Abstract
Purpose:
Segmentation of the hippocampus from magnetic resonance (MR) images is a key task in the evaluation of mesial temporal lobe epilepsy (mTLE) patients. Several automated algorithms have been proposed although manual segmentation remains the benchmark. Choosing a reliable algorithm is problematic since structural definition pertaining to multiple edges, missing and fuzzy boundaries, and shape changes varies among mTLE subjects. Lack of statistical references and guidance for quantifying the reliability and reproducibility of automated techniques has further detracted from automated approaches. The purpose of this study was to develop a systematic and statistical approach using a large dataset for the evaluation of automated methods and establish a method that would achieve results better approximating those attained by manual tracing in the epileptogenic hippocampus.
Methods:
A template database of 195 (81 males, 114 females; age range 32–67 yr, mean 49.16 yr) MR images of mTLE patients was used in this study. Hippocampal segmentation was accomplished manually and by two well-known tools (FreeSurfer and hammer) and two previously published methods developed at their institution [Automatic brain structure segmentation (ABSS) and LocalInfo]. To establish which method was better performing for mTLE cases, several voxel-based, distance-based, and volume-based performance metrics were considered. Statistical validations of the results using automated techniques were compared with the results of benchmark manual segmentation. Extracted metrics were analyzed to find the method that provided a more similar result relative to the benchmark.
Results:
Among the four automated methods, ABSS generated the most accurate results. For this method, the Dice coefficient was 5.13%, 14.10%, and 16.67% higher, Hausdorff was 22.65%, 86.73%, and 69.58% lower, precision was 4.94%, −4.94%, and 12.35% higher, and the root mean square (RMS) was 19.05%, 61.90%, and 65.08% lower than LocalInfo, FreeSurfer, and hammer, respectively. The Bland–Altman similarity analysis revealed a low bias for the ABSS and LocalInfo techniques compared to the others.
Conclusions:
The ABSS method for automated hippocampal segmentation outperformed other methods, best approximating what could be achieved by manual tracing. This study also shows that four categories of input data can cause automated segmentation methods to fail. They include incomplete studies, artifact, low signal-to-noise ratio, and inhomogeneity. Different scanner platforms and pulse sequences were considered as means by which to improve reliability of the automated methods. Other modifications were specially devised to enhance a particular method assessed in this study.
Keywords: medical imaging, image processing, segmentation, hippocampus, temporal lobe epilepsy, magnetic resonance imaging (MRI)
1. INTRODUCTION
Epilepsy is defined by the occurrence of at least two recurrent epileptic seizures in less than 24 h. The hippocampus is a seminal structure in the most common surgically treated form of epilepsy and it also factors highly in the pathology of Alzheimer’s disease and mild cognitive impairment. The prevalence of epilepsy in developing countries is 14–57 cases per 1000 individuals1,2 and in developed countries, 4–10 cases.3 Temporal lobe epilepsy (TLE) represents a group of disorders in which patients suffer from recurrent epileptic seizures arising in one or both temporal lobes of the brain. According to the International League Against Epilepsy (ILAE), TLE is categorized into two main groups: mesial TLE (mTLE) and lateral TLE. The most common form of epilepsy is mTLE, which is characterized by recurrent complex partial seizures.4 Hippocampal sclerosis is usually considered as its pathophysiological substrate.5
Surgical resection of the epileptogenic hippocampus and neighboring structures is considered optimal for long-term seizure freedom in drug-resistant mTLE patients. However, successful surgical outcome is dependent on an accurate analysis of pathoanatomical and functional changes in the hippocampus.6 Accurate segmentation of the hippocampus aids in establishing asymmetry regarding size and signal characteristics in order to disclose the likely site of epileptogenicity. With sufficient refinement, it may ultimately aid in the avoidance of invasive monitoring with its expense and risk for the patient. To this end, a reliable and consistent method for segmentation of the hippocampus from magnetic resonance imaging (MRI) is needed.
Subfields of the hippocampus are characterized by multiple edges of low contrast and low signal-to-noise ratio with discontinuous and missing boundaries. Moreover, both the size and shape of the hippocampus change along its longitudinal axis. These characteristics make the automated segmentation very challenging. Manual segmentation of the hippocampus, therefore, is regarded as the current gold standard7 for establishing dimension, although it involves experienced personnel in a time-consuming task not ideal in the current clinical practice. There are also both inter-rater and intrarater variabilities which produce inconsistencies in manual segmentation rendering it less than optimal; conversely, automated methods can be designed to address such variability analytically.
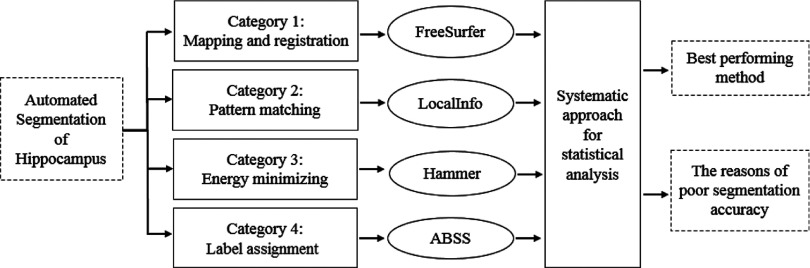
Over the past decade, new algorithms have been developed for automated segmentation of the hippocampus. Accuracy, reliability, and reproducibility have been the key elements in the evolution of these techniques. We have categorized automated techniques into four groups: (1) mapping and registration based on single or multiatlas; (2) pattern-matching based on information registration; (3) shape-fitting based on energy minimizing, and (4) label assignment based on pattern recognition and machine learning. In some studies, combinations of these categories have also been considered. For instance, both atlas registration and minimization of an energy function have been applied for segmentation purposes.8
In the first category, a reference image called an atlas mask is registered to the subject image. After registration, the coordinates of the region of interest (ROI) are mapped from the atlas image to the subject image.9 Many studies have been done on the atlas selection and its effect on segmentation accuracy in the hippocampal regions.10 A graph-cuts algorithm combined with atlas-based segmentation and morphological opening has been proposed for hippocampal extraction.11 By using a multiatlas segmentation approach, multiple atlases are registered to the target image and the deformed labels are combined.12 A multiatlas framework with autocontext models (ACMs) has been suggested for automated segmentation of 7T magnetic resonance (MR) images.13 Multiatlas methods are used as nonparametric regression models in the high-dimensional space of image patches for characterizing expected segmentation error.14 A multiatlas segmentation propagation technique has been adapted for performing label fusion using local similarity to involve pathology in TLE.15 FreeSurfer16 is a publicly available package with subcortical atlas-based features for automated segmentation of the brain structures. It includes volumetric segmentation, intersubject alignment, segmentation of hippocampal subfields, white matter fascicles segmentation, and construction of surface models of cerebral cortex. Nonlinear template matching is used in this tool for segmentation of brain structures such as the hippocampus. The atlas is registered to the target image and the atlas labels are then deformed to the coordinate framework of the target image.
Defined techniques in the second category segment a predefined ROI using a knowledge-based system. Pattern-matching has been shown as a robust and repeatable methodology for hippocampal segmentation in MR images.17 Prior knowledge about hippocampal position was used for volumetric diffeomorphic normalization and pattern-matching as a semiautomatic technique.18 Elastic pattern-matching and an evolutionary heuristic approach were combined with prior statistical information about ROI to control deformable patterns.19 LocalInfo is such an algorithm, with information-based registration for segmentation of brain structures.20 In this technique, the right and left hippocampi are segmented using information-based multiple atlases. It applies affine registration for representing the coordinates of the hippocampus in an atlas mask. Three- and ten-label fuzzy classification, tissue-type information extraction, and optimization of shape parameters are performed for the segmentation task. Extraction of principal and mean shapes by LocalInfo is accomplished through nonrigid registration of atlases and subject images, transformation to label maps with approximation to those that correspond ideally, affine registration, and principal component analysis (PCA).
In the third category, different energy-minimizing models guided by internal and external shape forces such as discrete contours, classic snakes, deformable contours, and level-set models have also been developed for automated segmentation of brain structures.21 Modified deformable counters with minimizing energy functions22,23 have been specifically suggested for medical image segmentation. A group of deformable templates, iteratively modified to minimize a hybrid local/global energy, have been used for fitting to the contours of target structures in MRI.24 hammer is an elastic registration method for MR images of the brain which is also publicly available. This method minimizes the energy function for deformable registration and segments brain structures from an atlas. It uses a hierarchical procedure for optimization of an energy function and a set of features to derive volumetric structures. Moreover, the concept of an attribute vector is used to characterize brain structures in the vicinity of each voxel. Geometric moment invariants (GMIs) are applied for representing the geometric structure of the underlying anatomy. This method includes a morphometric analysis for the segmentation of high-resolution images.25
The fourth category of automated segmentation techniques is the pattern-recognition algorithms such as artificial neural networks (ANNs) and other machine-learning techniques which assign labels to image patterns. The intensities of a MRI voxel and its neighboring voxels are applied as inputs to a three-layer ANN.26 Local label learning using both image intensity and texture features is proposed to estimate the target image’s segmentation label by support vector machine (SVM) to distinguish a hippocampal voxel from its neighboring voxels.27 A mask representing manual tracing is used as the search space and morphological operations are used to reduce extension of the region beyond the hippocampal boundaries. Automatic brain structure segmentation (ABSS)28 is based on ANNs and has been developed for the segmentation of the brain structures from a MRI. Shape and signed-distance functions of the hippocampal region are represented in different scales by GMIs and ANNs. For each scale, the GMIs, as well as voxel intensities and coordinates, are fed as input while the signed-distance function is considered as output of an ANN. Finally, ANN outputs of different stages are fed into another ANN. The output of the overall system consists of two different regions, inside and outside of the hippocampus.
