Abstract
Amino acid biosynthetic pathways that are absent in mammals are considered an attractive target for antifungal therapy. Leucine biosynthesis is one such target pathway, consisting of a five-step conversion process starting from the valine precursor 2-keto-isovalerate. Isopropylmalate dehydrogenase (Leu1) is an Fe-S cluster protein that is required for leucine biosynthesis in the model fungus Saccharomyces cerevisiae. The human pathogenic fungus Cryptococcus neoformans possesses an ortholog of S. cerevisiae Leu1, and our previous transcriptome data showed that the expression of LEU1 is regulated by iron availability. In this study, we characterized the role of Leu1 in iron homeostasis and the virulence of C. neoformans. We found that deletion of LEU1 caused leucine auxotrophy and that Leu1 may play a role in the mitochondrial-cytoplasmic Fe-S cluster balance. Whereas cytoplasmic Fe-S protein levels were not affected, mitochondrial Fe-S proteins were up- regulated in the leu1 mutant, suggesting that Leu1 mainly influences mitochondrial iron metabolism. The leu1 mutant also displayed increased sensitivity to oxidative stress and cell wall/membrane disrupting agents, which may have been caused by mitochondrial dysfunction. Furthermore, the leu1 mutant was deficient in capsule formation and showed attenuated virulence in a mouse inhalation model of cryptococcosis. Overall, our results indicate that Leu1 plays a role in iron metabolism and is required for virulence in C. neoformans.
Keywords: C. neoformans, Leu1, mitochondria, Fe-S cluster, virulence
1. Introduction
Amino acid biosynthetic pathways that are absent in mammals are considered an attractive target for antifungal therapy because several amino acid auxotrophs among various fungal pathogens show reduced virulence. The leucine biosynthetic pathway is an example of such a pathway, as it exists in fungi but is absent in humans. Particular attention has been paid to this pathway because of its characteristics in the model yeast Saccharomyces cerevisiae. In contrast to other amino acid biosynthetic pathways, leucine biosynthesis is partitioned between the mitochondria and cytoplasm, and its intermediate α-isopropylmalate plays a role as signal molecule within the cell (Kohlhaw, 2003). In S. cerevisiae, α-isopropylmalate binds to the homodimeric DNA binding protein Leu3 and positively regulates branched-chain amino acid biosynthesis, amino acid uptake, and nitrogen and carbohydrate metabolism. In the absence of α-isopropylmalate, Leu3 acts as a repressor together with the Gcn4 general amino acid control system (Friden and Schimmel, 1988; Sze et al., 1992; Zhou et al., 1987; Zhou et al., 1990). Moreover, studies have suggested a possible link between leucine biosynthesis and iron metabolism in S. cerevisiae, and isopropylmalate dehydrogenase encoded by LEU1 may play a key role. Leu1 is an Fe-S cluster protein with high homology to the iron regulatory protein Irp1 in mammalian cells, implying an influence on iron metabolism within the cell (Kaptain et al., 1991). Indeed, its transcript and protein levels were significantly reduced in iron deficiency, suggesting down-regulation of LEU1 is a strategy used by S. cerevisiae to adapt to iron scarcity (Bedekovics et al., 2011; Ihrig et al., 2010).
In this study, we identified and functionally characterized LEU1 in the opportunistic human fungal pathogen Cryptococcus neoformans, which causes life-threatening fungal meningitis in immunocompromised individuals such as HIV/AIDS patients. Every year, approximately one million cases of cryptococcal meningitis are reported globally, and the mortality rate has reached approximately 40% in sub-Saharan Africa (Hakim et al., 2000; Park et al., 2009). Amino acid biosynthetic pathways that are absent in the vertebrate host were also found to be important for the virulence of C. neoformans. For example, C. neoformans mutants lacking MET2, MET3, or MET6, which are essential for methionine biosynthesis, and ILV2, which is essential for isoleucine and valine biosynthesis, were avirulent in a murine inhalation model of cryptococcosis (Kingsbury, 2004; Nazi et al., 2007; Pascon et al., 2004; Yang et al., 2002). The contribution of the leucine biosynthetic pathway to virulence in C. neoformans has not yet been investigated, and the connection of Leu1 to iron regulation suggests that this enzyme may be doubly important for fungal proliferation in the host.
