Abstract
The proteasome has emerged as an intricate machine that has dynamic mechanisms to regulate the timing of its activity, its selection of substrates, and its processivity. The 19-subunit regulatory particle (RP) recognizes ubiquitinated proteins, removes ubiquitin, and injects the target protein into the proteolytic chamber of the core particle (CP) via a narrow channel. Translocation into the CP requires substrate unfolding, which is achieved through mechanical force applied by a hexameric ATPase ring of the RP. Recent cryo-electron microscopy studies have defined distinct conformational states of the RP, providing illustrative snapshots of what appear to be progressive steps of substrate engagement. Here, we bring together this new information with molecular analyses to describe the principles of proteasome activity and regulation.
Keywords: proteasome, ubiquitin, protein degradation, ATPase
The proteasome
The proteasome is the most complex protease known, and, with hundreds of substrates, the scope of its regulatory functions is likewise unparalleled. Its principal activity is to degrade proteins conjugated to ubiquitin. The proteolytic sites of the proteasome are sequestered within an internal chamber (1), which is essentially a topological compartment defined by the inner surfaces of the 28 subunits that form the proteasome core particle (CP, also known as the 20S particle) (Figure 1A–B). Cellular proteins have highly restricted access to this chamber and to the proteolytic sites within it, thus minimizing nonspecific proteolysis.
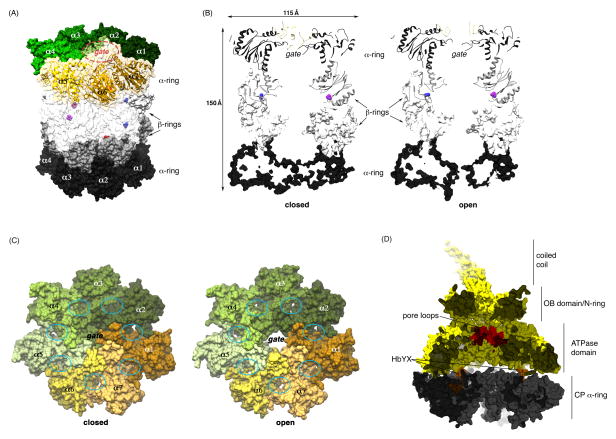
Figure 1. Substrate entry into the core particle (CP) is gated by N-terminal tails of the α subunits.
(A) A semi-transparent surface representation of the proteasome CP, generated using (PDB 1RYP) (1). For the top α-ring, ribbon diagrams are included for α-α7 and the gating residues circled. The top and bottom β-rings are distinguished by white and grey coloring respectively, and the N-terminal β1, β2, and β5 catalytic threonines rendered in blue, red, and purple, respectively. The surfaces of α-α7 and β-β7 of the top rings are transparent for visualization of the catalytic sites. (B) A cross-section of the CP in its closed (left) and open (right) state to illustrate the hollow interior and altered configuration of the N-terminal gating residues. The top α- and β-rings are rendered as ribbon diagrams whereas the bottom two rings are displayed as surfaces. The β1 and β5 catalytic sites are represented in blue and purple, respectively. Dimensions are provided for scale. (C) Top view of the α-rings for closed and open configurations of the CP. The inter-subunit clefts where C termini of many proteasome regulators dock, including the regulatory particle (RP), are circled and the gate labeled. PDB 1RYP (1) was used for the CP closed state represented in (A) – (C) whereas PDB 1FNT (2) was used for the open state in (B) and (C). The latter structure was solved in complex with the PA26 regulatory particle. (D) Surface diagram cut-away view of the CP α-ring (grey) with the RP ATPase ring (yellow) highlighting the pore loops (red), HbYX motifs (orange) and contiguous substrate entry channel. PDB 4CR4 (53) and 1FNT (2) were used to generate this image.
In the proteasome holoenzyme, the CP is complexed with the 19-subunit regulatory particle (RP, also known as the 19S particle and PA700) (Figure 2A). The RP contains receptors for ubiquitin and thus initiates substrate recognition. After conjugated ubiquitin is tethered to the RP, the attached substrate is translocated from the RP to the CP. Translocation requires ATP hydrolysis, which is effected by a heterohexameric ring of ATPases that are subunits of the RP. Here we describe our current understanding of the key steps in protein breakdown by the proteasome. Although it degrades hundreds of proteins, the proteasome is highly selective, and we will discuss the structural elements that restrict its activity to proper substrates. Finally we will review the results of recent cryoelectron microscopy studies that have highlighted the plasticity of the proteasome and revealed broad conformational switches that serve to orchestrate its diverse catalytic activities, including ATP hydrolysis, deubiquitination, and substrate proteolysis.
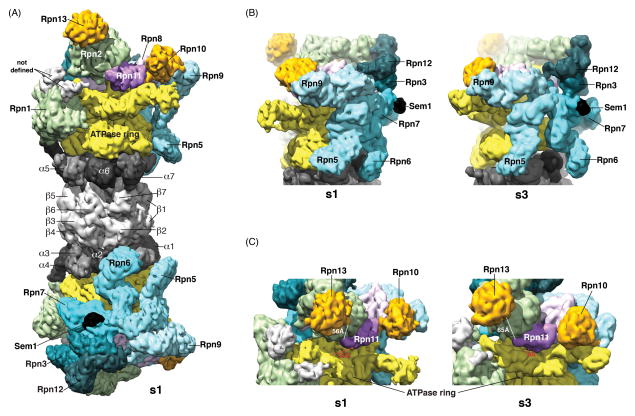
Figure 2. Cryo-electron microscopy reconstruction of the proteasome, highlighting differences in the s1 and s3 states.
(A) RPs stack against each end of the CP, rotated relative to each other by ~180°. The ATPase rings (yellow) abut the α-rings (grey) of the CP with six fanned out lid components (shades of blue) lining each side of the proteasome. Sem1 binds to this area and is shown in black. In this s1 state of the proteasome, Rpn11 (purple) is offset from the center of the ATPase core. Ubiquitin receptors Rpn10 and Rpn13 (orange) are at opposite sides of Rpn11. Above Rpn1 (green) is electron density that has not yet been defined (grey), as Rpn1 model structures do not map to incorporate these two regions. (B, C) Comparative views of the s1 (left) and s3 (right) states of the proteasome. In (B) major changes are observed for Rpn6, while in (C) Rpn11 is positioned more directly over the ATPase core in the s3 configuration. Distances are included between Rpn11 A183 and Rpn13 I8 (white) and between Rpn11 S120 and Rpt1 E170 (red) to highlight conformational changes. EMD 2594 and EMD 2596 (53) were used to generate the s1 and s3 images, respectively.
The catalytic core particle
In the CP, four stacked heteroheptameric rings of subunits are assembled into an α7β7β7α7 architecture (Figure 1A–B). Thus, the outer rings are formed by the α subunits and the inner rings by β subunits. Three of the seven β subunits are proteolytically active, cutting after hydrophobic (β5), basic (β2), or acidic (β1) residues (Figure 1A–B). The combination of limited specificity and multiple active sites ensures rapid degradation once substrates enter the chamber of the CP. In the crystal structure of the free CP–that is, with the RP absent–the hydrolytic chamber is essentially closed to its environment (1). However, a pathway for substrate entry can be formed axially at the faces of this barrel-like structure (2, 3). This translocation channel is gated, in that it exists in open and closed states that can be interconverted. In the free CP, the gate is closed (Figure 1A), as this site is occupied by a coalescence of the short, highly conserved N-terminal tails from the seven α subunits (3) (Figure 1A and 1B–C, left). When the RP binds the CP, it is positioned over the CP channel and opens the gate, relieving autoinhibition of the CP (Figure 1B–C, right and 1D).
The translocation channel is very narrow (2, 3), ensuring that, even in its open state, properly folded cellular proteins are excluded from the hydrolytic chamber of the CP. To traverse the channel, substrates must typically be unfolded by the ATPase ring of the RP (the Rpt ring), as is the case for ATP-dependent proteases in general (4–7). Globular domains of the substrate are denatured by being forced through the axial channel of the Rpt ring; unfolding is driven mechanically by translocation. The substrate translocation channel of the Rpt ring is coaxial with that of the CP (Figure 1D).
The regulatory particle
In a significant advance, the general architecture of the RP has been resolved (Figure 2A), with the geometry of the subunits assigned by the integration of cryo-electron microscopy (cryo-EM) with chemical cross-linking and other techniques (8–11). Two major subassemblies of the RP had been previously defined biochemically, the base and lid, each being a 9-subunit complex (12, 13). The RP dissociates readily into the base and lid in vitro and is now thought to assemble in cells through the same two complexes as intermediates to the fully formed 26S proteasome (14). The key component of the lid is Rpn11, a metalloprotease deubiquitinating enzyme that hovers above the substrate entry port of the Rpt ring (15–18) (Figure 2A). The base contains the above-mentioned Rpt ring complex (composed of the Rpt1-Rpt6 proteins), Rpn1, Rpn2, and Rpn13, the latter being a substrate receptor (19–21) that can dock ubiquitin at the proteasome both directly and indirectly. Its indirect mode of recognition involves reversible binding of shuttle receptors, which escort ubiquitin conjugates to the proteasome (22–26).