In this work, we present a systematic and statistical analysis approach for evaluation of automated segmentation methods in order to establish one that reliably approximates the results achieved by manual tracing of the hippocampus. As stated above, we have categorized the basic algorithms that may be used to develop an automated technique into four groups. Then, we have chosen a fully automated technique from each group and assessed accuracy, reliability, and reproducibility using a database of mTLE MRI studies. To this end, we have applied established methods of performance evaluation on the segmentation results. Next, based on the segmentation results, we have categorized the input conditions of those cases with unacceptable segmentation outcomes to find necessary conditions promoting optimal results. At the end, we have addressed the shortcomings of the automated segmentation methods and how to improve them.
The automatic methods chosen to evaluate each category were selected on the basis of availability and other criteria defining ease of use (Fig. 1). ABSS and LocalInfo were developed at our institution. FreeSurfer and hammer are public software and are readily accessible, whereas access to others is restricted. FreeSurfer and hammer are well-known tools and have been widely used for automatic segmentation. Comparison of these common methods with newer techniques (ABSS and LocalInfo) is useful to establish progress in methodology. The absence of any need for initial parameter settings or initial conditions in the chosen methods enhances reproducibility by remaining insensitive to user-defined parameters or variability in user setting. They considered in the literature to be of low complexity and expeditious, traits that are sought after in the imaging industry. Running time and complexity of methods are primary driving elements in clinical application.
FIG. 1.
The automatic techniques are categorized into four groups: (1) mapping and registration based on single or multiatlas, (2) pattern-matching based on information registration, (3) shape-fitting based on energy minimization, and (4) labeling assignment based on pattern recognition and machine learning. A fully automatic technique is chosen for each group. By statistical analysis, the accuracy, reliability, and reproducibility are assessed using a large database of mTLE MRI studies. The best performing method was selected and the input conditions of those cases with unacceptable segmentation outcomes were evaluated. Some modifications were specially devised to enhance a particular method assessed in this study.
The rest of the paper is organized as follows. In Sec. 2, the subjects and the imaging protocols are presented. In Sec. 3, the methods including skull stripping, manual and automatic segmentation, and performance measures are introduced. In Sec. 4, experimental results and statistical analysis of the validation methods are presented. The paper is concluded in Sec. 5 where a final discussion is presented.
2. MATERIALS
2.A. Subjects
An archival review of mTLE patients treated between June 1993 and June 2014 at Henry Ford Hospital, Detroit, MI was performed. The patients were evaluated by neurologic examination, video-electroencephalography (EEG), MR and nuclear medicine imaging, and neuropsychological testing. A template database of MR images of 195 mTLE patients (81 males, 114 females; age range of 32–67 yr, mean age of 49.16 yr) was used in the study. Analysis of the images was approved by the institutional IRB board. Individual patient consent was not required as the analysis used anonymized data that had been acquired previously for the patient care.
2.B. Imaging protocol
Preoperative MR images were obtained using a 1.5T or a 3.0T MRI system (GE Medical Systems, Milwaukee, USA) and included coronal T1-weighted images, using inversion recovery spoiled gradient echo, IRSPGR protocol. On the 1.5T MRI, T1-weighted imaging parameters were TR/TI/TE = 7.6/1.7/500 ms, flip angle = 20°, and voxel size = 0.781 × 0.781 × 2.0 mm3. On the 3.0T MRI, T1-weighted imaging parameters were TR/TI/TE = 10.4/4.5/300 ms, flip angle = 15°, and voxel size = 0.39 × 0.39 × 2.00 mm3.
3. METHODS
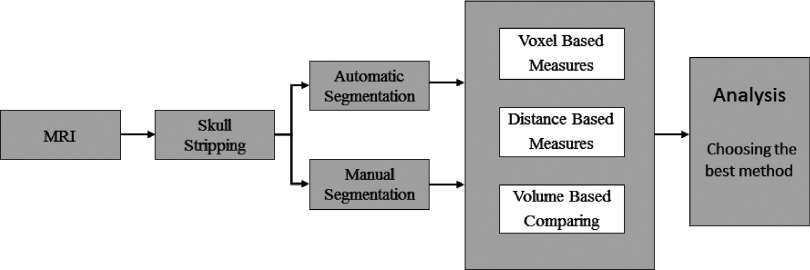
The following sequence of steps was carried out for each case: skull stripping, manual segmentation, automatic segmentation by four methods, statistical validation by voxel-, distance-, and volume-based measures, and analysis of the results to find out reasons for the failure of specific segmentation methods in certain cases. A flowchart of the steps carried out in this work is shown in Fig. 2. The steps are explained below.
FIG. 2.
Flowchart of the work: (1) skull stripping, (2) hippocampal segmentation by manual and automatic segmentation methods, (3) statistical validation by voxel, distance, and volume measures, and (4) analysis of the results based on the input data.
3.A. Skull stripping
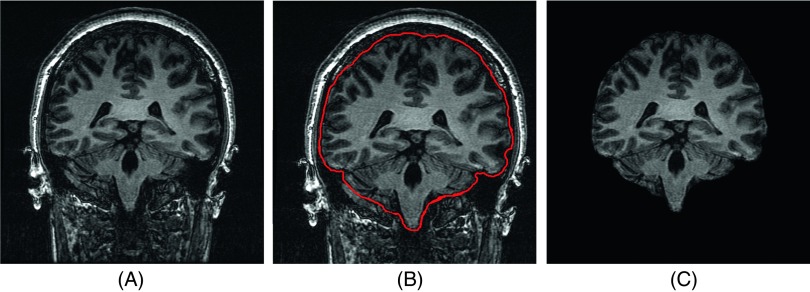
To enable further processing, scalp, skull, and dura were removed from the T1-weighted images by automated and manual skull stripping. Optimal elimination of nonbrain tissues provides for better subsequent volumetric analysis.29,30 The results of using the skull stripped images revealed that accuracy of some automatic segmentation algorithms like atlas-based methods and LocalInfo enhanced through skull stripping as other applications such as brain morphometry, cortical surface reconstruction, and presurgical planning. Image analysis software such as BrainSuite,31 MRIcro,32 spm,33 and label (Ref. 34) was used for this step. The stripping procedure was performed manually in the image slices that the results of automated methods were judged unacceptable. Figure 3 illustrates the procedure.
FIG. 3.
Skull-stripping steps: (A) input images, (B) brain contouring, and (C) removal of nonbrain tissues.
3.B. Manual segmentation
Using the coronal T1-weighted MR images, the hippocampi were outlined by a previously established protocol.35 To this end, the standard procedure applied in clinical trials was followed for all subjects. The ROIs encompassing the hippocampi were outlined in the coronal plane; then, fine-tuning steps were performed using the sagittal view. The hippocampal position was established using a MRI atlas as Refs. 36 and 37. Features of the MRI atlas and its distinction from other atlases have been previously reported.35 Separation of the anterior head of the hippocampus from the amygdala was facilitated by the linea alba and inferior limb of the lateral ventricle.38 For each subject, both of the right and left hippocampi were segmented using MRIcro by a research assistant who was trained on this task but was blind to the video-EEG monitoring and other clinical data. The hippocampus was traced posteriorly to include the gyrus fasciola.39 Figure 4 shows a sample result of the manual segmentation. Segmentation results were checked and corrected if needed by a neuroscientist with expertise in hippocampal segmentation (a co-author of the paper). Tracing of the right and left hippocampi boundaries per subject took approximately 5 h by an experienced research assistant and required optimal hand-eye coordination.9
FIG. 4.
Manual segmentation of the hippocampi using coronal T1-weighted MR images and a previously established segmentation protocol.
3.C. Automated segmentation
The automated methods were applied to the images in order to extract hippocampal volume. DICOM images were converted by MRIcro to NIfTI format; then, the following automated segmentation methods were applied to the images.
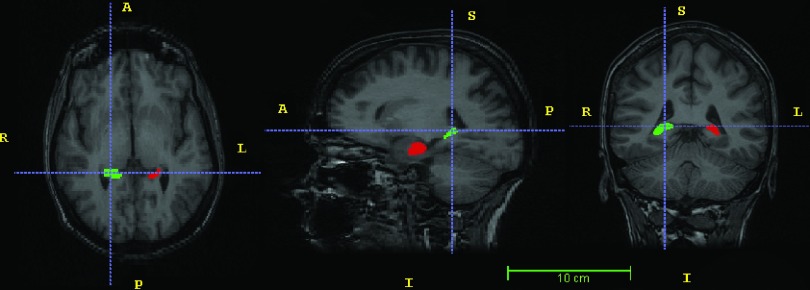
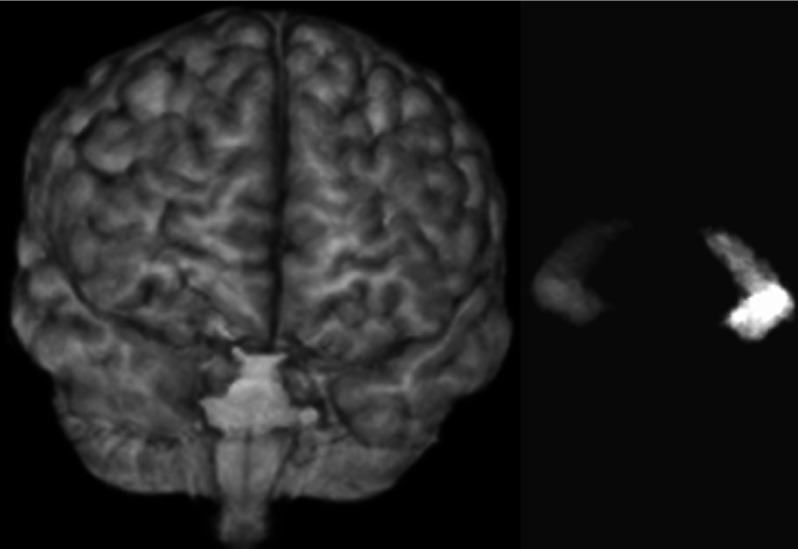
FreeSurfer version 5.3.0 was applied where the processing steps included transformation into Talairach coordinates, segmentation of the subcortical white matter and gray matter, and removal of nonbrain tissues. hammer, a technique based on the minimization of the energy function for deformable registration, was also applied. This method uses registration to warp a statistical model to the target subject’s coordinate framework. To compute the optimal segmentation, the information from the warped atlas is used in combination with a statistical intensity model.8 LocalInfo was applied next. This approach is specifically designed for hippocampal segmentation. Affine registration is applied in order to represent the coordinates of the hippocampus in the atlas mask. Finally, ABSS, a pattern-recognition algorithm was applied for hippocampal segmentation. In this approach, shape and signed-distance functions of the manually established hippocampus are represented in different scales to train ANNs. The trained ANNs are then applied to extract the right and left hippocampi. Figure 5 shows the surface-rendered cortex and the hippocampi of a 52 years old female with mTLE; the hippocampi are segmented using the ABSS method.