We have investigated iron uptake and regulation in C. neoformans because of the significant influence of iron on virulence factor expression and survival within the vertebrate host environment. Avirulence or attenuated virulence in a murine model of cryptococcosis is a typical consequence in mutants lacking any of the components of the iron uptake system or associated regulatory pathways in C. neoformans (Jung et al., 2009; Jung et al., 2010; Jung et al., 2008; Jung et al., 2006). Interestingly, our previous transcriptome data showed iron-responsive regulation of the gene encoding Leu1 in C. neoformans (Kim et al., 2012). Therefore, in this study, we aimed to study the role of Leu1 in iron metabolism and virulence in C. neoformans. Our data show that expression of Leu1 is influenced by iron levels and regulated by the iron regulatory transcription factors Cir1, Hap3 and HapX. We constructed strains lacking LEU1 and showed that the leu1 mutant has dysfunctional mitochondria resulting from deficient iron metabolism in the organelle, and is hypersensitive to oxidative stress and cell wall/membrane disturbing agents. Furthermore, we demonstrated that LEU1 is required for virulence in C. neoformans suggesting that the leucine biosynthetic pathway may be a potential target for antifungal drugs to treat cryptococcosis.
2. Materials and Methods
2.1 Strains and growth conditions
The strains used in this study are listed in Table S1. The strains were routinely cultured in yeast extract-Bacto peptone medium with 2.0% glucose (YPD; Difco) or yeast nitrogen base (YNB; Difco) with 2.0% glucose. YNB medium without amino acid supplementation was used to analyze amino acid auxotrophy. The low-iron medium was prepared as described elsewhere (Jung et al., 2009). Briefly, defined YNB medium was prepared and adjusted to pH 7.0 with 3- morpholinopropanesulfonic acid (MOPS), and the iron-limited condition was achieved by addition of 100 −M bathophenanthroline disulfonate (BPS). Iron-replete media were prepared by adding FeCl3 to the low-iron medium at a final concentration of 100 −M. To deplete intracellular iron, cells were precultured in low-iron medium at 30°C overnight for all experiments, as described previously (Jung et al., 2008).
2.2 Construction of strains
The sequence of the gene encoding LEU1 homolog (CNAG_00237) was obtained from the C. neoformans var. grubii serotype A genome database (http://www.broadinstitute.org/annotation/genome/cryptococcus_neoformans). The mutants, and the strains containing fusions with the green fluorescent protein (GFP) or the FLAG epitope tag, were constructed using the primers listed in Table S2. To construct the leu1 mutant, the gene-specific deletion cassette was prepared by overlapping PCR using primers LEU1_KO_1, LEU1_KO_2, LEU1_KO_3, LEU1_KO_4, LEU1_KO_5 and LEU1_KO_6, with the wild-type genomic DNA and the plasmid pCH233 as templates, and biolistically transformed into the wild-type strain as previously described (Toffaletti et al., 1993). The wild-type LEU1 coding region was replaced with the gene- specific deletion cassette containing the nourseothricin acetyltransferase (NAT) gene, and positive transformants were confirmed by PCR and Southern blot analysis (Fig. S1). To construct the reconstituted strain, the LEU1 gene was amplified with primers RE_1_F and RE_3_R using wild-type genomic DNA as the template. The amplified 4.1 kb DNA fragment was ligated into the plasmid pJAF1, which contains the neomycin resistance (NEO) gene, using the restriction enzymes SacI and SpeI. The resulting plasmid, named pWH126, was digested with BspHI and introduced into the leu1 mutant by biolistic transformation. Positive transformants containing the wild-type LEU1 gene at its original locus were identified by PCR. To construct the strain expressing a Leu1-GFP fusion protein, the plasmid pWH091, which contains the codon-optimized GFP gene, the GAL7 terminator and NEO, was used (Jung et al., 2009). The LEU1 gene was amplified by PCR from wild-type genomic DNA with primers GFP_F and GFP_R. The amplified DNA fragments were digested with BamHI and BglII, and cloned into the plasmid pWH091 digested with BamHI. The resulting plasmid pWH127 was digested with BamHI and introduced into the wild-type strain using biolistic transformation. Positive transformants were confirmed by PCR and western blot analysis. The sequences of S. cerevisiae Aco1 (NP_013407.1), Nfu1 (NP_012884.3) and Fra2 (NP_011295.1) were used to search for homologs in the C. neoformans genome. The identified homologs, CNAG_01137 (Aco1), CNAG_03395 (Nfu1) and CNAG_03927 (Fra2), were fused with the 3×FLAG epitope tag for protein expression analysis. To construct the Aco1-FLAG fusion protein, the ACO1 gene was amplified by PCR from the wild-type genomic DNA using primers ACO1_F and ACO1_R. The amplified DNA fragments were digested with BglII and cloned into the BamHI site of the plasmid pWH109, which contains the 3×FLAG epitope, the GAL7 terminator and the NEO gene. The resulting plasmid pWH133 was digested with HindIII, and introduced into the wild-type strain and the leu1 mutant by biolistic transformation. Positive transformants containing the recombinant gene at ACO1 locus were confirmed by PCR using primers Aco1confirm_F and NATterm2F, and western blot analysis. To construct the Nfu1-FLAG fusion protein, the NFU1 gene was amplified by PCR from the wild-type genomic DNA using primers NFU1_F and NFU1_R. The amplified DNA fragments were digested with BamHI and cloned into the BamHI site of the plasmid pWH109. The resulting plasmid pWH138 was digested with XbaI and introduced into the wild-type strain and the leu1 mutant by biolistic transformation. Positive transformants containing the recombinant gene at NFU1 locus were confirmed by PCR using primers Nfu1confirm_F and NATterm2F, and western blot analysis. To construct the Fra2-FLAG fusion protein, the FRA2 gene was amplified by PCR from wild-type genomic DNA using primers FRA2_F and FRA2_R. The amplified DNA fragments were digested with BglII and cloned into the BamHI site of the plasmid pWH109. The resulting plasmid pWH157 was digested with HindIII, and introduced into the wild-type strain and the leu1 mutant by biolistic transformation. Positive transformants containing the recombinant gene at NFU1 locus were confirmed by PCR using primers Fra2confirm_F and NATterm2F, and western blot analysis.
