Abstract
Cytoplasmic dynein, a member of the AAA family of ATPases, drives the processive movement of numerous intracellular cargos towards the minus end of microtubules. Here, we summarize the structural and motile properties of dynein and highlight features that distinguish this motor from kinesin-1 and myosin V, two well-studied transport motors. Integrating information from recent crystal and cryo-EM structures as well as high-resolution single molecule studies, we also discuss models for how dynein biases its movement in one direction along a microtubule track, and present a movie that illustrates these principles.
Overview of the Dynein Motor Protein
Eukaryotic cells use motor proteins to transport a variety of cargos, which include membrane-bounded organelles, mRNAs and proteins, along cellular “highways” of actin filaments and microtubules [1, 2]. The molecular motor myosin V moves cargos unidirectionally towards the barbed end of actin filaments. On microtubules, cytoplasmic dynein and kinesins are the two main classes of cargo-transporting motors. Microtubules have an intrinsic polarity, with a plus end and a minus end; in most cells, the plus ends are directed primarily towards the cell cortex, whereas most minus ends localize towards the cell center. Most kinesin family members transport cargoes towards the plus end, or cell cortex, whereas dynein transports cargo towards the minus end, or cell center. Kinesin-1 and myosin V are the best understood cargo-transporting molecular motors; their mechanisms of motility have been reviewed elsewhere [3–5] and are summarized in Box 1. In this review, we focus on how the dynein motor steps along a microtubule. Broadly, dyneins can be divided into two classes: axonemal and cytoplasmic. Axonemal dyneins regulate microtubule sliding in the axonemes of cilia and flagella, whereas cytoplasmic dynein facilitates movement of organelles and other cargos necessary for cellular function.
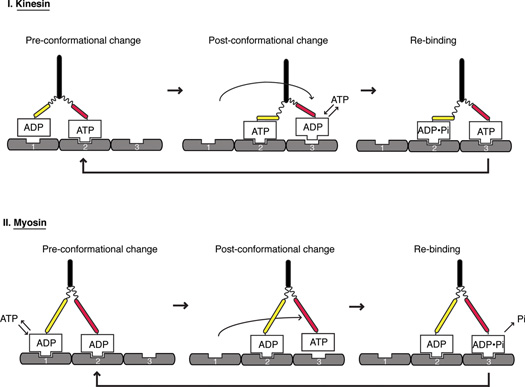
Box 1. Principles of how kinesin-1 and myosin V move processively along tracks.
Work on kinesin-1 and myosin V have revealed general principles of how these homodimeric motors move processively along tracks; schematics depicting the kinesin-1 (Figure I A) and myosin V (Figure IB) cycles are shown. In both cases, the two motor domains move in a handover-hand motor and one ATP is consumed per step [61,62]. Energy from the ATP hydrolysis cycle is transferred into structural changes in the motor that are needed for the following two critical events in the motility cycle.
Principles of how kinesin (A) and myosin (B) move along their tracks, microtubules and actin respectively. Nucleotide-dependent conformational changes are coordinated between the two heads to generate a uniform size of steps along the tracks.
1. Changing the strength of binding between the motor and its track
Just as G proteins use a GTPase cycle to change their affinity for effector proteins, motor proteins use ATP energy to change their strength of binding to their polymeric track. A strong binding state allows the motor to resist an opposing load. A weak binding state is needed to release the motor so that it can be displaced (see below) and then rebind to a forward binding site on the track and lock a “step” in place. Breaking a strong interaction between the motor and its track requires energy from the nucleotide hydrolysis cycle. For kinesin-1, the weak binding state occurs with ADP bound, whereas for myosin V, the weak binding state is ATP-bound. The binding energy from the motor-polymer interaction also can weaken the binding of ADP in the active site, thus advancing the chemical cycle of the motor by allowing ATP/ADP exchange.
2. Biasing the stepping of the motor in one direction along the polymer
If the motor dissociates and rebinds to the same position on the track, then no net motion will occur. For kinesin-1 and myosin-V, at least two mechanisms favor the rebinding of the dissociated motor domain to a forward binding site and help to coordinate the alternating actions of the two motor domains. First, a structural change in a mechanical element (two conformational states shown in red and yellow) in the polymer-bound motor displaces the detached partner motor domain and biases its Brownian search for a forward binding site [63,64]. In myosin-V, this conformational change is the swing of its long lever arm, whereas in kinesin-1, the docking of the neck linker to the catalytic core serves a similar role. A second mechanism for promoting directionality is through an asymmetry in ADP release kinetics of the two motor domains, which is regulated by the weak-to-strong binding transition of the motor to the polymer. In the case of kinesin, the transition to the strong binding state is disfavored if the detached motor domain attempts to bind at the rearward binding site but this transition is allowed at the forward binding site [65,66]. This asymmetry is likely mediated by the different neck linker position in the front and rear head. In the case of myosin V, the fully strongly bound actin state and concomitant ADP release are repressed in the front head of myosin-V motor domain; once rear head releases, then the lever arm swing and ADP release can occur in the front head [5,61,67].
Cytoplasmic dynein, a homodimer of two identical motor domains and several additional subunits attached to the non-motor “tail domain”, has numerous cargos and is involved in many cellular functions [1, 6, 7]. Cytoplasmic dynein 1 transports mRNA-protein complexes, viruses, membranous organelles, and certain proteins toward the cell center, whereas cytoplasmic dynein 2 transports cargo exclusively in the cilium. In metazoan cells, dynein 1 plays a critical role in mitosis, where it creates forces that slide microtubules to help build and maintain a bipolar spindle [8–10]. Dynein 1 also transports mitotic checkpoint proteins from the kinetochore towards the poles, which helps to regulate the timing of the metaphase-anaphase transition [11, 12]. During development, dynein 1 localized at the cell cortex positions the spindle during asymmetric cell division and transports critical mRNA complexes involved in developmental patterning [13–14]. The nucleus is also positioned by dynein 1, and errors in this process cause defects in brain development [15–17]. Many of the activities described above are carried out by dynein 1 working in conjunction with dynactin, a ~1.2 MDa, multi-subunit protein complex, and adaptor proteins that link dynein-dynactin to specific cargos [18]. In fact, mammalian cytoplasmic dynein 1 appears to exhibit poor motility in the absence of dynactin and an adapter [19, 20]. In contrast, budding yeast Saccharomyces cerevisiae dynein exhibits robust processive motility in the absence of dynactin [21]. In S. cerevisiae, dynein only appears to serve a non-essential function of pulling on microtubules from the cell cortex in order to position the mitotic spindle at the juncture of the mother-daughter bud neck; perhaps because of its limited functions, the regulation of yeast dynein appears to be less complex. Thus, yeast dynein has served as a good model system for studying the motility properties of dynein in vitro.
