YU, Egusa SF, Volgyi B, Inoue T, Bloomfield SA, Barres BA, Berson DM, Feldheim DA, Huberman AD. Cadherin-6 mediates axon-target matching in a non-image-forming visual circuit. Neuron. 2011;71:632–639. doi: 10.1016/j.neuron.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterhout JA, El-Danaf RN, Nguyen PL, Huberman AD. Birthdate and outgrowth timing predict cellular mechanisms of axon target matching in the developing visual pathway. Cell Reports. 2014;8:1006–1017. doi: 10.1016/j.celrep.2014.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak MW, Giolli RA, Pinto LH, Mangini NJ, Gregory KM, Vanable JW. Retinopretectal and accessory optic projections of normal mice and the OKN-defective mutant mice beige, beige-J, and pearl. J Comp Neurol. 1987;258:435–446. doi: 10.1002/cne.902580311. [DOI] [PubMed] [Google Scholar]
- Prusky GT, Silver BD, Tschetter WW, Alam NM, Douglas RM. Experience-dependent plasticity from eye opening enables lasting, visual cortex-dependent enhancement of motion vision. J Neurosci. 2008;28:9817–9827. doi: 10.1523/JNEUROSCI.1940-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard C, Hébert SS, De Strooper B. The amyloid-β precursor protein: integrating structure with biological function: APP: integrating structure with function. The EMBO Journal. 2005;24:3996–4006. doi: 10.1038/sj.emboj.7600860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringstedt T, Braisted JE, Brose K, Kidd T, Goodman C, Tessier-Lavigne M, O’Leary DD. Slit inhibition of retinal axon growth and its role in retinal axon pathfinding and innervation patterns in the diencephalon. J Neurosci. 2000;20:4983–91. doi: 10.1523/JNEUROSCI.20-13-04983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles E, Baier H. Assembly of synaptic laminae by axon guidance molecules. Curr Opin Neurobiol. 2012;22:799–804. doi: 10.1016/j.conb.2012.04.014. [DOI] [PubMed] [Google Scholar]
- Robles E, Laurell E, Baier H. The retinal projectome reveals brain-area-specific visual representations generated by ganglion cell diversity. Curr Biol. 2014;22:2085–96. doi: 10.1016/j.cub.2014.07.080. [DOI] [PubMed] [Google Scholar]
- Roohi J, Montagna C, Tegay DH, Palmer LE, DeVincent C, Pomeroy JC, Christian SL, Nowak N, Hatchwell E. Disruption of contactin 4 in three subjects with autism spectrum disorder. J Med Genet. 2009;46:176–182. doi: 10.1136/jmg.2008.057505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, Yamagata M. Many paths to synaptic specificity. Annu Rev Cell Dev Biol. 2009;25:161–195. doi: 10.1146/annurev.cellbio.24.110707.175402. [DOI] [PubMed] [Google Scholar]
- Schmidt ERE, Brignani S, Adolfs Y, Lemstra S, Demmers J, Vidaki M, Donahoo ALS, Lilleväli K, Vasar E, Richards LJ, et al. Subdomain-mediated axon-axon signaling and chemoattraction cooperate to regulate afferent innervation of the lateral habenula. Neuron. 2014;83:372–387. doi: 10.1016/j.neuron.2014.05.036. [DOI] [PubMed] [Google Scholar]
- Shimoda Y, Watanabe K. Contactins: emerging key roles in the development and function of the nervous system. Cell Adh Migr. 2009;3:64–70. doi: 10.4161/cam.3.1.7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JI. The accessory optic system. Annu Rev Neurosci. 1984;7:13–41. doi: 10.1146/annurev.ne.07.030184.000305. [DOI] [PubMed] [Google Scholar]
- Stafford BK, Park SJH, Wong KY, Demb JB. Developmental changes in NMDA receptor subunit composition at ON and OFF bipolar cell synapses onto direction-selective retinal ganglion cells. J Neurosci. 2014;34:1942–8. doi: 10.1523/JNEUROSCI.4461-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney NT, Tierney H, Feldheim DA. Tbr2 is required to generate a neural circuit mediating the pupillary light reflex. J Neurosci. 2014;34:5447–53. doi: 10.1523/JNEUROSCI.0035-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschetter WW, Douglas RM, Prusky GT. Experience-induced interocular plasticity of vision in infancy. Front Syst Neurosci. 2011;5:44. doi: 10.3389/fnsys.2011.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschetter WW, Alam NM, Yee CW, Gorz M, Douglas RM, Sagdullaev B, Prusky GT. Experience-enabled enhancement of adult visual cortex function. J Neurosci. 2013;33:5362–5366. doi: 10.1523/JNEUROSCI.5229-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh DM, Minogue AM, Sala Frigerio C, Fadeeva JV, Wasco W, Selkoe DJ. The APP family of proteins: similarities and differences. Biochem Soc Trans. 2007;35:416–420. doi: 10.1042/BST0350416. [DOI] [PubMed] [Google Scholar]
- Wang L, Rangarajan KV, Lawhn-Heath CA, Sarnaik R, Wang BS, Liu X, Cang J. Direction-specific disruption of subcortical visual behavior and receptive fields in mice lacking the beta2 subunit of nicotinic acetylcholine receptor. J Neurosci. 2009;29:12909–12918. doi: 10.1523/JNEUROSCI.2128-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Elstrott J, Feller MB. Two-photon targeted recording of GFP-expressing neurons for light responses and live-cell imaging in the mouse retina. Nature Protocols. 2010;5:1347–1352. doi: 10.1038/nprot.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata M, Sanes JR. Expanding the Ig superfamily code for laminar specificity in retina: expression and role of contactins. J Neurosci. 2012;32:14402–14414. doi: 10.1523/JNEUROSCI.3193-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonehara K, Ishikane H, Sakuta H, Shintani T, Nakamura-Yonehara K, Kamiji NL, Usui S, Noda M. Identification of retinal ganglion cells and their projections involved in central transmission of information about upward and downward image motion. PLoS ONE. 2009;4:e4320. doi: 10.1371/journal.pone.0004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara Y, Kawasaki M, Tamada A, Nagata S, Kagamiyama H, Mori K. Overlapping and differential expression of BIG-2, BIG-1, TAG-1, and F3: four members of an axon-associated cell adhesion molecule subgroup of the immunoglobulin superfamily. J Neurobiol. 1995;28:51–69. doi: 10.1002/neu.480280106. [DOI] [PubMed] [Google Scholar]
- Zuko A, Bouyain S, van der Zwaag B, Burbach JPH. Contactins: structural aspects in relation to developmental functions in brain disease. Adv Protein Chem Struct Biol. 2011;84:143–180. doi: 10.1016/B978-0-12-386483-3.00001-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuko A, Kleijer KTE, Oguro-Ando A, Kas MJH, van Daalen E, van der Zwaag B, Burbach JPH. Contactins in the neurobiology of autism. European Journal of Pharmacology. 2013;719:63–74. doi: 10.1016/j.ejphar.2013.07.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.