2.3 Northern blot analysis
Strains were grown in 50 mL of low-iron medium with or without 100 −M FeCl3 at 30°C for 12 hours. The cells were collected and washed twice with iron-chelated water. Total RNA was extracted using the RiboPure-Yeast Kit (Ambion), and the quantity of total RNA was measured by NanoDrop. Northern blot analysis was performed as described by Sambrook et al. using 20 −g of total RNA from each strain (Sambrook and Russell, 2001). The separated total RNA in the formaldehyde gels was transferred to an UltraBind transfer membrane (Gelman laboratory) and hybridized with a gene- specific DNA probe labeled with [32P]-dCTP. The membrane was exposed to a Phosphor screen (PerkinElmer) for 16 hours and scanned using a Packard cyclone phosphor imager (PerkinElmer).
2.4 Western blot analysis
Strains were grown in low-iron medium with or without 100 −M FeCl3 at 30°C for 12 hours and harvested. Pellets were resuspended in protein extraction buffer containing 50 mM HEPES KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycolate, 1 mM PMSF and a protease inhibitor cocktail (Sigma). Acid-washed glass-beads (0.4 mm; Sigma) were added, and the cells were disrupted using a bead-beater (BioSpec). The concentration of total protein was measured using the Bradford assay (Bradford, 1976). Equal amounts of protein were subjected to SDS- polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane (Amersham). Western blot analysis was performed using an anti-GFP monoclonal rabbit antibody (Sigma) and an anti-DDDDK polyclonal rabbit antibody (Abcam) as primary antibody, and a goat anti-rabbit IgG horseradish peroxidase conjugate (Santa Cruz) as a secondary antibody, followed by visualization using chemiluminescence.
2.5 Superoxide dismutase activity assay
The activity of superoxide dismutase was evaluated as previously described (Beauchamp and Fridovich, 1971). Briefly, the wild-type strain and the leu1 mutant were grown in YPD at 30°C overnight and pelleted by centrifugation at 13,000 rpm for 2 min. Cell pellets were resuspended with lysis buffer containing 50 mM Tris-Cl pH 7.5, 0.1 mM EDTA and 1 mM PMSF, and were disrupted as described above. The protein concentration of lysates was determined by the Bradford assay (Bradford, 1976). The total protein (40 −g) was subjected to the 10% native polyacrylamide gel electrophoresis at 120 V and 4°C for 2 hours. The superoxide dismutase activity was visualized by staining the gel with nitro blue tetrazolium (NBT) buffer containing 50 mM potassium phosphate buffer pH 7.4, 0.25 mM riboflavin, 1.23 mM NBT and 0.5 mM TEMED, with incubation in the dark for 30 min at room temperature. After staining, the gel was washed with distilled water and incubated on a light box for 15 min at room temperature before being photographed. The band intensity was analyzed using ImageJ software (http://imagej.nih.gov/ij/). Copper phthalocyanine-3,4′,4′,4′′- tetrasulfonic acid tetrasodium salt (CPTS) staining was used to demonstrate that all samples were loaded equally.
2.6 Microscopy
To visualize the Leu1-GFP fusion protein, the strain expressing the fusion protein was grown in YPD medium at 30°C overnight. Cells were washed twice with iron-chelated water and resuspended with YNB medium followed by incubation at 30°C for 6 hours. Mitotracker Red CMXRos (Molecular Probes) was added to YNB medium at a final concentration of 100 nM to stain mitochondria. The cells were incubated at 30°C for 30 min and visualized using an Axioplan 2 imaging system (Zeiss) with 100× magnification. Differential interference contrast (DIC) and fluorescent images were obtained using Metamorph imaging software (Universal Imaging Corp.). To evaluate capsule formation, cells were grown in defined low-iron medium at 30°C overnight and stained with India ink.