The ATPase ring of the regulatory particle
The Rpt proteins are members of the functionally diverse AAA (ATPases Associated with diverse cellular Activities) family of ATPases (7), which typically convert the energy of ATP hydrolysis into mechanical force. Current models of the Rpt ring have been shaped largely by analogy to other AAA rings, especially the elegantly dissected ClpXP, an ATP-dependent protease from E. (Escherichia) coli (6, 7). An archaeal protease, the PAN-20S complex, is representative of an ancient evolutionary precursor of the eukaryotic proteasome (27–29). A similar protease exists as well in the bacterial lineage–the actinobacterial ARC complex (AAA ATPase forming Ring-shaped Complexes) (30). The PAN-20S complex is a stripped-down version of the proteasome, containing only a 28-subunit 20S complex (orthologous to the eukaryotic CP) and a 6-subunit RP (a homomeric ATPase ring complex). The accretion of new RP subunits in the eukaryotic proteasome is evidently related to the introduction of ubiquitin as a signal for degradation; many of the subunits that are specific to eukaryotes are ubiquitin receptors, deubiquitinating enzymes, or scaffolding proteins that help to position and control these factors. Another feature that sets the proteasome apart from PAN and other prokaryotic complexes is the heteromeric nature of its AAA ring, which reflects specialization in the six Rpt subunits (31–34) and has allowed for innovation in the mechanisms of communication between these subunits (discussed later).
Studies of PAN highlighted the significance of the C-terminal tails of the ATPases. PAN subunits carry a signature HbYX (hydrophobic-tyrosine-X) element (28), which inserts into intersubunit pockets of the CP α subunits (Figure 1C). In yeast proteasomes, three of the six Rpt tails contain recognizable HbYX motifs, and these tails insert into specific α pockets in the holoenzyme (8, 9, 35, 36) (Figure 1C–D). However, significant evolutionary conservation is evident in the other tails, so they may also insert into α pockets conditionally or have a related function in the complex (37, 38). In any case, these tail-pocket interactions are the major, though probably not sole (35), means by which the RP and CP are joined. A peculiar aspect of RP-CP interface is that there is a symmetry mismatch between the heptameric Rpt and hexameric α rings; this precludes close complementarity between the rings and may underlie the highly dynamic nature of this interface (28, 36, 37). This mismatch is an ancient feature of ATP-dependent proteases, found in both archaea (PAN-20S) and bacteria (ClpXP and others).
The ATPase domains of the Rpt proteins remain only marginally understood. Individual Rpt proteins cooperate with each other extensively, as is generally the case in AAA ring complexes (6, 7, 32–34). ATPase activity involves the large and small AAA domains from each Rpt protein (Figure 3A), with nucleotide positioned between them for a given subunit (displayed for PAN in Figure 3B). In the course of the ATPase hydrolytic cycle, these two domains undergo rigid body rotations. However, the large AAA domain of one Rpt protein is fixed with respect to the small AAA domain of its neighbor, as shown previously for ClpX (6, 39). A second type of intersubunit communication is that ATP binding by one subunit is assisted by its neighbor’s “arginine finger” element (Figure 4), which contacts ATP and may be important for nucleotide hydrolysis (7, 39). Based on detailed mutational analysis (32–34), complex communication between Rpt proteins is achieved through these types of interactions, although the picture remains sketchy. For example, it is unclear whether the six subunits within a ring hydrolyze ATP in a random, sequential, or concerted manner (32, 33). To resolve this issue, it will be critical to obtain higher resolution structures trapped at different stages of substrate processing that include nucleotide within the AAA active sites.
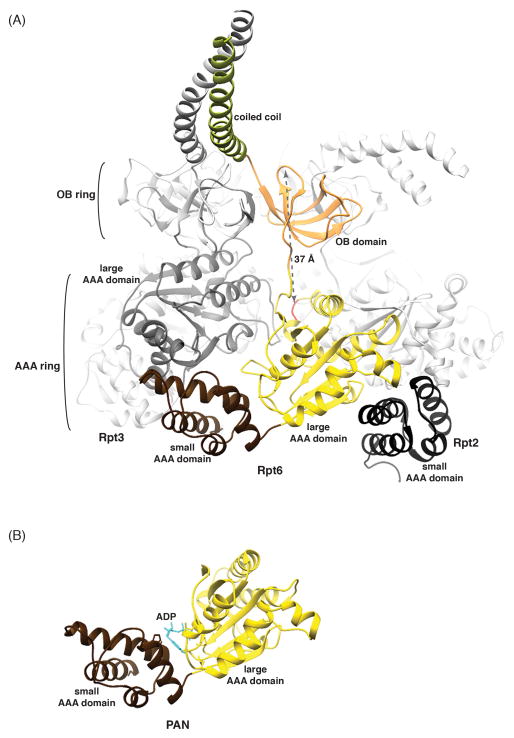
Figure 3. The RP ATPase ring components are comprised of four structural domains with similarity to archaeal PAN.
(A) The ATPase ring is displayed as a ribbon diagram highlighting the Rpt6 small (brown) and large (yellow with the pore loop red) AAA domains, OB domain (orange) and coiled coil (green). The Rpt6 small AAA domain packs against the Rpt3 large AAA domain (dark grey), while the Rpt6 large AAA domain packs against the Rpt2 small AAA domain (black). The distance between the Cα atoms of Rpt6 D118 and Y222 is included. PDB 4CR2 (53) was used for this image. (B) PAN ATPase domain with nucleotide (blue) bound between its large (yellow) and small (brown) AAA domains. The archaeal proteasome RP is formed from a PAN hexamer. PDB 3H4M (29) was used for this image.
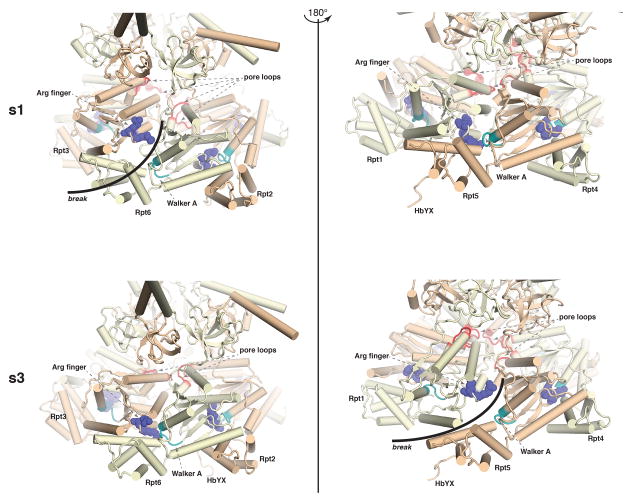
Figure 4. Conformational rearrangements that reconfigure the ATPase ring in the s1 and s3 states.
Expanded view in ribbon representation of the ATPase region that has a break (highlighted with a black arc) between the arginine finger (Arg finger, indigo) and Walker A motif (light blue) for the s1 (top, left) and s3 (bottom, right) configurations. These two views differ by a 180° rotation, and the corresponding view for each configuration is also included for comparison. The pore loops are highlighted in red to illustrate their transition from a spiral structure in s1 to a more planar structure in s3. HbYX motifs are labeled where visible. These images were generated from PDB accession codes 4CR2 and 4CR4 (53) for the s1 and s3 states, respectively.
The movements of the AAA domains serve primarily to drive substrate translocation, which is thought to reflect substrate-AAA domain contacts within the translocation channel. Specific loop elements, referred to as pore loops (Figures 1D, 3A, and 4, highlighted in red), may provide these contact sites (32, 33). The motion of the loops must be asymmetric in order to generate net movement towards the CP, although how these motions impart directionality is not understood at all.
The Rpt ring contains two other important types of elements, namely the OB domains and the coiled coil helices formed by heterodimerization of neighboring Rpt proteins (Figure 3A). The substrate entry port is defined by the Rpt OB domains, which form a distinct ring (the N-ring) adjacent to the catalytic AAA domains, at the side opposite to the CP (8–10, 29) (Figure 3A). The OB ring appears to restrict access through the entry port, while the Rpt coiled coil region promotes proper assembly of the RP (40) by making contacts with other regions of both the base and lid (8–10) (Figure 2). Other ATP-dependent proteases, whose ATPase complexes may resemble the proteasome closely, lack the OB ring, with the exception of PAN and ARC (29, 30). Its function is unknown, but one effect of the OB ring is to distance the AAA domain pore loops from the body of the substrate protein. For substrate to engage the axial pore loops of the AAA domain, it must span up to 40 Å through the center of the OB ring (Figure 3A). Thus, favorable substrates may typically arrive at the proteasome already unfolded in some region, most simply by possessing an intrinsically disordered segment. In some cases, a protein might unfold partially and transiently while bound to the proteasome by a mechanism that does not involve the AAA pore loops, most simply as a result of inherent conformational dynamics of the substrate.
The extended segments of the substrate that must reach to the pore loops are known as initiation sites (41). Even if ubiquitinated, a globular protein lacking initiation sites will be a poor proteasome substrate (41). Initiation sites have characteristic length requirements and sequence features, which can be used to fine-tune protein turnover rates, but they seem to lack defined motifs (42–45).