In the past few years, our understanding of dynein has advanced considerably; several X-ray and cryo-electron microscopy (cryo-EM) structures of various dyneins from different species have been reported, and the single molecule motility properties of S. cerevisiae dynein have been studied in exquisite detail. In this article, we review this recent progress and highlight similarities and differences between dynein and the other well-studied cargo transport motors kinesin-1 and myosin-V. We integrate structural data on cytoplasmic and axonemal dyneins from different species, and single molecule studies focused on yeast dynein, into a model for the biased stepping of dynein towards the minus end of the microtubule. This review does not cover the regulation of dynein motility by associated factors (e.g. Lis1, dynactin), for which considerable recent progress has also been made [19, 22, 23].
The dynein motor domain and its conformational changes
Architecture of the Dynein motor domain
The first low resolution crystal structures of the dynein motor domain were solved in 2011 [24, 25], shedding light on the architecture of the motor domain; subsequent crystal structures revealed details at higher resolution and in different nucleotide states [26–29]. The cytoplasmic dynein heavy chain is a homodimer comprising two identical motor domains. Each motor domain contains a hexameric AAA ring, with three appendages that protrude from the ring: the stalk, buttress (in some studies also called the “strut”), and linker (Figure 1). The building blocks for each AAA domain in the AAA ring are a large αβ domain (AAAL) and a small α-helical domain (AAAs). Together, AAAL and AAAs form one AAA domain. Most AAA proteins are homo-oligomeric rings, comprising multiple copies of an identical protomer [30]. Dynein is unusual in that its 6 AAA domains are synthesized as a single polypeptide, which folds to form an asymmetric AAA ring. Of the six AAA domains in dynein, AAA1–4 bind ATP [26, 27]; domains AAA1, AAA3 and AAA4 also are capable of hydrolyzing ATP. Hydrolysis at AAA1 is critical for motility, and most of this review focuses on how AAA1 drives dynein motility. Hydrolysis at AAA3, and perhaps AAA4, plays a regulatory role, which will be briefly discussed.
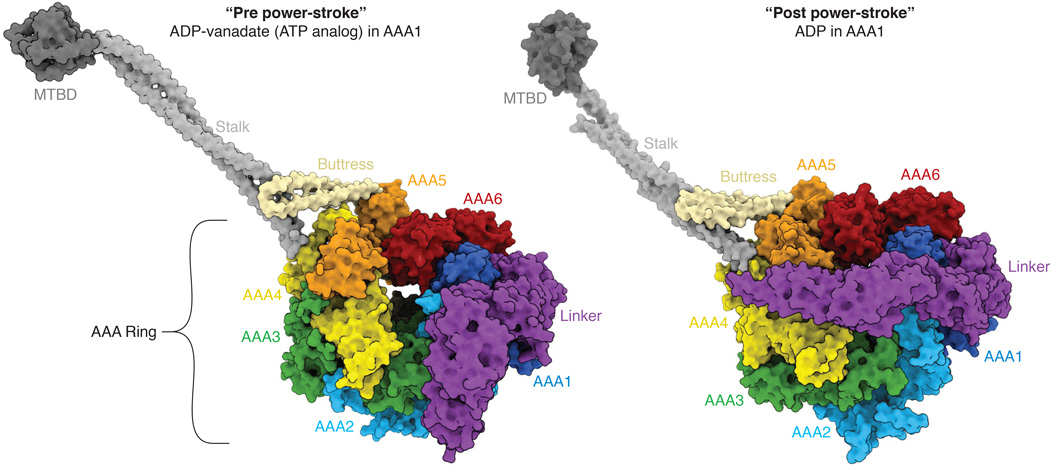
Figure 1.
Architecture of the dynein motor domain and the linker conformational change. Two crystal structures are shown: (Left panel) human cytoplasmic dynein 2 with ADP-vanadate, which is though to mimic ATP or ADP-Pi in AAA1 (PDB code: 4RH7) and (right panel) Dictyostelium cytoplasmic dynein with ADP bound in AAA1 (PDB code: 3VKH). AAA1 is the main hydrolytic site that drives the motility cycle. Transitions of the linker (magenta) between bent and straight conformations may be involved in driving dynein motility (see Figure 2). This figure was generated using ePMV [68].
In addition to the six AAA domains, dynein contains three appendages that protrude from the ring (Figure 1). The stalk and buttress are two coiled-coils that extend from the small domains of AAA4 and AAA5 respectively and interact with one another. The stalk is a long (~140 Å) coiled coil that contains a small, ~10 kDa microtubule binding domain (MTBD) at its distal tip. The buttress is a smaller coiled-coil that interacts with the stalk approximately one-third up the stalk from the ring. The stalk coiled-coil also provides a pathway of communication between the AAA ring and the MTBD. The two helices of the coiled coil can interact with one another in two possible ways, which results in two different registrations of interacting amino acids. Experiments by Gibbons et al. [31] first suggested that the “α-registry” is associated with a strong binding state of the MTBD and a four residue shifted “β+-registry” is associated with a weak binding state. Several subsequent experiments have provided additional support for this hypothesis [29, 32–35]. The sliding of one of the helices of the stalk between the α- and β-registries is thought to provide a mechanism for communicating between the AAA ring and the MTBD.
The third appendage of the ring is a predominantly helical “linker” domain that arches over one face of the ring. The N terminus of the linker domain extends into the cargo-binding tail region, while its C-terminal region continues into AAA1, the main ATPase site of the dynein ring. The linker is hypothesized to serve as a mechanical element. This hypothesis originally arose from 2D negative stain EM data of axonemal dynein [36] and later EM studies of cytoplasmic dynein [37]. In the ADP-vanadate trapped state, the linker is bent, and its N terminus is positioned at AAA2/3; in the ADP state, the linker extends straight across the ring towards AAA4/5 (Figure 1). The bent-to-straight “linker swing” was proposed to act as a power stroke, although its roles might be more complex as will be discussed later.
