Abstract
The lack of structural information on hepatitis C virus (HCV) surface proteins has so far hampered the development of effective vaccines. Recently, two crystallographic structures have described the core portion (E2c) of E2 surface glycoprotein, the primary mediator of HCV entry. Despite the importance of these studies, the E2 overall structure is still unknown and, most importantly, several biochemical and functional studies are in disagreement with E2c structures. Here, the main literature will be discussed and an alternative disulfide bridge pattern will be proposed, based on unpublished human monoclonal antibody reactivity. A modeling strategy aiming at recapitulating the available structural and functional studies of E2 will also be proposed.
Keywords: HCV, E2, X-ray structure, disulfide bridges, in silico modeling, monoclonal antibody, alanine scanning
Introduction
Hepatitis C virus (HCV) chronically infects approximately 180 million people worldwide and is a major cause of liver cirrhosis and hepatocellular carcinoma [1]. Until recently, the standard therapy has been limited to the use of nonspecific molecules [e.g. polyethylene glycol (PEG)-interferon (IFN)α and ribavirin], featuring variable efficacy depending on HCV genotype and being burdened by remarkable side-effects. The development of novel molecules specifically inhibiting HCV replicative stages has been driven by the structural characterization of internal viral proteins (e.g. protease and RNA polymerase) [2–4]. Conversely, only little direct structural information has been available for HCV surface glycoproteins (E1 and E2) responsible for viral attachment and entry. The lack of knowledge about E1 and E2 impairs the development of molecules inhibiting the early stages of infection and, most importantly, of immunogens capable of eliciting a protective immune response against HCV.
E1 and E2 are highly glycosylated transmembrane proteins that assemble in heterodimers, the functional unit necessary for viral attachment and entry. E2 mediates the viral attachment to host cells interacting with several host proteins such as CD81 (the main cellular receptor), the scavenger receptor class B type I (SR-BI) and members of the claudin/occludin family [5–7]. By contrast, the fusion step remains unclear, with E1 and E2 presenting a putative fusion peptide and probably co-participating in the conformational rearrangement necessary for viral entry [8,9]. Whereas the E1 structure remains largely unknown, that of the central portion [i.e. E2 core (E2c)] of E2 ectodomain (E2e) has been recently solved through X-ray crystallography [10,11]. These studies report for the first time the structure of E2 domains involved in the interaction with CD81, but significant structural information is still missing. Moreover, some structural features of E2c are in disagreement with previously crystallized short peptides and with biological data.
In this paper, we will discuss direct and indirect studies of E2 structure, highlighting the discrepancies between structural and functional data and proposing a novel E2 cysteine disulfide assignment based on unpublished immunological data that reconcile all previous evidence. We will then focus on possible computational and experimental methods that can be used to generate an E2e model comprising direct and indirect structural data that can be validated through biological features.
HCV: a heterogeneous and elusive pathogen
HCV is classified into the Flaviviridae family, which includes four genera of enveloped viruses sharing structural, genetic and replicative features. However, the hepacivirus genus, including HCV, is phylogenetically distant from the other main genera: flavivirus, pestivirus and pegivirus [12]. HCV is highly variable and is further classified into eight distinct genotypes, each comprising several subtypes [13]. This variability has greatly hampered the elucidation of HCV structure and replicative cycle; such studies are made even more difficult by the fact that HCV does not replicate in vitro and can be studied in only a few non-human primates (e.g. chimpanzees, gorillas). As a consequence, the vast majority of available data has been generated through surrogate systems such as virus-like particles (HCV-LP), viral pseudo-particles (HCVpp) and a laboratory-adapted strain (HCVcc).
Cryo-electron microscopy (cryoEM) experiments on the overall shape of HCV have led to different results. Two studies performed on HCVcc, HCV-LP and HCVpp demonstrated a smooth, regular surface that was analogous to viruses belonging to the Togaviridae family or to the flavivirus genus. However, HCVpp, HCVcc and HCV-LP showed variable particle diameter depending on the construct and the cell line used for production [14,15]. Recently, Catanese et al. analyzed the external morphology of HCV particles produced from primary human hepatocytes and showed that they do not adopt a prototypical icosahedral symmetry, different from what was observed with surrogate particles. Moreover, the external structure is highly heterogeneous, with a discontinuous envelope, high concentration of apolipoprotein (apo)E and low concentrations of apoB and apoA-I [16]. A thorough analysis of the HCV lipidome demonstrated a composition unique for enveloped viruses, with low cholesterol concentration and a marked enrichment of cholesteryl esters that destabilizes a regular lipid bilayer structure [17]. The heterogeneous structure of HCV envelope leads to an unusual assembly of surface proteins, with E1E2 heterodimers apparently not assembling in an ordered layer and with their transmembrane (TM) domains featuring a differential orientation depending on the local envelope structure [16]. The above results demonstrate the peculiarity of HCV surface structures, with their extreme heterogeneity posing a first major problem in their fine characterization.