An important feature of the proteasome is that it operates processively on most substrates. Successful initiation of substrate translocation, presumably by engagement between the initiation site and the AAA pore loops, will generally lead to processive degradation. However, engagement does not guarantee processive degradation, because the “grip” between the pore loops and the translocating substrate can be lost. Pore loop interaction is generally poor for low-complexity sequences in a substrate, and when such “slippery” elements are positioned adjacent to a folded domain, the mechanical force needed for unfolding of that domain cannot be generated and translocation is in turn stalled; thus, the structured domain is unable to traverse the constriction of substrate entry port. Although this outcome is rare, several proteasome substrates have specifically evolved to undergo this fate, such as p105 (46), Cubitus interruptus (47), Gli3 (48), Spt23 (49, 50), Mga2 (49, 51), and Svb (52). In some cases, partial degradation serves to regulate transcription factors by releasing them from anchoring domains that prevent their access to the nucleus. In the case of Cubitus interruptus (47) and Gli3 (48), degradation of a transcriptional activation domain spares a DNA-binding domain to convert the protein into a repressor.
Conformational switching of the proteasome holoenzyme
To date, three conformational states of the proteasome holoenzyme have been resolved: s1, s2, and s3 (53) (Figure 2). The ground state, s1, predominates under conditions of ATP hydrolysis and the absence of substrate (8–10). Interestingly, s3 can be induced by substrate engagement or chemically by replacing ATP with nonhydrolyzable nucleotide (ATPγS) (39, 54). S2 is a sparsely populated hybrid of s1 and s3 (discussed later; (53)). Presumably proteasomes dock substrate in the s1 state and proceed to the s3 state as the substrate is more intimately engaged, switching back to s1 when a given substrate is fully degraded. The relevant aspect of “engagement” is likely the formation of productive contacts between the initiation elements and the pore loops that are capable of driving substrate translocation. It is widely assumed that this is a pivotal point in the pathway to degradation, and it is sometimes referred to as a “commitment step” (15, 55). This step is not well defined experimentally, but appears to involve a higher-affinity interaction with the substrate, as expected from initiation site engagement (55).
The transition between s1 and s3 states involves sweeping adjustments to the structure of the RP (Figure 2B–C, and 4). So distinct are these states that it has been possible to resolve them within cells by electron cryotomography; the s1 state predominates, constituting 80% of RP in neurons, suggesting that only a minority of proteasomes are substrate-engaged in this cell type (56). In both states there is an interesting discontinuity in the AAA ring (Figure 4), but the site of this “break” switches across the ring in the transition (39). The break reflects an inherent screw axis to the ring that prevents stable closure. Proper alignment of the arginine finger between neighboring subunits cannot be achieved at the site of the break. As noted above, the large and small domains of neighboring ATPases are thought to move as a rigid body throughout the ATPase cycle; however, at the broken interface this connection is lost. Thus, in each state, one of the six Rpt proteins is expected to show compromised ATPase activity. Because of the break in the continuity, the ring is overall distinctly asymmetric, in striking contrast to the homomeric ATPase rings that often serve as model systems for the proteasome.
The motions of the Rpt proteins also affect RP-CP contacts (9, 36, 39, 53) and, in the s3 state, these motions bring the axis of the substrate translocation channel of the RP into close alignment with that of the CP (39, 54) (Figure 2C). The pore loops, although poorly visualized to date, are aligned in the s1 state in a kind of spiral staircase, with the pore loop of Rpt2 at the bottom and that of Rpt3 at the top (Figure 4, top). By contrast, the pore loops adopt a nearly planar configuration in the engaged s3 state (Figure 4, bottom). These data indicate that the pore loops migrate along the channel axis during the ATP cycle, consistent with a role in driving substrate translocation. Do the six pore loops grip substrate coordinately? This appears to be the case for the homomeric ClpX ATPase ring (57, 58). Most likely, the loops that engage substrate productively in a given power stroke will vary depending on the detailed sequence of the substrate at the site of the pore loop interaction. For the heteromeric Rpt ring of the proteasome, asymmetry in the positioning of the pore loops may be a key feature: when active-site mutants in Rpt proteins were compared, the strongest effects on degradation were seen from mutation of the three Rpt subunits with pore loops closest to the substrate entry port in the OB ring (Rpt3, Rpt4 and Rpt6 (33); but see (32)). These Rpt subunits all lie on one side of the ring.
Like the Rpt ring (Figure 4), the lid also shows notable differences between the s1 and s3 states (Figure 2B). The lid is modular: it has two MPN (Mpr1p and Pad1p N-terminus)-domain subunits (Rpn8 and Rpn11), which form a heterodimer, and 6 PCI (Proteasome, COP9, Initiation factor 3) domain subunits (Rpn3, Rpn5, Rpn6, Rpn7, Rpn9, and Rpn12). Each PCI subunit has a C-terminal helix, and these combine with two helices from Rpn8 and from Rpn11 to collectively form a bundle, from which the PCI domains emanate like a fan (59) (Figure 2A). This structure enables the PCI subunits to embrace base subunits at many points, in some cases (Rpn5 and Rpn6) extending as far as the CP (Figure 2A–B). Interestingly, these lid-base contact points differ substantially in the s1 and s3 configurations, particularly for Rpn6 (35) (Figure 2B).
In the most significant of many alterations of the lid in the s3 state, Rpn11 shifts with respect to the Rpt ring (39, 54), providing greater accessibility to its active site and closer alignment with the substrate entry port (Figure 2C). Rpn11 seems to be activated by this motion, which fits perfectly with its function of removing ubiquitin chains en bloc from substrates that are being translocated into the CP (15, 16, 60). Proteasomal Rpn11 activity is dependent on ATP hydrolysis by the Rpt proteins (15, 16), perhaps because the ubiquitin chain is brought to the active site of Rpn11 as a result of substrate translocation, and perhaps because Rpn11 is properly positioned to cleave chains from substrates when the proteasome is in the s3 state (Figure 2C). In summary, the different conformational states of the proteasome holoenzyme observed by cryoelectron microscopy could well represent different states along the enzymatic pathway for substrate degradation, with s1 well suited for receiving substrates and s3 in a state tailored for substrate deubiquitination and translocation through the CP gate.
Ubiquitin recognition
The initial stage of substrate recognition by the proteasome is mediated by a surprisingly complex set of ubiquitin receptors. Two receptors intrinsic to the RP are the aforementioned Rpn13, which is at an apical location, and Rpn10/S5a, which is seated close to the ATPases and Rpn11 (Figure 2A). Rpn10 recognizes ubiquitin chains via its helical UIM (Ubiquitin Interacting Motif) whereas subunit Rpn13 employs a globular Pru (Pleckstrin-like Receptor for Ubiquitin) domain (19, 20, 61–64). Other ubiquitin receptors–Rad23, Dsk2, and Ddi1–bind the proteasome reversibly, using a UBL (ubiquitin-like) domain that is recognized by Rpn1 and by the ubiquitin-binding sites of Rpn13 and Rpn10 (19, 23, 24, 65). Each of these “shuttle receptors” also has a UBA (ubiquitin-associated) domain that binds ubiquitin chains (25, 26). Mutations affecting UBQLN2, an apparent ortholog of Dsk2, are thought to lead to ALS (amyotrophic lateral sclerosis) (66). Genetic studies in yeast suggest the existence of additional ubiquitin receptors for the proteasome (19), not yet identified. Intrinsically disordered protein Sem1/Dss1/Rpn15, which functions in proteasome assembly (67, 68), has been proposed as an additional receptor (69), but has not yet been shown to bind ubiquitin in the context of an assembled proteasome.
Why the proteasome has such a variety of ubiquitin receptors is an open question. The ubiquitin-binding sites of most of the receptors—the shuttle receptors as well as Rpn10—are tethered to the proteasome via flexible linkers, which may provide for more robust chain recognition through avidity effects, as discussed later (70). Simultaneous binding of a chain to more than one receptor has been observed (62), but whether this is the norm remains to be established.
A tetraubiquitin chain linked via Lys48 in ubiquitin has long been considered the canonical recognition motif for the proteasome (71), but recent studies have indicated that a broader range of ubiquitination patterns on the substrate is compatible with efficient degradation, and not all substrates require ubiquitin groups to be arranged in a chain (72–75). Single-molecule studies revealed that the number of ubiquitin groups on a substrate affects substrate dwell time on the proteasome, and, for those substrates tested, dwell time increased exponentially up to three ubiquitins per substrate, then linearly from four to nine ubiquitins (60). In at least the one case tested, a substrate modified by two di-ubiquitin chains proved to be a superior proteasome substrate than one modified by a single tetraubiquitin chain (60). One suggested explanation for this is that an array of ubiquitin chains on the substrate is better suited to simultaneously engage multiple ubiquitin receptors at the proteasome and thus, through avidity, promotes a more stable interaction (60). Perhaps analogously, substrates carrying branched ubiquitin chains also appear to be degraded more readily (in a branched chain, at least one ubiquitin group is ubiquitinated at more than one site) (76).
It is also clear now that many ubiquitin-ubiquitin linkage types can target proteins to the proteasome to be degraded, rather than Lys48 exclusively (77–79). Lys63 chains of ubiquitin provide an interesting case in that they do not typically target proteins to proteasomes in vivo, yet they can target substrates to purified proteasomes (80–82) and, in at least one case, can target a protein for degradation in cells (81). Their weakness in targeting proteins for degradation has been attributed to competing ubiquitin receptors and alternatively, to Lys63-specific deubiquitination at the proteasome, but the contribution of these factors remains to be clearly understood (82, 83). In addition to ubiquitination, other mechanisms for targeting substrates to the proteasome also exist, although they are specialized (84–87).