Nucleotide-induced conformational changes
The conformational changes in different nucleotide states are described in detail in a recent review that compares crystal structures of the dynein motor domain bound to ADP, AMPPNP, ADP-vanadate or no nucleotide (“apo”, in which all nucleotide binding sites are empty except AAA2, which constitutively binds nucleotide) [29]. In brief, structural studies suggest that ATP binding at AAA1 triggers a series of conformational changes that propagate around the entire ring and reposition all of the AAA domains into a “closed” ring conformation (Figure 1). Additionally, the linker detaches from its docking site at AAA5, where it resides in the “apo” state. In the ATP-bound state, the linker can adopt a bent conformation, in which it contacts AAA3/2 (referred to in the dynein literature as the “pre-power stroke state”) [38]. In addition, it can although can explore various other conformations [28]. One important consequence of the AAA ring adopting a “closed” conformation is that AAA4 and AAA5 move relative to one another. This movement results in the buttress “pulling” on one of the helices of the stalk to which it is attached, thus shifting the coiled-coil to its β+ registry and inducing a weak binding state of the MTBD [29]. ATP hydrolysis and/or release of phosphate at AAA1 then allows the MTBD to return to a strong binding state (α-registry of the stalk), the linker to return to its straight conformation (“post-power stroke state”), and the AAA ring to adopt its “open” conformation.
Allosteric Roles for AAA3 and the linker
The nucleotide state of AAA3 plays a regulatory role in modulating the allosteric changes in the motor domain. When ADP is bound in AAA3, large conformational changes in the entire ring (AAA1–6) and linker occur, as described above. However, when ATP (modeled by AMPPNP) is bound in AAA3, only conformational changes in AAA1–4 are observed. In this case the linker remains straight and the MTBD remains locked in a tight binding state [28, 39, 40]. Consistent with the idea that the linker bending occurs when AAA3 is bound to ADP, a crystal structure of human dynein 2 in the presence of ADP-vanadate captures a bent conformation of the linker, and reveals electron density for ADP-vanadate for AAA1 but only ADP in AAA3 [29]. Single molecule studies agree with this structural data, showing that ATP binding at AAA3 results in strong microtubule binding, whereas ADP binding at AAA3 weakens the binding to microtubules [39]. However, when force is applied at the N-terminus of a linker, under high load, the binding and release from microtubules is independent of the nucleotide bound at AAA3 [40]. A more detailed understanding of how this AAA3-based regulatory mechanism is used to control dynein motility under low and high load in a cell remains to be elucidated.
The linker conformational change from straight to bent is likely key for promoting ATP hydrolysis in AAA1. In its straight conformation, the linker sterically inhibits the catalytic arginine finger in AAA2 from reaching its optimal position to promote hydrolysis of ATP in AAA1 [27, 28]. Linker bending removes this steric block to AAA1-AAA2 closure [29], allowing positioning of the arginine finger optimally for catalysis, and thus may facilitate ATP hydrolysis [28]. Conversely, docking of the N-terminal end of the straight linker onto AAA5, as in the “apo” state, may facilitate the opening of AAA1-AAA2 and the release of ADP to complete the ATPase cycle (Figure 1) [26]. The nucleotide-dependent allosteric changes of both the linker and AAA3 are important in regulating dynein’s chemo-mechanical cycle. The linker conformational change facilitates ATP hydrolysis at AAA1, the main site of hydrolysis, and AAA3 gates conformational changes around the ring and thus impacts binding of dynein to microtubules.
Comparison of Dynein with Kinesin and Myosin
In order to understand the mechanism of dynein, it is useful to compare and contrast dynein with the two best-studied processive motors- kinesin-1 and myosin V. Although they operate on different tracks (microtubules for kinesin-1; actin for myosin V), these motors use a similar mechanism for converting ATP energy into the hand-over-hand movement of their two identical motor domains along a filament (Box 1). First, while bound to the polymer, both motors use a large conformational change of a mechanical element (lever arm for myosin V and neck linker for kinesin-1) to displace a detached, partner motor domain, thereby biasing its reattachment to a forward binding site after a Brownian search. Second, as a result of having different orientations in the mechanical elements in the two-head-bound intermediate, the transition to the strong-binding state is favored in only one of the two motor domains (Box 1). This asymmetry helps to coordinate the chemical cycles of the two motor domains and helps to ensure that the motor domain, once detached, steps forward rather than revisits its last binding site. As will be discussed later, dynein may exploit similar principles in how it moves towards the microtubule minus end. However, as discussed next, dynein also differs from kinesin-1 and myosin V in several important ways, and these differences may provide clues into its mechanism. We focus our discussion on results from yeast dynein, which is the most extensively studied dynein motor to date and where there appears to be an emerging consensus with regard to its single molecule behavior. We note that mammalian dynein has not been studied in as much detail as yeast dynein, and may exhibit a different stepping behavior [41–43].
The two motor domains of dynein operate largely independently during motility
Single molecule fluorescence imaging of the individual motor domains of kinesin-1 and myosin V reveals an invariant hand-over-hand movement in which the rear head always passes the front head (Box 1). By contrast, imaging studies of the two dynein heads reveal that they step largely independently of one another, which gives rise to diverse types of steps [44, 45]. Like kinesin-1 and myosin V, the rear head of dynein can step past the front head, but these hand-over-hand steps account for less than 20% of the steps. In addition, dynein displays an inch-worm type of motion, which is never observed for kinesin-1 or myosin-V. Here, the front head can detach and then step forward, while the rear head remains fixed in position. The rear head also can take a step without necessarily passing the front head. Furthermore, the order of stepping of the two dynein heads appears to be largely stochastic; in some cases, one motor domain can step two or three times in a row before its partner head steps. This stepping pattern is very different from kinesin-1 and myosin V, which display a strict alternating pattern in which one motor domain steps and then the other. An exception to this stochastic stepping occurs when the two dynein motor domains become widely separated on the microtubule lattice. In this scenario, the probability of forward head stepping decreases, and either the rear head steps forward or the front head steps backward, both of which reduce the linker-mediated tension between the motor domains.
