Abstract
Exosomes are biological nanovesicles that are involved in cell-cell communication via the functionally-active cargo (such as miRNA, mRNA, DNA and proteins). Because of their nanosize, exosomes are explored as nanodevices for the development of new therapeutic applications. However, bulk, safe and cost-effective production of exosomes is not available. Here, we show that bovine milk can serve as a scalable source of exosomes that can act as a carrier for chemotherapeutic/chemopreventive agents. Drug-loaded exosomes showed significantly higher efficacy compared to free drug in cell culture studies and against lung tumor xenografts in vivo. Moreover, tumor targeting ligands such as folate increased cancer-cell targeting of the exosomes resulting in enhanced tumor reduction. Milk exosomes exhibited cross-species tolerance with no adverse immune and inflammatory response. Thus, we show the versatility of milk exosomes with respect to the cargo it can carry and ability to achieve tumor targetability. This is the first report to identify a biocompatible and cost-effective means of exosomes to enhance oral bioavailability, improve efficacy and safety of drugs.
Keywords: Milk-derived exosomes, drug delivery, tumor-targeting, chemotherapeutic drugs, chemopreventive agents
1 Introduction
Over the last three decades, a number of nanoparticle-delivery systems have been developed for cancer therapy, including natural and synthetic polymer-based, lipid-based, and organic and inorganic materials [1, 2]. However, due to inherent limitations, only a handful of them have been studied in clinics. The encapsulation of doxorubicin in liposomes (DoxilR) and paclitaxel in protein-based nanoparticles (AbraxaneR) represent two of the successful applications [3-5]. Factors that have stalled the clinical introduction of other nanoparticles include high cost, difficulty in reproducibly synthesizing them in sufficient quantities, and/or toxicity issues [2, 6]. The development of ideal nanoparticles with attributes such as long circulation time, evasion of the host immune system, and ability to target specific cells, minimum off site toxicity, and ability to carry versatile therapeutics remains elusive [7, 8].
Nature-derived nanoparticles could potentially overcome some of the limitations of synthetic liposomes. Among the different secreted membrane vesicles, exosomes intrinsically possess many attributes of a drug delivery vehicle [9, 10], such as they: i) are well tolerated in the body, as evidenced by their wide distribution in various biological fluids (including milk) [11-13], ii) have longer circulating half-life, iii) are internalized by other cells, iv) carry small molecules and cargoes (such as miRNA, mRNA, and DNA), that make these vesicles as delivery vehicles of therapeutics [14-16], and v) are amenable to ligand attachment for tumor targetability.
Although the field of exosome-based therapeutics is in its infancy, the ability to engineer exosomes to display proteins, incorporate specific nucleic acid and protein cargos, load therapeutic agents, its targeted uptake and tolerance in vivo have been demonstrated to some extent [14, 15]. However, before exosomes are accepted as a delivery vehicle in clinics, the development of biocompatible, economically-viable source and methods for harvesting exosomes, which are effective and well-tolerated in vivo, must be demonstrated.
We report here the suitability of bovine milk as a potentially scalable source of exosomes that could serve as a drug delivery vehicle. Bovine milk consumption is generally considered to be safe and to provide important nutritional benefits [17]. Thus, availability, cost and toxicity considerations make bovine milk a suitable natural source for large-scale production of exosomes. We demonstrate that milk-derived exosomes can serve as a vehicle to deliver both hydrophilic and lipophilic small molecules, including chemotherapeutic (chemo) drugs. Using in vitro and in vivo models, we show enhanced biological efficacy of the exosomal formulations. This effect was further increased by the addition of tumor-targeting ligand, folic acid (FA). Therefore, milk exosomes represent a potentially scalable, biocompatible and cost-effective means to potentially enhance oral bioavailability, improve efficacy and safety of drugs.
2 Materials and Methods
2.1 Isolation of exosomes
Milk from pasture-fed Holstein and Jersey cows during the mid-lactation period was obtained from a local dairy; colostrum was from 2-3 days postpartum. Exosomes were isolated by differential centrifugation. Briefly, milk was centrifuged at 13,000×g in 250 ml centrifuge bottles (Nalgene, Thermofisher Scientific, Holtsville, NY) using TA-10.250 rotor and Allegra 25R centrifuge (Beckman Coulter, Brea, California) at 4 °C for 30 min to remove fat globules, casein aggregates and other debris. The whey was collected by passing through a cheese cloth and subsequently transferred into 70 ml polycarbonate tubes and centrifuged at 100,000×g in Type 45 Ti fixed angle rotor using Optima LE-80K Ultracentrifuge (Beckman Coulter, Brea, California) at 4 °C for 60 min to remove large particles and microvesicles. Forty-five ml of the supernatant was carefully removed from the top and the lower slush portion along with pellet was discarded. This supernatant (70 ml/tube) was finally centrifuged at 135,000×g for 90 min at 4 °C in a Type 45 Ti fixed angle rotor using Optima LE-80K Ultracentrifuge. The supernatant was discarded and the exosome pellet thus obtained was washed thrice with PBS. The exosome pellets were pooled and re-suspended in PBS to give homogenous suspension and filtered through 0.22 μm for sterilization. The total protein content of exosomes was determined and adjusted to get 6 mg/ml and stored in aliquots at −80 °C until use.
2.2 Protein determination
An aliquot of milk exosome preparation was used for protein estimation using the BCA kit (Thermo Scientific, Rockford, IL). Exosome preparations, usually diluted by 10-fold, were compared in triplicates against serially diluted BSA as standard according to manufactures instructions. Values were extrapolated from this curve, using a third-order polynomial equation, with r2>0.98 for each assay.
2.3 NanoSight and Zetasizer
The size distribution of the isolated exosomes was measured by NanoSight and Zetasizer (Malvern Instruments Ltd, Malvern, Worcestershire, UK). A monochromatic laser beam at 405 nm was applied to the diluted suspension of vesicles. Filtered PBS was used as a negative control. A video of 30s was taken with a frame rate of 30 frames/s and particle movement was analyzed by NTA software (version 2.2, NanoSight). The NTA software is optimized first to identify and then track each particle on a frame-by-frame basis, and its Brownian movement is tracked and measured. The velocity of particle movement was used to calculate particle size by applying the two-dimensional Stokes–Einstein equation [18]. All samples were evaluated in 4 replicates. Size determination of isolated exosomes was also performed using a Zetasizer Nano ZS (Malvern Instruments). Exosomes were diluted in 1 ml PBS, and parameters such as zeta potential (electronegativity) and size distribution were analyzed at 37 °C according to the manufacturer's instructions.
2.4 Scanning electron microscopy (SEM)
Exosomes (6 mg/ml) were filtered through 0.22 μm syringe filter (Corning Incorporated, Manassas, VA) and diluted to 1000-fold using deionized water. Diluted exosomes (5 μl) were added onto a clean silica (~300 nm SiO2) wafers and air-dried for 30 min. A conductive layer of platinum metal was coated for 30 secs at a current of 20 mA and grounded with copper tape. Exosomes were imaged in Zeiss Supra 35 VP SEM (Thornwood, NY) under low accelerated voltage (5KV) using secondary electron detectors.
2.5 Atomic force microscopy (AFM)
Exosomes (6 mg/ml) were filtered through 0.2 μm syringe filter (Corning Incorporated, Manassas, VA) and diluted to 600-fold using deionized water. Then 5 μl of the diluted exosomes was added on to a cleaned silica (~300 nm SiO2) wafers and air-dried for 30 min. Asylum MF-3D (Asylum Research, Oxford Instruments, Goleta, CA) atomic force microscope in tapping mode, and silicon probes coated with aluminum (Force Constant = 40 Nm-1; Resonant Frequency = 300 kHz, BudgetSensors.com) was used for imaging. Topographic height, amplitude and phase retraces were imaged concurrently with a fixed force (< 1 nN) with a scanning rate of 1Hz. The images were recorded at 256 × 256 pixels and processed using IGOR software.
2.6 Opti-prep density gradient
Buoyant density of the milk exosomes and the drug-loaded exosomes was determined by layering on top of an Opti-prep density gradient (10-60%; w/v) medium (Sigma-Aldrich, St. Louis, MO) at 150,000×g and 4 °C for 16 h in a swing bucket rotor (SW 41Ti, Beckman Coulter Inc, Fullerton, CA). Distinct bands were collected from the tube, 10 ml of PBS was added to each sample, and exosomes were collected by centrifugation for 2 h at 135,000×g.
2.7 Isolation of total RNA
mirVana miRNA Isolation kit (Applied Biosystems, Foster City, CA) was used to isolate total RNA for mRNA expression studies. Small RNA was further enriched from total RNA for qPCR analysis of miRNAs according to the manufacturer's protocol. Trace genomic DNA in the crude total RNA samples was removed by incubation with 10 units of DNase I per 100 μg RNA (Ambion, Austin, TX) at 37 °C for 30 min. The concentration of the total and small RNA was determined with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) and RNA integrity was verified with a Bioanalyzer 2100 (Agilent, Palo Alto, CA).
2.8 qRT-PCR for miRNA and mRNA expression
For miRNA analysis of the 5 selected miRNAs (miR-21, -181a, -155, -223 and -146a), the individual TaqMan human MicroRNA Assays were used. Briefly, 25 ng of total RNA was reverse-transcribed in a final volume of 20 μl with 12.5 nM of each RT primer using the TaqMan MicroRNA Reverse Transcription Kit. TaqMan miRNA PCR kit was used to perform PCR reactions on the ABI 7900 Real-Time PCR System (Applied Biosystems, Foster City, CA). The reactions were initiated in a 96-well optical plate at 95 °C for 5 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Relative miRNA expression was assessed using the differences in normalized Ct (ΔΔCt) method after normalization to 5S rRNA.
To determine the exosome-associated mRNA expression levels, one-Step SYBR green qRT-PCR Kit (Quanta Biosciences, Gaithersburg, MD) was used to perform cDNA synthesis and PCR amplification simultaneously from 100 ng of total RNA according to the manufacturer's instructions. Bovine specific primers for CD36, CD63, E1α, FAS, MFG-E8, MHC-II, PIGR and XDH were designed using primer 3 express software and synthesized (IDT DNA Technologies, Coralville, IA,). Reactions were run under the following conditions: hold at 50 °C for 10 min, 95 °C for 5 min, then 40 cycles at 95 °C for 10 s and 60 °C for 30 s. Relative gene expression was assessed using the differences in normalized Ct (ΔΔCt) method after normalization to 5S rRNA.