HCV E2: a heterogeneous and elusive protein
E2 is a highly glycosylated protein encompassing 362 residues (numbering starting from residue 384 of the genotype 1a H77 isolate polyprotein); it is formed by a 334-residue-long ectodomain and a C-terminal TM domain. The ectodomain (E2e) can be further divided into a receptor-binding domain and a short C-terminal membrane-proximal stem region. E2 is the most variable HCV protein, with high sequence variability focused in two hypervariable regions (HVR1, residues 384–410; and HVR2, residues 460–485) and an inter-genotypic variable region (IgVR, residues 569–581) [18].
All attempts to determine the structure of complete E2e have failed previously and, before the solution of E2c domain, the only available structures were of fragments spanning residues 412–423 and 430–446, solved through X-ray crystallography, and fragment 684–719, derived using NMR [19–25]. The difficulty in solving the E2e structure is at least partially related to its glycosylation state; E2 has 11 N-glycosylation and five O-glycosylation sites with variable degrees of micro-heterogeneity, crucial in maintaining its proper conformation and aggregation state [26,27]. Circular dichroism and infrared spectroscopy experiments estimated that the E2e comprises ~35% β-sheets and ~5% α-helices, with a high degree of disorder [28]. Moreover, E2 tertiary folding is maintained by 18 completely conserved cysteines involved in disulfide bridges; as described later, cysteine coupling can vary based on the expression system and, most importantly, on the E2 form (e.g. soluble, truncated). Finally, E2 must be expressed together with E1 to achieve its correct conformation [29].
The variability of E2 disulfide bridges and its glycosylation state highlight the need to express it in eukaryotic cells using a protein as close as possible to the wild type (WT). Furthermore, the high degree of E2 structural flexibility clearly impairs homogeneous crystal packing and effects the reconstruction of E2e density maps obtained through small-angle X-ray scattering (SAXS) and single-particle electron cryomicroscopy (cryoEM), leading to low-resolution maps [10,11].
E2c: a structural comparison
Notwithstanding all of these obstacles, two E2c structures have been recently derived from synthetic constructs comprising the central part of E2e [residues 412–645 of E2 from H77 isolate (genotype 1a) in Kong et al. (PDB ID: 4MWF) and residues 456–656 of E2 from J6 isolate (genotype 2a) in Khan et al. (PDB ID: 4NX3)] [10,11]. The construct used by Kong and colleagues also replaces HVR2 with a short GSSG linker and removes two N-glycosylation sites (N448 and N576). Furthermore, 4MWF construct has standardized N-glycosylations obtained growing E2c-expressing cells in presence of kifunensine, whereas 4NX3 N-glycosylations were enzymatically removed before crystallization and present a N542E mutation. These proteins are thus truncated, expressed in soluble form, folded in the absence of E1 and present different glycosylation patterns, necessities for their crystallization but also potentially introducing artifacts.
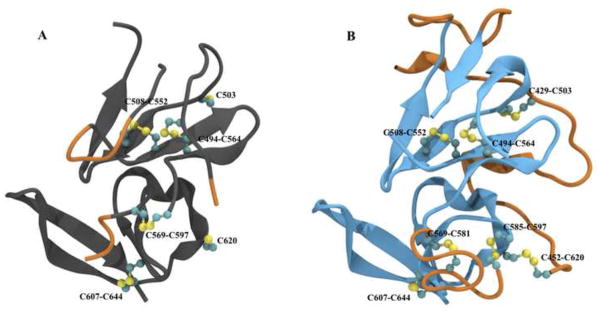
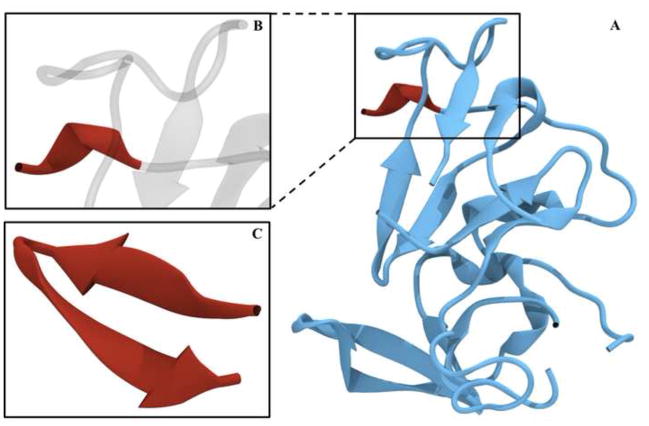
Portions of both structures are highly disordered, with approximately one-third of the protein remaining unsolved. In both E2c structures, two ordered tertiary arrangements can be defined: a central Ig-like domain formed by a four-stranded lower β-sheet and a two-stranded upper β-sheet; and a C-terminal four-stranded β-sheet. Short α-helices are dispersed in loops surrounding these domains. Structural superimposition indicates an α-carbon root-mean-square deviation below 1 Å, suggesting overall similarity. However, the structures differ significantly in the Ig-like domain upper layer, which in 4MWF is formed by two antiparallel β-strands, whereas in 4NX3 it is unstructured (Figure 1). The E2c structures also diverge in the outer loops, as evidenced by discrepancies in disulfide bridges. Another notable structural discrepancy is evident for a region spanning residues 412–423, a neutralizing epitope and part of the CD81 binding site (CD81bs); in all available co-crystals with different monoclonal antibodies (mAbs) this peptide has a well-defined β-hairpin arrangement, whereas it is unstructured in 4MWF and not included in 4NX3 (Figure 2) [21,22,24,25]. Importantly, this linear epitope keeps the same β-hairpin structure when bound to antibodies with completely different paratope structure and contacting different epitopic residues (e.g. mAbs AP33 and HCV1), making less probable a mAb-induced conformational change [25]. Taken together, these pieces of evidence highlight the variations in E2 folding and cysteine coupling in the E2c crystal structures.