FIG. 5.
Surface rendering of the cortex (left) and segmented hippocampi (right) of a 52 years old female with mTLE. The hippocampal segmentation is performed using the ABSS method.
3.D. Performance measures
The accuracy of different automated segmentation techniques was assessed by comparison of their segmentation to manual segmentation. Intraexpert and interexpert variabilities of segmentation and quality evaluation were considered.40 Several performance metrics, widely used in the literature, were considered to assess correlation and overlap between automatic and manual segmentation results. Three groups of measurements were implemented. The first involved voxel-based metrics such as Dice coefficient, similarity, precision, average symmetric surface distance (ASSD), sensitivity, specificity, accuracy, negative predictive value, and rational absolute value degree (RAVD). The second group was composed of metrics based on distance evaluation such as Hausdorff, Hausdorff 95, root mean square (RMS), and mean distance (MD). In the third group, volume-based comparisons were used to find the overlap between automated and manual techniques in 3D. Among the extracted measures, those judged most efficient in computing intuitively reasonable quantities with a firm underlying theoretical basis were analyzed.
3.D.1. Measurements based on voxel
The similarity between automated and manual segmentation results can be assessed using overlap measures. One of the most popular methods, used for comparing each of the four automated methods against the gold standard, was based on the Dice coefficient, defined as
| (1) |
where A and R represent the set of segmented hippocampal voxels by automatic and manual methods and N, the relative number of elements. In the binary case, each voxel can rate as 1 for belonging to the structure or 0 as a background voxel. The Dice coefficient ranges from 0 to 1, where 1 refers to complete overlap. The volumes are measured by voxel counts.
By definition, true positive (TP), the voxel proportion that correctly determines the hippocampus; true negative (TN), the voxel proportion of the background that segments as background; false positive (FP), the voxel proportion of the background that segments as hippocampus, and false negative (FN), the voxel proportion of the hippocampus that segments as background. Precision or positive predictive value was defined as the number of true positive voxels for hippocampal segmentation divided by the sum of both true positives and false positives for voxel segmentation of the hippocampus,
| (2) |
Another measure that is analogous to precision is similarity. In this metric, FN, the voxel proportion of the hippocampus that segments as background was added to thedenominator,
| (3) |
Other metrics used include
| (4) |
| (5) |
| (6) |
| (7) |
3.D.2. Measurements based on distance
The degree of resemblance of two images that are superimposed on each other is determined by the Hausdorff distance,41 which is the distance between two compact nonempty subsets of a metric space42 and is calculated here to find the similarity of automatic and manual segmentation results. Given two closed and bounded subsets, A and R, of a metric space M, the Hausdorff distance is defined as
| (8) |
where
| (9) |
and ‖ ⋅ ‖ is some underlying norm on the points of A and R. As H(A, R) diminishes, the overlap between A and R increases. Due to outliers, the 95th percentile can be used for comparison as Hausdorff 95.
RMS uses surface-to-surface geometrics to define a distance metric. It is commonly used as a statistical measure to show the magnitude of a varying quantity,
| (10) |
where SA denotes the set of surface voxels of A (automatic), SR denotes the set of surface voxels of R (manual), and is the squared nearest Euclidian distance from a surface point “a” to the surface SA. This metric is based on the distances between hippocampal surfaces extracted by two segmentation algorithms. RMS is used to evaluate whether the quantity of segmentation varies, so that the smaller its value, the higher its similarity.
ASSD, calculated as the average distance of all contour points which are segmented automatically to the closest contour point of manual tracing, is as follows:
| (11) |
where the notations are the same as for RMS.
For each voxel on the boundary of the automatic segmented region, the distance to the closest voxel on the boundary of the true label (manual) is found as dA. Using this vector, MD is defined by
| (12) |
where NA denotes the total number of voxels on the boundary of the hippocampus which are segmented automatically.
3.D.3. Measurement based on volume
Reliability of automated techniques was evaluated by volumetric assessment of the resulting hippocampal volumes. For this, the volume of hippocampal structures was estimated using each of the above techniques and compared to the results obtained by the manual approach. To study the strength of the correlation and similarity of the extracted volumes, the Pearson cross correlation of two datasets as a function of subject number was found using
| (13) |
where VA and VR are the volume generated by the automatic and manual (reference) approaches, E is the expected value operator, and σ is the standard deviation of the data. Figure 6 shows the extracted volume waveform per subject for the manual and FreeSurfer techniques, separated for the right and left hippocampi. For modeling the relationship between scalar arrays of extracted volumes by automated and manual approaches, a linear regression was found. To analyze the “agreement” between automated and gold standard (i.e., manual) results, the Bland–Altman plot was used. To this end, the horizontal and vertical axes were defined as
| (14) |
where VR is the volume segmented manually and VA is the volume segmented by an automatic approach.
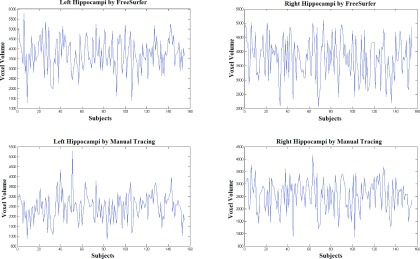
FIG. 6.
Volume waveforms vs subjects for the right and left hippocampi (patients with Dice > 4.0), for the automated and manual methods.
Finally, by the same notation, the RAVD is defined as
| (15) |
4. RESULTS AND ANALYSIS
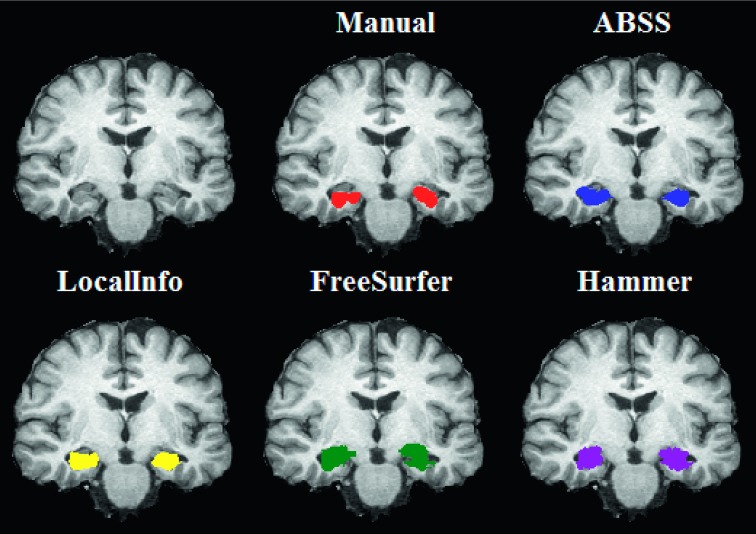
A comparison of manual and automatic hippocampal segmentation methods from a representative mTLE case is presented in Fig. 7. All methods (software) were applied in the manner suggested by the software developer, i.e., without any initialization by the user; the inputs to the software were the anatomical T1-weighetd images only. As a visual demonstration of good and poor performances, it represents a typical case where ABSS outperforms the other methods.
FIG. 7.
A comparison of manual and automatic hippocampal segmentation methods. The figure shows an intermediate (first row, first column) section of a skull-stripped T1 image of a mTLE patient. The first overlay (first row, second column) shows the manual segmentation, the second overlay (first row, third column) shows the ABSS segmentation, the third overlay (second row, first column) shows the LocalInfo segmentation, the fourth overlay (second row, second column) shows the FreeSurfer segmentation, and the last overlay (second row, third column) shows the hammer segmentation.
Different performance measurements were extracted and calculated for each of the segmentation methods from the T1-weighted images of the mTLE subjects. The resulting Hausdorff, Hausdorff 95, Dice, similarity, precision, RMS, mean distance, ASSD, sensitivity, specificity, accuracy, NPV, and RAVD values are presented in Table I. Dice, Hausdorff, precision, and RMS produced values that were felt to fulfill a reasonable basis for analysis.
TABLE I.