Deubiquitination
Once at the proteasome, deubiquitinating enzymes (DUBs) act coordinately with the Rpt proteins to prepare substrates for translocation into the proteolytic chamber of the CP. Removing ubiquitin allows substrates to pass through the narrow CP gate (Figure 1); however, premature deubiquitination of substrates can lead to their release prior to proteolysis. In addition to Rpn11, mammalian proteasomes contain two other DUBs, Usp14 and Uch37 (also known as Uch-L5), which differ fundamentally from Rpn11 in that their activity is ATP-independent. Studies in yeast showed that the Usp14 ortholog, Ubp6, can inhibit substrate degradation by the proteasome and that this inhibition occurs through both deubiquitination and a distinct noncatalytic mechanism (88, 89). What fraction of proteasome substrates is susceptible to these effects is not clear. Consistent with these observations, a small-molecule inhibitor of Usp14 was found to accelerate the degradation of some, albeit not all, proteasome substrates in mammalian cells (90). Proteasomes are not ordinarily saturated with Usp14 or Ubp6 (89, 90), so proteasome output can be regulated by induction of these enzymes. In the case of Ubp6, a deficiency of activity leads to hyper-degradation and depletion of ubiquitin (91), triggering elevated synthesis of Ubp6 in a feedback mechanism that helps to provide a set-point for cellular ubiquitin levels (89).
Ubiquitin can be trapped in the active site of most deubiquitinating enzymes, including Ubp6 and Usp14, through the use of probes such as ubiquitin-aldehyde (UbAl) and ubiquitin vinylsulfone (UbVS), which form covalent adducts with the active-site cysteines of these enzymes (92, 93). Proteasomes associated with Ubp6-UbVS or Usp14-UbAl show elevated ATPase and CP peptidase activity (94, 95). Although these might obviously be considered signatures of an activated state of the proteasome, such proteasomes are, on the contrary, inhibited in their capacity to degrade true ubiquitin-conjugated substrates (95). UbVS strengthens or elicits what is evidently the same noncatalytic effect of Ubp6 as discovered using the substrate cyclin B (88, 89). Another key effect of active-site occupancy of Ubp6 is that the catalytic domain of the enzyme, which is normally difficult to resolve by cryo-EM, becomes fixed in the neighborhood of subunit Rpt1 (95, 96) (Figure 5). This finding suggests that the Ubp6 catalytic domain may interact dynamically with the proteasome until it is occupied by a ubiquitinated substrate.
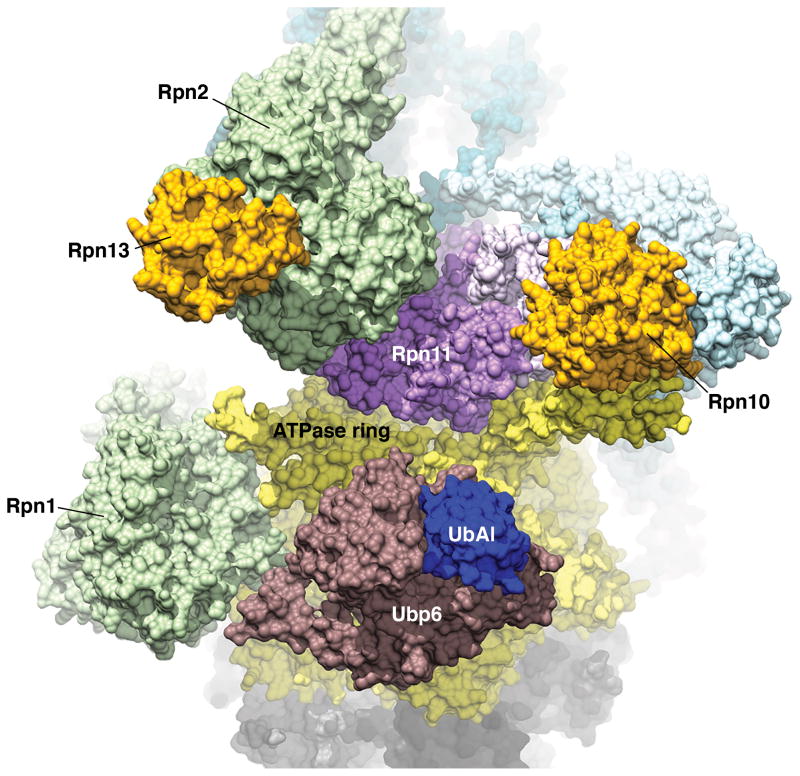
Figure 5. Cryo-electron microscopy-based structure of the proteasome RP with deubiquitinating enzyme Ubp6-UbAl.
Structure of the proteasome s2 state induced by Ubp6 (burgundy) covalently bound to ubiquitin-aldehyde (UbAl, indigo). PDB 5A5B (96) was used to generate this figure, which is colored according to Figure 2.
Unexpectedly, Ubp6-UbAl dramatically perturbed the dynamics of the overall structure of the proteasome, in that it promoted the s2 state (96) (Figure 5), which is otherwise a rare state of the complex (53). S2 is a hybrid state with features of both s1, the substrate-free, engagement-competent state, and s3, the substrate-engaged state associated with translocation (53). Perhaps the elevated ATPase and peptidase activity of the Ubp6-UbAl form of the proteasome reflects its being driven to the s2 state (96). It may be that this leads to a block in degradation via the noncatalytic effect of Ubp6 described earlier. Although Ubp6-UbAl promotes the s2 state in the absence of substrate, it may instead stabilize the s3 state when the proteasome has engaged a substrate (95). Indeed, Ubp6-UbAl does not seem to impair degradation when the proteasome is in the s3 state, but rather, appears to hinder proteasome transition back to the s1 state (95). It is likely that only the s1 state is competent to initiate substrate engagement by the ATPases and thus, Ubp6-UbAl holds the proteasome in a state that cannot support a new round of substrate translocation into the CP (95).
Uch37/Uch-L5 is, like Usp14, a proteasome-associated DUB whose activity is ATP-independent (97). It has long been thought to work in concert with Usp14 and Rpn11 in preparing substrates for entry through the CP gate by removing ubiquitin and thus to promote ubiquitin homeostasis. Uch37 is also, like Usp14, hypothesized to antagonize degradation of at least some proteasome substrates by premature release of conjugated ubiquitin (97). However, when ubiquitin-protein conjugates are used as substrates in vitro, the kinetics of Uch37 activity is slow, which would limit its capacity to compete effectively against substrate degradation. To resolve this conundrum, it will be important to identify preferred substrates of the enzyme.
Uch37 is a component of both the proteasome and the chromatin remodeling factor Ino80 (98); it binds Rpn13 at the proteasome (99–102) and NFRKB (Nuclear Factor Related to KappaB binding protein) at Ino80 (98). Rpn13 and NFRKB both employ DEUBAD (DEUBiquitinase Adaptor) domains to bind Uch37 via its unique C-terminal ULD (Uch37-Like Domain). Whereas the Rpn13 DEUBAD domain activates Uch37, that of Ino80 suppresses its activity, reflecting their divergent effects on the interaction of the Uch37 catalytic domain with ubiquitin (103–105). In the case of NFRKB, ubiquitin interaction is sterically blocked, whereas interaction with Rpn13 seems to direct an activation loop in Uch37 away from its catalytic site, thus increasing catalytic site accessibility (103, 104). To better understand Uch37 function, it will be important to identify how its suppression by NFRKB may be reversed and what its specific targets might be when associated with Ino80. As to proteasomal Uch37, it will be interesting to test whether the docking of ubiquitin chains on Rpn13 leads to optimal presentation of substrates to Uch37. In summary, the three deubiquitinating enzymes of the proteasome are strikingly distinct from one another in function and to date only partially understood. One key consequence of deubiquitination is that if it occurs before the substrate is committed to degradation, the substrate may dissociate from the proteasome instead of being degraded.
Activators of the core particle
The degradation of ubiquitinated proteins relies on RP-CP and RP2-CP proteasomes. However, a set of factors can compete with the RP for occupancy of the ends of the CP cylinder, including PA28αβ, PA28γ, PA200/Blm10, and possibly p97/Cdc48/VCP (106, 107). PA28αβ, PA28γ, and p97 are oligomeric ring complexes, whereas Blm10 is a large monomer that folds into a toroidal shape; nonetheless, these activators have in common the ability to open the CP channel by docking at the α-ring inter-subunit pockets (Figure 1C), as discussed earlier for the RP. Like the RP, PA28αβ and PA28γ have axial channels that align with that of the CP in the assembled complex, and an unobstructed path for substrate entry is perhaps present in p97 and Blm10 as well (107–111).
PA28αβ, PA28γ, and Blm10 do not have ATPase activity so they cannot drive substrate translocation into the CP, as does the RP. This does not exclude a role in promoting protein degradation however, particularly for substrates that have little or no tertiary structure, such as the CDK inhibitors p16, p19, and p21 (112, 113), which can potentially gain access to the CP by diffusion if the gate is open. Interestingly, these activators do not appear to recognize ubiquitin and may provide a ubiquitin-independent avenue for CP-dependent protein degradation. In lieu of ubiquitin, direct interactions between the activator and the substrate may be critical for their selectivity, which may extend even to specific post-translationally modified forms of the substrate (114). Substrates for PA28γ include steroid receptor coactivator 3 (115), the core protein of hepatitis C virus (116), and CDK inhibitors (112, 113), as mentioned above; for PA200/Blm10, the transcriptional activator Sfp1 (117), the GTPase dynamin (110), and acetylated histones (114) are reported targets.
