Abstract
Over the past decade, it became evident that proteins perform critical functions as components of specialized macromolecular complexes. Here, we discuss a recent study by Wan and colleagues, which highlights the significance of protein complexes by studying their conservation in organisms separated by up to a billion years of evolution.
Keywords: Protein interactions, protein-protein interaction, evolution, macromolecular complex, mass spectrometry, interactome
Charles Darwin originally hypothesized in his seminal work On the Origin of Species that natural selection, more colloquially known as `survival of the fittest,' is the primary driver of evolution [1]. Since Darwin's era, this tenet has been supported by evidence from multiple scientific disciplines. The use of radiometric dating techniques and expansion of the fossil record has clarified the structural and ontological relationship between present-day animals and their ancestors. Yet, we do not have a complete picture of the molecular underpinnings of biochemical evolution.
One goal at the heart of biochemical evolution is to understand the interplay among central dogma components and the mechanisms by which new cellular functions have arisen in species of greater complexity. The development of genome sequencing approaches has enabled more detailed insight into the molecular `history' of natural selection and DNA-protein relationships. Born from this work is the idea that genetic mutations resulting in non-conservative amino acid substitutions would often be deleterious to protein structure and thus disfavored by natural selection. Yet, paradoxically, this type of amino acid substitution would have significant potential to create new protein functions. Recent work in bacteria has shown that one strategy to overcome the inherent negative selectivity against mutations that disrupt protein structure/function is to increase chaperone activity, albeit at an additional cellular energy burden [2]. These studies underscore the broader importance of interpreting a gain or loss of function in the context of protein-protein interactions.
This is not a trivial task, yet the advent of mass spectrometry-based proteomics, paired with biochemical fractionation and affinity purification techniques, has rapidly generated large-scale datasets on intracellular protein-protein relationships [3–5]. These studies have convincingly supported the idea that proteins rarely function autonomously, but rather as components of multiprotein complexes and interdependent signaling cascades. So, while the guiding hand of natural selection preserves positive adaptations to protein structure, it must also accommodate the secondary effects on physical interacting partners and regulatory relationships.
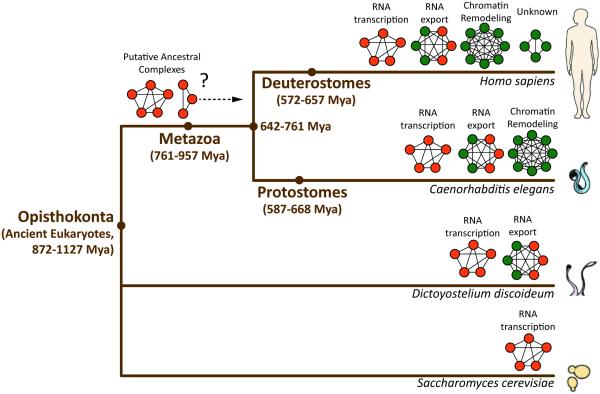
Comparative analyses of protein interaction networks within whole organisms have shown that physical associations (protein complexes) and functional relationships can be conserved between orthologous groups [6]. Yet, there are many unanswered questions regarding the scope of protein network conservation across phyla separated by hundreds of millions of years of evolution. A recent work published in Nature from Wan and colleagues [7] elegantly tackles this essential aspect of biochemical evolution. To gain insight into the biochemical evolution of eukaryotes, they studied the protein association networks across nine unique extant species, ranging from the unicellular slime mold, Dictyostelium discoideum, to Homo sapiens. The proteome of each organism was biochemically separated, totaling nearly 6,400 fractions, which were then analyzed by quantitative mass spectrometry, resulting in the co-fractionation of 13,386 protein orthologue profiles. Using an integrated computational platform to determine high-confidence intra-species protein associations and between species conservation, 981 putative conserved multiprotein groups were identified. In evolutionary terms, the protein components comprising these conserved groups spanned from 500 million to over 1 billion years old. Strikingly, 490 complexes contained ancient components (> 1 billion years old) that were ubiquitously expressed in different cell and tissue types, were frequently abundant, and had low domain complexity. Several core metazoan macromolecular complexes, such as those involved in metabolic regulation and RNA transcription, displayed exquisite evolutionary stability. This suggests that once the physical and functional coordination of complex components have been set by natural selection, these primordial complexes are rarely re-designed, but refined through the incorporation of additional components that add specialized functionality (Figure 1).
Figure 1.