Experiments by the Yildiz laboratory with recombinant dynein heterodimers further underscore the conclusion that an individual dynein motor domain acts as an autonomous stepper. In these experiments, a wildtype dynein motor domain was heterodimerized either with a “dead” motor domain bearing a crippling mutation in the AAA1 active site [44] or with only a “crutch” consisting of the dynein coiled-coil stalk and MTBD without the ring [46]. Remarkably, both types of active-inactive heterodimeric motors step at close to wildtype velocities along the microtubule. The active motor is usually (~75%) in the lead, and its movement forces its dysfunctional partner also to step along the microtubule. This finding contrasts results obtained from a comparable kinesin wildtype plus hydrolysis-dead heterodimer, which moves 10–20-fold slower than native homodimeric kinesin [47]. These experiments indicate that one dynein head can step along the microtubule very effectively, provided that it has a “tether” (a partner motor domain or even a microtubule-binding “crutch”) that prevents it from diffusing away from the microtubule surface when it is searching for a new tubulin binding site after releasing from the microtubule. Thus, the walking mechanism of dynein does not require communication between two actively cycling ATPase motor domains, as is true of kinesin-1 and myosin V.
Dynein exhibits a weak directional bias and variable step sizes
Kinesin-1, myosin V, and yeast cytoplasmic dynein display comparable processivity, moving at ~1–2 microns per encounter with the track. During such an excursion under zero load, kinesin-1 and myosin-V very rarely take backwards steps. By contrast, dynein frequently (20% of the time) takes backward steps [21, 44, 45]. This finding indicates that the mechanisms that dynein uses to produce directional bias are less efficient than those of kinesin-1 or myosin V.
Single molecule data show that the step sizes of yeast dynein (measured by marking the position of the motor domain ring and not the MTBD) are highly variable: 8 to 32 nm in both the forward and backward directions, with an average of 10–12 nm [21, 44, 45, 48]. Sideways stepping to adjacent protofilaments is also common. These results suggest that the detached dynein motor domain can access many possible tubulin binding sites on the lattice. By contrast, kinesin only takes 8.2 nm steps [49] (the size of the tubulin dimer), and the step sizes of myosin V are close to 36 nm [50], which is the helical pitch of the actin filament.
The main catalytic site of dynein is spatially distant from the microtubule-binding domain
In kinesin and myosin, the polymer-binding and catalytic sites are integrated within the same domain. Thus, during a step, the polymer-binding and catalytic sites advance by the same distance. By contrast, the microtubule-binding and catalytic AAA ring of dynein are separated by a long (~140 Å). The main role of the large AAA ring is to displace the small (~15 kD) MTBD along the microtubule towards the minus end. The MTBD of dynein must move by a unitary distance corresponding to the spacing of tubulin binding sites; however, because the stalk is flexible and can potentially bend or alter its angle with respect to the ring, the ring itself may not undergo the same net displacement as the MTBD. The length of the stalk coiled-coil is highly conserved throughout the dynein superfamily. Thus, the separation between the microtubule and catalytic sites is likely to be an important feature in the dynein mechanism.
How Dynein Steps Towards the Microtubule Minus End
How is dynein stepping biased towards the minus end of a microtubule? The answer to this central question is not entirely clear, but here we synthesize data and models from the literature, and discuss how several mechanisms might synergize with one another to promote the weak directional preference of dynein.
How does the leading head step towards the minus end?
Figure 2 shows a model in which the leading motor domain of dynein takes a step forward to a new tubulin binding site, while the lagging head remains bound in place. As described earlier, neither kinesin-1 nor myosin V can execute this type of step. Two potential mechanisms might enable the front motor domain of dynein to step without any active assistance from the lagging head. It is also important to keep in mind that these biasing mechanisms are relatively weak, because the leading head frequently takes backward as well as forward steps [44, 45] (Figure 2).
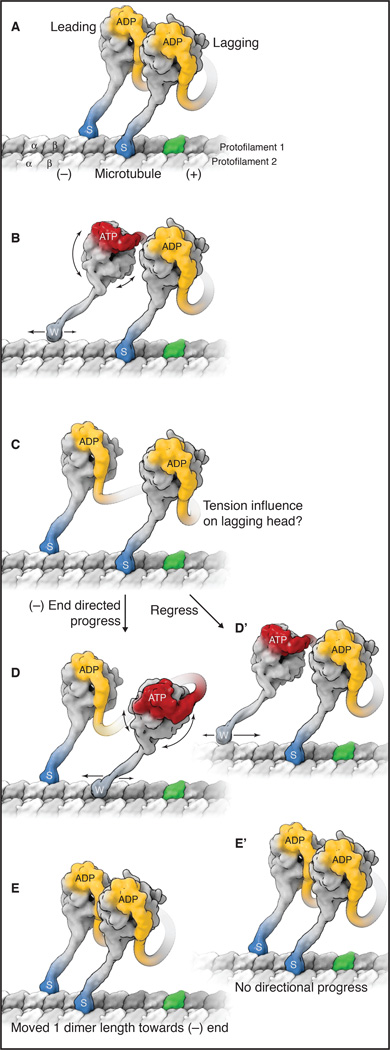
Figure 2.
A model for dynein stepping along a microtubule. See also Movie S1. A) The sequence starts with a cytoplasmic dynein dimer with ADP bound in both motor domains (yellow) and both microtubule-binding domains (MTBDs) tightly bound to the microtubule, in the strong-binding state (indicated by “s” and darker blue color). One tubulin dimer in the microtubule is highlighted in green to allow easier identification of dynein progression towards the minus end. B) When ADP is exchanged for ATP in the leading head (red), conformational changes occur in the ring, which are communicated to the MTBD via a change in registry of the coiled-coil stalk, thus releasing the MTBD from the microtubule. This sequence triggers the reposition of the N terminus of the linker over AAA2/3 and a resultant rotation of the ring/MTBD and a slight forward re-positioning of the MTBD. The weakly bound motor domain (indicated by “w” and lighter blue color) is also subject to Brownian motion, which causes a two-dimensional search of the MTBD on top of the microtubule lattice. The weak-to-strong binding transition is favored with the stalk pointing backwards, and this locks a forward step in place by the front head (see also Figure 3). This asymmetry in the binding properties of the MTBD provides an additional proof-reading mechanism that favors a forward versus a backward step. C) This strong rebinding triggers the movement of the linker from AAA2/3 to AAA4/5. This minus-end directed shift of the N terminus of the linker reduces the slack in the connection between the motor domains and applies tension to the lagging motor domain. At this point, two potential subsequent steps are shown (D,E and D',E'). D and E) The rear head now detaches after ADP/ATP exchange and undergoes the same sequence of events described for the front head, again resulting in a step forward towards the front head and relieving the tension between the two motor domains. D’ and E’) The leading motor domain undergoes ADP/ATP exchange and detaches, but tension between the motor domains overrides the forward biasing mechanisms and the leading MTBD steps backward. In this instance, the homodimer does not undergo a net displacement from A to E’.