2.7 Virulence assay
Virulence was assayed in an inhalation model of cryptococcosis using female BALB/c mice (4 to 6 weeks old) from Charles River Laboratories (Ontario, Canada) as previously described (Hu and Kronstad, 2010). Briefly, C. neoformans strains were grown in 5 mL of YPD at 30oC overnight, washed twice with PBS (Invitrogen Canada), and resuspended in PBS. BALB/c mice, in groups of 10, were intranasally inoculated with a suspension of 2 ×105 cells in 50 −L. The health status of the mice was monitored daily post-inoculation. Mice reaching the humane endpoint were euthanized by CO2 anoxia. Statistical analyses of survival differences were performed by log rank tests using GraphPad Prism 5 for Windows (GraphPad software). The fungal load on lungs, brain, kidney, liver, spleen and blood on three mice was assessed at time of death. Organs were removed aseptically, weighted, homogenized and diluted in PBS prior to inoculation on YPD plates. Blood was retrieved from the heart and directly inoculated onto YPD plates. Plates were incubated at 30°C for 48 hours and CFU counts were determined. The protocols for the virulence assay (protocol A13-0093) were approved by the University of British Columbia Committee on Animal Care.
3. Results
3.1. LEU1 expression is regulated by iron in C. neoformans
Our previous transcriptome analysis with a mutant lacking the CFO1 gene encoding a ferrioxidase for high affinity iron uptake revealed that several genes are differentially regulated by intracellular iron levels in C. neoformans. These differentially regulated genes included the gene for a protein highly homologous to Leu1 of the model yeast S. cerevisiae, with 73% similarity and 60% identity. S. cerevisiae Leu1 catalyzes the conversion of α-isopropylmalate into β-isopropylmalate during leucine biosynthesis. Similar to S. cerevisiae Leu1, the amino acid sequence of C. neoformans Leu1 contains a conserved aconitase family signature that binds the 4Fe-4S cluster (data not shown). As mentioned, Cfo1 is required for high-affinity iron uptake, and a strain lacking the gene displayed significantly reduced intracellular iron levels. Therefore, our previous observation of significant down-regulation of LEU1 transcripts in cfo1 mutants implied a positive influence of iron on LEU1 expression. To confirm this, we measured transcript levels of LEU1 in wild-type cells grown in either high- or low- iron conditions and found that the mRNA levels were indeed reduced in response to iron deficiency in the medium (Fig. 1A).
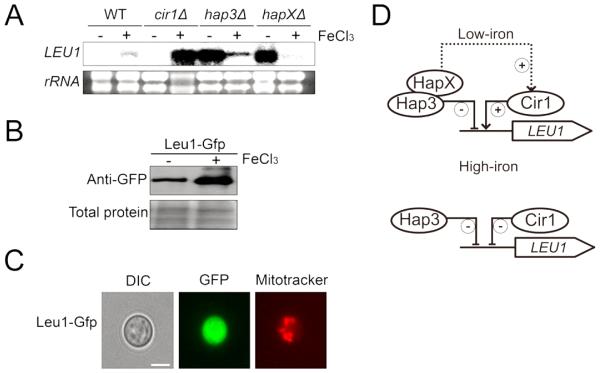
Fig. 1. Iron-dependent regulation of LEU1.
(A) Transcript levels of LEU1 in the wild-type and the mutants lacking CIR1, HAP3 or HAPX grown in low- (0 −M FeCl3) or high- (+100 −M FeCl3) iron medium were evaluated by Northern blot analysis. (B) Leu1 was fused with GFP, and the abundance of the protein in the cells grown in low- (0 −M FeCl3) or high- (+100 −M FeCl3) iron medium was evaluated by Western blot analysis using an anti-GFP antibody. (C) Localization of Leu1-GFP by fluorescent microscopy. Mitotracker was used to stain mitochondria. (D) Proposed model of regulation of LEU1 expression by Cir1, Hap3 and HapX.
The genes required for iron uptake and metabolism are regulated by several transcription factors including Cir1, Hap3 and HapX in C. neoformans. In this study, we found that all three transcription factors influenced the expression of Leu1. In the mutant lacking CIR1, LEU1 transcripts were almost undetectable in cells grown in low-iron medium, whereas very low levels of the LEU1 transcript were produced in the wild-type cells grown in the same medium. In contrast, LEU1 showed the opposite pattern of expression in the cir1 mutant grown in high-iron medium, as it was transcribed more abundantly than in the wild type. These results suggest dual regulatory roles of Cir1 on LEU1, i.e., positive and negative regulation in low- and high-iron conditions, respectively. The transcript levels of LEU1 were also dramatically increased in both hap3 and hapX mutants grown in low-iron medium, which suggested that Hap3 and HapX may coordinately act to negatively regulate LEU1 in iron- deficient conditions. The hap3 mutant, but not the hapX mutant, showed increased levels of LEU1 transcripts compared to the wild type upon growth in high-iron medium, implying that the regulatory effect of Hap3 on LEU1 may be slightly different from that of HapX in C. neoformans. Distinct regulation by Hap3 and HapX was suggested previously (Jung et al., 2010). The Leu1 protein was also fused with green fluorescence protein (GFP) to investigate whether the levels of the protein reflected the altered transcript levels. The Leu1-GFP fusion protein restored the growth of the leu1 mutant in the absence of leucine in the medium suggesting that the protein is functional (Fig. S2). The western blot analysis shown in Fig. 1B revealed that Leu1-GFP protein levels are also influenced by iron levels in the medium, with results similar to those from the Northern blot analysis. Furthermore, fluorescence microscopy showed that Leu1 is localized in the cytosol (Fig. 1C). Together, our data suggest that the expression of Leu1 is influenced by iron levels and regulated by the iron regulatory transcription factors Cir1, Hap3 and HapX.