2.9 Western blot analysis
Milk exosomes were analyzed for exosomal surface proteins by western blot as described [19] and blots were probed for CD63, CD81, Tsg101 and Alix (Cell-Signaling, Danvers, MA). To rule out presence of contaminating mutlivesicular bodies (MVBs) and endoplasmic reticulum (ER) exosomes preparations were also probed with MVB markers such as integrin β1, p-selectin and CD40 and ER marker calnexin (Cell-Signaling, Danvers, MA). Appropriate secondary antibodies were used and detection carried out using enhanced chemiluminescence reagent (Thermo Scientific, Waltham, MA). Equal loading of the proteins was confirmed using β-actin (Sigma-Aldrich, St. Louis, MO).
2.10 In vivo biodistribution of exosomes
Athymic nude mice (n=4 per group) were employed to study biodistribution of exosomes administered via oral and intravenous routes. Animals were fed with purified AIN-93M diet and water ad libitum. Milk exosomes were labeled with near-infrared fluorescent dye DiR (20 μM) by incubation at 37 °C for 30 min, followed by centrifugation at 10,000×g for 30 min to remove unbound dye. Labeled exosomes were concentrated with vivaspin 500 centrifugal filter devices (10,000K MWCO, Sartorius Stedim, Bohemia, New York) and washed thrice with PBS. Exosome pellets were suspended in PBS and sterilized by passing through 0.22 μm filter. Animals were administered with a single dose of 100 μl DiR-labeled exosomes (60 mg/kg Exo protein). Animals were euthanized after 4 days of treatment; different organs were collected and imaged ex vivo using Photon Imager Optima (Biospace lab, Paris, France). The relative intensities were measured and compared with untreated control. For in vivo stability study, after administration of DiR-labeled exosomes by oral gavage as described above, blood were collected at different time points (1, 4, 24, 48, 72 and 144 h) and imaged for fluorescent intensity.
2.11 Drug encapsulation and in vitro release
The drug loading of chemopreventive [withaferin A (WFA), bilberry-derived anthocyanidins (Anthos) and curcumin (Cur)] and chemotherapeutic drugs [paclitaxel (PAC) and docetaxel (DOC)] agents was achieved by mixing the test agent (dissolved in ethanol or 1:1 mixture of ethanol and acetonitrile) with exosome suspension in the proportion of 1:9 at room temperature (22 °C). In a separate experiment, we determined that these solvents had no effect on the particle size, coagulation, etc. Unbound drug was removed by a low-speed centrifugation (10,000×g) for 10 min, and the drug-loaded exosomes were collected by centrifugation at 135,000×g for 2 h. The pellet was suspended in PBS and stored at −80 °C. Drug loading was determined by analysis of drug spectrophotometrically and/or by ultra-performance liquid chromatography (UPLC and protein in the sample and percent drug load was calculated.
FA was loaded similar to other small drug molecules, except that it was mixed together with WFA and dissolved in dimethyl sulfoxide and the final concentration was maintained at 2.5% (v/v). Vehicle-treated exosomes were processed in parallel to serve as a control. We determined the effect of solvents on the exosomes, based on the size and molecular markers and anti-proliferative activity. The presence of up to 10% of the solvents did not affect the exosomes properties in any manner and also confirmed the activity of all formulations in cell culture.
For in vitro release, exosomes loaded with WFA, PAC, and DOC were added to the dialysis tubes and left in the PBS (pH 7.4) containing 0.2% Tween-80, in order to provide sink conditions. Aliquots (50 μl) were collected from the dialysis tubes after 1, 2, 4, 8, 24, 48 and 72 h, and the residual drug levels were measured spectrophotometrically or by UPLC. The amount of drug released was determined by subtracting from the initial amount.
2.12 UPLC analysis
Percent drug load was determined by UPLC (Shimadzu, Kyoto, Japan). Briefly, 0.95 ml acetonitrile was added to 50 μl of drug loaded exosomes to extract the drug and precipitate the exosomal proteins. The precipitated proteins were separated by centrifugation at 10,000×g for 10 min, supernatant was separated and 5 μl samples were analyzed on UPLC system using a Shim-Pack XR-ODS II reverse phase column (Shimadzu; 150 × 3.0 mm i.d., 2.2 μm). Acetonitrile and water were used at a flow rate of 0.7 ml/min with a linear gradient elution in which acetonitrile concentration was increased from 5 to 60% from 1.3 to 5.1 min, followed by an increase to 80% from 5.1 to 7.7 min and finally, to 100% in 10 min, the latter ratio was maintained till 10.9 min and finally decreased to 5% in 12 min. The WFA was detected at 215 nm by PDA-UV and total WFA concentration was calculated against the standard curves of WFA. Similarly, other drugs like PAC were extracted using acetonitrile, supernatant was separated by centrifugation and samples were analyzed spectrophotometrically and drug concentration was calculated against a standard curve of reference compounds.
2.13 Cell culture
Human lung cancer (A549 and H1299), breast cancer (MDA-MB-231 and T47D) cell lines and human normal bronchial epithelial cells (Beas-2B) were obtained from American Type Cell Culture (ATCC, Manassas, VA). All cancer cell lines were supplemented with 10% fetal bovine serum and antibiotics. The A549 and H1299 cells were grown in F-12K and DMEM media, respectively. The T47D cells were grown in RPMI supplemented with 0.2 units/ml insulin and MDA-MB-231 cells were grown in L-15 media. The Beas-2B cells were grown in Bronchial Epithelial Medium (BEBM) media supplemented with all the additives of BEGM, Kit (Lonza/Clonetics Corporation, San Diego, CA). All cell lines except MDA-MB-231 were grown at 5% CO2. All cell lines were maintained at 37 °C in a humidified chamber.
2.14 Confocal microscopy
Milk exosomes were labeled with PHK-67 as per manufacturer's instructions and labeled exosomes were washed twice with PBS and concentrated with VIVASPN 500 centrifugal filter devices (10,000 MWCO; Sartorius Stedim, Bohemia, New York). Human lung cancer H1299 cells were seeded into BD Falcon 8-well chamber Culture Slides (BD Biosciences, San Jose, CA) and labeled exosomes were added at various concentrations and incubated variable times. For confocal microscopy, samples were fixed with 4% methanol-free paraformaldehyde for 10 min, permeabilized in 0.1% Triton-X 100 in PBS for 3 min. After fixation and permeabilization, cells were washed thrice with PBS, blocked in 1% bovine serum albumin in PBS for 10 min, and stained with a solution containing 0.5 unit/μl Alexa Fluor 488 -conjugated phalloidin (Molecular Probes, Life Technologies, Grand Island, NY) for 20 min at room temperature. DNA was visualized through DAPI staining (Molecular Probes, Life Technologies, Grand Island, NY).
2.152.15 Electrophoretic Mobility Shift Assay (EMSA)
DNA binding of NF-κB was measured by EMSA as described [20]. Briefly, human lung cancer A549 cells pretreated with milk exosomes at 37 °C for 6h and then challenged with tumor necrosis factor- α (TNF-α) (1 ng/ml) or LPS (1 μg/ml) for 30 min and nuclear extracts were prepare. Nuclear protein (10 μg in each assay) was incubated at room temperature for 30 min with 0.2 μg of 32P-end-labeled double-stranded oligonucleotide containing the NF-κB binding motif (Promega, Madison, WI, USA) and 1 μg of poly (dI-dC) as an inhibitor of nonspecific binding, in binding buffer containing 20 mM N-2-hydroxyethylpiperazine-N'-2 ethanesulfonic acid (HEPES; pH 7.4), 60 mM KCl, 5 mM MgCl2, 0.2 mM EDTA, 0.5 mM dithiothreitol, 0.5 mM phenylmehylsulfonyl fluoride, 1% Nonidet P-40 and 8% glycerol. The sequence of the double-stranded oligomers used for EMSA was 5'-AGT TGA GGG GAC TTT CCC AGG C-3'. In a cold-competition experiment, unlabeled oligonucleotide was incubated with extracts for 30 min at room temperature prior to the addition of the radiolabeled probe. The reaction mixtures were electrophoresed through 7.5% native polyacrylamide gels. After the gels were dried, DNA-protein complexes were visualized and quantified by Packard InstantImager (Downers Grove, IL).
2.16 Antiproliferative activity
The antiproliferative activity of the exosomal formulations of the test agents against normal bronchial epithelial cells (Beas-2B), breast (MDA-MB-231, T47D), and lung (A549, H1299) cancer cells was assessed by MTT assay, as described elsewhere [19]. Briefly, cells were seeded in a 96-well plate for overnight, and treated with either drug alone, exosomal formulation of the drug or exosomes alone at various concentrations for 72 h. At the end of treatment period the medium was changed and incubated with MTT (0.5 mg/ml) for 2 h, followed by solubilization in dimethyl sulfoxide and spectrophotometric measurement at 570 nm.
2.17 Antitumor activity of drugs delivered as exosomal formulations
Female athymic nude mice (4-5 week old) were purchased from Harlan (Harlan Labs, Indianapolis, IN) and provided with AIN-93M diet and water ad libitum. Human lung cancer A549 cells in serum-free media (F-12K) were mixed with matrigel matrix in 1:1 ratio and a suspension of 2.5 × 106 cells (100 μl) was injected subcutaneously into the right flank of each mouse. When tumors grew to ~80 mm3, animals were randomized into five groups (n = 6-8). The treatment group received intraperitoneal (i.p) injections of WFA or Exo-WFA (4 mg WFA/kg; 25 mg protein/kg b. wt.) on alternate days (3 doses a week). Two control groups received exosomes alone (25 mg protein/kg b. wt.) or PBS alone. Tumor size, animal weights and diet intake were monitored weekly. All animals were euthanized after 7 weeks of treatment. Animals were also examined for any visible signs of toxicity.