Repositioning of the stalk/MTBD by a rotation of the AAA ring
Several studies [25–27, 38, 51] have shown that the N-terminus of the linker is positioned primarily over AAA4/AAA5 in ADP/apo states and over AAA2/3 in ATP/ADP-Pi states of AAA1 (note: other positions of the linker between AAA2–4 are also observed with ATP or ATP analogues [28, 38]). How does this conformational change impact movement on the microtubule? The answer will likely depend upon whether dynein is strongly-bound or detached from the microtubule. If strongly-bound, then the ring will be held in place by the docked MTBD/stalk and the linker will be “swing” relative to the microtubule, analogous to the power stroke of myosin (Box 1). However, if detached from the microtubule in the ATP state, then the ring can potentially rotate relative to the microtubule to allow AAA2/3 to make contact with the N-terminal domains of the linker [37] (Figure 2). Indeed, “ring rotation” has been recently observed in EM reconstructions for in situ axonemal dynein in an ATP-like state, while the linker was more or less stationary perhaps due to connections with the tail domain or with the partner head [52]. Importantly, the angular change of the stalk associated with ring rotation would shift the position of the MTBD further towards the microtubule minus end, and thereby bias its rebinding to a new tubulin site towards the minus end (Figure 2). Because of the 140 Å length of the stalk, a small rotation of the ring would lead to a much larger displacement of the MTBD. This type of biasing mechanism could explain how a dynein motor domain can step without a functional partner motor domain [46].
Brownian motion and rebinding of the MTBD
To further bias the directionality of dynein, the ring rotation described above may be augmented by an inherent asymmetry in the microtubule rebinding by the MTBD during a Brownian search. A mechanism of a random diffusional search combined with the asymmetric binding of a motor to a polymer was first suggested by Huxley in 1957 as a model for muscle contraction (Figure 3). Similar to this model, dynein has been shown to undergo one-dimensional diffusion along the microtubule surface in its weakly bound (ATP) state. This result suggests that the MTBD can hold on to the microtubule weakly and that thermal energy can displace the MTBD from one tubulin subunit to the next along the microtubule [35, 53, 54]. Furthermore, optical trap studies have uncovered a directional asymmetry in the binding of dynein to the microtubules. Specifically, when dynein is pulled backward, it holds on more tightly to the MT compared to when it is pulled forward [40, 46, 55]. The angle between the MTBD and the stalk, where there appears to be a flexible joint [56, 57], may play a role in governing this binding asymmetry. Based upon these results and others [32], we speculate that the detached MTBD, exploring space by thermal motion, will more likely undergo a weak-to-strong binding transition if it reaches a forward tubulin and has a preferred geometry of its stalk pointing backward (Figure 3).
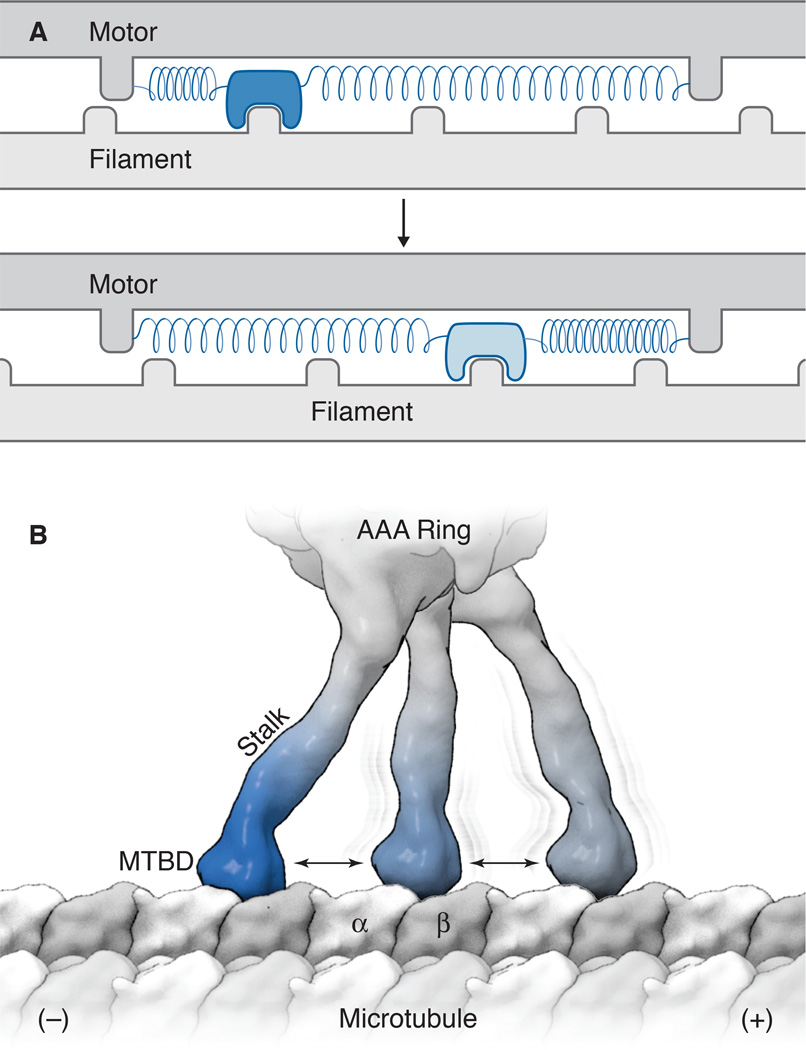
Figure 3.
Asymmetry in polymer binding can create a directional bias. A) A depiction of a model by Andrew Huxley for muscle contraction [69]. In this model, which was developed before the myosin structure was known, the motor contains a small actin-binding element linked to a core by a spring-like element, which moves back-and-forth by thermal motion relative to the actin filament. The binding of this fluctuating element to the actin filament increases when displaced by thermal fluctuations in one direction (top panel, dark blue reveals a state with a high association rate). Conversely, its association rate is lower (bottom panel, light blue) when stretched in the opposite direction. Binding to the filament occurs spontaneously, but ATP energy is needed to dissociate the actomyosin bond, thus creating a mechanism that drives multiple force-generating cycles. B) A model for the binding of the MTBD to the microtubule. The stalk/MTBD can explore different binding sites by Brownian motion; the weak-to-strong transition of the MTBD (dark blue) is enhanced when displaced towards the minus end (left in this figure) but less likely when interacting with other binding sites (lighter blue).