3.2. LEU1 is required for leucine biosynthesis
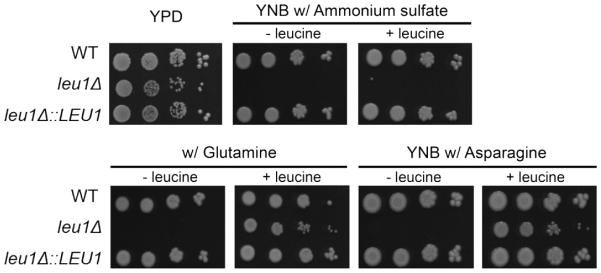
To characterize the function of LEU1 in C. neoformans, a leu1 mutant was constructed by a targeted gene deletion method using biolistic transformation. A LEU1 reconstituted strain was also constructed by integration of the wild-type gene into its original locus in the leu1 mutant, and this strain was used throughout the study (see Materials and Methods). We first confirmed loss of the function of LEU1 in the mutant based on its amino acid auxotrophy. Cells were grown in YNB media with or without leucine in the presence of ammonium sulfate as a sole nitrogen source. As expected, the leu1 mutants were not able to grow without leucine, confirming a defect in leucine biosynthesis. However, addition of leucine did not restore growth of the mutant (Fig. 2). We speculated that nitrogen catabolite repression may interfere with leucine uptake in C. neoformans in the presence of ammonium sulfate, which is generally considered a primary nitrogen source. We therefore replaced ammonium sulfate with glutamine or asparagine and re-tested leucine supplementation. As expected, the replacement of the nitrogen source and addition of leucine to the media led to restoration of the growth of the leu1 mutant. Nitrogen catabolite repression is well studied in fungi. In general, the uptake of poor nitrogen sources such as amino acids and nitrate is inhibited by high quality primary nitrogen sources such as ammonium (Hofman-Bang, 1999; Marzluf, 1997). For example, a negative effect of ammonium on leucine uptake was shown in S. pombe yeast, and a preference for ammonium over leucine as a nitrogen source was also shown in a plant pathogenic fungus Geotrichum candidum (Adour et al., 2010; Weisman et al., 2005). Nitrogen metabolic repression has also been studied in C. neoformans. It was suggested that a GATA transcription factor Gat1 transcriptionally regulates expression of nitrogen catabolism associated genes together with Tar1, which is an Nmr family repressor of nitrogen utilization (Kmetzsch et al., 2011; Lee et al., 2011; Lee et al., 2012). Taken together, our results suggested that LEU1 is required for leucine biosynthesis in C. neoformans, and that leucine uptake is likely under the control of nitrogen catabolite repression.
Fig. 2. LEU1 is required for leucine biosynthesis.
The growth of the leu1 mutant in YNB media containing various nitrogen sources with (+) or without (-) leucine (2 mM) was monitored. Ten-fold serial dilutions of cells (starting at 104 cells) were spotted onto the plates and incubated at 30°C for 2 days.