PA28αβ is strongly induced by interferon and promotes effective class I antigen presentation for certain epitopes (118). Class I antigens must be 8–10 residues in length and are produced mainly through the breakdown of intracellular proteins by the proteasome, which releases oligopeptide products of varied length. Interestingly, the enhancement of antigen presentation by PA28αβ need not involve accelerating the degradation of the proteins that contain these epitopes. For example, PA28αβ could function to preserve cleavage products at a length suitable for antigen presentation by promoting their rapid exit from the internal chamber of the CP. This model assumes a hybrid RP:CP:PA28αβ proteasome in which substrate is delivered to one end of the CP by the RP, and exits as peptides at the other end of the CP via a channel held open by PA28αβ. Such hybrid proteasomes have been found in human cells (119).
P97/Cdc48/VCP was suggested to be a CP activator only recently (108, 109), and more work is needed to define how it cooperates with the proteasome. It is clearly capable of activating the CP in a reconstituted system, consistent with its homohexameric structure and C-terminal HbYX motifs. P97 is similar to the RP, rather than PA28 or PA200, in that it processes ubiquitin conjugates and has ATPase activity. It performs myriad functions in the cell through the use of a rich network of adaptor proteins, including cofactors that bind ubiquitin (120). A unifying feature of p97 is that it extracts ubiquitinated substrates from membranes and protein complexes. The extracted proteins may then be degraded by the proteasome, although in some cases extraction is not followed by proteolysis. Proteasomal degradation of such ubiquitin conjugates has generally been envisaged to occur through a process separate from the extraction step (121, 122), with p97 relaying the substrate to the RP. However, a p97-CP complex could bypass the RP, to function as an alternative proteasome. This is an exciting idea, but evidence of a p97-CP complex that functions in cells is still needed (56).
In summary, the CP associates with a variety of activator complexes to form an ensemble of proteolytic species. Most of these activators probably confer to the CP the ability to degrade nonubiquitinated proteins, but the range of substrates for these pathways may be limited by the lack of unfoldase activity of PA28αβ, PA28γ, and PA200/Blm10.
Regulation of proteasome activity
For years, the activity of the proteasome in cells was thought to be subject to very little control. It now appears that global regulation of the ubiquitin-proteasome system is mediated primarily by direct modulation of the proteasome, and the variety of known proteasome regulators is impressive: some regulate the proteasome by associating with it, others modify the proteasome post-translationally, and still others promote the transcription of genes for proteasome subunits.
A conserved ubiquitin ligase (123, 124), known in yeast as Hul5 and in mammals as Ube3C, is associated more strongly with the proteasome under conditions of reduced proteasome function, or “proteasome stress” (125–127). Hul5 also promotes resistance to heat stress and is a major mediator of ubiquitination under these conditions (128). Hul5 and Ube3c exert a positive effect on degradation, particularly its processivity; in its absence, some substrates are degraded incompletely, resulting in the accumulation of protein fragments (129–132). The basis for these effects may be the capacity of Hul5 to add ubiquitin promiscuously to proteasome-bound ubiquitinated proteins (124). This activity might promote effective degradation of stalled substrates, as implied by the link between Hul5/Ube3C and processivity. Hul5 and Ube3C can also modify ubiquitin receptors of the proteasome with ubiquitin (124–126), though the importance of this type of modification in comparison to substrate modification remains to be clarified.
A second factor that is recruited to proteasomes during proteasome stress is Ecm29 (123, 127, 133–135). Ecm29 is a large protein that contacts both the RP and the CP (123, 133), suppressing ATPase, peptidase, and substrate degradation activity by the proteasome. The broad effects of Ecm29 may suggest that it drives proteasomes into the s1 state, or traps this state. Such a mode of action, if validated, would be interestingly related to the noncatalytic effect of Ubp6, which, as discussed earlier, is proposed to inhibit the proteasome by stabilizing the s2 or s3 state (95, 96). The example of Ecm29 illustrates that the regulatory response to physiological stress is not always to enhance proteasome activity, as Ecm29 is a suppressive factor. Similarly, under oxidative stress, proteasome activity is diminished, in this case through reversible dissociation of the CP and RP (134, 136). Interestingly, proteasome inhibition leads to ubiquitination of proteasome subunits and selective degradation of the complex by autophagy in Arabodospsis thaliana (137).
Perhaps the dominant form of stress-dependent regulation of the proteasome is through induction of its synthesis. Elegant studies in yeast uncovered a simple negative feedback loop involving Rpn4 as a constitutive substrate of the proteasome that also promotes transcription from every gene encoding a proteasome subunit (138). When Rpn4 degradation is compromised, its accumulation leads to elevated levels of proteasome synthesis. Mammals lack an Rpn4 ortholog, but transcription factor Nrf1 (Nuclear factor erythroid derived 2-related factor 1) provides a comparable feedback mechanism, which is notably also elicited by proteasome inhibitors such as bortezomib (139–142). Nrf1 is targeted biosynthetically to the endoplasmic reticulum (ER) membrane; to be active in transcription, a fragment of Nrf1 must be released from the membrane by proteolysis. The identity of the protease that releases Nrf1 from the membrane is under debate and the proteasome itself has been proposed. Nrf1 is an ortholog of the well-studied SKN-1 of C. elegans, which similarly controls proteasome levels (143). It should be noted that neither Rpn4, Nrf1, nor SKN-1 is dedicated exclusively to the proteasome, but rather the induction of elevated rates of proteasome synthesis is part of a general stress response. Interestingly, activation of the TORC1 (Target of Rapamycin Complex) kinase complex also leads to Nrf1-dependent elevation of proteasome synthesis, although TORC1 does not influence the rate of Nrf1 processing but rather its transcript levels (144).
Elevation of proteasome levels can be achieved not only through coordinate induction of all genes encoding proteasome subunits, but also through induction of a single subunit, if that subunit is limiting for assembly. Remarkably, specific modulation of the lid subunit Rpn6 can exert a major influence on both stem cell function and organismal longevity; its synthesis is under the control of the Foxo transcription factors Daf-16 in C. elegans and FOXO4 in mammals (145, 146).
Proteasome activity is regulated not only through modulation of its cellular level, but also through post-translational modifications, such as ADP-ribosylation (147), of components of proteasome subpopulations. This has been studied most extensively for phosphorylation, as neuronal-activity-dependent phosphorylation of Rpt6 controls proteasome recruitment into dendritic spines (148). Other phosphorylation events control the stability of RP-CP association (149).
Other aspects of proteasome function
The complexity of the proteasome is daunting, and due to space constraints, we are unable to cover all of the important topics associated with its function. The reader is referred to previous work on proteasome inhibitors as anti-cancer drugs (150, 151), the importance of proteasomes in health and disease (152, 153), mechanisms of proteasome assembly (154–156), and tissue-specific proteasome variants (157), such as the remarkable thymoproteasome (158).
Concluding remarks
Is there another enzyme that has to negotiate such a vast and diverse set of substrates as the proteasome? Not only does the proteasome degrade hundreds of proteins, it must also recognize substrate-bound ubiquitin chains in a range of topologies. Although the initial binding of ubiquitin chains by the proteasome is not as discriminating as once thought, the architecture of ubiquitin modification may exert a stronger influence on downstream events such as degradation and deubiquitination (60), for reasons not yet determined. Might these downstream events depend on which of the many ubiquitin receptors are engaged by the substrate? If a ubiquitinated substrate is initially docked at the proteasome via shuttle factors, do subsequent processing events depend on the substrate being transferred to intrinsic receptors?
Although the questions above remain to be answered, substrate processing has been observed for years to rely on complex dynamics throughout the proteasome. With the recent resolution of discrete conformational states, these diverse events can be seen as broadly concerted and indeed, more readily interpretable. A key feature of the conformational states of the proteasome is that they are governed by associated molecules: substrate, nucleotide, and cofactors such as Ubp6. However, the inherent functional differences between the s1 and s3 forms of the proteasome are still largely unclear. Moreover the choreography the proteasome as it transitions from substrate capture to translocation and unfolding, and finally peptide and ubiquitin release, remains based on still images. As structural approaches, single-molecule studies, and mutational analyses are increasingly refined, the pathway of substrates through the landscape of the proteasome may soon be traceable.
Trends Box.
Docking of a substrate at the proteasome RP complex is mediated by ubiquitin recognition, but to be degraded the substrate must be translocated through a channel leading from the RP the proteolytic CP complex.
Because the channel from the RP to the CP is narrow, translocation generally requires unfolding of the substrate. Hydrolysis of ATP supplies the mechanical force required for substrate unfolding and translocation.
Protein loops that line the channel within the RP interact with substrate and move axially to direct vectorial motion of the substrate towards the CP.
Ubiquitin promotes degradation, but if not removed can impede translocation because it resists unfolding.
Ubiquitin is removed from the substrate either prior to substrate entry into the translocation channel or contemporaneously with this event, depending on the deubiquitinating enzyme. Rapid deubiquitination can preempt substrate degradation.
Recent cryoelectron microscopy studies indicate that the proteasome adopts distinct conformational states, which appear to be distinguishable as substrate receiving or substrate engaged states.