Performance measurements estimated for evaluating automatic segmentation methods.
| Method | Hausdorff | Hausdorff 95 | Dice | Similarity | Precision | RMS | Mean distance | ASSD | Sensitivity | Specificity | Accuracy | NPV | RAVD | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subject 1 | ABSS | 3.00 | 1.00 | 0.86 | 0.76 | 0.89 | 0.56 | 0.27 | 0.29 | 0.84 | 1.00 | 1.00 | 1.00 | −0.06 |
| LocalInfo | 4.12 | 1.00 | 0.85 | 0.73 | 0.83 | 0.63 | 0.35 | 0.33 | 0.86 | 1.00 | 1.00 | 1.00 | 0.05 | |
| Freesurfer | 7.55 | 3.74 | 0.70 | 0.53 | 0.93 | 1.45 | 0.46 | 0.89 | 0.56 | 1.00 | 1.00 | 1.00 | −0.40 | |
| hammer | 12.21 | 2.45 | 0.71 | 0.55 | 0.78 | 1.31 | 0.72 | 0.77 | 0.65 | 1.00 | 1.00 | 1.00 | −0.16 | |
| Subject 2 | ABSS | 4.24 | 1.41 | 0.83 | 0.72 | 0.78 | 0.69 | 0.44 | 0.38 | 0.89 | 1.00 | 1.00 | 1.00 | 0.13 |
| LocalInfo | 4.58 | 1.41 | 0.81 | 0.67 | 0.75 | 0.75 | 0.49 | 0.44 | 0.87 | 1.00 | 1.00 | 1.00 | 0.17 | |
| Freesurfer | 7.07 | 4.24 | 0.66 | 0.50 | 0.79 | 1.51 | 0.61 | 0.97 | 0.57 | 1.00 | 1.00 | 1.00 | −0.27 | |
| hammer | 11.00 | 4.00 | 0.67 | 0.50 | 0.68 | 1.52 | 0.99 | 0.90 | 0.66 | 1.00 | 1.00 | 1.00 | −0.03 | |
| Subject 3 | ABSS | 7.62 | 2.24 | 0.73 | 0.58 | 0.79 | 1.00 | 0.50 | 0.59 | 0.69 | 1.00 | 1.00 | 1.00 | −0.13 |
| LocalInfo | 31.89 | 27.31 | 0.06 | 0.03 | 0.04 | 14.74 | 14.32 | 12.07 | 0.11 | 1.00 | 1.00 | 1.00 | 1.36 | |
| Freesurfer | 11.18 | 6.08 | 0.58 | 0.41 | 0.75 | 2.16 | 0.71 | 1.34 | 0.48 | 1.00 | 1.00 | 1.00 | −0.37 | |
| hammer | 11.49 | 6.00 | 0.42 | 0.27 | 0.47 | 2.71 | 1.78 | 1.92 | 0.38 | 1.00 | 1.00 | 1.00 | −0.19 | |
| Subject 4 | ABSS | 7.14 | 4.24 | 0.73 | 0.58 | 0.74 | 1.48 | 0.70 | 0.87 | 0.72 | 1.00 | 1.00 | 1.00 | −0.03 |
| LocalInfo | 6.71 | 3.74 | 0.69 | 0.53 | 0.68 | 1.45 | 0.83 | 0.95 | 0.71 | 1.00 | 1.00 | 1.00 | 0.05 | |
| Freesurfer | 11.22 | 7.28 | 0.63 | 0.46 | 0.75 | 2.49 | 0.69 | 1.53 | 0.54 | 1.00 | 1.00 | 1.00 | −0.28 | |
| hammer | 11.22 | 4.69 | 0.64 | 0.47 | 0.66 | 2.00 | 1.10 | 1.27 | 0.62 | 1.00 | 1.00 | 1.00 | −0.07 |
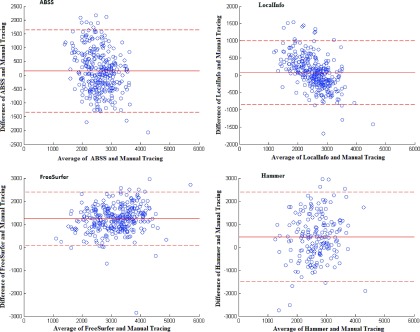
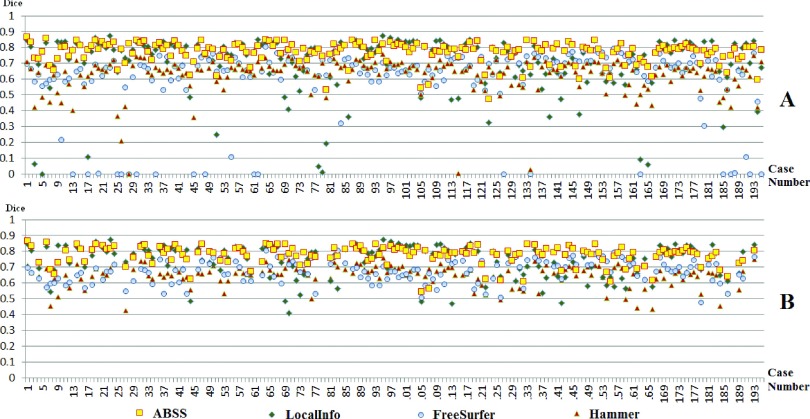
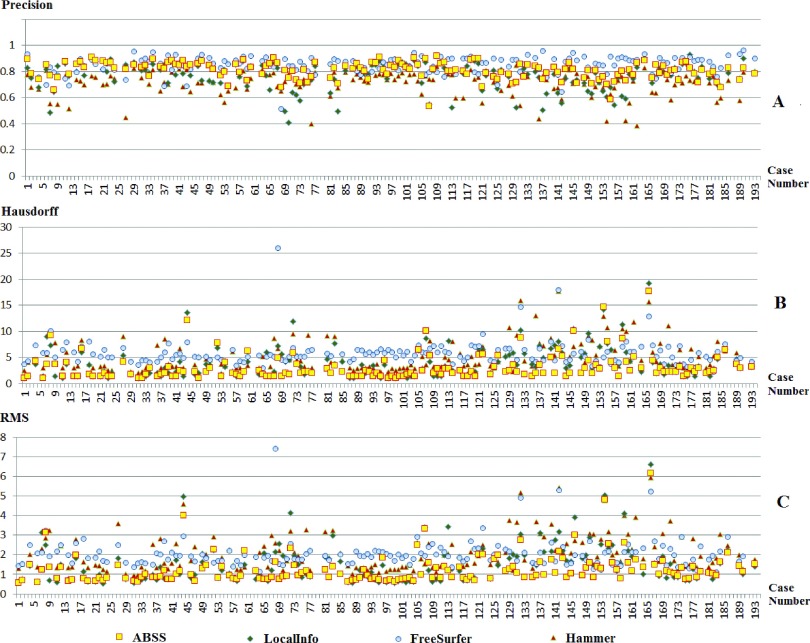
Figure 8 compares the extracted volumes by linear regression per subject and Fig. 9 shows the resulting Bland–Altman plots. Table II summarizes the results in terms of the mean and standard error of the Dice coefficient, Hausdorff distance, precision, and RMS. The Dice coefficient of the evaluated methods by case number is shown in Fig. 10(A). A value less than 0.4 represents an unacceptable overlap with the established gold standard. Such cases were removed from analysis [Fig. 10(B)]. Figure 11 shows precision, Hausdorff, and RMS values for each of the methods. Overall analysis of the voxel and distance-based measures, excluding precision, confirms that ABSS provides more correlation, overlap, and reproducibility than the other methods.
FIG. 8.
Hippocampal volumes (mm3) estimated by the manual and automated methods for the left and right hippocampi and their linear regression.
FIG. 9.
Bland–Altman plots comparing manual results versus the following: ABSS (first row, first column), LocalInfo (first row, second column), FreeSurfer (second row, first column), and hammer (second row, second column) results for the left and right hippocampi.
TABLE II.
Mean and standard error of different measures.
| ABSS | LocalInfo | FreeSurfer | hammer | |
|---|---|---|---|---|
| Dice | 0.78 ± 0.01 | 0.74 ± 0.01 | 0.67 ± 0.01 | 0.65 ± 0.01 |
| Hausdorff | 3.09 ± 0.20 | 3.79 ± 0.23 | 5.77 ± 0.20 | 5.24 ± 0.23 |
| Precision | 0.81 ± 0.01 | 0.77 ± 0.01 | 0.85 ± 0.01 | 0.71 ± 0.01 |
| RMS | 1.26 ± 0.06 | 1.50 ± 0.08 | 2.04 ± 0.06 | 2.08 ± 0.07 |
FIG. 10.
Dice coefficient of the four automatic segmentation methods (FreeSurfer, hammer, LocalInfo, ABSS) vs case number for the following: (A) all 195 mTLE patients and (B) 157 mTLE patients with Dice > 0.4. Since those cases with unacceptable automated segmentation outcomes were evaluated separately, they were removed in (B) of this figure and the subsequent figures.
FIG. 11.
Precision (A), Hausdorff distance (B), and RMS (C) of the four automatic segmentation methods (ABSS, LocalInfo, Freesurfer, hammer) vs case number for 157 mTLE patients with Dice >0.4, sorted by case numbers.