How does the lagging head step towards the minus end?
Each dynein motor domain can step in an independent and autonomous manner. Thus, the lagging motor domain can detach and bias its forward motion using the same mechanism described for the leading motor domain. However, the forward movement of the lagging motor domain might also be aided through tension created by the leading head. How is this tension generated? If the leading head succeeds in taking several successive forward steps, then the inter-head distance increases and the connection between the two heads becomes increasingly taut. This tension appears to increase the probability of the lagging head taking a forward step [44, 48]. In addition, the bent-to-straight conformational change of the linker in the strongly bound leading head also would be expected to increase tension and thus potentially pull the lagging motor domain towards the microtubule minus end [6] (Figure 2). However, the degree to which the lagging head “feels” the tension from the linker conformational change and shifts its position towards the minus end depends upon the compliance of the elements holding the two heads together, which is still poorly understood for native dynein. Unlike kinesin-1 and myosin V, the dynein dimerization domain is located near the N terminus of the polypeptide chain and thus is very distal from the motor domains [23, 58].
Dynein stepping: an animation
To illustrate how dynein’s structural properties and conformational changes described above might lead to the complex stepping behavior of yeast dynein, we have generated an animation based upon the crystal structures of dynein in different nucleotide states and its single molecule stepping behavior. Specifically, this animation highlights the following features of dynein stepping, most of which have been described earlier in this review: 1) The “inch-worm” step (the leading head takes a step followed by the lagging head. 2) An occasional “hand-over-hand” step (20% of the time according to Qiu et al. [45]). 3) Both alternating and consecutive steps of the two dynein motor domains. 4) Backwards steps (approximately 20% of the steps for yeast dynein. 5). Different sized steps (8–32 nm for the AAA ring). 6) Switching of a motor domain to an adjacent protofilament track [21]. 7) An inherent asymmetry in positioning of the motor domains, such that the leading motor domain is most often positioned to the right of lagging motor domain, when viewed down the axis of the microtubule towards the minus end [44, 45]. The animation also features the conformational changes of the linker and MTBD as well as the Brownian motion of the entire motor and its subdomains, and illustrates how such dynamics might enable the motor to step along the microtubule.
In generating this animation, gaps in information or understanding also become more apparent; indeed, many aspects of this animated model are speculative. First, the structure and compliance of the element connecting the two motor domains is not known, and its modeling in the animation is somewhat arbitrary. These details are important for understanding how the two motors domains might communicate with one another through tension. However, unlike kinesin-1, the dynein dimer is able to move processively with diverse structural elements connecting the two motors domains, including a short connection to an anti-parallel GST dimer [21, 24], DNA-based dimerization [59], or a longer flexible linker, as exists in native dynein [58]. We currently do not understand how dynein can accommodate such diverse connections between its two motor domains and the role of this region in the native dynein molecule. Second, defining hinge points that allow relative motions of dynein subdomains is important in generating the animation; these frames of reference and relative motion impact the degree of ring rotation (leading head stepping), how the MTBD can undergo a Brownian search along the microtubule lattice, and how one motor domain can pass its partner. While some information is known, many of these details are educated guesses at the moment. Third, the relative contributions to dynein stepping of linker conformational changes, asymmetric binding of the MTBD, and perhaps other biasing mechanisms are not well-defined at present. Fourth, while we have “snap shots” of dynein in different nucleotide states, the temporal sequence of conformational changes within this large motor is not known. For example, the exact order of events of ATP binding to the motor, microtubule release, and the linker bending remains speculative. Fifth, the conformational states of the AAA ring are derived from crystal structures, and we do not know how microtubule binding might impact these conformational states. Sixth, current information on stepping is based upon high resolution tracking data from a fluorescent a dye in the AAA ring, not in the MTBD. As discussed earlier, the positions of the ring and the MTBD may differ due to the flexibility of the stalk and rotations of the ring.
Concluding Remarks
Recent crystal and EM structures have focused the dynein motor domain and have illuminated how transitions in the nucleotide state of AAA1 and AAA3 in the asymmetric, hexameric AAA ring trigger allosteric changes over long distances in the motor domain (reviewed in Schmidt et al. [60]). Single molecule and mutagenesis studies also have provided exquisite details on the stepping behavior of yeast dynein and how it differs from kinesin-1 and myosin V. Despite this recent deluge of information, the structural basis of unidirectional motion of dynein remains incompletely understood. Thus the challenge remains to develop more direct evidence for or against the models discussed here. Part of this challenge will be in understanding the dynamics of the dynein molecule. We have snapshots of dynein from X-ray crystallography, but dynein is likely to be a very dynamic protein machine that explores many conformations. To help to understand such dynamics, new efforts will be needed to develop conformation-sensitive probes so that changes in the structure of dynein can be observed in real-time and ascertain how these changes relate to specific steps that dynein takes on a microtubule. It also will be important to obtain higher resolution structures of the entire motor domain when it is in microtubule bound states, particularly in the dynein dimer. In addition, new advances in cryo-EM allow identification of different conformational states even within a single nucleotide-bound chemical state; such conformational heterogeneity will provide information on the dynamics of the motor domain. Finally, future single molecule studies of mammalian dynein-dynactin and axonemal dynein will provide a broader understanding of dynein stepping and extend the current extensive work on yeast dynein. Collectively, such experimental information will help to provide a clearer understanding of the dynein mechanism.
Supplementary Material
Outstanding Questions Box.
-
-
How does dynein move toward the microtubule minus end? We discuss several ideas in this review, but are they correct? What are the relative contributions of the linker versus the microtubule-binding domain in directional motion?
-
-
How are the two dynein motor domains connected to one another in a homodimer? Can a linker conformation in one dynein motor domain increase tension and bias the position of the partner motor domain?
-
-
Why does dynein use a long coiled coil to separate the polymer-binding domain from the ATPase ring? Why is its length so highly conserved?