In another study, nude mice carrying the A549 lung tumor xenograft (n = 8-10) were treated with PBS, exosomes alone, Exo-WFA and FA-loaded Exo-WFA (Exo-WFA-FA). The dose of WFA was 8 mg/kg b. wt. Animals received the intervention when tumors grew to ~80 mm3. Agents were administered by oral gavage on alternate days (3 doses a week). Animals were euthanized after 8 weeks and tumors were exercised. All other conditions were as in the previous study. Since FA + drug and exosomal formulations of FA did not provide any higher antiproliferative activity compared to the free drug based on cell culture studies, we did not include this group in the in vivo xenograft study. All the treatments were performed using PBS as control.
2.18 Toxicity assessment
Potential short-term and long-term toxicity of milk exosomes was assessed in female Sprague-Dawley rats (5-6 week old) (Harlan Labs, Indianapolis, IN). Wild-type rats were selected over mice to assess toxicity for the need of more blood to perform serum chemistry and hematological profiles. Animals were provided with AIN-93M diet and water ad libitum. After acclimation, animals were randomized into 4 groups (n=4) for short-term toxicity assessment and treated with a single dose of either PBS or exosomes (25 mg exosomal protein/kg b. wt.) by oral gavage, and animal were euthanized after 1, 3 and 6 h. For long-term toxicity assessment animals were randomized into 2 groups (n = 4) and treated daily with either PBS or exosomes (25 mg exosomal protein/kg b. wt.) by oral gavage. After 15 days, animals were euthanized by CO2 asphyxiation. Blood was collected at the time of euthanasia, and hematological parameters were analyzed using whole blood by Cell Dyn 3500 hematology analyzer (Abbott laboratories, Santa Clara, CA) as described earlier [21]. Serum was used to measure various biochemical parameters of liver and kidney function by an automated AU640® Chemistry Analyzer (Beckman Coulter, Inc., Brea, CA, USA) that uses an ion selective electrode to measure various electrolytes and in-built automated spectrophotometric techniques for all other assays.
2.19 Cytokine measurement
To assess cytokine levels in response to milk exosomes, serum samples from toxicity studies in vivo using Sprague-Dawley rats were utilized. Prior to use, serum samples were diluted 1:4 in sample diluent and cytokines were analyzed using a Bio-Plex Pro rat cytokine Th1/Th2 12-plex immunoassay (Bio-Rad, Hercules, CA) according to manufacturer's instructions. The panel of cytokines included IL-1α, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-10, IL-12p70, IL-13, GM-CSF, IFN-γ and TNF-α. After collection, all the samples from each time point were run in single Luminex experiment. Samples were read on a Bio-Plex 200 system with Bio-Plex Manager software (Bio-Rad, Hercules, CA). Data are expressed as pg of cytokine/ml (mean ± SEM), with n = 3 in duplicate for each group.
3 Results
3.1 Isolation and biological characterization of milk-derived exosomes
We isolated milk exosomes from bovine raw milk by differential centrifugation (Fig. 1A, Supplementary Fig. S1). The average yield of the exosomes was 335 ± 48 mg per liter of the milk prepared at different times. These exosomes were largely 40-100 nm in diameter, as measured by NanoSight (Fig. 1B), and exhibited a polydispersity index (PDI) of 0.22 ± 0.06, calculated at 37 °C using Zetasizer (Fig. 1C). The exosome size was confirmed to be < 80 nm by atomic force microscopy (AFM) and scanning electron microscopy (SEM) (Fig. 1D, 1E and Supplementary Fig. S2). Buoyant density of the milk exosomes was determined using Opti-prep density-gradient centrifugation. We observed that exosomes resulted in a major band at 30% of Opti-prep concentration which corresponded to approximately 1.18 g/ml density (Fig. 1F), in agreement with reported values [13, 22]. Exosome protein lysates were prepared and verified with western blot analysis for vital exosomal membrane markers CD63, CD81, transpanins, Tsg101 and alix [23, 24] (Fig. 2A). The absence of microvesicle surface markers β-integrin β1, p-selectin and CD40 [25] and endoplasmic reticulum (ER) marker calnexin [26] confirmed that the vesicles isolated were not contaminated with other multivesicular bodies. Exosomes were also isolated from bovine colostrum and observed a significantly higher yield (1.5 times compared to mature milk) with a buoyant density similar to that of mature milk.
Fig. 1. Isolation and characterization of milk-exosomes.
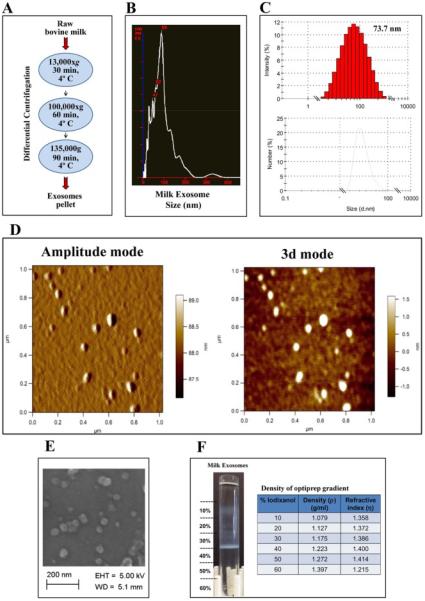
(A) Schematic representation of the major steps involved in the isolation of exosomes from bovine raw milk. (B) The exosome suspension (6 mg/ml protein concentration) was diluted 20-50 fold in phosphate-buffer-saline (PBS), pH 7.4 and a total of 200 μl was analyzed by NanoSight. (C) Bovine milk-derived exosomes were analyzed using 1 ml of the diluted suspension (1 mg/ml) in disposable cuvettes and milk exosomes size distribution was measured by Zetasizer. (D) Diluted exosomal suspension was loaded on a cleaned silicon wafers and air-dried for 30 min. Asylum MF-3D (Asylum Research, Oxford Instruments) AFM in 3d and tapping mode, and silicon probes coated with aluminum (Force Constant = 40 Nm-1; Resonant Frequency = 300 kHz, Budget Sensors.com) were used for imaging. Topographic and amplitude images were captured concurrently with a fixed force (<1 nN) with a scanning rate of 1Hz. (E) Exosomal suspension was filtered through 0.22 μm and loaded over clean silicon wafers and air-dried for 30 min. Silicon wafers were grounded using copper adhesive tape for conductivity. Exosomes were imaged in Zeiss Supra 35 SEM under beam energies (5kV). 142,000 × magnification. (F) Milk exosomes were layered onto Opti-prep gradient (10-60%) and centrifuged at 150,000×g for 16 h using 41 Ti swing rotor. The densities and refractive index values for the corresponding fractions are indicated in the table.
Fig. 2. Exosomal markers and cargo.
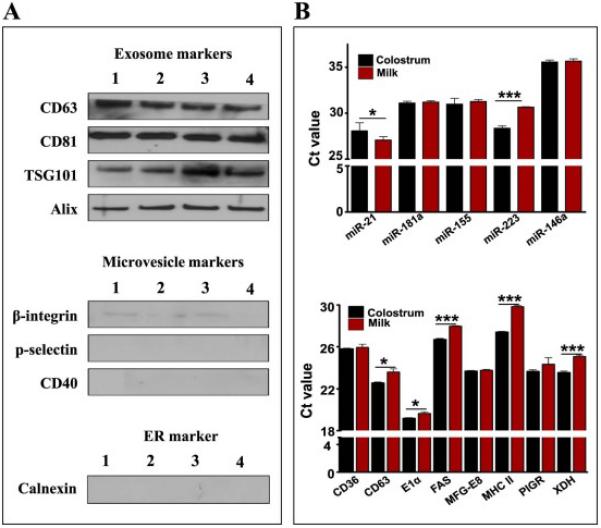
(A) Exosomes isolated in four different batches (numbered 1 - 4) were analyzed for the indicated exosomal and plasma membrane proteins. (B) Total RNA was isolated from 1 ml exosomal suspension (6 mg exosomal protein) by Trizol method (n=3), and reverse transcription and PCR were performed using bovine specific Taqman probes and primers (Applied Biosystems) for indicated immune-related miRNAs (Top) and exosome-related mRNAs (Bottom). Ct (threshold cycle) is plotted on y-axis and comparison between milk and colostrum exosomes were made. Student's t-test was performed to determine statistical significance; p<0.05=*, p<0.01= ** & p<0.001= ***.
In view of reports indicating the presence of RNA components in exosomes derived from various cells and secretory fluids including milk, we analyzed select miRNAs and mRNAs in the milk- and colostrum-derived exosomes by RT-PCR. As shown in Fig. 2B, a number of immune-related miRNAs –miR-155, -181a, -146a and -223, with the suggestive role in human health [27, 28], were detected in the milk and colostrum exosomes. Presence of exosomal mRNAs such as CD36, CD63, Eα1, FAS, MFG-E8, MHC-II, PIGR and XDH in exosomes isolated from milk and colostrum was also confirmed by RT-PCR (Fig. 2B).
We observed that when milk exosomes were stored at ≤6 mg exosomal protein/ml at −80 °C, they remain largely free of coagulation for several months. Exosomes had initial particle size of 95.5 ± 14.8 nm which was insignificantly (104.5 ± 12.0 nm) altered after 18 months of storage. Effect of storage on biological activity of exosomes was assessed by anti-proliferation activity and the results indicated no loss of activity upon storage. These findings suggested that milk-derived exosomes can be stored for prolonged periods without causing significant changes in their physical and biological properties.
3.2 Uptake, distribution and toxicity of milk exosomes in vitro and in vivo
To investigate the in vitro cell uptake of exosomes, human lung cancer H1299 cells were treated with PKH67-labeled milk exosomes at various concentrations (0-500 μg/ml) and durations (1-16 h). We observed a concentration-dependent increase in uptake of exosomes until 50 μg/ml concentration; at higher concentrations, the effect was non-linear (Fig. 3A). Similarly, a time-dependent increase in uptake of exosomes was observed for 8 h, which reached a plateau by 16 h (Fig. 3B). Thus, our results indicated concentration- and time-dependent uptake of exosomes by the cells. The cellular internalization of PKH67-labeled milk exosomes was examined in H1299 cells by confocal microscopy. The images revealed that the PHK-67-labeled exosomes co-localize uniformly inside the cells (Fig. 3C), which was confirmed by depth Z-scanning (Supplementary Fig. S3).
Fig. 3. Uptake of milk exosomes.