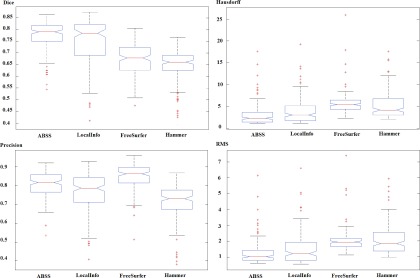
The Dice coefficient for ABSS is 5.13% (p-value < 2 × 10−3), 14.10% (p-value < 5 × 10−33), and 16.67% (p-value < 2 × 10−47) higher compared to LocalInfo, FreeSurfer, and hammer, respectively. The ABSS method of segmentation, therefore, is shown to have more overlap with the gold standard than the other methods. The Hausdorff distance for ABSS is 22.65% (p-value < 3 × 10−2), 86.73% (p-value < 7 × 10−19), and 69.58% (p-value < 2 × 10−11) lower compared to LocalInfo, FreeSurfer, and hammer, respectively, also confirming that the ABSS automated segmentation method better approximates the gold standard. The precision for ABSS is 4.94% (p-value < 3 × 10−10), −4.94% (p-value < 3 × 10−8), and 12.35% (p-value < 2 × 10−21) higher compared to LocalInfo, FreeSurfer, and hammer, respectively. That obtained using Freesurfer ranked the highest. The RMS distance for ABSS is 19.05% (p-value < 2 × 10−2), 61.90% (p-value < 6 × 10−18), and 65.08% (p-value < 5 × 10−16) lower compared to LocalInfo, FreeSurfer, and hammer, respectively, demonstrating less variation in values than the other competing methods. Since all methods were paired for comparison, the differences among groups were analyzed by ANOVA. The standard ANOVA is illustrated in Table III and a graphical depiction of groups for each measurement in 157 subjects by quartile is shown in Fig. 12. The statistical significance of the model which is shown by a large F and small values of p correspond to a large difference in the center lines of the boxplots.
TABLE III.
The standard ANOVA table. The source of variability is indicated in the second column. The third column comprises the sum of squares (SS) due to each source and the fourth column, the degrees of freedom (df) associated with each source. The mean squares (MS), which is the ratio SS/df, is shown in the fifth column. The ratio of the mean squares is shown as the F-statistic in column number six, and finally, the last column is the p-value derived from the cumulative distribution function of F.
| Measurements | Source | Sum of squares (SS) | Degree of freedom (df) | Mean squares (MS) | F-statistic | p-value |
|---|---|---|---|---|---|---|
| Dice | Columns | 1.66 | 3 | 0.55 | 94.13 | 2.91 × 10−50 |
| Error | 3.67 | 624 | 5.89 × 10−3 | — | — | |
| Total | 5.33 | 627 | — | — | — | |
| Hausdorff | Columns | 730.41 | 3 | 243.47 | 32.83 | 1.03 × 10−19 |
| Error | 4627.01 | 624 | 7.415 | — | — | |
| Total | 5357.42 | 627 | — | — | — | |
| Precision | Columns | 1.62 | 3 | 0.54 | 76.75 | 2.89 × 10−42 |
| Error | 4.39 | 624 | 7.04 × 10−3 | — | — | |
| Total | 6.01 | 627 | — | — | — | |
| RMS | Columns | 76.89 | 3 | 25.63 | 34.92 | 7.11 × 10−21 |
| Error | 457.96 | 624 | 0.73 | — | — | |
| Total | 534.85 | 627 | — | — | — |
FIG. 12.
Graphical depiction of groups using Dice (first row, first column), Hausdorff (first row, second column), precision (second row, first column), and RMS (second row, second column) on 157 subjects by quartile. Outliers are plotted by a plus sign.
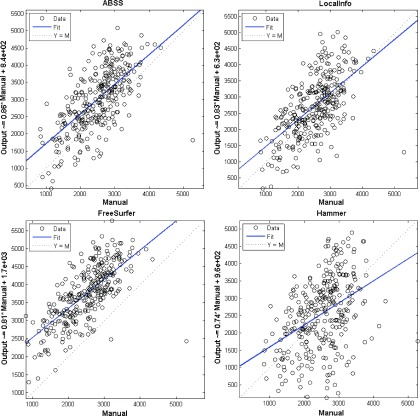
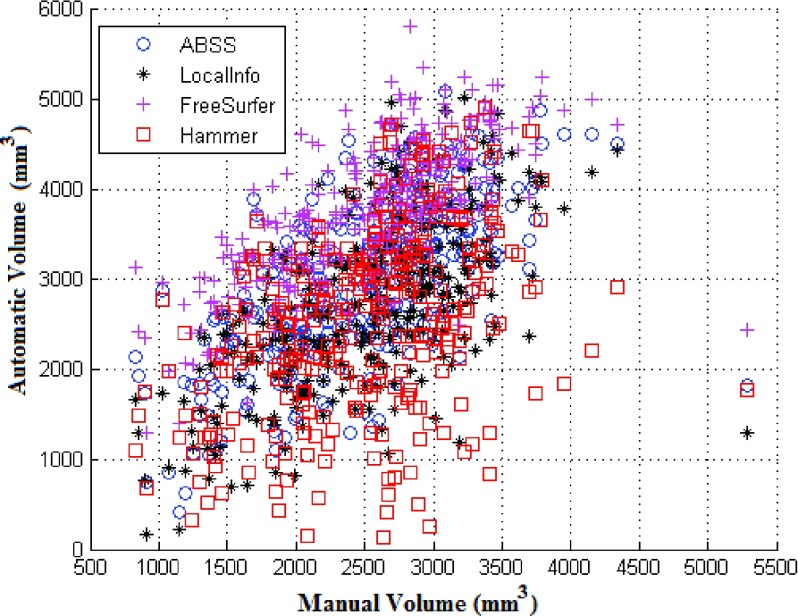
Table IV shows the mean and standard deviation of the segmented volumes, correlation coefficients for each hippocampus (left-to-right ratio), and the limits of agreement with a benchmark for each of the methods. Bias as a source of systematic difference between two methods is identified. The volumes generated by each of the automatic methods compared to those obtained by manual means are shown in Fig. 13. This scatterplot shows FreeSurfer to have extracted larger volumes than the other methods.
TABLE IV.
Summary of the means and standard deviations of hippocampal volumes, cross correlation with the manual method, left-to-right ratios, limits of agreement, and bias between automatic and manual methods.
| Hippocampal volume (voxel) | Cross-correlation | ||||||
|---|---|---|---|---|---|---|---|
| Left | Right | Left | Right | Left-to-right hippocampal ratio | Limits of agreement with manual (voxel) | Bias (voxel) | |
| Manual | 2499.37 ± 798.35 | 2577.43 ± 742.58 | — | — | 0.96 | — | — |
| ABSS | 2552.16 ± 521.25 | 2672.40 ± 542.56 | 0.97 | 0.98 | 0.95 | [−842.37; 1001.21] | 79.42 |
| LocalInfo | 2728.33 ± 610.43 | 2713.90 ± 595.13 | 0.96 | 0.97 | 1.00 | [−842.37; 1001.21] | 153.19 |
| FreeSurfer | 3727.91 ± 1011.63 | 3763.47 ± 854.27 | 0.97 | 0.98 | 0.99 | [87.09; 2387.15] | 1237.12 |
| hammer | 2601.63 ± 606.47 | 2501.97 ± 789.94 | 0.93 | 0.94 | 1.04 | [−1343.07; 1649.45] | 445.19 |
FIG. 13.
Visual comparison of the hippocampus volumes (mm3) estimated by the automated segmentation methods vs ground truth (manual segmentation).
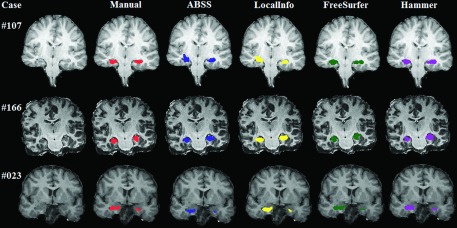
Although ABSS is shown here to be superior to other segmentation methods by the statistical methods applied, it may not be the best method in all cases. From the numerical data, we know that there are a few cases in which other methods outperformed ABSS. Figure 10(B) and 11 reveal that for subject numbers 107, 166, and 23, hammer, FreeSurfer, and LocalInfo, respectively, produced better segmentation than ABSS. The segmentation of these cases is shown in Fig. 14. Evaluations of these cases reveal features that create flaws in the ABSS segmentation methodology. Incomplete imaging in the coronal view for case number 23 rendered a truncated image of the posterior hippocampal region. Reliance on precise skull-stripping for the ABSS method created the suboptimal result for case number 166 where poor skull-stripping resulted in a dark region in the inferior aspect of the brain. In case number 107, unusual shapes of the left and right temporal lobes rendered inaccurate segmentation because of a reliance upon a relatively normal morphology.
FIG. 14.
Segmentation results of other methods in cases that they outperformed ABSS. For subject numbers 107, 166, and 23, hammer, FreeSurfer, and LocalInfo, respectively, produced better segmentation than ABSS.
5. DISCUSSION
Automated techniques for hippocampal segmentation have been developed in the research community to reduce the time-consuming workload and improve upon reproducibility because of the variability encountered in the manual method. There has been consensus that manual measurement of the hippocampal volume is, however, more accurate than the automated techniques because of the better visual definition of the hippocampal margins37 and that it remains the ground truth (gold standard) currently.43 Automated methods, on the other hand, promote operator independence, higher reproducibility, and improved clinical applicability.15 Their application will be judged valid if they provide results comparable to those obtained by the manual contouring of the structure.44 A reliable, objective, and reproducible technique for automated hippocampal segmentation, in particular, would expedite the confident processing of absolute volumes in clinical cases in order to judge the degree of bihemispheric asymmetry and establish the degree of atrophy over time.
Relatively few studies have compared results of the automated hippocampal segmentation methods with those of manual tracing in epilepsy cases.32,39 Several automated hippocampal segmentation methods have been proposed in the literature; however, most have been tested only in nonepileptic subjects. FreeSurfer was shown to have greater accuracy than FSL/FIRST especially in the head and tail portions of the hippocampus when compared to manual tracing in nonepileptic subjects.45 Its feasibility in distinguishing hippocampal volumes in the young adult was also confirmed.46 Both the volume and shape of the hippocampi change in several neuropsychiatric conditions and in pathologic brains such as mTLE. Difficulties arise in the accurate delineation of the hippocampal boundaries under some of these circumstances. Further estimation of the efficacy of the automated methods must, therefore, be obtained to undertake a proper assessment of their applicability. Few reports of automated hippocampal segmentation in cases affected by mTLE have been published.20,39,47,48 In this study, four automated methods were compared against the manual approach using 195 mTLE cases to provide a robust comparative analysis of their respective performances.