-
-
Does dynein use dynactin or other associated proteins to improve the coupling between the two motor domains or change the efficiency of forward stepping?
Trends Box.
-
-
The architecture and nucleotide-dependent conformational changes of the dynein motor domain were recently resolved in several crystal and cryo-EM structures.
-
-
Dynein displays conspicuous differences from kinesin and myosin, including the independent stepping behavior of the two motor domains in the homodimer, its much weaker directional bias, and the long separation between the polymer-binding domain and the catalytic body of the enzyme.
-
-
The dynein linker domain plays an important role in the mechanics of movement but also regulates specific transitions in the ATPase cycle.
-
-
Dynein may use several mechanisms to bias its movement towards the minus end, including conformational changes of its linker domain, Brownian search and an asymmetric binding mechanism of its microtubule-binding domain.
Acknowledgments
We are grateful to members of the Vale lab for their comments on this review. Our work on dynein is funded by the NIH (R01GM09731 to RDV and a K99 to GB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Allan VJ. Cytoplasmic dynein. Biochem. Soc. Trans. 2011;39:1169–1178. doi: 10.1042/BST0391169. [DOI] [PubMed] [Google Scholar]
- 2.Verhey KJ, et al. Kinesin assembly and movement in cells. Annu. Rev. Biophys. 2011;40:267–288. doi: 10.1146/annurev-biophys-042910-155310. [DOI] [PubMed] [Google Scholar]
- 3.Block SM. Kinesin motor mechanics: binding, stepping, tracking, gating, and limping. Biophys. J. 2007;92:2986–2995. doi: 10.1529/biophysj.106.100677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vale RD, Milligan RA. The way things move: looking under the hood of molecular motor proteins. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- 5.Hammer JA, Sellers JR. Walking to work: roles for class V myosins as cargo transporters. Nat. Rev. Mol. Cell Biol. 2012;13:13–26. doi: 10.1038/nrm3248. [DOI] [PubMed] [Google Scholar]
- 6.Roberts AJ, et al. Functions and mechanics of dynein motor proteins. Nat. Rev. Mol. Cell Biol. 2013;14:713–726. doi: 10.1038/nrm3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raaijmakers JA, Medema RH. Function and regulation of dynein in mitotic chromosome segregation. Chromosoma. 2014;123:407–422. doi: 10.1007/s00412-014-0468-7. [DOI] [PubMed] [Google Scholar]
- 8.Tanenbaum ME, et al. Cytoplasmic dynein crosslinks and slides anti-parallel microtubules using its two motor domains. eLife. 2013;2:e00943. doi: 10.7554/eLife.00943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heald R, et al. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- 10.Rusan NM, et al. Reorganization of the microtubule array in prophase/prometaphase requires cytoplasmic dynein-dependent microtubule transport. J. Cell Biol. 2002;158:997–1003. doi: 10.1083/jcb.200204109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howell BJ, et al. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 2001;155:1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varma D, et al. Direct role of dynein motor in stable kinetochore-microtubule attachment, orientation, and alignment. J. Cell Biol. 2008;182:1045–1054. doi: 10.1083/jcb.200710106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carminati JL, Stearns T. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niedner A, et al. Of social molecules: The interactive assembly of ASH1 mRNA-transport complexes in yeast. RNA Biol. 2014;11:998–1009. doi: 10.4161/rna.29946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai J-W, et al. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat. Neurosci. 2007;10:970–979. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- 16.Burke B, Roux KJ. Nuclei take a position: managing nuclear location. Dev. Cell. 2009;17:587–597. doi: 10.1016/j.devcel.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 17.Wynshaw-Boris A, et al. Lissencephaly: mechanistic insights from animal models and potential therapeutic strategies. Semin. Cell Dev. Biol. 2010;21:823–830. doi: 10.1016/j.semcdb.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schroer TA. Dynactin. Annu. Rev. Cell Dev. Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- 19.McKenney RJ, et al. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science. 2014;345:337–341. doi: 10.1126/science.1254198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlager MA, et al. In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J. 2014;33:1855–1868. doi: 10.15252/embj.201488792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reck-Peterson SL, et al. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toropova K, et al. Lis1 regulates dynein by sterically blocking its mechanochemical cycle. eLife. 2014;3:e03372. doi: 10.7554/eLife.03372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Urnavicius L, et al. The structure of the dynactin complex and its interaction with dynein. Science. 2015;347:1441–1446. doi: 10.1126/science.aaa4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carter AP, et al. Crystal structure of the dynein motor domain. Science. 2011;331:1159–1165. doi: 10.1126/science.1202393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kon T, et al. X-ray structure of a functional full-length dynein motor domain. Nat. Struct. Mol. Biol. 2011;18:638–642. doi: 10.1038/nsmb.2074. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt H, et al. Insights into dynein motor domain function from a 3.3-\AA crystal structure. Nat. Struct. Mol. Biol. 2012;19 doi: 10.1038/nsmb.2272. 492-7-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kon T, et al. The 2.8 \AA crystal structure of the dynein motor domain. Nature. 2012;484:345–350. doi: 10.1038/nature10955. [DOI] [PubMed] [Google Scholar]
- 28.Bhabha G, et al. Allosteric communication in the dynein motor domain. Cell. 2014;159:857–868. doi: 10.1016/j.cell.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt H, et al. Structure of human cytoplasmic dynein-2 primed for its power stroke. Nature. 2015;518:435–438. doi: 10.1038/nature14023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 31.Gibbons IR, et al. The affinity of the dynein microtubule-binding domain is modulated by the conformation of its coiled-coil stalk. J. Biol. Chem. 2005;280:23960–23965. doi: 10.1074/jbc.M501636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carter AP, et al. Structure and functional role of dynein’s microtubule-binding domain. Science. 2008;322:1691–1695. doi: 10.1126/science.1164424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kon T, et al. Helix sliding in the stalk coiled coil of dynein couples ATPase and microtubule binding. Nat. Struct. Mol. Biol. 2009;16:325–333. doi: 10.1038/nsmb.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Redwine WB, et al. Structural basis for microtubule binding and release by dynein. Science. 2012;337:1532–1536. doi: 10.1126/science.1224151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uchimura S, et al. A flipped ion pair at the dynein-microtubule interface is critical for dynein motility and ATPase activation. J. Cell Biol. 2015;208:211–222. doi: 10.1083/jcb.201407039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burgess SA, et al. Dynein structure and power stroke. Nature. 2003;421:715–718. doi: 10.1038/nature01377. [DOI] [PubMed] [Google Scholar]