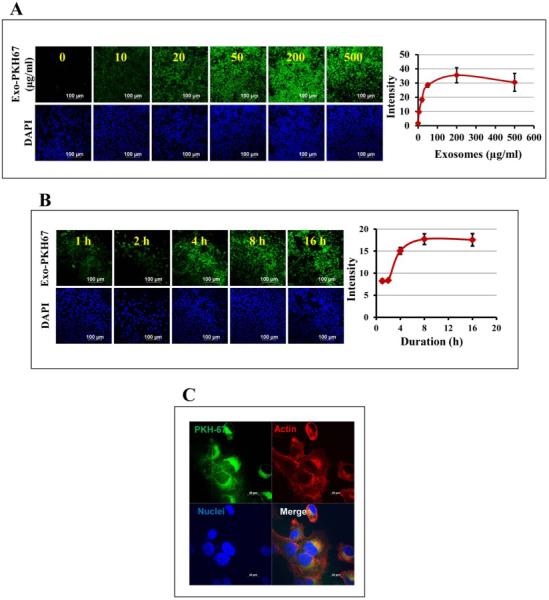
(A) Dose-dependent uptake of milk exosomes by human lung cancer H1299 cells in vitro. Left: PKH-67-labeled milk exosomes were added to H1299 cells in 8-well chamber slide at 0-500 μg/ml and the uptake was monitored after 6 h. Cells were fixed and visualized under confocal microscope. Nuclei were visualized by staining with DAPI. Representative confocal images are shown (left panel). Scale bar, 100 μm. Fluorescent intensity of the PKH-67-labelled exosomes was quantified using ImageJ software (right panel). (B) PKH-67-labeled milk exosomes were added to H1299 in 8-well chamber slide at 50 μg/ml and the uptake was monitored after 1, 2, 4, 8 and 16 h. Cells were fixed and visualized. Confocal images are show in left panel and quantification of fluorescent images in right panel. (C) 500 μg/ml of the PKH67-labelled milk exosomes were added per 40,000 cells and incubated at 37 °C for 4 h. The uptake of the labeled exosomes was detected by confocal microscopy. Alexa fluor-phalloidin 594 (red) was used to detect actin filaments and DAPI for nucleus (blue) and PKH-67 to label the exosomes (green). Scale bar, 20 μm.
To determine their stability and distribution in vivo, exosomes were labelled with lipophilic near-IR fluorescent dye DiR. The stability of exosomes was quantified by the fluorescent signal intensity in blood at different time intervals (1, 4, 24, 48, 72 and 144 h). Our results indicated highest fluorescent signal after 24 h and a steady decrease in the intensity levels was observed until day 6 suggesting clearance from circulation (Fig. 4A).
Fig. 4. Biodistribution, stability and immunological response of milk exosomes.
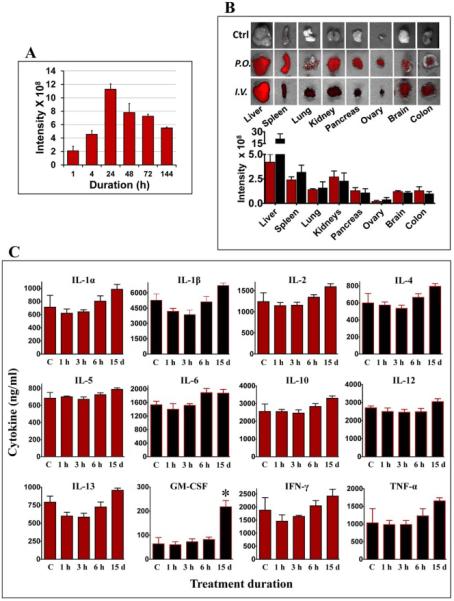
(A) Blood from nude mice treated with a single dose of 60 mg DiR-labeled exosomes by gavage (p.o.) was drawn and imaged at indicated times. Untreated control mice had baseline intensity value of < 1.25 × 108 units. (B) Female nude mice were administered a single dose of 60 mg DiR-labeled exosomes/kg body weight by gavage (p.o.) or intravenously (i.v.). Top: Ex vivo-imaging of the tissues after 4 days of exosome administration were performed using Biospace lab Photon Imager Optima. Representative images are shown. Bottom: Fluorescent intensity of exosomes distributed in various tissues quantified using photon optima software. Figure shows average ± standard deviation of four animals. (C) Female Sprague-Dawley rats were treated with a single dose (short-term toxicity) of exosomes (25 mg exosomal protein/kg, b.wt.) by oral gavage or once daily for 15 days (long-term toxicity) and the serum was analyzed for cytokine levels using bio-plex cytokine Th1/Th2 assay. Acute toxicity observations were made at 1h, 3h and 6h post treatment. Vehicle-treated control (vehicle) animals were used as reference for baseline cytokine levels.
The in vivo biodistribution was examined after administration of DiR-labelled milk exosomes by oral gavage or intravenous injection (i.v.) in female athymic nude mice. Our findings suggested that route of administration had a significant influence on the tissue distribution of DiR-labeled exosomes (Fig. 4B). Various organs imaged ex vivo, indicated that the tissue distribution of the exosomes within the liver, lung, kidney, pancreas, spleen, ovaries, colon and brain was somewhat uniform with the gavage route; however, with the i.v. route, the exosome concentration predominated in the liver. In the other organs, the distribution of exosomes was similar between the oral and i.v. routes.
To determine any systemic toxicity, the milk exosomes were administered to female Sprague-Dawley rats by oral gavage at 25 mg/kg b. wt. A single dose was monitored over 1, 3, and 6 h to study short-term toxicity and daily doses were administered for 15 d to study long-term toxicity. All animals survived the duration of the study and had no noticeable changes in clinical signs, body weight gains and diet intake etc. Administration of exosomes did not result in any significant changes in biochemical parameters such as serum aspartate transaminase (AST) and alanine transferase (ALT), creatinine and blood urea nitrogen (BUN) compared to untreated animals, suggesting that exosomes treatment did not adversely affect liver and kidney function at all the time points examined. All other biochemical and hematological parameters remained unaltered except triglycerides, indicating no short-term or long-termtoxicity due to milk exosomes. Table I-II represents biochemical and hematological parameters values at 6 h and 15 day after administration of milk exosomes. Decrease in triglycerides was observed at both 6 h and 15 d treatment with exosomes compared to vehicle treated animals. In addition, cytokine profiling indicated no significant changes in pro- and anti-inflammatory cytokines and chemokines, except for the anti-inflammatory cytokine GM-CSF which was significantly elevated (Fig. 4C) suggesting possible beneficial effects due to the milk exosomes. These initial biocompatibility results indicate that the milk exosomes may be used as drug delivery vehicle in vivo.
Table I.
Effect on biochemical profile (systemic toxicity) following 6 h or 15 day exposure to milk exosomes in Sprague-Dawley rats
| Biochemical profile | 6h Exposure | 15 day Exposure | ||
|---|---|---|---|---|
| Control | Milk Exo | Control | Milk Exo | |
| Liver profile | ||||
| AST (SGOT) | 202.8 ± 49.3 | 204.8 ± 43.1 | 235 ± 85.7 | 220.5 ± 58.7 |
| ALT (SGPT) | 65.3 ± 12.8 | 56.8 ± 13.4 | 66.0 ± 3.2 | 64.2 ± 9.7 |
| Alk Phosphatase | 192 ± 18.3 | 162.3 ± 23.8 | 243.8 ± 58.9 | 151.8 ± 55.7 |
| GGT | 1.3 ± 0.5 | 2.8 ± 2.1 | 5.3 ± 1.0 | 3.3 ± 1.7 |
| Amylase | 529.5 ± 107.2 | 585.5 ± 162.0 | 529.8 ± 55.3 | 496.0 ± 42.9 |
| CPK | 873.5 ± 238.6 | 915.8 ± 388.0 | 25.0 ± 0.0 | 25.0 ± 0.0 |
| Kidney profile | ||||
| BUN | 19.8 ± 0.5 | 17.5 ± 2.6 | 23.8 ± 3.1 | 18.8 ± 1.9 |
| BUN/Creatinine Ratio | 36 ± 3.6 | 33.5 ± 5.0 | 98.8 ± 42.1 | 65.5 ± 30.3 |
| Phosphorus | 18.4 ± 3.0 | 13.1 ± 0.9 | 15.9 ± 1.7 | 15.1 ± 2.4 |
| Calcium | 12.4 ± 1.0 | 11.7 ± 0.9 | 10.6 ± 1.1 | 11.1 ± 1.1 |
| Total Protein | 6.9 ± 0.4 | 6.5 ± 0.5 | 6.8 ± 0.2 | 6.7 ± 0.5 |
| Albumin | 3.8 ± 0.2 | 3.7 ± 0.3 | 3.9 ± 0.2 | 4.0 ± 0.3 |
| Globulin | 3.1 ± 0.2 | 2.8 ± 0.2 | 2.9 ± 0.1 | 2.7 ± 0.2 |
| A/G Ratio | 1.2 ± 0.1 | 1.3 ± 0.0 | 1.3 ± 0.1 | 1.5 ± 0.1 |
| Glucose | 193.8 ± 55.8 | 208.3 ± 28.6 | 205.5 ± 31.0 | 193.5 ± 34.8 |
| Cholesterol | 94 ± 10.1 | 81.5 ± 9.3 | 105.8 ± 14.6 | 105.5 ± 12.9 |
| Triglyceride | 134.5 ± 26.6 | 86.0 ± 26.8* | 117.3 ± 9.4 | 69.3 ± 13.8** |
Female Sprague Dawley rats (6-7) weeks old were provided control diet (AIN 93M) and water ad libitum and treated with milk-derived exosomes (25 mg/kg, b. wt.) by oral gavage for 6 h or 15 days, once daily. At euthanasia blood was collected and analyzed using an automated AU640® Chemistry Analyzer (Beckman Coulter, Inc., Brea, CA, USA) by Antech diagnostics. Data represent average and Sprague-Dawley of four animals. Statistical analysis was performed by student t-test.
p-value <0.05
p-value <0.01.
Table II.