Figure 1.
Comparison between 4NX3 and 4MWF E2 crystallographic structures. Shared structures (Ig-like domain and the C-terminal β-sheet) are colored in gray (4NX3) and cyan (4MWF). Orange segments indicate discrepancies between structures. Cysteines are represented in ball-and-stick configuration.
Figure 2.
Structural comparison of an E2 segment spanning residues 412–423 in 4MWF and co-crystallized with neutralizing murine mAb AP33 (PDB ID: 4GAJ). (a) The 4MWF structure in cyan, solved portion of residues 412–423 in red. (b) A zoomed in representation of these residues in red, the remaining is transparent. (c) E2 residues 412–423 in 4GAJ structure.
Biochemical characterization: the importance of disulfide bridges
E2 has 18 cysteines completely conserved across all genotypes, with a multifaceted role; they are involved in maintaining the correct protein topology and are crucial players in the conformational rearrangement upon viral attachment that leads to the fusion stage [30,31]. The 4NX3 construct includes 14 cysteines but only ten are present in the crystallographic structure, eight of which are involved in disulfide bridges; 4MWF includes 16 cysteines and all those that have been resolved are involved in seven disulfide bridges. Only three disulfide bridges (C494-C564, C508-C552 and C607-C644) are shared between E2c structures, and are probably crucial in maintaining the fundamental Ig-like domain and the C-terminal β-sheet conformation. Moreover, these three disulfide bridges are necessary for the capability of soluble E2, HCVpp and HCVcc to bind the CD81 large extracellular loop (LEL) [18,31,32]. Of the four additional cysteines present in the 4NX3 structure, two are involved in a disulfide bridge (C569-C597) and two are unbound (C503 and C620). The C569-C597 bridge is not present in 4MWF, where C569 bonds with C581 and C597 bonds with C585. In 4NX3, C503 and C620 are lacking their partners, which are not included in the 4NX3 construct. Conversely, in 4MWF C503 is coupled to C429 and C620 is coupled to C452.
The experimental setup-dependent variability of the E2 disulfide bridge pattern has been highlighted in several biochemical and functional studies. Krey et al. analyzed cysteine connectivity through mass spectrometry of E2e expressed as a soluble, secreted protein, and suggested that all cysteines are involved in disulfide bridges [28]. Other functional studies using HCVpp and HCVcc analyzed the impact of cysteine to alanine (C→A) mutations on the interaction with CD81, viral infectivity and E1E2 incorporation in virions [18,31,32]. These experiments could not confirm Krey and colleagues’ data on disulfide bridges, suggesting that information generated with soluble E2 might not reflect the native form. Fraser et al. added another missing piece investigating the role of E2 cysteines in viral attachment and entry. Through alkylation experiments they demonstrated that E2 on the virion surface has at least one free cysteine that is crucial in post-attachment E2 rearrangement necessary for viral entry [30].
Taken together, these studies suggest that cysteine connectivity in native E2 differs from E2 that is truncated, expressed in soluble form or folded in absence of E1, leading to a non-native tertiary arrangement [33]. Furthermore, discrepancies in the two E2c crystallographic structures suggest that even small differences in the truncated forms can lead to a different disulfide bridge pattern.
Experimental data: from immunology to structure and back
The biochemical and functional characterization of E2 suggests that the E2c structures reflect, at least in some areas, non-native folding and cannot be used to assign unambiguously the native cysteine coupling of E2. We reasoned that additional information might be obtained by assessing E2 single-residue mutants for the binding to mAbs to identify residues crucial in keeping E2 in its correct conformation. To this end, we have previously cloned from an HCV-infected patient a panel of human mAbs recognizing highly divergent viral genotypes, suggesting the conserved nature of their epitopes [34–37]. Moreover, all of the mAbs recognize conformational epitopes and exhibit different neutralizing activity, suggesting that their binding could provide information on the structural features of native E2.