Acknowledgments
Proteasome studies in the authors’ laboratories are supported by grants from the National Institutes of Health (R37-GM043601 to D.F., R01-GM65592 to D.F.) and by the Intramural Research Program of the NIH, NCI, CCR to K.J.W. We thank A. Matouschek for valuable comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Groll M, et al. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 2.Whitby FG, et al. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature. 2000;408:115–120. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]
- 3.Groll M, et al. A gated channel into the proteasome core particle. Nat Struct Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 4.Weber-Ban EU, Reid BG, Miranker AD, Horwich AL. Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature. 1999;401:90–93. doi: 10.1038/43481. [DOI] [PubMed] [Google Scholar]
- 5.Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- 6.Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- 7.Nyquist K, Martin A. Marching to the beat of the ring: polypeptide translocation by AAA+ proteases. Trends Biochem Sci. 2014;39:53–60. doi: 10.1016/j.tibs.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lasker K, et al. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc Natl Acad Sci U S A. 2012;109:1380–1387. doi: 10.1073/pnas.1120559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beck F, et al. Near-atomic resolution structural model of the yeast 26S proteasome. Proc Natl Acad Sci U S A. 2012;109:14870–14875. doi: 10.1073/pnas.1213333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lander GC, et al. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.da Fonseca PC, He J, Morris EP. Molecular model of the human 26S proteasome. Mol Cell. 2012;46:54–66. doi: 10.1016/j.molcel.2012.03.026. [DOI] [PubMed] [Google Scholar]
- 12.Glickman MH, et al. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 13.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomko RJ, Jr, Hochstrasser M. Molecular architecture and assembly of the eukaryotic proteasome. Annu Rev Biochem. 2013;82:415–445. doi: 10.1146/annurev-biochem-060410-150257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verma R, et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 16.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 17.Worden EJ, Padovani C, Martin A. Structure of the Rpn11-Rpn8 dimer reveals mechanisms of substrate deubiquitination during proteasomal degradation. Nat Struct Mol Biol. 2014;21:220–227. doi: 10.1038/nsmb.2771. [DOI] [PubMed] [Google Scholar]
- 18.Pathare GR, et al. Crystal structure of the proteasomal deubiquitylation module Rpn8–Rpn11. Proc Natl Acad Sci U S A. 2014;111:2984–2989. doi: 10.1073/pnas.1400546111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Husnjak K, et al. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schreiner P, et al. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature. 2008;453:548–552. doi: 10.1038/nature06924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamazaki J, Hirayama S, Murata S. Redundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis. PLoS Genet. 2015;11:e1005401. doi: 10.1371/journal.pgen.1005401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elsasser S, Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat Cell Biol. 2005;7:742–749. doi: 10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- 23.Hiyama H, et al. Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26 S proteasome. J Biol Chem. 1999;274:28019–28025. doi: 10.1074/jbc.274.39.28019. [DOI] [PubMed] [Google Scholar]
- 24.Walters KJ, Kleijnen MF, Goh AM, Wagner G, Howley PM. Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry. 2002;41:1767–1777. doi: 10.1021/bi011892y. [DOI] [PubMed] [Google Scholar]
- 25.Bertolaet BL, et al. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat Struct Biol. 2001;8:417–422. doi: 10.1038/87575. [DOI] [PubMed] [Google Scholar]
- 26.Chen L, Shinde U, Ortolan TG, Madura K. Ubiquitin-associated (UBA) domains in Rad23 bind ubiquitin and promote inhibition of multi-ubiquitin chain assembly. EMBO Rep. 2001;2:933–938. doi: 10.1093/embo-reports/kve203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benaroudj N, Zwickl P, Seemuller E, Baumeister W, Goldberg AL. ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Mol Cell. 2003;11:69–78. doi: 10.1016/s1097-2765(02)00775-x. [DOI] [PubMed] [Google Scholar]
- 28.Smith DM, et al. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang F, et al. Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell. 2009;34:473–484. doi: 10.1016/j.molcel.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Djuranovic S, et al. Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases. Mol Cell. 2009;34:580–590. doi: 10.1016/j.molcel.2009.04.030. [DOI] [PubMed] [Google Scholar]
- 31.Rubin DM, Glickman MH, Larsen CN, Dhruvakumar S, Finley D. Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J. 1998;17:4909–4919. doi: 10.1093/emboj/17.17.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erales J, Hoyt MA, Troll F, Coffino P. Functional asymmetries of proteasome translocase pore. J Biol Chem. 2012;287:18535–18543. doi: 10.1074/jbc.M112.357327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beckwith R, Estrin E, Worden EJ, Martin A. Reconstitution of the 26S proteasome reveals functional asymmetries in its AAA+ unfoldase. Nat Struct Mol Biol. 2013;20:1164–1172. doi: 10.1038/nsmb.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peth A, Nathan JA, Goldberg AL. The ATP costs and time required to degrade ubiquitinated proteins by the 26 S proteasome. J Biol Chem. 2013;288:29215–29222. doi: 10.1074/jbc.M113.482570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pathare GR, et al. The proteasomal subunit Rpn6 is a molecular clamp holding the core and regulatory subcomplexes together. Proc Natl Acad Sci U S A. 2012;109:149–154. doi: 10.1073/pnas.1117648108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian G, et al. An asymmetric interface between the regulatory and core particles of the proteasome. Nat Struct Mol Biol. 2011;18:1259–1267. doi: 10.1038/nsmb.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park S, et al. Reconfiguration of the proteasome during chaperone-mediated assembly. Nature. 2013;497:512–516. doi: 10.1038/nature12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sokolova V, Li F, Polovin G, Park S. Proteasome Activation is Mediated via a Functional Switch of the Rpt6 C-terminal Tail Following Chaperone-dependent Assembly. Sci Rep. 2015;5:14909. doi: 10.1038/srep14909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sledz P, et al. Structure of the 26S proteasome with ATP-gammaS bound provides insights into the mechanism of nucleotide-dependent substrate translocation. Proc Natl Acad Sci U S A. 2013;110:7264–7269. doi: 10.1073/pnas.1305782110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inobe T, Genmei R. N-Terminal Coiled-Coil Structure of ATPase Subunits of 26S Proteasome Is Crucial for Proteasome Function. PLoS One. 2015;10:e0134056. doi: 10.1371/journal.pone.0134056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat Struct Mol Biol. 2004;11:830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- 42.Fishbain S, et al. Sequence composition of disordered regions fine-tunes protein half-life. Nat Struct Mol Biol. 2015;22:214–221. doi: 10.1038/nsmb.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Lee R, et al. Intrinsically disordered segments affect protein half-life in the cell and during evolution. Cell Rep. 2014;8:1832–1844. doi: 10.1016/j.celrep.2014.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fishbain S, Prakash S, Herrig A, Elsasser S, Matouschek A. Rad23 escapes degradation because it lacks a proteasome initiation region. Nat Commun. 2011;2:192. doi: 10.1038/ncomms1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inobe T, Fishbain S, Prakash S, Matouschek A. Defining the geometry of the two-component proteasome degron. Nat Chem Biol. 2011;7:161–167. doi: 10.1038/nchembio.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 47.Tian L, Holmgren RA, Matouschek A. A conserved processing mechanism regulates the activity of transcription factors Cubitus interruptus and NF-kappaB. Nat Struct Mol Biol. 2005;12:1045–1053. doi: 10.1038/nsmb1018. [DOI] [PubMed] [Google Scholar]
- 48.Schrader EK, Harstad KG, Holmgren RA, Matouschek A. A three-part signal governs differential processing of Gli1 and Gli3 proteins by the proteasome. J Biol Chem. 2011;286:39051–39058. doi: 10.1074/jbc.M111.274993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoppe T, et al. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- 50.Rape M, et al. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell. 2001;107:667–677. doi: 10.1016/s0092-8674(01)00595-5. [DOI] [PubMed] [Google Scholar]
- 51.Rape M, Jentsch S. Productive RUPture: activation of transcription factors by proteasomal processing. Biochim Biophys Acta. 2004;1695:209–213. doi: 10.1016/j.bbamcr.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 52.Zanet J, et al. Pri sORF peptides induce selective proteasome-mediated protein processing. Science. 2015;349:1356–1358. doi: 10.1126/science.aac5677. [DOI] [PubMed] [Google Scholar]
- 53.Unverdorben P, et al. Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome. Proc Natl Acad Sci U S A. 2014;111:5544–5549. doi: 10.1073/pnas.1403409111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matyskiela ME, Lander GC, Martin A. Conformational switching of the 26S proteasome enables substrate degradation. Nat Struct Mol Biol. 2013;20:781–788. doi: 10.1038/nsmb.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peth A, Uchiki T, Goldberg AL. ATP-dependent steps in the binding of ubiquitin conjugates to the 26S proteasome that commit to degradation. Mol Cell. 2010;40:671–681. doi: 10.1016/j.molcel.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Asano S, et al. Proteasomes. A molecular census of 26S proteasomes in intact neurons. Science. 2015;347:439–442. doi: 10.1126/science.1261197. [DOI] [PubMed] [Google Scholar]