Effect on the hematological parameters (systemic toxicity) following 6h or 15 day exposure to milk exosomes in Sprague-Dawley rats.
| Hematological profile | 6h Exposure | 15 day Exposure | ||
|---|---|---|---|---|
| Control | Milk Exo | Control | Milk Exo | |
| WBC | 5.7 ± 2.5 | 6.5 ± 1.6 | 5.7 ± 1.7 | 7.0 ± 2.0 |
| HGB | 14.0 ± 1.2 | 13.5 ± 0.9 | 14.0 ± 0.3 | 13.8 ± 0.6 |
| HCT | 44.0 ± 3.5 | 41.5 ± 3.4 | 43.3 ± 2.1 | 42.3 ± 1.9 |
| MCV | 60.8 ± 1.0 | 59.0 ± 1.8 | 45.5 ± 29.0 | 59.3 ± 1.7 |
| MCHC | 31.8 ± 0.5 | 32.4 ± 1.0 | 32.3 ± 2.3 | 32.8 ± 0.5 |
| Platelet Count | 689.0 ± 145.8 | 605.8 ± 433.2 | 755.7 ± 134.9 | 772.8 ± 85.8 |
| Neutrophils | 12.0 ± 3.5 | 32.7 ± 35.8 | 11.5 ± 3.0 | 13.5 ± 8.4 |
| Lymphocytes | 84.5 ± 4.5 | 50.8 ± 40.4 | 86.0 ± 2.8 | 81.0 ± 7.4 |
Female Sprague Dawley rats (6-7) weeks old were provided control diet (AIN 93M) and water ad libitum and treated with milk-derived exosomes (25 mg/kg, b. wt.) by oral gavage for 6 h or 15 days, once daily. At euthanasia blood was collected and analyzed using an automated AU640® Chemistry Analyzer (Beckman Coulter, Inc., Brea, CA, USA) by Antech diagnostics. Data represent average and Sprague-Dawley of four animals. Statistical analysis was performed by student t-test and no significant change was observed in milk exosome treated animals compared to control group.
3.3 Milk exosomes as nanocarriers of small drug molecules
Because exosomes are lipid bilayer nanovesicles with embedded proteins, they possibly have various binding sites on their surfaces. To exploit the possibility of using milk exosomes as a drug carrier, different hydrophilic and lipophilic agents, including chemotherapeutic drugs, were loaded onto exosomes using appropriate solvents. A wide variety of compounds [CUR, WFA, anthocyanidins (Anthos), PAC and docetaxel (DOC)] that vary in their lipophilicity, molecular weights, and functional groups were loaded onto the exosomes. Buoyant densities of the drug-loaded exosomes remained unaltered compared to unloaded or vehicle-treated exosomes (Fig 5A). The drug loading was assessed after solvent extraction of the drug formulations, followed by analysis of the drug by spectrophotometry or UPLC and protein by standard BCA assay. The results suggested that the drug loading of chemotherapeutic and chemopreventive agents varied from 10% - 40% based on the test agent.
Fig. 5. in vitro release profile and stability of drug-loaded exosomes.
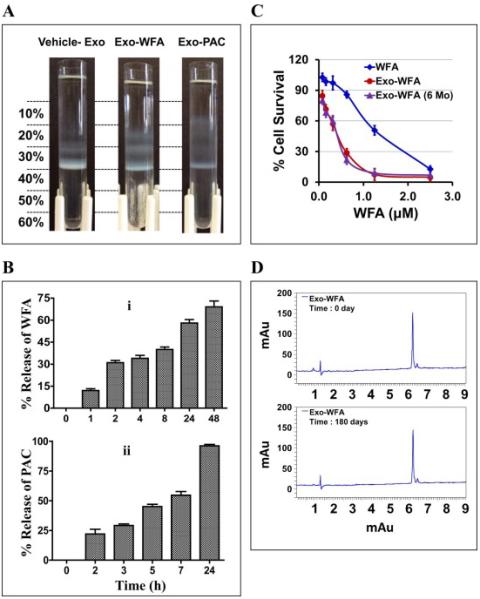
(A) Separation of vehicle- and drug-loaded milk exosomes by Opti-prep density gradient. Indicated agents were mixed with exosome in the presence of 10% ethanol. Unbound drug was removed by centrifugation at 10,000×g for 10 min. The drug loaded exosomes were layered onto Opti-prep gradient (10 - 60%) and separated by centrifugation at 150,000×g for 16 h using SW 41 Ti swing rotor. (B) The release study was done of Exo-WFA (top) and Exo- PAC (bottom) formulations was using dialysis tubes against buffer containing the surfactant, Tween-80 (0.02%) at 37°C. (C) Anti-proliferative activity of exosomal-WFA formulation of fresh preparation (red) and after 6 months of storage (purple) against A549 cells. Free WFA was included for reference (blue) (mean ± SD). (D) UPLC profile of WFA extracted from exosomal formulation at day 0 and after 180 days.
The drug release kinetics from the exosomal formulations were determined in vitro using tube-o-dialyzer tubes in the presence of Tween-80. Being a detergent Tween-80 helps in the extraction of drug from the exosomes by dissociating drug-exosomes interaction. The results indicated that Exo-WFA exhibited a time-dependent release. The cumulative release of WFA was 12, 31, 34, 40, and 58% after 1, 2, 4, 8 and 24 h, respectively (Fig. 5B). WFA extracted from the residual material was found to be stable during the workup, based on UPLC analysis. Similarly, exosomal formulations of the chemo drugs, PAC (Fig. 5B) exhibited time-dependent release. The cumulative release of PAC was 22, 29, 45 and 55% after 2, 3, 5, and 7 h, respectively, with almost the entire drug (96%) released after 24 h. The cumulative release of DOC was over 50% after 23 h (Supplementary Fig. S4).
The effect of long-term storage on the efficacy and stability of Exo-WFA was determined by anti-proliferative activity assay and solvent extraction of WFA followed by UPLC analysis. The efficacy results of the formulation as tested against lung cancer A549 cells in vitro indicated efficacy similar to at the time of its preparation (Fig. 5C). Furthermore, after 6 months of storage at −80 °C, the levels of WFA were found to be essentially unaltered (Fig. 5D). These findings suggest that exosomes can be effectivity loaded with small molecules to increase drug stability and the drug formulations and be stored for long-term without losing drug efficacy.
3.4 Enhanced anti-cancer and anti-inflammatory effects by exosomal delivery
We examined the anti-proliferative activity of drug-loaded exosomes against human lung (A549 and H1299) and breast (MDA-MB-231 and T47D) cancer cell lines Exosomal formulations show enhanced efficacy as determined by reduction in IC50 values of Exo-WFA compared to WFA alone (0.4 μM vs 1.25 μM) and Exo-PAC compared to PAC alone (1.56 nM vs 3.12 nM) in A549 lung cancer cells after 72 h of treatment (Fig. 6A). We further observed that milk exosomes per se (in the absence of any drug) exhibited significant growth-inhibitory effects against lung (A549 and H1299) and breast (MDA-MB-231 and T47D) cancer cells that ranged from 15-45%, depending on the dose and cell line (Fig. 6B). These data indicate that the enhanced effects observed with the exosomal formulations could be partly due to milk exosome per se, in addition to increased stability of the drug and higher cellular uptake. Normal lung (Beas-2b) cells on other hand exhibited no toxicity to the milk exosomes and were well tolerated up to a concentration of 50 μg/ml dose for 72 h compared to A549 lung cancer cells (Fig. 6C).
Fig. 6. Increased drug bioavailability and enhanced anti-cancer effects.
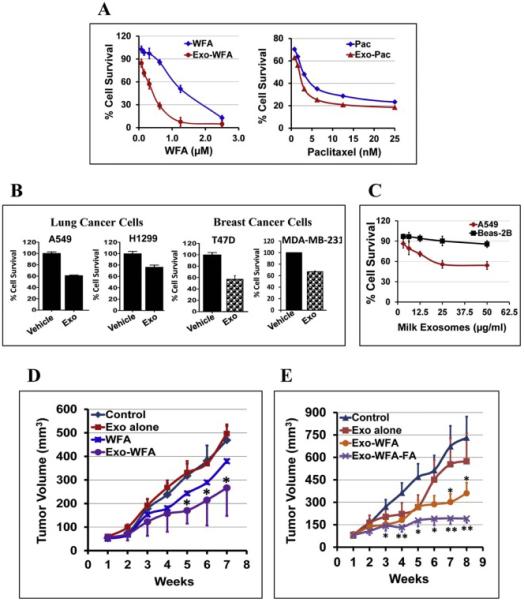
(A) Antiproliferative activity of drug-loaded milk exosomes versus free drugs [withaferin A (WFA) and paclitaxel (PAC)] in human lung cancer A549 cells. For Exo-drug treatments, exosomal protein concentration was maintained constant (50 μg/ml). (B) Anti-proliferative activity of milk exosomes per se against human lung (A549 and H1299), and breast (T47D and MDA-MB-231) cancer cells. Cells were treated with 50 μg/ml exosomal protein for 72 h. The percent cell survival was analyzed by MTT assay. Data represent average ± SD (n=3). (C) Anti-proliferative activity of milk exosomes per se at concentrations 0- 50 μg/ml for 72 h against human normal lung Beas-2b and lung cancer (A549) cells. Data represent average ± SD (n=3). (D) Following inoculation with human lung cancer A549 cells (2.5 × 106 cells), when tumor xenografts grew to over 80 mm3, animals were treated i.p. three times a week with Exo-WFA (4 mg/kg WFA and 25 mg/kg b. wt.). Two other groups were treated i.p. with Exo alone (25 mg/kg b. wt.) or WFA (4 mg/kg). Data represent average ± SE (n = 6–8); SE is not shown in WFA alone for clarity. Statistical analysis was done using student's t-test; *, p < 0.05; **, p < 0.005. (E) Animals bearing A549 xenografts were treated with oral gavage three times a week with FA-Exo-WFA (8 mg/kg WFA and 25 mg/kg b. wt. exo protein) to achieve tumor targeting. Two other groups were treated with Exo alone (25 mg/kg b. wt.) or vehicle. Data represent average ± SE (n = 8–10). Statistical analysis was done using student's t-test; *, p < 0.05; **, p < 0.005.