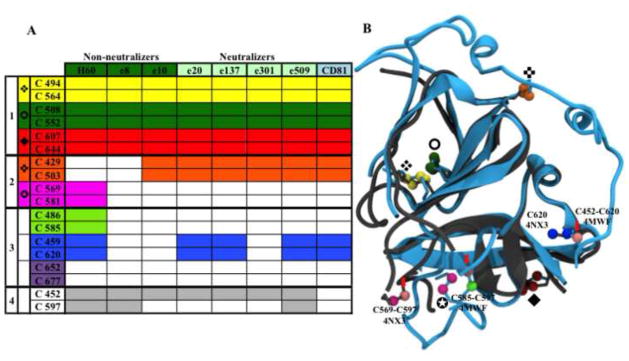
Here, we report the testing of this panel of mAbs for their binding to each E2 C→A variant generated by shotgun mutagenesis, following previously described protocols [38,39]. A conformation-sensitive murine antibody (H60 – a generous gift from Dr Jean Dubuisson) and an anti-V5 tag linear antibody used as expression control were also tested [40,41]. The construct used to generate all tested E2 variants was based on E1E2 WT sequence spanning residues 192–746 of H77 isolate polyprotein (UniProt ID: F5BWY6), with the V5 tag located C-terminally to E2 to minimize its impact on E1E2 folding. Each E2 variant was co-expressed with E1 on human HEK-293 cells, and the effect of each mutation on a mAb reactivity was directly evaluated by cytofluorimetry on fixed and permeabilized cells. Considering mean cellular fluorescence, antibody reactivity against each mutant E2 was calculated relative to the WT by subtracting the signal from mock-transfected controls and normalizing it to the signal from WT-transfected controls. A C→A mutation was considered as abrogating the binding when the reactivity was below 30% compared with WT (Figure 3) [40–42].
Figure 3.
Proposed disulfide bond pairs for E2. (a) Monoclonal antibody (mAb) and CD81 large extracellular loop (LEL) reactivity patterns against C→A mutants of full-length E2 (H77 strain) co-expressed with E1 in human HEK293 cells [31]. Single C→A mutants showing a binding below 30% compared with the wild type (WT) are marked with colored blocks. Cysteine pairs are grouped to highlight (1) disulfide bridges confirmed by 4NX3, 4MWF and mAb binding, (2) disulfide bridges confirmed by 4MWF and mAb binding, (3) disulfide bridges proposed according to mAb reactivity patterns and (4) free cysteines. Disulfide bridges belonging to groups (1) and (2) are marked with different symbols in (a) and (b). (b) 4NX3 (in gray) and 4MWF (in cyan) superimposed structures. Cysteines are colored as in (a); E2c cysteines that are inconsistent with mAb reactivity are reported in light pink, marked with red arrows and labeled. In 4MWF C652 and C677 are outside the crystallized constructs, whereas C459 and C486 are not solved; in 4NX3, C429, C452, C652 and C677 are not included in the construct, whereas C459, C486, C581 and C585 are not solved. Consequently, all of them are not shown in (b).
The expression of full-length E1 and E2 on human cells allowed us to study E2 in conditions that promote the formation of its native structure. Interestingly, the differential reactivity pattern of mAb recognition of the C→A mutants suggests an alternative disulfide bridge pattern necessary for E2 in its native tertiary conformation (Figure 3). Specifically, our data suggest that seven disulfide bridges are formed by C494-C564, C508-C552, C607-C644, C429-C503, C569-C581, C486-C585 and C459-C620, whereas C452 and C597 are in a free state.
The proposed pattern of cysteine disulfide pairs includes three disulfide bridges present in both E2c structures (C494-C564, C508-C552 and C607-C644). Mutation to alanine of any one of these six cysteines abrogates the binding of neutralizing and non-neutralizing mAbs and CD81 LEL, highlighting the crucial part played by these bridges in maintaining not only the CD81bs but also the overall E2 conformation [31]. The other four disulfides suggested by our mAb reactivity patterns could help resolve the discrepancy between the two published E2c structures. Our data also highlight the presence of two free cysteines (C452 and C597), as previously predicted by Fraser et al [30].
Our mAb reactivity pattern is in agreement with 4MWF for two cysteine pairs, C429-C503 and C569-C581, in addition to the three consistent between the structures. The C429-C503 pair plays a fundamental role in keeping together CD81bs residues located in the Ig-like domain and those lying in the 412–423 and 434–446 regions [43–45]. The disruption of C429-C503 abrogated the binding of all tested neutralizing mAbs, whereas the binding of e8 and H60, two non-neutralizing antibodies, were not affected. Disruption of the C569-C581 disulfide abrogated the binding of H60 only; these cysteines are immediately flanking the IgVR domain and induce the formation of an exposed loop.