- 57.Iosefson O, Nager AR, Baker TA, Sauer RT. Coordinated gripping of substrate by subunits of a AAA+ proteolytic machine. Nat Chem Biol. 2015;11:201–206. doi: 10.1038/nchembio.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iosefson O, Olivares AO, Baker TA, Sauer RT. Dissection of Axial-Pore Loop Function during Unfolding and Translocation by a AAA+ Proteolytic Machine. Cell Rep. 2015;12:1032–1041. doi: 10.1016/j.celrep.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Estrin E, Lopez-Blanco JR, Chacon P, Martin A. Formation of an intricate helical bundle dictates the assembly of the 26S proteasome lid. Structure. 2013;21:1624–1635. doi: 10.1016/j.str.2013.06.023. [DOI] [PubMed] [Google Scholar]
- 60.Lu Y, Lee BH, King RW, Finley D, Kirschner MW. Substrate degradation by the proteasome: a single-molecule kinetic analysis. Science. 2015;348:1250834. doi: 10.1126/science.1250834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Q, Young P, Walters KJ. Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition. J Mol Biol. 2005;348:727–739. doi: 10.1016/j.jmb.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 62.Zhang N, et al. Structure of the s5a:k48-linked diubiquitin complex and its interactions with rpn13. Mol Cell. 2009;35:280–290. doi: 10.1016/j.molcel.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang D, et al. Together, Rpn10 and Dsk2 can serve as a polyubiquitin chain-length sensor. Mol Cell. 2009;36:1018–1033. doi: 10.1016/j.molcel.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J Biol Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- 65.Elsasser S, et al. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- 66.Deng HX, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tomko RJ, Jr, Hochstrasser M. The intrinsically disordered Sem1 protein functions as a molecular tether during proteasome lid biogenesis. Mol Cell. 2014;53:433–443. doi: 10.1016/j.molcel.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bohn S, et al. Localization of the regulatory particle subunit Sem1 in the 26S proteasome. Biochem Biophys Res Commun. 2013;435:250–254. doi: 10.1016/j.bbrc.2013.04.069. [DOI] [PubMed] [Google Scholar]
- 69.Paraskevopoulos K, et al. Dss1 is a 26S proteasome ubiquitin receptor. Mol Cell. 2014;56:453–461. doi: 10.1016/j.molcel.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aufderheide A, Unverdorben P, Baumeister W, Forster F. Structural disorder and its role in proteasomal degradation. FEBS Lett. 2015 doi: 10.1016/j.febslet.2015.07.034. [DOI] [PubMed] [Google Scholar]
- 71.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kravtsova-Ivantsiv Y, Cohen S, Ciechanover A. Modification by single ubiquitin moieties rather than polyubiquitination is sufficient for proteasomal processing of the p105 NF-kappaB precursor. Adv Exp Med Biol. 2011;691:95–106. doi: 10.1007/978-1-4419-6612-4_10. [DOI] [PubMed] [Google Scholar]
- 73.Dimova NV, et al. APC/C-mediated multiple monoubiquitylation provides an alternative degradation signal for cyclin B1. Nat Cell Biol. 2012;14:168–176. doi: 10.1038/ncb2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shabek N, et al. The size of the proteasomal substrate determines whether its degradation will be mediated by mono- or polyubiquitylation. Mol Cell. 2012;48:87–97. doi: 10.1016/j.molcel.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 75.Kulathu Y, Komander D. Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol. 2012;13:508–523. doi: 10.1038/nrm3394. [DOI] [PubMed] [Google Scholar]
- 76.Meyer HJ, Rape M. Enhanced protein degradation by branched ubiquitin chains. Cell. 2014;157:910–921. doi: 10.1016/j.cell.2014.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kirkpatrick DS, et al. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 78.Jin L, Williamson A, Banerjee S, Philipp I, Rape M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell. 2008;133:653–665. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu P, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hofmann RM, Pickart CM. In vitro assembly and recognition of Lys-63 polyubiquitin chains. J Biol Chem. 2001;276:27936–27943. doi: 10.1074/jbc.M103378200. [DOI] [PubMed] [Google Scholar]
- 81.Saeki Y, et al. Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J. 2009;28:359–371. doi: 10.1038/emboj.2008.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jacobson AD, et al. The lysine 48 and lysine 63 ubiquitin conjugates are processed differently by the 26 s proteasome. J Biol Chem. 2009;284:35485–35494. doi: 10.1074/jbc.M109.052928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nathan JA, Kim HT, Ting L, Gygi SP, Goldberg AL. Why do cellular proteins linked to K63-polyubiquitin chains not associate with proteasomes? EMBO J. 2013;32:552–565. doi: 10.1038/emboj.2012.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schmidtke G, Aichem A, Groettrup M. FAT10ylation as a signal for proteasomal degradation. Biochim Biophys Acta. 2014;1843:97–102. doi: 10.1016/j.bbamcr.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 85.Godderz D, Schafer E, Palanimurugan R, Dohmen RJ. The N-terminal unstructured domain of yeast ODC functions as a transplantable and replaceable ubiquitin-independent degron. J Mol Biol. 2011;407:354–367. doi: 10.1016/j.jmb.2011.01.051. [DOI] [PubMed] [Google Scholar]
- 86.Rani N, Aichem A, Schmidtke G, Kreft SG, Groettrup M. FAT10 and NUB1L bind to the VWA domain of Rpn10 and Rpn1 to enable proteasome-mediated proteolysis. Nat Commun. 2012;3:749. doi: 10.1038/ncomms1752. [DOI] [PubMed] [Google Scholar]
- 87.Tanji K, Tanaka T, Kamitani T. Interaction of NUB1 with the proteasome subunit S5a. Biochem Biophys Res Commun. 2005;337:116–120. doi: 10.1016/j.bbrc.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 88.Hanna J, et al. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 89.Hanna J, Meides A, Zhang DP, Finley D. A ubiquitin stress response induces altered proteasome composition. Cell. 2007;129:747–759. doi: 10.1016/j.cell.2007.03.042. [DOI] [PubMed] [Google Scholar]
- 90.Lee BH, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hanna J, Leggett DS, Finley D. Ubiquitin depletion as a key mediator of toxicity by translational inhibitors. Mol Cell Biol. 2003;23:9251–9261. doi: 10.1128/MCB.23.24.9251-9261.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hershko A, Rose IA. Ubiquitin-aldehyde: a general inhibitor of ubiquitin-recycling processes. Proc Natl Acad Sci U S A. 1987;84:1829–1833. doi: 10.1073/pnas.84.7.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ekkebus R, Flierman D, Geurink PP, Ovaa H. Catching a DUB in the act: novel ubiquitin-based active site directed probes. Curr Opin Chem Biol. 2014;23:63–70. doi: 10.1016/j.cbpa.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Peth A, Besche HC, Goldberg AL. Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Mol Cell. 2009;36:794–804. doi: 10.1016/j.molcel.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bashore C, et al. Ubp6 deubiquitinase controls conformational dynamics and substrate degradation of the 26S proteasome. Nat Struct Mol Biol. 2015;22:712–719. doi: 10.1038/nsmb.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Aufderheide A, et al. Structural characterization of the interaction of Ubp6 with the 26S proteasome. Proc Natl Acad Sci U S A. 2015;112:8626–8631. doi: 10.1073/pnas.1510449112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- 98.Yao T, et al. Distinct modes of regulation of the Uch37 deubiquitinating enzyme in the proteasome and in the Ino80 chromatin-remodeling complex. Mol Cell. 2008;31:909–917. doi: 10.1016/j.molcel.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yao T, et al. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat Cell Biol. 2006;8:994–1002. doi: 10.1038/ncb1460. [DOI] [PubMed] [Google Scholar]
- 100.Hamazaki J, et al. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 2006;25:4524–4536. doi: 10.1038/sj.emboj.7601338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Qiu XB, et al. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 2006;25:5742–5753. doi: 10.1038/sj.emboj.7601450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen X, Lee BH, Finley D, Walters KJ. Structure of proteasome ubiquitin receptor hRpn13 and its activation by the scaffolding protein hRpn2. Mol Cell. 2010;38:404–415. doi: 10.1016/j.molcel.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sahtoe DD, et al. Mechanism of UCH-L5 activation and inhibition by DEUBAD domains in RPN13 and INO80G. Mol Cell. 2015;57:887–900. doi: 10.1016/j.molcel.2014.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.VanderLinden RT, et al. Structural basis for the activation and inhibition of the UCH37 deubiquitylase. Mol Cell. 2015;57:901–911. doi: 10.1016/j.molcel.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen X, Walters KJ. Structural plasticity allows UCH37 to be primed by RPN13 or locked down by INO80G. Mol Cell. 2015;57:767–768. doi: 10.1016/j.molcel.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fort P, Kajava AV, Delsuc F, Coux O. Evolution of proteasome regulators in eukaryotes. Genome Biol Evol. 2015;7:1363–1379. doi: 10.1093/gbe/evv068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kish-Trier E, Hill CP. Structural biology of the proteasome. Annu Rev Biophys. 2013;42:29–49. doi: 10.1146/annurev-biophys-083012-130417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Barthelme D, Sauer RT. Identification of the Cdc48*20S proteasome as an ancient AAA+ proteolytic machine. Science. 2012;337:843–846. doi: 10.1126/science.1224352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Barthelme D, Jauregui R, Sauer RT. An ALS disease mutation in Cdc48/p97 impairs 20S proteasome binding and proteolytic communication. Protein Sci. 2015;24:1521–1527. doi: 10.1002/pro.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tar K, et al. Proteasomes associated with the Blm10 activator protein antagonize mitochondrial fission through degradation of the fission protein Dnm1. J Biol Chem. 2014;289:12145–12156. doi: 10.1074/jbc.M114.554105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dange T, et al. Blm10 protein promotes proteasomal substrate turnover by an active gating mechanism. J Biol Chem. 2011;286:42830–42839. doi: 10.1074/jbc.M111.300178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li X, et al. Ubiquitin- and ATP-independent proteolytic turnover of p21 by the REGgamma-proteasome pathway. Mol Cell. 2007;26:831–842. doi: 10.1016/j.molcel.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 113.Chen X, Barton LF, Chi Y, Clurman BE, Roberts JM. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGgamma proteasome. Mol Cell. 2007;26:843–852. doi: 10.1016/j.molcel.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Qian MX, et al. Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell. 2013;153:1012–1024. doi: 10.1016/j.cell.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li X, et al. The SRC-3/AIB1 coactivator is degraded in a ubiquitin- and ATP-independent manner by the REGgamma proteasome. Cell. 2006;124:381–392. doi: 10.1016/j.cell.2005.11.037. [DOI] [PubMed] [Google Scholar]