Phylogenetic Relationships and Protein Complex Conservation Among Yeast (Saccharomyces cerevisiae), Slime Mold (Dictyostelium discoideum), Worms (Caenorhabditis elegans), and Humans (Homo sapiens). Conserved protein complexes were determined by biochemical co-fractionation, mass spectrometry, and computational scoring [7]. Representative complexes were selected to highlight the increasing diversity from lower to higher order present-day eukaryotes. Lower order eukaryotes tend to possess highly conserved complexes containing ancient components (orange nodes) that can be traced through hundreds of millions of years (Mya) of evolution, or a mixture of old and new components (green nodes). Higher order eukaryotes often retain these protein complexes, while also possessing complexes that have acquired newer components, reflecting more-specialized functions. The evolution of ancestral complexes to present-day complexes remains to be fully understood.
Importantly, the conserved complexes clustered by Wan and colleagues have statistically significant overlap with findings from other complementary affinity purification-mass spectrometry studies, which provide an orthogonal validation of the identified protein complexes [5]. Yet, a striking aspect of their study was that about two-thirds of the conserved complexes had not been annotated in protein interaction databases. This includes a novel `Commander' complex, whose co-fractionation profiles supported a 500 kDa composition. The identification of each component by mass spectrometry allowed the authors to design functional studies that uncovered a role for the Commander complex in embryonic neural development. Using antisense morpholinos to individually knockdown the COMMD2 and COMMD3 subunits in tadpoles caused impaired head and eye development and neural patterning. Further genetic and mechanistic investigation of the members of the Commander complex is warranted, given the recent reports that linked CCDC22, another Commander complex component, to human brain developmental disabilities [8].
Placing this evolutionary conservation in a broader context, the authors noted that the number of conserved complexes represent an estimated 10 – 25% of all predicted eukaryotic complexes [9]. Therefore, greater mechanistic understanding of the role for complexes unique to multicellular organisms in processes such as cell differentiation and adhesion, and by extension tumorigenesis, will likely come as evolutionarily younger and lower abundance complexes are characterized. The identification of mixed complexes, containing ancient components revised with modern gene products during evolution, provides intriguing targets for understanding protein complex 'speciation' events. Understanding how and why these modern components were integrated into ancestral protein complexes would provide the molecular basis for adaptation of a species to its changing environment. A particularly relevant example is the continued co-evolution of host-pathogen interactions. The dynamic assembly of protein complexes plays a critical role in host responses to viral and bacterial pathogens (e.g., [10]). Correlating the biochemical evolution of protein complexes to the biological evolution of host-pathogen relationships could aid in the identification of critical anti-viral host factors. Additionally, this study highlights the need to investigate other aspects of protein evolution. The increasing depth of analysis afforded by proteomic technologies has now cataloged hundreds of thousands of post-translational modifications across species. Classifying post-translational modifications by their degree of conservation will accelerate the discovery of specific modification sites with critical regulatory roles. Ultimately, as science continues to reveal the effects of natural selection at the amino acid, protein, and protein assembly level, a clearer picture of the relationship between biochemical and biological evolution will emerge.
Acknowledgments
Many scientists have contributed to protein interaction studies, and we apologize for not being able to give credit because of citation limits. We are grateful for funding from NIH (GM114141 and HL127640) to IMC and NJCCR to TMG.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Darwin CR. On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life. John Murray; London: 1859. [PMC free article] [PubMed] [Google Scholar]
- 2.Pechmann S, Frydman J. Interplay between chaperones and protein disorder promotes the evolution of protein networks. PLoS Comput. Biol. 2014;10:e1003674. doi: 10.1371/journal.pcbi.1003674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guruharsha KG, et al. A Protein Complex Network of Drosophila melanogaster. Cell. 2011;147:690–703. doi: 10.1016/j.cell.2011.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hein MY, et al. A Human Interactome in Three Quantitative Dimensions Organized by Stoichiometries and Abundances. Cell. 2015;163:712–723. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 5.Huttlin EL, et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharan R, et al. Conserved patterns of protein interaction in multiple species. Proc. Natl. Acad. Sci. U. S. A. 2005;102:1974–1979. doi: 10.1073/pnas.0409522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wan C, et al. Panorama of ancient metazoan macromolecular complexes. Nature. 2015;525:339–344. doi: 10.1038/nature14877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voineagu I, et al. CCDC22: a novel candidate gene for syndromic X-linked intellectual disability. Mol. Psychiatry. 2012;17:4–7. doi: 10.1038/mp.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stumpf MP, et al. Estimating the size of the human interactome. Proc. Natl. Acad. Sci. U. S. A. 2008;105:6959–6964. doi: 10.1073/pnas.0708078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diner BA, et al. Interactions of the Antiviral Factor Interferon Gamma-Inducible Protein 16 (IFI16) Mediate Immune Signaling and Herpes Simplex Virus-1 Immunosuppression. Mol. Cell. Proteomics. 2015;14:2341–2356. doi: 10.1074/mcp.M114.047068. [DOI] [PMC free article] [PubMed] [Google Scholar]