Mutation of C452 (not included in the 4NX3 construct, and coupled to C620 in 4MWF) and C597 (coupled to C569 in 4NX3 and to C585 in 4MWF) demonstrated unique mAb reactivity patterns that do not match the mAb reactivity data for any other cysteine mutations; consequently, we hypothesize that C452 and C597 remain unbound in native E2. The C585A mutant affected only H60 binding and, considering mAb reactivity, we propose a novel C486-C585 coupling; this long-range bond in native E2 could participate in maintaining the structure of the unsolved E2c region spanning residues 453–491.
Our mAb reactivity patterns cannot confirm the C452-C620 disulfide proposed by 4MWF; the C620A mutant has a reactivity profile that uniquely matches with C459A, strongly suggesting an alternative C459-C620 coupling. Finally, two C-terminal cysteines (C652 and C677) that are not included in the E2c constructs when mutated to alanine do not have any effect on mAb or CD81 LEL binding; however, they both cause decreased E1E2 heterodimerization, suggesting their involvement in a disulfide bridge [31].
In conclusion, assessing the binding properties of all C→A E2 mutants expressed in a near-native environment to a large panel of mAbs with diverse conformational epitopes allowed us to study the local and general impact of each cysteine on E2 native conformation. The obtained results confirm five disulfide bridges already identified in E2c (three in each of 4NX3 and 4MWF, and two only in 4MWF) as necessary to maintain the overall E2 structure but also suggesting novel couplings, not present in the E2c truncated structures, that are required for E2 native folding.
Concluding remarks: in silico merging of structural and immunological data
HCV E2 represents a clear example of how use of a ‘gold-standard’ technique, such as X-ray crystallography, to obtain the atomic structure of a protein could require additional biochemical evidence to provide a comprehensive picture of the native structure. Moreover, biases introduced by crystallographic conditions (e.g. protein modifications or the crystallization protocol) can affect the native structure [46,47].
HCV glycoproteins are challenging targets for structural studies, because they are flexible, highly glycosylated transmembrane proteins that require multi-subunit assembly and oligomerization to acquire their native folding. To date, these characteristics have hampered all attempts to obtain their full-length structure through X-ray crystallography or lower-resolution techniques such as cryoEM and SAXS. Considering this, the recently solved E2c structures represent a groundbreaking improvement in comprehending its interaction with CD81 and the structure of neutralizing epitopes. However, the discrepancies with biological data and the lack of structural information for approximately half of E2e indicate the need to refine and extend these structures.
Monoclonal antibodies recognizing conformational epitopes represent a potent tool to validate a structure and obtain indirect structural information. In fact, considering the buried area at the paratope–epitope interface, residues recognized as fundamental for mAb–antigen binding must lie in close proximity; the characterization of conformational epitopes can therefore lead to the identification of sequentially distant domains that are close in the tertiary structure [48].
Many techniques exploiting experimental and/or computational methods are available to locate a mAb epitope, but the low yield of E2 expression and method-dependent variability make many of them unfeasible [49–51]. Here, we have applied the alanine-scanning method because it allows the rapid identification of epitopic residues on proteins expressed in near-native conditions, a fundamental requirement to reduce possible biases resulting from non-native E2 conformations. Alanine scanning consists of individually substituting each residue in the target protein with alanine and subsequently assessing the ability of each variant to bind mAbs compared with the WT protein. Although this technique does not provide direct structural data and indirect and allosteric effects must also be considered, it presents several advantages. Alanine scanning can be performed on the full-length protein expressed in native conditions, reducing the risk of introducing structural biases. Also, its usage is not restricted to epitope evaluation; as described here, it can be used to assess the role of key residues such as cysteines in correct protein folding. Moreover, single alanine point mutations can be introduced in the context of whole viral particles; several publications report the role of E2 cysteine or histidine single mutations in HCVcc and HCVpp to study their impact in viral attachment, entry and assembly [31,32,52]. Considering all of these aspects, alanine scanning can provide useful and reliable information to be used for protein modeling and structure validation.
In addition to experimental methods, several in silico approaches have been developed to predict ab initio protein folding based only on linear sequence, which could be applied to E2 in future studies. Given the high sequence variability of E2 and the availability of a vast number of sequences, it is an ideal target for algorithms analyzing large multiple sequence alignments that infer residue pair proximity based on their co-evolution. Recently, several algorithms have been developed, most of them implementing a direct coupling analysis method that allows direct and indirect contributions to be disentangled, resulting in a more precise identification of residue couples in close proximity, for examples see [53–56]. Evolutionary coupling analysis has proven itself to predict protein folding correctly when co-evolving contacts are merged with secondary structure information and, although all test cases were structurally stable proteins (e.g. globular, channels), useful distance constraints can be derived to reconstruct missing domains of a partially solved protein structure such as HCV E2 [56,57].