- 116.Moriishi K, et al. Proteasome activator PA28gamma-dependent nuclear retention and degradation of hepatitis C virus core protein. J Virol. 2003;77:10237–10249. doi: 10.1128/JVI.77.19.10237-10249.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lopez AD, et al. Proteasomal degradation of Sfp1 contributes to the repression of ribosome biogenesis during starvation and is mediated by the proteasome activator Blm10. Mol Biol Cell. 2011;22:528–540. doi: 10.1091/mbc.E10-04-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cascio P. PA28alphabeta: the enigmatic magic ring of the proteasome? Biomolecules. 2014;4:566–584. doi: 10.3390/biom4020566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tanahashi N, et al. Hybrid proteasomes. Induction by interferon-gamma and contribution to ATP-dependent proteolysis. J Biol Chem. 2000;275:14336–14345. doi: 10.1074/jbc.275.19.14336. [DOI] [PubMed] [Google Scholar]
- 120.Buchberger A, Schindelin H, Hanzelmann P. Control of p97 function by cofactor binding. FEBS Lett. 2015 doi: 10.1016/j.febslet.2015.08.028. [DOI] [PubMed] [Google Scholar]
- 121.Richly H, et al. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 122.Kim I, Mi K, Rao H. Multiple interactions of rad23 suggest a mechanism for ubiquitylated substrate delivery important in proteolysis. Mol Biol Cell. 2004;15:3357–3365. doi: 10.1091/mbc.E03-11-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Leggett DS, et al. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- 124.Crosas B, et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–1413. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- 125.Besche HC, et al. Autoubiquitination of the 26S proteasome on Rpn13 regulates breakdown of ubiquitin conjugates. EMBO J. 2014;33:1159–1176. doi: 10.1002/embj.201386906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jacobson AD, MacFadden A, Wu Z, Peng J, Liu CW. Autoregulation of the 26S proteasome by in situ ubiquitination. Mol Biol Cell. 2014;25:1824–1835. doi: 10.1091/mbc.E13-10-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Park S, Kim W, Tian G, Gygi SP, Finley D. Structural defects in the regulatory particle-core particle interface of the proteasome induce a novel proteasome stress response. J Biol Chem. 2011;286:36652–36666. doi: 10.1074/jbc.M111.285924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fang NN, Ng AH, Measday V, Mayor T. Hul5 HECT ubiquitin ligase plays a major role in the ubiquitylation and turnover of cytosolic misfolded proteins. Nat Cell Biol. 2011;13:1344–1352. doi: 10.1038/ncb2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kohlmann S, Schafer A, Wolf DH. Ubiquitin ligase Hul5 is required for fragment-specific substrate degradation in endoplasmic reticulum-associated degradation. J Biol Chem. 2008;283:16374–16383. doi: 10.1074/jbc.M801702200. [DOI] [PubMed] [Google Scholar]
- 130.Aviram S, Kornitzer D. The ubiquitin ligase Hul5 promotes proteasomal processivity. Mol Cell Biol. 2010;30:985–994. doi: 10.1128/MCB.00909-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Martinez-Noel G, et al. Identification and proteomic analysis of distinct UBE3A/E6AP protein complexes. Mol Cell Biol. 2012;32:3095–3106. doi: 10.1128/MCB.00201-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chu BW, et al. The E3 ubiquitin ligase UBE3C enhances proteasome processivity by ubiquitinating partially proteolyzed substrates. J Biol Chem. 2013;288:34575–34587. doi: 10.1074/jbc.M113.499350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.De La Mota-Peynado A, et al. The proteasome-associated protein Ecm29 inhibits proteasomal ATPase activity and in vivo protein degradation by the proteasome. J Biol Chem. 2013;288:29467–29481. doi: 10.1074/jbc.M113.491662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang X, Yen J, Kaiser P, Huang L. Regulation of the 26S proteasome complex during oxidative stress. Sci Signal. 2010;3:ra88. doi: 10.1126/scisignal.2001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lee SY, De la Mota-Peynado A, Roelofs J. Loss of Rpt5 protein interactions with the core particle and Nas2 protein causes the formation of faulty proteasomes that are inhibited by Ecm29 protein. J Biol Chem. 2011;286:36641–36651. doi: 10.1074/jbc.M111.280875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Livnat-Levanon N, et al. Reversible 26S proteasome disassembly upon mitochondrial stress. Cell Rep. 2014;7:1371–1380. doi: 10.1016/j.celrep.2014.04.030. [DOI] [PubMed] [Google Scholar]
- 137.Marshall RS, Li F, Gemperline DC, Book AJ, Vierstra RD. Autophagic Degradation of the 26S Proteasome Is Mediated by the Dual ATG8/Ubiquitin Receptor RPN10 in Arabidopsis. Mol Cell. 2015;58:1053–66. doi: 10.1016/j.molcel.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Xie Y, Varshavsky A. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc Natl Acad Sci U S A. 2001;98:3056–3061. doi: 10.1073/pnas.071022298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Radhakrishnan SK, et al. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol Cell. 2010;38:17–28. doi: 10.1016/j.molcel.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Steffen J, Seeger M, Koch A, Kruger E. Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Mol Cell. 2010;40:147–158. doi: 10.1016/j.molcel.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 141.Radhakrishnan SK, den Besten W, Deshaies RJ. p97-dependent retrotranslocation and proteolytic processing govern formation of active Nrf1 upon proteasome inhibition. Elife. 2014;3:e01856. doi: 10.7554/eLife.01856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sha Z, Goldberg AL. Proteasome-mediated processing of Nrf1 is essential for coordinate induction of all proteasome subunits and p97. Curr Biol. 2014;24:1573–1583. doi: 10.1016/j.cub.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Li X, et al. Specific SKN-1/Nrf stress responses to perturbations in translation elongation and proteasome activity. PLoS Genet. 2011;7:e1002119. doi: 10.1371/journal.pgen.1002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhang Y, Manning BD. mTORC1 signaling activates NRF1 to increase cellular proteasome levels. Cell Cycle. 2015;14:2011–2017. doi: 10.1080/15384101.2015.1044188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Vilchez D, et al. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature. 2012;489:304–308. doi: 10.1038/nature11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Vilchez D, et al. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature. 2012;489:263–268. doi: 10.1038/nature11315. [DOI] [PubMed] [Google Scholar]
- 147.Cho-Park PF, Steller H. Proteasome regulation by ADP-ribosylation. Cell. 2013;153:614–627. doi: 10.1016/j.cell.2013.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Djakovic SN, et al. Phosphorylation of Rpt6 regulates synaptic strength in hippocampal neurons. J Neurosci. 2012;32:5126–5131. doi: 10.1523/JNEUROSCI.4427-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Guo X, et al. UBLCP1 is a 26S proteasome phosphatase that regulates nuclear proteasome activity. Proc Natl Acad Sci U S A. 2011;108:18649–18654. doi: 10.1073/pnas.1113170108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Goldberg AL. Development of proteasome inhibitors as research tools and cancer drugs. J Cell Biol. 2012;199:583–588. doi: 10.1083/jcb.201210077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Huber EM, Groll M. Inhibitors for the immuno- and constitutive proteasome: current and future trends in drug development. Angew Chem Int Ed Engl. 2012;51:8708–8720. doi: 10.1002/anie.201201616. [DOI] [PubMed] [Google Scholar]
- 152.Schmidt M, Finley D. Regulation of proteasome activity in health and disease. Biochim Biophys Acta. 2014;1843:13–25. doi: 10.1016/j.bbamcr.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Brehm A, et al. Additive loss-of-function proteasome subunit mutations in CANDLE/PRAAS patients promote type I IFN production. J Clin Invest. 2015;125 doi: 10.1172/JCI81260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sahara K, Kogleck L, Yashiroda H, Murata S. The mechanism for molecular assembly of the proteasome. Adv Biol Regul. 2014;54:51–58. doi: 10.1016/j.jbior.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 155.Kunjappu MJ, Hochstrasser M. Assembly of the 20S proteasome. Biochim Biophys Acta. 2014;1843:2–12. doi: 10.1016/j.bbamcr.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Saeki Y, Tanaka K. Assembly and function of the proteasome. Methods Mol Biol. 2012;832:315–337. doi: 10.1007/978-1-61779-474-2_22. [DOI] [PubMed] [Google Scholar]
- 157.Kniepert A, Groettrup M. The unique functions of tissue-specific proteasomes. Trends Biochem Sci. 2014;39:17–24. doi: 10.1016/j.tibs.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 158.Murata S, et al. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science. 2007;316:1349–1353. doi: 10.1126/science.1141915. [DOI] [PubMed] [Google Scholar]