We then determined if exosomal formulation could be effective in inhibiting tumor growth than free drug using in vivo tumor models. Mice bearing human lung cancer (A549) xenografts were treated with i.p injection of suboptimal doses of WFA at 4 mg/kg b. wt., either as free WFA or Exo-WFA. The suboptimal dose was opted in order to appreciate the anticipated higher efficacy of the exosomal formulation. Additional groups received vehicle or exosomes alone at the same doses used for the exosomal formulation. A significantly greater tumor inhibitory effect was observed with Exo-WFA compared to free WFA (46% vs 23%) (Fig. 6D), thus, confirming the enhanced efficacy of WFA in exosomal formulation.
We then investigated if oral delivery of exosomal formulation carrying tumor-targeting ligand FA would further enhance the anti-tumor effect in mice bearing human lung cancer (A549) xenografts. For this study, milk exosomes were co-loaded with FA and WFA (i.e., Exo-WFA-FA), and all the test agents (vehicle, Exo, Exo-WFA and Exo-WFA-FA) were administered by oral gavage using a WFA dose that was 2-fold higher (8 mg/kg) than that used in our previous study in view of oral administration. The results presented in Fig. 6E indicate that the FA-tagged exosomal formulation (Exo-WFA-FA) led to significantly higher growth inhibition than Exo-WFA (74% vs. 50%; p = 0.016) after 8 weeks when compared to untreated control. Exosomes per se exhibited 21% inhibition vs. vehicle-treated animals. These data indicate tumor targeting feasibility of milk exosomes for enhanced anti-tumor activity.
Since human and bovine milk exosomes can potentially modulate immune cell function and influence the immune system [29], we determined the anti-inflammatory effects of milk exosomes in vitro in H1299 cells challenged with either TNF-α or LPS to induce NF-κB activation. Cells pre-treated with milk exosomes exhibited signification inhibition of both constitutive and induced NF-κB levels (Fig. 7A) indicating anti-inflammatory effects of milk-derived exosomes. To validate the in vitro anti-inflammatory findings of milk exosomes, Sprague-Dawley rats were treated intraperitoneally with LPS (10 mg/kg b.wt.) and milk exosomes (25 mg exosomal protein/kg b.wt.), alone and in combination. After 6 h, animals were euthanized and the lung and liver NF-κB levels were measured. An evidence of a modest (30%-40%) reduction in the NF-κB levels with milk exosomes was observed (Fig. 7B). These protective effects of the milk exosomes presumably are derived from cargo it carries, such as immune factors, miRNAs and proteins [12, 13]. Additionally, our findings show that treatment of wild-type Sprague-Dawley rats with milk exosomes (25 mg/kg exosome protein, once daily) for 15 days did not induce inflammation marker NF-κB in the lung and liver (Fig. 7C) compared to positive control for inflammation LPS suggesting a lack of inflammatory reaction due to the cross-species origin of the milk exosomes.
Fig. 7. Anti-inflammatory effects of milk exosomes.
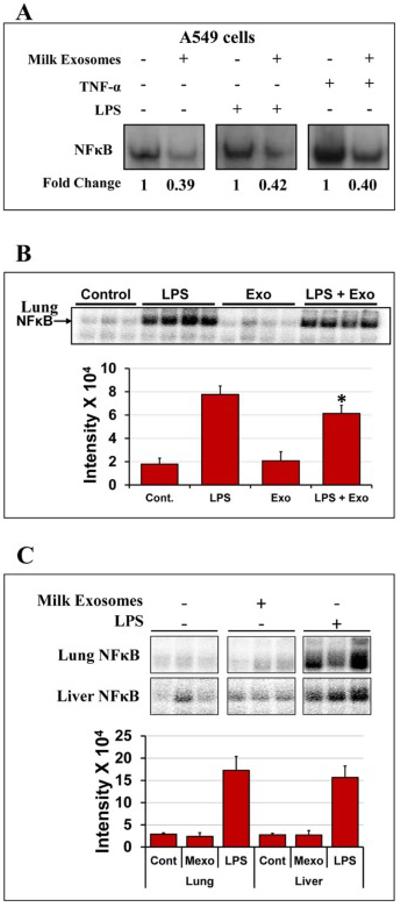
(A) Human lung A549 cancer cells were pre-treated with Exo (90 μg/ml Exo protein), for 6 h followed by treatment with or without tumor necrosis factor (TNF)-α (10 ng/ml) or LPS (1 μg/ml) to induce NF-κB activation. NF-κB levels were determined by electrophoretic mobility shift assay (EMSA). (B) Female Sprague-Dawley rats were treated with milk exosomes daily (25 mg Exo protein/kg b.wt.) by gavage for 15 days. Top: Lung and liver tissues were analyzed for NF-κB by EMSA. Animals treated with LPS served as a positive control. Data shown are from 3 individual animals. Bottom: Bar graph represents intensity quantification of NF-κB levels. Cont- vehicle control; Mexo- milk exosomes and LPS- Lipopolysaccharide. (C) Anti-inflammatory activity of milk exosomes in the lung tissue of female Sprague-Dawley rats treated with LPS as measured NF-κB activation by EMSA. Rats were treated with LPS (10 mg/kg b.wt.) and milk exosomes (25 mg Exo protein/kg b. wt.), both intraperitoneally, alone or in combination for 6 h. Data from 3-4 animals are shown for each group. Free probe was in excess in all samples and not shown for clarity. Bar graph represents intensity quantification, p<0.01.
4 Discussion
The focus of this study was to develop a biocompatible, exosome-based drug delivery technology. Secreted membrane vesicles such as exosomes inherently possess many characteristics of a drug delivery vehicle and could be accepted in clinics [30]. Here, we show bovine milk as a viable source for isolation of exosomes. Exosomes have been purified from biological fluids and cell culture media using a variety of strategies such as ultracentrifugation, density-based separation (sucrose and Opti-prep), precipitation (ExoQuick), ultrafiltration, and immunoaffinity (reviewed in [31]). However, all isolation procedures unvaryingly have some disadvantages and contain proportions of other membranous vesicles as contaminants [32]. We employed differential centrifugation method to isolate exosomes from bovine milk that was essentially free of any contamination from microvesicles. We acknowledge the fact that the method of vesicle isolation by differential centrifugation is unlikely to completely eliminate the presence of other vesicular bodies. Therefore, our protocol involved discarding the pellet collected at 100,000×g, thus, largely eliminating microvesicles contamination from the supernatant used to harvest the exosomes at 135,000×g. The 100,000×g centrifugation step is usually exercised to collect microvesicles [33] and sometimes exosomes from biological fluids [11]. This approach significantly improved the purity of the exosomes at the expense of somewhat lower yield. Our method is also cost-effective as it does not require any filtrations or sucrose density gradient purifications.
Although there are several reports on exosomes isolation from bovine milk using differential centrifugation alone [34, 35] or in combination with density gradient fractionation [13, 36] or precipitation [37, 38] methods, none of these describe quantitative yield of exosomes. When this manuscript was in preparation, Arntz et al., [39] reported differential centrifugation and filtration methods to isolate exosomes from commercial semi-skimmed milk and collected exosomes pellet at 110,000×g, with a reported yield of 200 mg protein per liter of milk. Our protocol on other hand resulted in higher yield at 335 ± 49 mg protein per liter of milk. Compared to milk typically one liter of cell culture supernatant yields about 0.5-2.0 mg exosomal protein depending of the cell type from which exosomes are harvested [40-43]. Thus, for drug delivery purposes isolation of exosomes from milk is extremely desirable as the yield is over 200 fold higher than those derived from cell culture supernatants. Although differential ultracentrifugation has been the gold standard for exosome isolation and purification, and large-scale ultracentrifugation is possible and has been used for production of other biologics, concerns relating to the relatively low yield prevail and remain to be evaluated for exosome isolation. Alternatively, filtration-based methods that can process thousands of liters are projected as viable alternative for industrial scale production of intact extracellular vesicles for therapeutic applications [44].
Characterization of the isolated particles by NanoSight and Zetasizer indicated an average particle size of <100 nm. The particle size measured by SEM and AFM further confirmed the size to be <80 nm. The buyout density of milk exosomes was estimated as 1.18 g/ml. Immunoblot analysis of the milk exosomes confirmed the presence of endocytic vesicle markers (CD63, CD81, Tsg101 and Alix) and absence of microvesicle (integrin β1 and p-selectin and CD40) and ER (calnexin) markers. Despite reports in the literature, it is arguable whether universal markers for exosomes and microvesicles can be established. Nevertheless, these physical and molecular attributes were in accordance with reports for classification of the secreted vesicle as exosomes [11, 31]. The size and polydispersity index of exosomes further suggested that the material was less prone to excessive aggregation and precipitation, which is a favorable attribute for drug delivery system. The isolated milk exosomes could be stored for prolonged periods at optimal conditions such as at concentration < 6 mg/ml exosomal protein and at −80° C to minimize coagulation or loss of activity.
Immune-regulatory miRNAs and mRNAs are found in both human and bovine milk and play a role in thymic Treg differentiation [27, 45, 46]. Reports have also indicated presence of the RNA moieties in the exosomes isolated from milk [12, 45, 46]. Our findings show presence of several mammary gland- and immune function-related genes and miRNAs. Colostrum contained higher levels of immune-related miRNAs (namely, miR-223), and gene transcripts, a finding that correlates with the higher immune-boosting effects of colostrum compared with matured milk [12]. Recently, Melnik et al. reviewed the role of milk exosomal miRNAs in promoting thymic regulatory T cell maturation and in controlling pivotal target genes [47].
We demonstrated the uptake and biocompatibility of milk exosomes in human cancer cell lines and rodent models. A dose and time dependent uptake of milk exosomes by cancer cells with a non-linear increase beyond 50 μg/ ml and after 8 h of incubation. Similar uptake of bovine colostrum/milk and human breast milk exosomes by cells have been reported [12, 13, 42]. In vivo stability and distribution studies showed that DiR-labeled exosomes were stable in circulation for long periods (up to 6 days). Therefore, exosomal drug delivery is likely to increase drug efficacy due to prolong circulation time and facilitate uptake by cells. It was noted that route of administration had a considerable impact on the tissue distribution of DiR-labeled exosomes. Administration of exosomes by oral gavage route had somewhat uniform tissue distribution while predominant hepatic accumulation of exosomes was observed by i.v. route. Milk derived exosomes did not elicit any systemic toxicity or adverse immune reaction during short-term (1-6 h) or long-term (15 d) exposure in wild type rats. All biochemical, hematological parameters and cytokine profile, except triglycerides and GM-CSF remained unaltered indicating cross-species tolerance of milk exosomes following oral administration. A decrease in triglycerides level and increase in anti-inflammatory cytokine GM-CSF was noted in milk exosome treated animals. Although, these findings indicate potential health benefits of milk exosomes, it needs to be confirmed using a more appropriate animal model with larger sample size.