3.3. Leu1 influences expression of mitochondrial Fe-S proteins and may interfere with mitochondrial iron homeostasis
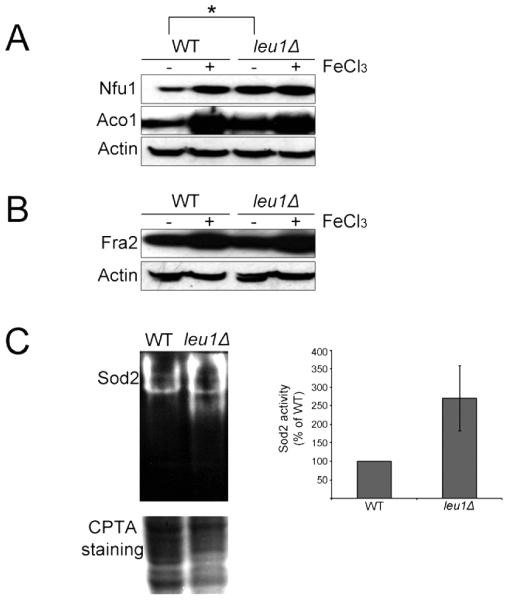
As mentioned above, Leu1 is an Fe-S cluster-containing protein localized in the cytosol. In S. cerevisiae, Leu1 plays a key role in the mitochondrial-cytoplasmic Fe-S cluster balance, and a mutant lacking LEU1 displays up-regulation of the components required for mitochondrial Fe-S cluster synthesis (Bedekovics et al., 2011). These observations led us to investigate whether Leu1 plays a similar role in C. neoformans. Two putative mitochondrial proteins, a homolog (CNAG_03395) of S. cerevisiae Nfu1, which is a mitochondrial protein involved in Fe-S cluster synthesis (Schilke et al., 1999), and a homolog (CNAG_01137) of S. cerevisiae aconitase (Aco1), which is often used as a marker for the level of mitochondrial Fe-S cluster synthesis (Bedekovics et al., 2011; Kispal et al., 1997), were selected and fused with a FLAG epitope tag. The abundance of these proteins was then evaluated in the leu1 mutant and the wild-type strain grown in low- and high-iron media. The homolog (CNAG_03927) of S. cerevisiae Fra2, which is a cytosolic Fe-S containing protein that is involved in regulation of iron uptake and homeostasis, was also selected and fused with the same epitope tag to determine whether Leu1 influences levels of a cytosolic Fe-S cluster protein. The results of Western blot analysis showed that the levels of both Nfu1 and Aco1 were increased in the leu1 mutant compared to the wild-type cells (Fig. 3A). Furthermore, the increased levels of the proteins were only observed in cells from low-iron media but not from high-iron media suggesting that the role of Leu1 in mitochondrial iron metabolism may only be critical in iron deficient conditions. No change was observed when the protein level of Fra2 was compared between the leu1 mutant and wild-type cells suggesting that Leu1 mainly influences mitochondrial but not cytosolic iron metabolism (Fig. 3B). As further evidence of perturbed iron status, we also observed increased activity for the mitochondrial manganese dependent superoxide dismutase (Sod2) in the leu1 mutant compared to the wild-type strain. This result indicated that mitochondria in the leu1 mutant suffered from oxidative stress, which may be caused by up-regulation of iron metabolism within the organelle (Fig. 3C).
Fig. 3. Deletion of LEU1 increased the levels of mitochondrial Fe-S cluster proteins and increased oxidative stress within mitochondria.
(A) Strains carrying the mitochondrial Fe-S cluster proteins Nfu1 and Aco1 fused with the FLAG epitope tag were used to investigate the influence of Leu1 on expression of the proteins. Cells were grown in low- (0 −M FeCl3) or high- (+100 −M FeCl3) iron medium, and the protein levels were analyzed by Western blot using an anti-FLAG antibody. Asterisk indicates increased protein levels in the leu1 mutant compared to the wild type. (B) The strain carrying the cytosolic Fe-S cluster protein Fra2 fused with the FLAG epitope tag was used to investigate the influence of Leu1 on the expression of the protein. Cells were grown in low- (0 −M FeCl3) or high- (+100 −M FeCl3) iron medium, and the protein levels were analyzed by Western blot using the anti-FLAG antibody. (C) Mitochondrial superoxide dismutase (Sod2) activity was analyzed by native gel electrophoresis and nitroblue tetrazolium staining. Sod2 activity on the same gel was quantified and expressed in the graph (right panel). Similar results were obtained in three independent experiments (p = 0.0226 by t-test). Proteins were transferred to the nitrocellulose membrane and stained with CPTA (copper phthalocyanine-3,4′,4′,4′′-tetrasulfonic acid tetrasodium) to show equal loading of each sample.