Analysis of voxel-, distance-, and volume-based metrics shows that ABSS and LocalInfo are more accurate segmentation methods in the case of mTLE. According to the Dice coefficient, Hausdorff distance, and root mean square distance, hammer and FreeSurfer were discovered to be less accurate. FreeSurfer was more precise than LocalInfo and hammer; however, when other parameters were engaged, this advantage was not sustained. This result was predictable because FreeSurfer segments larger regions than other automated methods and manual segmentation. True positive values and, ultimately, precision increase under these circumstances. Consequently, precision is not as meaningful as other measures in the evaluation of the hippocampal segmentation results. A combination of several metrics would be a better overall measure of performance.
Bland–Altman plots were used to compare the automated segmentation results with the gold standard and assess agreement among the automated methods by investigating the existence of any systematic differences with a fixed bias. The mean value of the difference of the evaluated techniques from the gold standard shows the presence of a fixed bias on the basis of the one-sample t-test. A high correlation would not always indicate that there was a good agreement with the gold standard. hammer and ABSS showed a very weak linear relationship and cross-correlation with the manual results. Some limitation in agreement and a fixed bias in this circumstance must be considered. Greater agreement between ABSS and LocalInfo is evident than with the manual technique when the mean difference is considered as the estimated bias. The left/right hippocampal ratios show a large bias in several mTLE cases, although they approximate unity in control cases. Left to right hippocampal ratios remove considerations of the brain size through normalization.49 These were found not to change significantly among the automated methods compared to those of the manual method. Therefore, the automated methods are thought to work well with ratio metrics in the analysis of asymmetry.
ABSS as a label assignment technique provides a robust segmentation in comparison to other techniques. Such machine-learning techniques search to choose the best labels as ROIs, minimizing disagreement with true segmentations of example images.50 ABSS segments are based on an interconnected group of artificial neurons and it models complex relationships between inputs and outputs by supervised learning. The generalization capability of machine-learning algorithms offers some theoretical insight into the reasons underlying the superior performance of ABSS. This machine-learning approach provides for training by several different atlases, comprised of both normal subjects and patients, and finds hypotheses that explain past observations to make accurate predictions. A weak point for some methods, such as FreeSurfer and LocalInfo, which require registration, will be certain details that may be lost as a high degree of variability is not allowed because of the regularization used in registration.51 Moreover, in these techniques, where one-to-one mapping between the input image and all anatomical images of atlases is not possible, a registration-based labeling framework inevitably will create incorrect labels. Also, due to the inherent registration errors, local incorrect matching can lead to segmentation errors.52 Another shortcoming of methods using label fusion techniques, such as LocalInfo, may arise through the same weighting applied to all samples wherein the criterion-based voting procedure is sensitive to registration error.53 These are limitations not shared by ABSS which will have reduced segmentation errors relative to the other methods.
In the literature, segmentation techniques have often been tested with few subjects and a small database. Therefore, such studies may have been biased toward good results. The present study employs a large dataset and has evaluated the segmentation methods with both nonpathological material and real patient data. This approach has been supported in the literature.48,54 In comparison to manual segmentation, the results of automatic segmentation remain relatively weak but, with continuing improvement, their applicability supports growing acceptance.48
Among 195 subjects, 38 could not be segmented efficiently at least by one of the automated methods as their Dice measure was less than 0.4. We evaluated the input MR images slice-by-slice to find the reason for this inability. The in-plane resolutions and slice thicknesses were evaluated. Their orientation using ImageJ and MRICro was checked and, in some cases, the vendors contacted to discuss possible sources of error. The ABSS method showed acceptable segmentation performance for all subjects, always demonstrating more than 40% coverage agreement between manual and automated segmentations while other methods failed adequate agreement in some subjects. The results show that ABSS, as a pattern-recognition technique, has the ability of learning and adapting to the manual segmentation and to produce acceptable results. Since there has been controversy over the manual segmentation method to be used in particular imaging studies,55 the automatic methods that have learning ability would be able to adjust to specific definitions of a structure and thus would greatly benefit the neuroimaging community.56
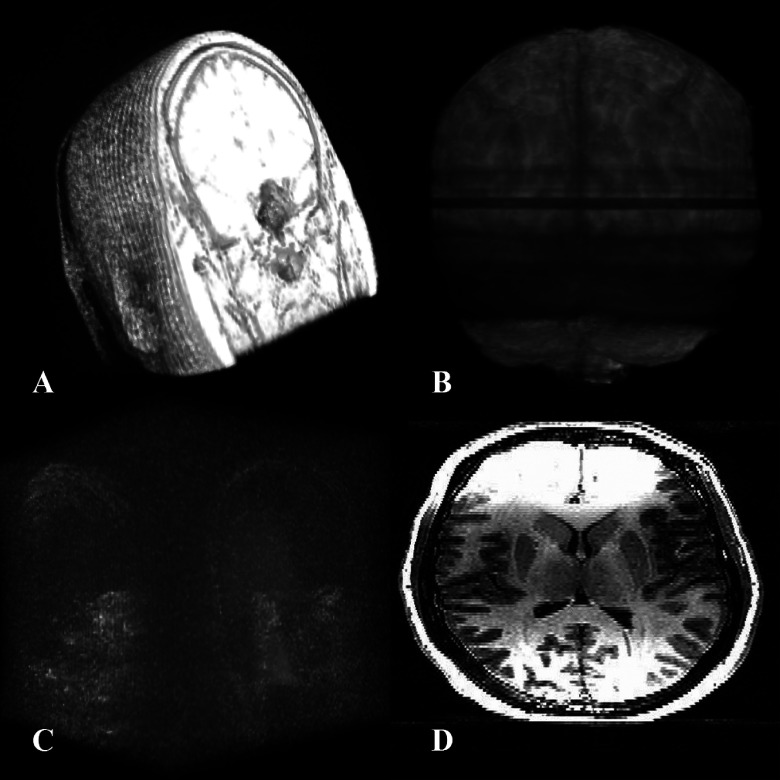
The input conditions of those cases with unacceptable segmentation results (i.e., Dice < 0.4) were addressed and categorized into four groups (see Fig. 15): (1) incomplete imaging that affected all automated methods, (2) an artifact with an interference pattern posed further difficulties as addressed in Ref. 57. Among the known MRI artifacts in the case of hippocampal segmentation are motion, vascular pulsation, susceptibility, chemical shift, Gibbs ringing, RF overflow, partial volume, cross-excitation, shading, zebra stripes, and zipper artifacts,58 (3) poor image quality further added to the problem. Subjects with an extensive childhood stroke, a large dysplastic hippocampus, or those with extensive subependymal heterotopia are examples of conditions contributing to the latter. In this category, some of the heterotopic gray matter could be misclassified as hippocampus. An extensive stroke affecting the hippocampus can result in significant change with resulting poor image quality as was noted in one of our cases that proved to be an outlier, (4) field inhomogeneity or image distortion with signal loss, created by magnetic material inside or outside the patient or scanning at the edge of the field. Apart from the above technical issues, methods of skull stripping also vary in their performance. The adjacent membranes or meninges surrounding the brain are found to be variably stripped and may contribute to volumetric alterations. Minor field inhomogeneities may be corrected before the application of the segmentation algorithms.
FIG. 15.
Four categories of input images in which segmentation output is unfavorable: (A) incomplete image set, (B) image with artifact, (C) noisy, low-quality image, and (D) inhomogeneous image.
Some limitations of this study require accounting. FreeSurfer and hammer use atlases which differ from those used with LocalInfo and ABSS. This may account for some difference in the average hippocampal volume obtained by each method. Such differences, however, do not hinder an assessment of performance of each method as it compares to the results obtained by the manual method. The comparisons drawn with the dice coefficients between the automated and manual methods are meant to declare which method should be preferred. Another limitation concerned manual tracing itself as the gold standard for segmentation given the inherent difficulties with inconsistent labeling across the slice direction.8 This matter was addressed by the selection of 195 high-quality manually segmented cases where clearly defined anatomical landmarks could be ascertained.
After analyzing the structure and functionality of the automated methods, we divided them to four general groups: (1) mapping and registration based on single or multiatlas; (2) pattern-matching based on information registration; (3) shape-fitting based on energy minimizing; and (4) label assignment based on pattern recognition and machine learning. By considering the input conditions of those cases with unacceptable segmentation results, some modifications can be devised and suggested that would enhance performance through improved reliability and reproducibility. The accuracy of atlas-based segmentation methods is dependent upon the scanner platform and pulse sequence that vary with the chosen atlas. Intensity normalization can be used to adjust the atlas intensity model of segmentation to new input data.59 Furthermore, a specific protocol can be established to define the hippocampus anatomically in vivo using a multitemplate fusion approach driven by expert manual labels.55 In the pattern-matching approaches, prior statistical information regarding the target shape can be used to guide the dynamic wrapping behavior of the deformable structures. A hybrid method to combine an evolutionary heuristic and a general elastic template-matching approach has been developed to improve these algorithms.19 Energy minimizing techniques would benefit by defining a new term in their energy function to address a bias field for intensity inhomogeneities. A local clustering criterion function in the level set functions has been suggested to address intensity properties with respect to the neighborhood center in MR images.60 Finally, there have been new ideas to improve pattern recognition techniques of segmentation based on image conditions. These have included computational models of fuzzy clustering, possibilistic clustering, geo-statistics, and knowledge combination.61 Supervised frameworks for segmentation using a probabilistic boosting tree classifier62 and autocontext information, combining multiple dynamic models and deep learning architecture,63 are some examples.