Milk exosomes exhibited versatility in carrying a wide range of small drug molecules of varied lipophilicity. Similar to liposomes, exosomes have a bi-lipid membrane and an aqueous core, therefore they could be potentially loaded with both hydrophilic and lipophilic drugs [31]. Although the exact mechanism of drug loading onto or into exosomes needs to explored, we hypothesize that the surface lipid and protein nature of the exosomes could, in part, facilitate the drug interaction, and thus loading. However, exosomal drug uptake by diffusion could also be a possible mechanism. Recent studies have exploited the idea of both passive methods such as incubation [48] and active encapsulation techniques such as electroporation, saponin treatment, extrusion and hypotonic dialysis [49, 50] for drug loading in cell-derived exosomes. These techniques could be adapted to milk exosomes to increase drug encapsulation that will result in further improvement in drugs therapeutic effect. In vitro release rates of different drugs ranged between 50-90% in 24 h, suggesting that rapid extraction of drug from exosomes is unlikely to be the case. Drug release from exosomes could be a combination of release of surface bound moieties and by diffusion of encapsulated drug. Slow-release of drugs from exosomes over a period of 24 - 48 h combined with longer circulation time are favorable properties implying that drugs can efficiently reach the target site with higher stability for enhanced therapeutic efficacy.
The enhanced in vitro growth inhibitory and in vivo anti-tumor activity that was observed with exosomal formulation could possibly be due to increased stability of the drugs formulated with exosomes. In addition, modest to significant protective effects such as anti-inflammatory, anti-proliferative and anti-tumor were observed by exosomes alone in the absence of any drug with minimum or no toxicity to normal cells. It is speculated that milk exosomal macromolecules such as miRNAs and mRNAs can play a critical role in the development and modulation of the immune system [35, 46]. Several studies have shown that one of the most remarkable characteristics of malignant cells is the alteration of biologic systems located at the plasma membrane level. Thus, it is plausible that there is a higher uptake of exosomes by cancer cells compared to the normal cells due to the leakiness of cancer cells. Moreover, milk exosomes might be involved in inhibiting cancer-related pathways (e.g., inhibition of NF-κB) for more cell kill against the cancer cells than normal cells. Therefore, the enhanced effects of the exosomal formulations could be ascribed, in part, to increased drug stability in the formulation, longer circulation time, higher uptake and to the intrinsic properties of exosomes.
FA and other vitamin receptors are overexpressed in many cancers and have been extensively exploited to achieve tumor targeting [51]. Tumor targeting of drugs is much sought to reduce off-target toxicity, reduce dose and improve drug efficacy. To this end, we show the feasibility of drug loaded milk exosomes functionalized with tumor targeting ligand such as FA for enhanced anti-tumor activity. Moreover, this higher anti-tumor effects was achieved by oral delivery of the exosomal formulation, indicating the potential of effective oral anti-cancer therapy by exosomal drug delivery. Further, tissue targeting or site-specific delivery of drug loaded exosomes, can be exploited by adding a wide variety of tumor-targeting ligands such as antibodies (e.g., VEGF, EGFR), peptides (e.g., transferrin, integrins, Her2), or receptor-targets (e.g., FA, biotin and hyaluronic acid) to the milk exosomes. Milk exosomes could also be explored to load and deliver potentially other macromolecules such as siRNA, miRNAs, plasmid DNA, cDNA, and proteins (antioxidant enzymes, etc.). Additionally, the protective effects of milk exosomes per se are very intriguing and suggest utility of these nanovesicles against many inflammation-based diseases. For example, exosomes from bovine milk and colostrum could be exploited as additives in formula milk thus potentially serve as immune booster in young children and could also be used for immune-compromised cancer patients undergoing chemotherapy.
5 Conclusions
In summary, we demonstrated that raw mature bovine milk can serve as a biocompatible and cost-effective source for harvesting bulk quantities of exosomes and that milk exosomes have tremendous potential as a drug carrier for hydrophilic and lipophilic agents, including chemo drugs. This nanodevice technology can overcome the limitations associated with the poor oral bioavailability of chemopreventives and chemotherapeutics and lower the total administered dose, thus minimizing or eliminating toxicity generally associated with high doses. Exosomal formulation of drugs can not only enhance biological efficacy and but exosomes functionalized with ligands for tumor targeting can further improve specificity and eliminate off-target side effects of drugs. Milk-derived exosomes has several properties such as physical and biological stability, tolerability, scalability of manufacturing process, versatility of agents it can carry, and ability to functionalize with ligands for targeting, that make it an ideal candidate for drug delivery with wide therapeutic applications. However, further studies are warranted to rule out any potential toxicity with long-term use of milk exosomes.
Supplementary Material
Highlights.
Bovine milk is a biocompatible and scalable source of exosomes.
Milk exosomes can serve as a vehicle to deliver both hydrophilic and lipophilic small drug molecules.
Exosomal formulations provide enhanced biological efficacy.
Milk exosomes can be functionalized with ligands such as folic acid to achieve tumor targeting.
Milk exosomes per se exhibit protective effects presumably due to the endogenous cargo of macromolecules.
Acknowledgements
This work was supported from the USPHS grants CA-118114 and CA-125152, Kentucky Lung Cancer Research Program, Agnes Brown Duggan Endowment, and Helmsley Funds. Dr. Tereza Paronyan is gratefully acknowledged for her assistance in SEM and AFM analyses and Dr. Manicka Vadhanam for useful discussions.
Abbreviations
- miRNA
microRNA
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide
- PBS
phosphate-buffered saline
- qRT-PCR
quantitative reverse transcription -polymerase chain reaction
- cDNA
complementary deoxynucleic acid
- SEM
scanning electron microscopy
- AFM
atomic force microscopy
- CUR
curcumin
- WFA
withaferin A
- Anthos
anthocyanidins
- FA
folic acid
- PAC
paclitaxel
- DOC
docetaxel
- EMSA
Electrophoretic Mobility Shift Assay (EMSA)
- UPLC
ultra-performance liquid chromatography
- TNF-α
tumor necrosis factor- α
- LPS
Lipopolysaccharide
- MVB
mutlivesicular bodies
- VEGF
Vascular endothelial growth factor
- EGFR
Epidermal growth factor receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jabir NR, Tabrez S, Ashraf GM, Shakil S, Damanhouri GA, Kamal MA. Nanotechnology-based approaches in anticancer research. International journal of nanomedicine. 2012;7:4391–4408. doi: 10.2147/IJN.S33838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lim EK, Jang E, Lee K, Haam S, Huh YM. Delivery of cancer therapeutics using nanotechnology. Pharmaceutics. 2013;5:294–317. doi: 10.3390/pharmaceutics5020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barenholz Y. Doxil(R)--the first FDA-approved nano-drug: lessons learned. Journal of controlled release : official journal of the Controlled Release Society. 2012;160:117–134. doi: 10.1016/j.jconrel.2012.03.020. [DOI] [PubMed] [Google Scholar]
- 4.Ciruelos E, Jackisch C. Evaluating the role of nab-paclitaxel (Abraxane) in women with aggressive metastatic breast cancer. Expert review of anticancer therapy. 2014;14:511–521. doi: 10.1586/14737140.2014.883922. [DOI] [PubMed] [Google Scholar]
- 5.Saif MW. U.S. Food and Drug Administration approves paclitaxel protein-bound particles (Abraxane(R)) in combination with gemcitabine as first-line treatment of patients with metastatic pancreatic cancer. JOP : Journal of the pancreas. 2013;14:686–688. doi: 10.6092/1590-8577/2028. [DOI] [PubMed] [Google Scholar]
- 6.Bamrungsap S, Zhao Z, Chen T, Wang L, Li C, Fu T, Tan W. Nanotechnology in therapeutics: a focus on nanoparticles as a drug delivery system. Nanomedicine. 2012;7:1253–1271. doi: 10.2217/nnm.12.87. [DOI] [PubMed] [Google Scholar]
- 7.Desai N. Challenges in development of nanoparticle-based therapeutics. The AAPS journal. 2012;14:282–295. doi: 10.1208/s12248-012-9339-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lai RC, Yeo RW, Tan KH, Lim SK. Exosomes for drug delivery - a novel application for the mesenchymal stem cell. Biotechnology advances. 2013;31:543–551. doi: 10.1016/j.biotechadv.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 9.Kooijmans SA, Vader P, van Dommelen SM, van Solinge WW, Schiffelers RM. Exosome mimetics: a novel class of drug delivery systems. International journal of nanomedicine. 2012;7:1525–1541. doi: 10.2147/IJN.S29661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lakhal S, Wood MJ. Exosome nanotechnology: an emerging paradigm shift in drug delivery: exploitation of exosome nanovesicles for systemic in vivo delivery of RNAi heralds new horizons for drug delivery across biological barriers. Bioessays. 2011;33:737–741. doi: 10.1002/bies.201100076. [DOI] [PubMed] [Google Scholar]
- 11.Thery C, Amigorena S, Raposo G, Clayton A. Bonifacino Juan S., editor. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Current protocols in cell biology / editorial board. 2006 doi: 10.1002/0471143030.cb0322s30. Chapter 3 Unit 3 22. [DOI] [PubMed] [Google Scholar]
- 12.Admyre C, Johansson SM, Qazi KR, Filen JJ, Lahesmaa R, Norman M, Neve EP, Scheynius A, Gabrielsson S. Exosomes with immune modulatory features are present in human breast milk. J Immunol. 2007;179:1969–1978. doi: 10.4049/jimmunol.179.3.1969. [DOI] [PubMed] [Google Scholar]