3.4. Leu1 plays roles in oxidative stress responses and in maintaining cell wall and membrane integrity
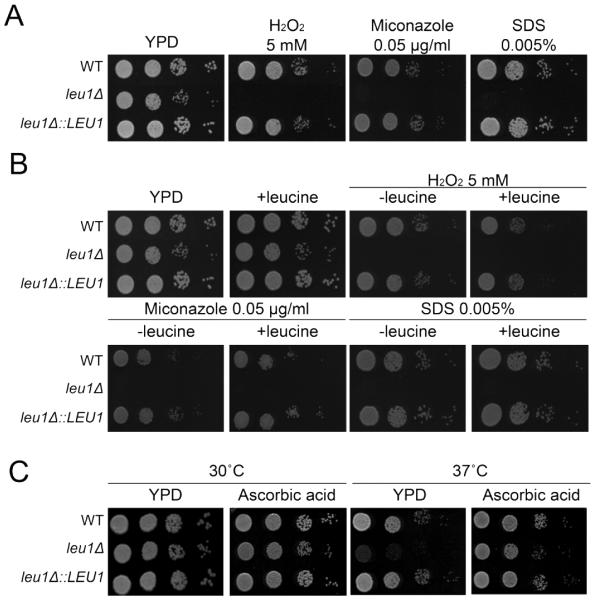
Mitochondrial dysfunction leads to cell wall and membrane defects in several fungi including Candida albicans, C. glabrata and S. cerevisiae (Batova et al., 2008; Brun et al., 2005; Dagley et al., 2011). Furthermore, fungal cells with abnormal mitochondrial iron metabolism display increased sensitivity to oxidative stress (Foury and Cazzalini, 1997; Gakh et al., 2008; Thomas et al., 2013). Therefore, we reasoned that altered mitochondrial iron metabolism in the leu1 mutant may change cell wall and membrane integrity as well as sensitivity to oxidative stress in C. neoformans. Significant growth defects were observed when the leu1 mutant was grown in the presence of oxidative stress-inducing agent hydrogen peroxide (H2O2) (Fig. 4A). Moreover, the leu1 mutant showed increased sensitivity to the antifungal drug miconazole, which is known to trigger oxidative damage in fungal cells (Kobayashi et al., 2002). Increased sensitivity was also observed when the leu1 mutants were cultured in media containing the cell wall and membrane-disrupting agent sodium dodecyl sulfate (SDS). Together, these results indicated that deletion of LEU1 influences oxidative stress responses, and cell wall and membrane integrity in C. neoformans. Moreover, the growth defect of leu1 mutants in the presence of H2O2 and SDS was not restored by exogenously added leucine. Thus, the additional roles of Leu1 in the response to oxidative stress and in cell wall integrity are independent of leucine biosynthesis (Fig. 4B). Similar data were obtained from the experiments using YNB minimal media with asparagine as a sole nitrogen source (Fig. S3). Temperature stress is also related to oxidative stress and mitochondrial function in S. cerevisiae (Davidson and Schiestl, 2001; Davidson et al., 1996). Therefore, we also tested the growth of the leu1 mutant at the host body temperature of 37°C and found significantly reduced growth of the mutant cells. This temperature-sensitive phenotype of the leu1 mutant was ameliorated by addition of the reducing agent ascorbic acid, indicating that oxidative stress contributed to the inhibition of the growth of the mutant cells at 37°C (Fig. 4C). Overall, the hypersensitivity of the leu1 mutant to oxidative stress, the cell wall and membrane disrupting agent, and host body temperature most likely resulted from altered mitochondrial iron homeostasis.
Fig. 4. Deletion of LEU1 increased sensitivity to oxidative stress and altered cell wall and membrane integrity.
(A) The growth of the leu1 mutant in YPD media containing H2O2, miconazole and SDS was monitored. (B) The growth of the leu1 mutant in the same media but with (+) or without (-) leucine (2 mM) was monitored. (C) The growth of the leu1 mutant in YPD media in the absence or presence of ascorbic acid (20 mM) at 30°C or 37°C was monitored. For all experiments, ten-fold serial dilutions of cells (starting at 104 cells) were spotted onto the plates indicated and incubated at 30°C for 2 days.
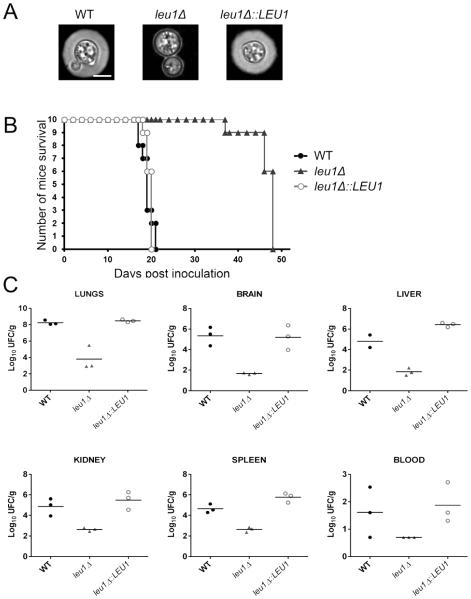
3.5. Deletion of LEU1 causes a defect in capsule formation and attenuated virulence in a mouse inhalation model of cryptococcosis
Melanin synthesis and capsule formation are major virulence traits that contribute to the survival of C. neoformans within the host environment by mediating resistance to oxidative stress and protecting fungal cells from phagocytes, respectively (Kronstad et al., 2008; Kronstad et al., 2011). We evaluated melanin synthesis and capsule formation in the leu1 mutant and found no change in melanin synthesis, but markedly reduced capsule formation compared to the wild-type and reconstituted strains (data not shown, Fig. 5A). We speculate that reduced cell wall integrity caused by altered mitochondrial iron homeostasis may account for the acapsular phenotype of the leu1 mutant. We next investigated the virulence of the leu1 mutant in comparison with the wild type and reconstitute strain using a mouse inhalation model of cryptococcosis. In total, 10 BALB/c mice were infected with each cryptococcal strain, and their survival was monitored daily. We found that the leu1 mutant showed significantly attenuated virulence compared to the wild-type and reconstituted strains (Fig. 5B). The fungal loads in systemic organs (liver, spleen, kidney, lungs and brain) as well as in blood were determined in three mice per group when they reached 15% weigh loss. The results showed at least a 100-fold decrease in leu1 mutant CFU’s compared to the wild type, while the CFUs for the reconstituted mutant were similar to wild type (Fig. 5C). These data suggest that LEU1 is required for the full virulence of C. neoformans, and this is consistent with the increased sensitivity to oxidative stress and high temperature, the reduced cell wall integrity, and the acapsular phenotype of the leu1 mutant.