ACKNOWLEDGMENT
This work was supported in part by NIH Grant No. R01-EB013227.
REFERENCES
- 1.Burneo J. G., Tellez-Zenteno J., and Wiebe S., “Understanding the burden of epilepsy in Latin America: A systematic review of its prevalence and incidence,” Epilepsy Res. 66, 63–74 (2005). 10.1016/j.eplepsyres.2005.07.002 [DOI] [PubMed] [Google Scholar]
- 2.Carpio A. and Hauser W. A., “Epilepsy in the developing world,” Curr. Neurol. Neurosci. Rep. 9, 319–326 (2009). 10.1007/s11910-009-0048-z [DOI] [PubMed] [Google Scholar]
- 3.Bell G. and Sander J., “CPD—Education and self-assessment the epidemiology of epilepsy: The size of the problem,” Seizure 10, 306–316 (2001). 10.1053/seiz.2001.0584 [DOI] [PubMed] [Google Scholar]
- 4.Fritschy J.-M., Kiener T., Bouilleret V., and Loup F., “GABAergic neurons and GABA-receptors in temporal lobe epilepsy,” Neurochem. Int. 34, 435–445 (1999). 10.1016/S0197-0186(99)00040-6 [DOI] [PubMed] [Google Scholar]
- 5.Engel J., “Mesial temporal lobe epilepsy: What have we learned?,” Neuroscientist 7, 340–352 (2001). 10.1177/107385840100700410 [DOI] [PubMed] [Google Scholar]
- 6.Hosseini M.-P., Nazem-Zadeh M. R., Mahmoudi F., Ying H., and Soltanian-Zadeh H., “Support vector machine with nonlinear-kernel optimization for lateralization of epileptogenic hippocampus in MR images,” in 36th Annual International Conference of the IEEE on Engineering in Medicine and Biology Society (EMBC) (IEEE, Chicago, IL, 2014), pp. 1047–1050. 10.1109/EMBC.2014.6943773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheenstra A. E., van de Ven R. C., van der Weerd L., van den Maagdenberg A. M., Dijkstra J., and Reiber J. H., “Automated segmentation of in vivo and ex vivo mouse brain magnetic resonance images,” Mol. Imaging 8(1), 35–44 (2009). [PubMed] [Google Scholar]
- 8.van der Lijn F., den Heijer T., Breteler M., and Niessen W. J., “Hippocampus segmentation in MR images using atlas registration, voxel classification, and graph cuts,” NeuroImage 43, 708–720 (2008). 10.1016/j.neuroimage.2008.07.058 [DOI] [PubMed] [Google Scholar]
- 9.Carmichael O. T., Aizenstein H. A., Davis S. W., Becker J. T., Thompson P. M., Meltzer C. C., and Liu Y., “Atlas-based hippocampus segmentation in Alzheimer’s disease and mild cognitive impairment,” NeuroImage 27, 979–990 (2005). 10.1016/j.neuroimage.2005.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aljabar P., Heckemann R. A., Hammers A., Hajnal J. V., and Rueckert D., “Multi-atlas based segmentation of brain images: Atlas selection and its effect on accuracy,” NeuroImage 46, 726–738 (2009). 10.1016/j.neuroimage.2009.02.018 [DOI] [PubMed] [Google Scholar]
- 11.Kwak K., Yoon U., Lee D.-K., Kim G. H., Seo S. W., Na D. L., Shim H.-J., and Lee J.-M., “Fully-automated approach to hippocampus segmentation using a graph-cuts algorithm combined with atlas-based segmentation and morphological opening,” Magn. Reson. Imaging 31, 1190–1196 (2013). 10.1016/j.mri.2013.04.008 [DOI] [PubMed] [Google Scholar]
- 12.Heckemann R. A., Hajnal J. V., Aljabar P., Rueckert D., and Hammers A., “Automatic anatomical brain MRI segmentation combining label propagation and decision fusion,” NeuroImage 33, 115–126 (2006). 10.1016/j.neuroimage.2006.05.061 [DOI] [PubMed] [Google Scholar]
- 13.Kim M., Wu G., Li W., Wang L., Son Y.-D., Cho Z.-H., and Shen D., “Automatic hippocampus segmentation of 7.0 Tesla MR images by combining multiple atlases and auto-context models,” NeuroImage 83, 335–345 (2013). 10.1016/j.neuroimage.2013.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Awate S. P. and Whitaker R. T., “Multiatlas segmentation as nonparametric regression,” IEEE Trans. Med. Imaging 33, 1803–1817 (2014). 10.1109/TMI.2014.2321281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winston G. P., Cardoso M. J., Williams E. J., Burdett J. L., Bartlett P. A., Espak M., Behr C., Duncan J. S., and Ourselin S., “Automated hippocampal segmentation in patients with epilepsy: Available free online,” Epilepsia 54, 2166–2173 (2013). 10.1111/epi.12408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischl B., “FreeSurfer,” NeuroImage 62, 774–781 (2012). 10.1016/j.neuroimage.2012.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haller J. W., Christensen G. E., Joshi S. C., Newcomer J. W., Miller M. I., Csernansky J. G., and Vannier M. W., “Hippocampal MR imaging morphometry by means of general pattern matching,” Radiology 199, 787–791 (1996). 10.1148/radiology.199.3.8638006 [DOI] [PubMed] [Google Scholar]
- 18.Pluta J., Avants B. B., Glynn S., Awate S., Gee J. C., and Detre J. A., “Appearance and incomplete label matching for diffeomorphic template based hippocampus segmentation,” Hippocampus 19, 565–571 (2009). 10.1002/hipo.20619 [DOI] [PubMed] [Google Scholar]
- 19.Pitiot A., Toga A. W., and Thompson P. M., “Adaptive elastic segmentation of brain MRI via shape-model-guided evolutionary programming,” IEEE Trans. Med. Imaging 21, 910–923 (2002). 10.1109/TMI.2002.803124 [DOI] [PubMed] [Google Scholar]
- 20.Akhondi-Asl A., Jafari-Khouzani K., Elisevich K., and Soltanian-Zadeh H., “Hippocampal volumetry for lateralization of temporal lobe epilepsy: Automated versus manual methods,” NeuroImage 54, S218–S226 (2011). 10.1016/j.neuroimage.2010.03.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen L., Saykin A. J., Kim S., Firpi H. A., West J. D., Risacher S. L., McDonald B. C., McHugh T. L., Wishart H. A., and Flashman L. A., “Comparison of manual and automated determination of hippocampal volumes in MCI and early AD,” Brain Imaging Behav. 4, 86–95 (2010). 10.1007/s11682-010-9088-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hosseini M. P., Soltanian-Zadeh H., and Akhlaghpoor S., “Three cuts method for identification of COPD,” Acta Med. Iran. 51, 771–778 (2013), see http://www.ncbi.nlm.nih.gov/pubmed/24390946. [PubMed] [Google Scholar]
- 23.Hosseini M. P., Soltanian-Zadeh H., and Akhlaghpoor S., “Detection and severity scoring of chronic obstructive pulmonary disease using volumetric analysis of lung CT images,” Iran. J. Radiol. 9, 22–27 (2012). 10.5812/iranjradiol.6759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pitiot A., Delingette H., Thompson P. M., and Ayache N., “Expert knowledge-guided segmentation system for brain MRI,” NeuroImage 23, S85–S96 (2004). 10.1016/j.neuroimage.2004.07.040 [DOI] [PubMed] [Google Scholar]
- 25.Shen D. and Davatzikos C., “ hammer: Hierarchical attribute matching mechanism for elastic registration,” IEEE Trans. Med. Imaging 21, 1421–1439 (2002). 10.1109/tmi.2002.803111 [DOI] [PubMed] [Google Scholar]
- 26.Hult R. and Agartz I., “Segmentation of multimodal MRI of hippocampus using 3d grey-level morphology combined with artificial neural networks,” in Image Analysis: Proceedings of the 14th Scandinavian Conference, SCIA 2005, Joensuu, Finland, 19–22 June 2005 (Springer, 2005), pp. 272–281. [Google Scholar]
- 27.Hao Y., Wang T., Zhang X., Duan Y., Yu C., Jiang T., and Fan Y., “Local label learning (LLL) for subcortical structure segmentation: Application to hippocampus segmentation,” Hum. Brain Mapp. 35, 2674–2697 (2014). 10.1002/hbm.22359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moghaddam M. J. and Soltanian-Zadeh H., “Automatic segmentation of brain structures using geometric moment invariants and artificial neural networks,” in Information Processing in Medical Imaging (Springer, Berlin, Heidelberg, 2009), pp. 326–337. [DOI] [PubMed] [Google Scholar]
- 29.Sadananthan S. A., Zheng W., Chee M. W., and Zagorodnov V., “Skull stripping using graph cuts,” NeuroImage 49, 225–239 (2010). 10.1016/j.neuroimage.2009.08.050 [DOI] [PubMed] [Google Scholar]
- 30.Galdames F. J., Jaillet F., and Perez C. A., “An accurate skull stripping method based on simplex meshes and histogram analysis for magnetic resonance images,” J. Neurosci. Methods 206, 103–119 (2012). 10.1016/j.jneumeth.2012.02.017 [DOI] [PubMed] [Google Scholar]
- 31.Shattuck D. W. and Leahy R. M., “BrainSuite: An automated cortical surface identification tool,” Med. Image Anal. 6, 129–142 (2002). 10.1016/S1361-8415(02)00054-3 [DOI] [PubMed] [Google Scholar]
- 32.Rorden C. and Brett M., MRIcro (2005), availabile at http://www.sph.sc.edu/comd/rorden/mricro.html. [Google Scholar]
- 33.Speier W., Iglesias J. E., El-Kara L., Tu Z., and Arnold C., “Robust skull stripping of clinical glioblastoma multiforme data,” in Medical Image Computing and Computer-Assisted Intervention–MICCAI (Springer, Toronto, Canada, 2011), pp. 659–666. [DOI] [PubMed] [Google Scholar]