- 13.Hata T, Murakami K, Nakatani H, Yamamoto Y, Matsuda T, Aoki N. Isolation of bovine milk-derived microvesicles carrying mRNAs and microRNAs. Biochem Biophys Res Commun. 2010;396:528–533. doi: 10.1016/j.bbrc.2010.04.135. [DOI] [PubMed] [Google Scholar]
- 14.El-Andaloussi S, Lee Y, Lakhal-Littleton S, Li J, Seow Y, Gardiner C, Alvarez-Erviti L, Sargent IL, Wood MJ. Exosome-mediated delivery of siRNA in vitro and in vivo. Nature protocols. 2012;7:2112–2126. doi: 10.1038/nprot.2012.131. [DOI] [PubMed] [Google Scholar]
- 15.van den Boorn JG, Schlee M, Coch C, Hartmann G. SiRNA delivery with exosome nanoparticles. Nature biotechnology. 2011;29:325–326. doi: 10.1038/nbt.1830. [DOI] [PubMed] [Google Scholar]
- 16.Sun DM, Zhuang XY, Xiang XY, Liu YL, Zhang SY, Liu CR, Barnes S, Grizzle W, Miller D, Zhang HG. A Novel Nanoparticle Drug Delivery System: The Anti-inflammatory Activity of Curcumin Is Enhanced When Encapsulated in Exosomes. Mol Ther. 2010;18:1606–1614. doi: 10.1038/mt.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haug A, Hostmark AT, Harstad OM. Bovine milk in human nutrition--a review. Lipids in health and disease. 2007;6:25. doi: 10.1186/1476-511X-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dragovic RA, Gardiner C, Brooks AS, Tannetta DS, Ferguson DJP, Hole P, Carr B, Redman CWG, Harris AL, Dobson PJ, Harrison P, Sargent IL. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomed-Nanotechnol. 2011;7:780–788. doi: 10.1016/j.nano.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munagala R, Kausar H, Munjal C, Gupta RC. Withaferin A induces p53-dependent apoptosis by repression of HPV oncogenes and upregulation of tumor suppressor proteins in human cervical cancer cells. Carcinogenesis. 2011;32:1697–1705. doi: 10.1093/carcin/bgr192. [DOI] [PubMed] [Google Scholar]
- 20.Paul PK, Gupta SK, Bhatnagar S, Panguluri SK, Darnay BG, Choi Y, Kumar A. Targeted ablation of TRAF6 inhibits skeletal muscle wasting in mice. The Journal of cell biology. 2010;191:1395–1411. doi: 10.1083/jcb.201006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bansal SS, Vadhanam MV, Gupta RC. Development and in vitro-in vivo evaluation of polymeric implants for continuous systemic delivery of curcumin. Pharmaceutical research. 2011;28:1121–1130. doi: 10.1007/s11095-011-0375-z. [DOI] [PubMed] [Google Scholar]
- 22.Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012;40:D1241–1244. doi: 10.1093/nar/gkr828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee Y, El Andaloussi S, Wood MJ. Exosomes and microvesicles: extracellular vesicles for genetic information transfer and gene therapy. Hum Mol Genet. 2012;21:R125–134. doi: 10.1093/hmg/dds317. [DOI] [PubMed] [Google Scholar]
- 24.Mathivanan S, Simpson RJ. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics. 2009;9:4997–5000. doi: 10.1002/pmic.200900351. [DOI] [PubMed] [Google Scholar]
- 25.Taylor DD, Gercel-Taylor C. The origin, function, and diagnostic potential of RNA within extracellular vesicles present in human biological fluids. Frontiers in genetics. 2013;4:142. doi: 10.3389/fgene.2013.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, Zimmermann P, David G. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nature cell biology. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 27.Kosaka N, Izumi H, Sekine K, Ochiya T. microRNA as a new immune-regulatory agent in breast milk. Silence. 2010;1:7. doi: 10.1186/1758-907X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu LF, Liston A. MicroRNA in the immune system, microRNA as an immune system. Immunology. 2009;127:291–298. doi: 10.1111/j.1365-2567.2009.03092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corrado C, Raimondo S, Chiesi A, Ciccia F, De Leo G, Alessandro R. Exosomes as Intercellular Signaling Organelles Involved in Health and Disease: Basic Science and Clinical Applications. Int J Mol Sci. 2013;14:5338–5366. doi: 10.3390/ijms14035338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnsen KB, Gudbergsson JM, Skov MN, Pilgaard L, Moos T, Duroux M. A comprehensive overview of exosomes as drug delivery vehicles - Endogenous nanocarriers for targeted cancer therapy. Bba-Rev Cancer. 2014;1846:75–87. doi: 10.1016/j.bbcan.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 31.Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Bba-Gen Subjects. 2012;1820:940–948. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 32.Tauro BJ, Greening DW, Mathias RA, Ji H, Mathivanan S, Scott AM, Simpson RJ. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56:293–304. doi: 10.1016/j.ymeth.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 33.Batista BS, Eng WS, Pilobello KT, Hendricks-Munoz KD, Mahal LK. Identification of a conserved glycan signature for microvesicles. Journal of proteome research. 2011;10:4624–4633. doi: 10.1021/pr200434y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolf T, Baier SR, Zempleni J. The Intestinal Transport of Bovine Milk Exosomes Is Mediated by Endocytosis in Human Colon Carcinoma Caco-2 Cells and Rat Small Intestinal IEC-6 Cells. The Journal of nutrition. 2015;145:2201–2206. doi: 10.3945/jn.115.218586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Izumi H, Tsuda M, Sato Y, Kosaka N, Ochiya T, Iwamoto H, Namba K, Takeda Y. Bovine milk exosomes contain microRNA and mRNA and are taken up by human macrophages. Journal of dairy science. 2015;98:2920–2933. doi: 10.3168/jds.2014-9076. [DOI] [PubMed] [Google Scholar]
- 36.Reinhardt TA, Lippolis JD, Nonnecke BJ, Sacco RE. Bovine milk exosome proteome. Journal of proteomics. 2012;75:1486–1492. doi: 10.1016/j.jprot.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 37.Pieters BC, Arntz OJ, Bennink MB, Broeren MG, van Caam AP, Koenders MI, van Lent PL, van den Berg WB, de Vries M, van der Kraan PM, van de Loo FA. Commercial cow milk contains physically stable extracellular vesicles expressing immunoregulatory TGF-beta. PloS one. 2015;10:e0121123. doi: 10.1371/journal.pone.0121123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamada T, Inoshima Y, Matsuda T, Ishiguro N. Comparison of methods for isolating exosomes from bovine milk. The Journal of veterinary medical science / the Japanese Society of Veterinary Science. 2012;74:1523–1525. doi: 10.1292/jvms.12-0032. [DOI] [PubMed] [Google Scholar]
- 39.Arntz OJ, Pieters BC, Oliveira MC, Broeren MG, Bennink MB, de Vries M, van Lent PL, Koenders MI, van den Berg WB, van der Kraan PM, van de Loo FA. Oral administration of bovine milk derived extracellular vesicles attenuates arthritis in two mouse models. Molecular nutrition & food research. 2015;59:1701–1712. doi: 10.1002/mnfr.201500222. [DOI] [PubMed] [Google Scholar]
- 40.Sheng H, Hassanali S, Nugent C, Wen L, Hamilton-Williams E, Dias P, Dai YD. Insulinoma-released exosomes or microparticles are immunostimulatory and can activate autoreactive T cells spontaneously developed in nonobese diabetic mice. J Immunol. 2011;187:1591–1600. doi: 10.4049/jimmunol.1100231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Estelles A, Sperinde J, Roulon T, Aguilar B, Bonner C, LePecq JB, Delcayre A. Exosome nanovesicles displaying G protein-coupled receptors for drug discovery. International journal of nanomedicine. 2007;2:751–760. [PMC free article] [PubMed] [Google Scholar]
- 42.Rahman MJ, Regn D, Bashratyan R, Dai YD. Exosomes released by islet-derived mesenchymal stem cells trigger autoimmune responses in NOD mice. Diabetes. 2014;63:1008–1020. doi: 10.2337/db13-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitchell JP, Court J, Mason MD, Tabi Z, Clayton A. Increased exosome production from tumour cell cultures using the Integra CELLine culture system. J Immunol Methods. 2008;335:98–105. doi: 10.1016/j.jim.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 44.E.L.A. S, Mager I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nature reviews. Drug discovery. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 45.Sun Q, Chen X, Yu J, Zen K, Zhang CY, Li L. Immune modulatory function of abundant immune-related microRNAs in microvesicles from bovine colostrum. Protein Cell. 2013;4:197–210. doi: 10.1007/s13238-013-2119-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou Q, Li M, Wang X, Li Q, Wang T, Zhu Q, Zhou X, Gao X, Li X. Immune-related microRNAs are abundant in breast milk exosomes. Int J Biol Sci. 2012;8:118–123. doi: 10.7150/ijbs.8.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Melnik BC, John SM, Schmitz G. Milk: an exosomal microRNA transmitter promoting thymic regulatory T cell maturation preventing the development of atopy? J Transl Med. 2014;12:43. doi: 10.1186/1479-5876-12-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang T, Martin P, Fogarty B, Brown A, Schurman K, Phipps R, Yin VP, Lockman P, Bai S. Exosome delivered anticancer drugs across the blood-brain barrier for brain cancer therapy in Danio rerio. Pharmaceutical research. 2015;32:2003–2014. doi: 10.1007/s11095-014-1593-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tian YH, Li SP, Song J, Ji TJ, Zhu MT, Anderson GJ, Wei JY, Nie GJ. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials. 2014;35:2383–2390. doi: 10.1016/j.biomaterials.2013.11.083. [DOI] [PubMed] [Google Scholar]
- 50.Fuhrmann G, Serio A, Mazo M, Nair R, Stevens MM. Active loading into extracellular vesicles significantly improves the cellular uptake and photodynamic effect of porphyrins. Journal of Controlled Release. 2015;205:35–44. doi: 10.1016/j.jconrel.2014.11.029. [DOI] [PubMed] [Google Scholar]
- 51.Chen S, Zhao X, Chen J, Chen J, Kuznetsova L, Wong SS, Ojima I. Mechanism-based tumor-targeting drug delivery system. Validation of efficient vitamin receptor-mediated endocytosis and drug release. Bioconjugate chemistry. 2010;21:979–987. doi: 10.1021/bc9005656. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