An in silico model of the entire E2e could be obtained by merging the data from all available direct and indirect structural studies with novel functional data obtained through alanine scanning and mAb reactivity patterns. Because there are no homologous structures, a ‘divide-and-conquer’ approach could be used to predict ab initio the folding of those domains that remain unsolved [58,59]. Proximity information can be subsequently applied to merge crystallographic and NMR structures with predicted fragments to obtain a comprehensive model of the whole E2e. For this purpose, a possible strategy could be based on distance geometry algorithms, extensively used to generate NMR structural ensembles and recently applied also to reconstruct a 3D protein structure from evolutionary coupling data [60–62].
The obtained models could then be structurally confirmed fitting them into the low-resolution cryoEM and SAXS maps available for E2e [10,11]. Furthermore, a strong biological validation could be performed mapping functional and immunogenic regions. As an example, two of the mAbs herein described (i.e. e137 and e8) could be used to assess a model validity mapping their epitopes on a modeled E2 3D structure; e8 epitope, a non-neutralizing antibody with no competing activity against the CD81 LEL, should map at a distance from the CD81bs compatible with their steric hindrance. Conversely, a broad-neutralizing mAb that competes for CD81 binding, such as e137, should map in close proximity of the CD81bs [36,37]. Therefore, a thorough mAb characterization in terms of biological activity and epitope recognition could provide fundamental information about the structure–function relationship and confirm a model structure.
To conclude, HCV E2 is a challenging target to study through classic structural biological approaches; recent E2c structures shed light on the structure of E2 but discrepancies with biological data, together with missing information about some E2 domains, highlight the need to generate a more complete model. Considering the characteristics of E2, we believe that a multidisciplinary approach will be required to achieve such a complex goal.
Highlights.
HCV/E2 core (E2c) crystal structures present structural discrepancies
HCV/E2 conformation and cysteine coupling relies on the experimental setup
HCV/E2c structures are in partial disagreement with functional data
Monoclonal antibody reactivity suggests an alternative E2 cysteine coupling
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lavanchy D. Evolving epidemiology of hepatitis C virus. Clin Microbiol Infect. 2011;17:107–115. doi: 10.1111/j.1469-0691.2010.03432.x. [DOI] [PubMed] [Google Scholar]
- 2.Delemos AS, Chung RT. Hepatitis C treatment: an incipient therapeutic revolution. Trends Mol Med. 2014;20:315–321. doi: 10.1016/j.molmed.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Rose L, et al. Sofosbuvir: a nucleotide NS5B inhibitor for the treatment of chronic hepatitis C infection. Ann Pharmacother. 2014;48:1019–1029. doi: 10.1177/1060028014534194. [DOI] [PubMed] [Google Scholar]
- 4.Scheel TK, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med. 2013;19:837–849. doi: 10.1038/nm.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu S, et al. Tight junction proteins claudin-1 and occludin control hepatitis C virus entry and are downregulated during infection to prevent superinfection. J Virol. 2009;83:2011–2014. doi: 10.1128/JVI.01888-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pileri P, et al. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 7.Scarselli E, et al. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 2002;21:5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lavillette D, et al. Characterization of fusion determinants points to the involvement of three discrete regions of both E1 and E2 glycoproteins in the membrane fusion process of hepatitis C virus. J Virol. 2007;81:8752–8765. doi: 10.1128/JVI.02642-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu YZ, et al. How hepatitis C virus invades hepatocytes: the mystery of viral entry. World J Gastroenterol. 2014;20:3457–3467. doi: 10.3748/wjg.v20.i13.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan AG, et al. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature. 2014;509:381–384. doi: 10.1038/nature13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kong L, et al. Hepatitis C virus E2 envelope glycoprotein core structure. Science. 2013;342:1090–1094. doi: 10.1126/science.1243876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapoor A, et al. Identification of rodent homologs of hepatitis C virus and pegiviruses. MBio. 2013;4:e00216–00213. doi: 10.1128/mBio.00216-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Argentini C, et al. HCV genetic variability: from quasispecies evolution to genotype classification. Future Microbiol. 2009;4:359–373. doi: 10.2217/fmb.09.8. [DOI] [PubMed] [Google Scholar]
- 14.Bonnafous P, et al. Characterization of hepatitis C virus pseudoparticles by cryo-transmission electron microscopy using functionalized magnetic nanobeads. J Gen Virol. 2010;91:1919–1930. doi: 10.1099/vir.0.021071-0. [DOI] [PubMed] [Google Scholar]
- 15.Yu X, et al. Cryo-electron microscopy and three-dimensional reconstructions of hepatitis C virus particles. Virology. 2007;367:126–134. doi: 10.1016/j.virol.2007.05.038. [DOI] [PubMed] [Google Scholar]