- 34.Shi F., Wang L., Dai Y., Gilmore J. H., Lin W., and Shen D., “ label: Pediatric brain extraction using learning-based meta-algorithm,” NeuroImage 62, 1975–1986 (2012). 10.1016/j.neuroimage.2012.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jafari-Khouzani K., Elisevich K. V., Patel S., and Soltanian-Zadeh H., “Dataset of magnetic resonance images of nonepileptic subjects and temporal lobe epilepsy patients for validation of hippocampal segmentation techniques,” Neuroinformatics 9, 335–346 (2011). 10.1007/s12021-010-9096-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duvernoy H. M., Cattin F., and Risold P.-Y., The Human Hippocampus: Functional Anatomy, Vascularization and Serial Sections with MRI (Springer-Verlag Berlin Heidelberg, 2013). [Google Scholar]
- 37.Boccardi M., Ganzola R., Bocchetta M., Pievani M., Redolfi A., Bartzokis G., Camicioli R., Csernansky J. G., de Leon M. J., and deToledo-Morrell L., “Survey of protocols for the manual segmentation of the hippocampus: Preparatory steps towards a joint EADC-ADNI harmonized protocol,” J. Alzheimer’s Dis. 26, 61–75 (2011). 10.3233/JAD-2011-0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Téllez-Zenteno J. F., Dhar R., and Wiebe S., “Long-term seizure outcomes following epilepsy surgery: A systematic review and meta-analysis,” Brain 128, 1188–1198 (2005). 10.1093/brain/awh449 [DOI] [PubMed] [Google Scholar]
- 39.Germeyan S. C., Kalikhman D., Jones L., and Theodore W. H., “Automated versus manual hippocampal segmentation in preoperative and postoperative patients with epilepsy,” Epilepsia 55, 1374–1379 (2014). 10.1111/epi.12694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Commowick O. and Warfield S. K., “Estimation of inferential uncertainty in assessing expert segmentation performance from STAPLE,” IEEE Trans. Med. Imaging 29, 771–780 (2010). 10.1109/TMI.2009.2036011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huttenlocher D. P., Klanderman G. A., and Rucklidge W. J., “Comparing images using the Hausdorff distance,” IEEE Trans. Pattern Anal. Mach. Intell. 15, 850–863 (1993). 10.1109/34.232073 [DOI] [Google Scholar]
- 42.Andreev A. and Kirov N., “Hausdorff distances for searching in binary text images,” Serdica J. Comput. 3, 23–46 (2009). [Google Scholar]
- 43.Doring T. M., Kubo T. T., Cruz L. C. H., Juruena M. F., Fainberg J., Domingues R. C., and Gasparetto E. L., “Evaluation of hippocampal volume based on MR imaging in patients with bipolar affective disorder applying manual and automatic segmentation techniques,” J. Magn. Reson. Imaging 33, 565–572 (2011). 10.1002/jmri.22473 [DOI] [PubMed] [Google Scholar]
- 44.Dewey J., Hana G., Russell T., Price J., McCaffrey D., Harezlak J., Sem E., Anyanwu J. C., Guttmann C. R., and Navia B., “Reliability and validity of MRI-based automated volumetry software relative to auto-assisted manual measurement of subcortical structures in HIV-infected patients from a multisite study,” NeuroImage 51, 1334–1344 (2010). 10.1016/j.neuroimage.2010.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morey R. A., Petty C. M., Xu Y., Hayes J. P., Wagner H. R., Lewis D. V., LaBar K. S., Styner M., and McCarthy G., “A comparison of automated segmentation and manual tracing for quantifying hippocampal and amygdala volumes,” NeuroImage 45, 855–866 (2009). 10.1016/j.neuroimage.2008.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wenger E., Mårtensson J., Noack H., Bodammer N. C., Kühn S., Schaefer S., Heinze H. J., Düzel E., Bäckman L., and Lindenberger U., “Comparing manual and automatic segmentation of hippocampal volumes: Reliability and validity issues in younger and older brains,” Hum. Brain Mapp. 35, 4236–4248 (2014). 10.1002/hbm.22473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hosseini M.-P., Nazem-Zadeh M. R., Pompili D., and Soltanian-Zadeh H., “Statistical validation of automatic methods for hippocampus segmentation in MR images of epileptic patients,” in 36th Annual International Conference of the IEEE on Engineering in Medicine and Biology Society (EMBC) (IEEE, Chicago, IL, 2014), pp. 4707–4710. 10.1109/EMBC.2014.6944675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pardoe H. R., Pell G. S., Abbott D. F., and Jackson G. D., “Hippocampal volume assessment in temporal lobe epilepsy: How good is automated segmentation?,” Epilepsia 50, 2586–2592 (2009). 10.1111/j.1528-1167.2009.02243.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McDonald C. R., D. J. Hagler, Jr., Ahmadi M. E., Tecoma E., Iragui V., Dale A. M., and Halgren E., “Subcortical and cerebellar atrophy in mesial temporal lobe epilepsy revealed by automatic segmentation,” Epilepsy Res. 79, 130–138 (2008). 10.1016/j.eplepsyres.2008.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jain V., Seung H. S., and Turaga S. C., “Machines that learn to segment images: A crucial technology for connectomics,” Curr. Opin. Neurobiol. 20, 653–666 (2010). 10.1016/j.conb.2010.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coupé P., Manjón J. V., Fonov V., Pruessner J., Robles M., and Collins D. L., “Patch-based segmentation using expert priors: Application to hippocampus and ventricle segmentation,” NeuroImage 54, 940–954 (2011). 10.1016/j.neuroimage.2010.09.018 [DOI] [PubMed] [Google Scholar]
- 52.Rousseau F., Habas P. A., and Studholme C., “A supervised patch-based approach for human brain labeling,” IEEE Trans. Med. Imaging 30, 1852–1862 (2011). 10.1109/TMI.2011.2156806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lötjönen J. M., Wolz R., Koikkalainen J. R., Thurfjell L., Waldemar G., Soininen H., Rueckert D., and Initiative A. S. D. N., “Fast and robust multi-atlas segmentation of brain magnetic resonance images,” NeuroImage 49, 2352–2365 (2010). 10.1016/j.neuroimage.2009.10.026 [DOI] [PubMed] [Google Scholar]
- 54.Merkel B., Steward C., Vivash L., Malpas C. B., Phal P., Moffat B. A., Cox K. L., Ellis K. A., Ames D. J., and Cyarto E. V., “Semi-automated hippocampal segmentation in people with cognitive impairment using an age appropriate template for registration,” J. Magn. Reson. Imaging 42, 1631–1638 (2015). 10.1002/jmri.24966 [DOI] [PubMed] [Google Scholar]
- 55.Nestor S. M., Gibson E., Gao F.-Q., Kiss A., and Black S. E., “A direct morphometric comparison of five labeling protocols for multi-atlas driven automatic segmentation of the hippocampus in Alzheimer’s disease,” NeuroImage 66, 50–70 (2013). 10.1016/j.neuroimage.2012.10.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pipitone J., Park M. T. M., Winterburn J., Lett T. A., Lerch J. P., Pruessner J. C., Lepage M., Voineskos A. N., and Mallar Chakravarty M., “Multi-atlas segmentation of the whole hippocampus and subfields using multiple automatically generated templates,” NeuroImage 101, 494–512 (2014). 10.1016/j.neuroimage.2014.04.054 [DOI] [PubMed] [Google Scholar]
- 57.Teitelbaum G., W. Bradley, Jr., and Klein B., “MR imaging artifacts, ferromagnetism, and magnetic torque of intravascular filters, stents, and coils,” Radiology 166, 657–664 (1988). 10.1148/radiology.166.3.3340759 [DOI] [PubMed] [Google Scholar]
- 58.Mirowitz S. A., “MR imaging artifacts. Challenges solutions,” Magn. Reson. Imaging Clin. North Am. 7, 717–732 (1999). [PubMed] [Google Scholar]
- 59.Han X. and Fischl B., “Atlas renormalization for improved brain MR image segmentation across scanner platforms,” IEEE Trans. Med. Imaging 26, 479–486 (2007). 10.1109/TMI.2007.893282 [DOI] [PubMed] [Google Scholar]
- 60.Li C., Huang R., Ding Z., Gatenby J., Metaxas D. N., and Gore J. C., “A level set method for image segmentation in the presence of intensity inhomogeneities with application to MRI,” IEEE Trans. Image Process. 20, 2007–2016 (2011). 10.1109/tip.2011.2146190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pham T. D. and Berger K., “Automated detection of white matter changes in elderly people using fuzzy, geostatistical, and information combining models,” IEEE Trans. Inf. Technol. Biomed. 15, 242–250 (2011). 10.1109/TITB.2010.2081996 [DOI] [PubMed] [Google Scholar]
- 62.Mayer A., Zimmerman-Moreno G., Shadmi R., Batikoff A., and Greenspan H., “A supervised framework for the registration and segmentation of white matter fiber tracts,” IEEE Trans. Med. Imaging 30, 131–145 (2011). 10.1109/TMI.2010.2067222 [DOI] [PubMed] [Google Scholar]
- 63.Carneiro G. and Nascimento J. C., “Combining multiple dynamic models and deep learning architectures for tracking the left ventricle endocardium in ultrasound data,” IEEE Trans. Pattern Anal. Mach. Intell. 35, 2592–2607 (2013). 10.1109/TPAMI.2013.96 [DOI] [PubMed] [Google Scholar]