- 37.Roberts AJ, et al. AAA+ Ring and linker swing mechanism in the dynein motor. Cell. 2009;136:485–495. doi: 10.1016/j.cell.2008.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roberts AJ, et al. ATP-Driven Remodeling of the Linker Domain in the Dynein Motor. Structure. 2012 doi: 10.1016/j.str.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeWitt MA, et al. The AAA3 domain of cytoplasmic dynein acts as a switch to facilitate microtubule release. Nat. Struct. Mol. Biol. 2015;22:73–80. doi: 10.1038/nsmb.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nicholas MP, et al. Cytoplasmic dynein regulates its attachment to microtubules via nucleotide state-switched mechanosensing at multiple AAA domains. Proc. Natl. Acad. Sci. U. S. A. 2015;112:6371–6376. doi: 10.1073/pnas.1417422112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mallik R, et al. Cytoplasmic dynein functions as a gear in response to load. Nature. 2004;427:649–652. doi: 10.1038/nature02293. [DOI] [PubMed] [Google Scholar]
- 42.Toba S, et al. Overlapping hand-over-hand mechanism of single molecular motility of cytoplasmic dynein. Proc. Natl. Acad. Sci. 2006;103:5741–5745. doi: 10.1073/pnas.0508511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nicholas MP, et al. Control of cytoplasmic dynein force production and processivity by its C-terminal domain. Nat. Commun. 2015;6:6206. doi: 10.1038/ncomms7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeWitt MA, et al. Cytoplasmic dynein moves through uncoordinated stepping of the AAA+ ring domains. Science. 2012;335:221–225. doi: 10.1126/science.1215804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qiu W, et al. Dynein achieves processive motion using both stochastic and coordinated stepping. Nat. Struct. Mol. Biol. 2012;19:193–200. doi: 10.1038/nsmb.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cleary FB, et al. Tension on the linker gates the ATP-dependent release of dynein from microtubules. Nat. Commun. 2014;5:4587. doi: 10.1038/ncomms5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thoresen T, Gelles J. Processive movement by a kinesin heterodimer with an inactivating mutation in one head. Biochemistry (Mosc.) 2008;47:9514–9521. doi: 10.1021/bi800747e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belyy V, et al. Cytoplasmic dynein transports cargos via load-sharing between the heads. Nat. Commun. 2014;5:5544. doi: 10.1038/ncomms6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Visscher K, et al. Single kinesin molecules studied with a molecular force clamp. Nature. 1999;400:184–189. doi: 10.1038/22146. [DOI] [PubMed] [Google Scholar]
- 50.Mehta AD, et al. Myosin-V is a processive actin-based motor. Nature. 1999;400:590–593. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- 51.Burgess SA, et al. Dynein structure and power stroke. Nature. 2003;421:715–718. doi: 10.1038/nature01377. [DOI] [PubMed] [Google Scholar]
- 52.Lin J, et al. Structural mechanism of the dynein power stroke. Nat. Cell Biol. 2014;16:479–485. doi: 10.1038/ncb2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vale RD, et al. One-dimensional diffusion of microtubules bound to flagellar dynein. Cell. 1989;59:915–925. doi: 10.1016/0092-8674(89)90614-4. [DOI] [PubMed] [Google Scholar]
- 54.Ross JL, et al. Processive bidirectional motion of dynein-dynactin complexes in vitro. Nat. Cell Biol. 2006;8:562–570. doi: 10.1038/ncb1421. [DOI] [PubMed] [Google Scholar]
- 55.Gennerich A, et al. Force-induced bidirectional stepping of cytoplasmic dynein. Cell. 2007;131:952–965. doi: 10.1016/j.cell.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Imai H, et al. Direct observation shows superposition and large scale flexibility within cytoplasmic dynein motors moving along microtubules. Nat. Commun. 2015;6:8179. doi: 10.1038/ncomms9179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nishikawa Y, et al. Structure of the Entire Stalk Region of the Dynein Motor Domain. J. Mol. Biol. 2014;426:3232–3245. doi: 10.1016/j.jmb.2014.06.023. [DOI] [PubMed] [Google Scholar]
- 58.Chowdhury S, et al. Structural organization of the dynein-dynactin complex bound to microtubules. Nat. Struct. Mol. Biol. 2015;22:345–347. doi: 10.1038/nsmb.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Derr ND, et al. Tug-of-war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science. 2012;338:662–665. doi: 10.1126/science.1226734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmidt H. Dynein motors: How AAA+ ring opening and closing coordinates microtubule binding and linker movement. BioEssays. 2015;37:532–543. doi: 10.1002/bies.201400215. [DOI] [PubMed] [Google Scholar]
- 61.Sakamoto T, et al. Direct observation of the mechanochemical coupling in myosin Va during processive movement. Nature. 2008;455:128–132. doi: 10.1038/nature07188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Visscher K, et al. Single kinesin molecules studied with a molecular force clamp. Nature. 1999;400:184–189. doi: 10.1038/22146. [DOI] [PubMed] [Google Scholar]
- 63.Rice S, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 64.Fujita K, et al. Switching of myosin-V motion between the lever-arm swing and brownian search-and-catch. Nat. Commun. 2012;3:956. doi: 10.1038/ncomms1934. [DOI] [PubMed] [Google Scholar]
- 65.Hackney DD. Evidence for alternating head catalysis by kinesin during microtubule-stimulated ATP hydrolysis. Proc. Natl. Acad. Sci. U. S. A. 1994;91:6865–6869. doi: 10.1073/pnas.91.15.6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hackney DD. The tethered motor domain of a kinesin-microtubule complex catalyzes reversible synthesis of bound ATP. Proc. Natl. Acad. Sci. U. S. A. 2005;102:18338–18343. doi: 10.1073/pnas.0505288102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Purcell TJ, et al. A force-dependent state controls the coordination of processive myosin V. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13873–13878. doi: 10.1073/pnas.0506441102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Johnson GT, et al. ePMV embeds molecular modeling into professional animation software environments. Struct. Lond. Engl. 1993. 2011;19:293–303. doi: 10.1016/j.str.2010.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huxley AF. Muscle structure and theories of contraction. Prog. Biophys. Biophys. Chem. 1957;7:255–318. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.