- 16.Catanese MT, et al. Ultrastructural analysis of hepatitis C virus particles. Proc Natl Acad Sci U S A. 2013;110:9505–9510. doi: 10.1073/pnas.1307527110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merz A, et al. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J Biol Chem. 2011;286:3018–3032. doi: 10.1074/jbc.M110.175018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCaffrey K, et al. Expression and characterization of a minimal hepatitis C virus glycoprotein E2 core domain that retains CD81 binding. J Virol. 2007;81:9584–9590. doi: 10.1128/JVI.02782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Albecka A, et al. Identification of new functional regions in hepatitis C virus envelope glycoprotein E2. J Virol. 2011;85:1777–1792. doi: 10.1128/JVI.02170-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng L, et al. Structural evidence for a bifurcated mode of action in the antibody-mediated neutralization of hepatitis C virus. Proc Natl Acad Sci U S A. 2013;110:7418–7422. doi: 10.1073/pnas.1305306110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong L, et al. Structure of hepatitis C virus envelope glycoprotein E2 antigenic site 412 to 423 in complex with antibody AP33. J Virol. 2012;86:13085–13088. doi: 10.1128/JVI.01939-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong L, et al. Structural basis of hepatitis C virus neutralization by broadly neutralizing antibody HCV1. Proc Natl Acad Sci U S A. 2012;109:9499–9504. doi: 10.1073/pnas.1202924109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krey T, et al. Structural basis of HCV neutralization by human monoclonal antibodies resistant to viral neutralization escape. PLoS Pathog. 2013;9:e1003364. doi: 10.1371/journal.ppat.1003364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pantua H, et al. Glycan shifting on hepatitis C virus (HCV) E2 glycoprotein is a mechanism for escape from broadly neutralizing antibodies. J Mol Biol. 2013;425:1899–1914. doi: 10.1016/j.jmb.2013.02.025. [DOI] [PubMed] [Google Scholar]
- 25.Potter JA, et al. Toward a hepatitis C virus vaccine: the structural basis of hepatitis C virus neutralization by AP33, a broadly neutralizing antibody. J Virol. 2012;86:12923–12932. doi: 10.1128/JVI.02052-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brautigam J, et al. Mass spectrometric analysis of hepatitis C viral envelope protein E2 reveals extended microheterogeneity of mucin-type O-linked glycosylation. Glycobiology. 2013;23:453–474. doi: 10.1093/glycob/cws171. [DOI] [PubMed] [Google Scholar]
- 27.Iacob RE, et al. Mass spectrometric characterization of glycosylation of hepatitis C virus E2 envelope glycoprotein reveals extended microheterogeneity of N-glycans. J Am Soc Mass Spectrom. 2008;19:428–444. doi: 10.1016/j.jasms.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krey T, et al. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. PLoS Pathog. 2010;6:e1000762. doi: 10.1371/journal.ppat.1000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brazzoli M, et al. Folding and dimerization of hepatitis C virus E1 and E2 glycoproteins in stably transfected CHO cells. Virology. 2005;332:438–453. doi: 10.1016/j.virol.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 30.Fraser J, et al. Hepatitis C virus (HCV) envelope glycoproteins E1 and E2 contain reduced cysteine residues essential for virus entry. J Biol Chem. 2011;286:31984–31992. doi: 10.1074/jbc.M111.269605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCaffrey K, et al. Role of conserved cysteine residues in hepatitis C virus glycoprotein e2 folding and function. J Virol. 2012;86:3961–3974. doi: 10.1128/JVI.05396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W, et al. Alanine scanning mutagenesis of hepatitis C virus E2 cysteine residues: insights into E2 biogenesis and antigenicity. Virology. 2014;448:229–237. doi: 10.1016/j.virol.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 33.Owsianka A, et al. Functional analysis of hepatitis C virus E2 glycoproteins and virus-like particles reveals structural dissimilarities between different forms of E2. J Gen Virol. 2001;82:1877–1883. doi: 10.1099/0022-1317-82-8-1877. [DOI] [PubMed] [Google Scholar]
- 34.Bugli F, et al. Mapping B-cell epitopes of hepatitis C virus E2 glycoprotein using human monoclonal antibodies from phage display libraries. J Virol. 2001;75:9986–9990. doi: 10.1128/JVI.75.20.9986-9990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mancini N, et al. Hepatitis C virus (HCV) infection may elicit neutralizing antibodies targeting epitopes conserved in all viral genotypes. PLoS One. 2009;4:e8254. doi: 10.1371/journal.pone.0008254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perotti M, et al. Identification of a broadly cross-reacting and neutralizing human monoclonal antibody directed against the hepatitis C virus E2 protein. J Virol. 2008;82:1047–1052. doi: 10.1128/JVI.01986-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sautto G, et al. Anti-hepatitis C virus E2 (HCV/E2) glycoprotein monoclonal antibodies and neutralization interference. Antiviral Res. 2012;96:82–89. doi: 10.1016/j.antiviral.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 38.Paes C, et al. Atomic-level mapping of antibody epitopes on a GPCR. J Am Chem Soc. 2009;131:6952–6954. doi: 10.1021/ja900186n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sukupolvi-Petty S, et al. Functional analysis of antibodies against dengue virus type 4 reveals strain-dependent epitope exposure that impacts neutralization and protection. J Virol. 2013;87:8826–8842. doi: 10.1128/JVI.01314-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deleersnyder V, et al. Formation of native hepatitis C virus glycoprotein complexes. J Virol. 1997;71:697–704. doi: 10.1128/jvi.71.1.697-704.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flint M, et al. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J Virol. 1999;73:6235–6244. doi: 10.1128/jvi.73.8.6235-6244.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davidson E, Doranz BJ. A high-throughput shotgun mutagenesis approach to mapping B-cell antibody epitopes. Immunology. 2014 doi: 10.1111/imm.12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Drummer HE, et al. A conserved Gly436-Trp-Leu-Ala-Gly-Leu-Phe-Tyr motif in hepatitis C virus glycoprotein E2 is a determinant of CD81 binding and viral entry. J Virol. 2006;80:7844–7853. doi: 10.1128/JVI.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Owsianka AM, et al. Identification of conserved residues in the E2 envelope glycoprotein of the hepatitis C virus that are critical for CD81 binding. J Virol. 2006;80:8695–8704. doi: 10.1128/JVI.00271-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rothwangl KB, et al. Dissecting the role of putative CD81 binding regions of E2 in mediating HCV entry: putative CD81 binding region 1 is not involved in CD81 binding. Virol J. 2008;5:46. doi: 10.1186/1743-422X-5-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quick M, et al. Experimental conditions can obscure the second high-affinity site in LeuT. Nat Struct Mol Biol. 2012;19:207–211. doi: 10.1038/nsmb.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rashin AA, et al. Factors correlating with significant differences between X-ray structures of myoglobin. Acta Crystallogr D Biol Crystallogr. 2014;70:481–491. doi: 10.1107/S1399004713028812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sautto G, et al. Structural and antigenic definition of hepatitis C virus E2 glycoprotein epitopes targeted by monoclonal antibodies. Clin Dev Immunol. 2013;2013:450963. doi: 10.1155/2013/450963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Castelli M, et al. Peptide-based vaccinology: experimental and computational approaches to target hypervariable viruses through the fine characterization of protective epitopes recognized by monoclonal antibodies and the identification of T-cell-activating peptides. Clin Dev Immunol. 2013;2013:521231. doi: 10.1155/2013/521231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clementi N, et al. Characterization of epitopes recognized by monoclonal antibodies: experimental approaches supported by freely accessible bioinformatic tools. Drug Discov Today. 2013;18:464–471. doi: 10.1016/j.drudis.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 51.Clementi N, et al. Epitope mapping by epitope excision, hydrogen/deuterium exchange, and peptide-panning techniques combined with in silico analysis. Methods Mol Biol. 2014;1131:427–446. doi: 10.1007/978-1-62703-992-5_26. [DOI] [PubMed] [Google Scholar]
- 52.Boo I, et al. Distinct roles in folding, CD81 receptor binding and viral entry for conserved histidine residues of hepatitis C virus glycoprotein E1 and E2. Biochem J. 2012;443:85–94. doi: 10.1042/BJ20110868. [DOI] [PubMed] [Google Scholar]
- 53.Baldassi C, et al. Fast and accurate multivariate gaussian modeling of protein families: predicting residue contacts and protein-interaction partners. PLoS One. 2014;9:e92721. doi: 10.1371/journal.pone.0092721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Juan D, et al. Emerging methods in protein co-evolution. Nat Rev Genet. 2013;14:249–261. doi: 10.1038/nrg3414. [DOI] [PubMed] [Google Scholar]
- 55.Jones DT, et al. PSICOV: precise structural contact prediction using sparse inverse covariance estimation on large multiple sequence alignments. Bioinformatics. 2012;28:184–190. doi: 10.1093/bioinformatics/btr638. [DOI] [PubMed] [Google Scholar]
- 56.Marks DS, et al. Protein 3D structure computed from evolutionary sequence variation. PLoS One. 2011;6:e28766. doi: 10.1371/journal.pone.0028766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hopf TA, et al. Three-dimensional structures of membrane proteins from genomic sequencing. Cell. 2012;149:1607–1621. doi: 10.1016/j.cell.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maupetit J, et al. PEP-FOLD: an online resource for de novo peptide structure prediction. Nucleic Acids Res. 2009;37:W498–503. doi: 10.1093/nar/gkp323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu D, Zhang Y. Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins. 2012;80:1715–1735. doi: 10.1002/prot.24065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brunger AT. Version 1.2 of the crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 61.Brunger AT, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 62.Hodsdon ME, et al. The NMR solution structure of intestinal fatty acid-binding protein complexed with palmitate: application of a novel distance geometry algorithm. J Mol Biol. 1996;264:585–602. doi: 10.1006/jmbi.1996.0663. [DOI] [PubMed] [Google Scholar]