Fig. 5. The leu1 mutant is deficient in capsule formation and has attenuated virulence.
(A) Strains were grown in defined low-iron medium at 30°C. Photographs were taken after 24-h incubation and after negative staining using India ink to visualize the capsule. The scale bar indicates 5 −m. (B) Ten female BALB/c mice were infected intranasally with each of the strains indicated, and the survival of the mice was monitored twice per day. The results from the assays indicate that LEU1 is required for full virulence. (C) Distribution of fungal cells in the organs of mice infected by the inhalation method. Organs from mice infected with the wild-type strain, the leu1 mutant or the LEU1 reconstituted strain were collected at time of death, and fungal burdens were monitored in organs by determining colony-forming units (CFU) upon plating on YPD medium. Three mice for each strain were used for the experiments, and horizontal bars in each graph represent an average CFUs. In all organs, differences of fungal burdens between the leu1 mutant and the wild-type, and between the leu1 mutant and the reconstitute strain were statistical significance (p<0.05) except that in blood (p × 0.1599, the leu1 mutant vs. the wild-type; p × 0.052, the leu1 mutant vs. the reconstitute strain).
4. Discussion
The branched amino acids include leucine, isoleucine and valine, and their synthesis is partitioned into the mitochondrial matrix and the cytoplasm in fungi. The biosynthetic pathways for the branched amino acids are commonly found in eubacteria, archaebacteria, fungi and plants. However, these pathways are absent in mammals and have thus been considered a target for herbicides and fungicides (Kohlhaw, 2003; McCourt and Duggleby, 2006). The potential of targeting this pathway for antifungal treatment has been suggested in studies with C. albicans and Aspergillus fumigatus (Kingsbury and McCusker, 2010; Oliver et al., 2012). In C. neoformans, the Ilv2 protein encoding acetolactate synthase in the branched amino acid biosynthesis pathway was disrupted to study its potential as an antifungal drug target, and the ilv2 mutant showed significantly reduced growth at 37°C and was avirulent in the murine inhalation model of cryptococcosis (Kingsbury et al., 2004).
Here we studied the iron-responsive regulation of LEU1 and its functional characteristics in C. neoformans. Our study demonstrated that the transcript levels of the gene were decreased upon iron deprivation. Similar down-regulation of LEU1 in iron-deprived conditions was previously observed in S. cerevisiae (Hausmann et al., 2008; Ihrig et al., 2010; Puig et al., 2005). In this fungus, expression of LEU1 is controlled by the Leu3 transcription activator in combination with the iron-responsive metabolite α-isopropylmalate and posttranscriptional mRNA degradation mediated by the mRNA binding proteins Cth1 and Cth2 (Hausmann et al., 2008; Ihrig et al., 2010; Puig et al., 2005). This transcriptional regulation may also be associated with mitochondrial Fe-S cluster synthesis because intracellular iron deprivation resulted in low activity of Fe-S cluster-containing proteins including Leu1 and triggered a decrease in the expression of the LEU1 gene. Atm1 and Yah1 are proteins required for mitochondrial Fe-S cluster synthesis and export in S. cerevisiae, and mutants lacking the genes encoding these proteins displayed significantly reduced LEU1 expression (Ihrig et al., 2010). Glutamate synthase encoded by GLT1 is another Fe-S cluster-containing protein involved in amino acid synthesis, and its transcript and protein levels were also reduced by iron deficiency in S. cerevisiae (Hausmann et al., 2008). These previous observations in S. cerevisiae and our current findings support the hypothesis that C. neoformans down-regulates the genes encoding Fe-S- containing proteins in response to iron deficiency, in turn reducing the biosynthesis of amino acids including leucine as an adaptation strategy.
Our study revealed regulatory mechanisms for the iron-responsive expression of LEU1. Significant de-repression of LEU1 expression was observed in hap3 and hapX mutants grown in the low-iron condition. These results suggest that the Hap complex plays a negative regulatory role in LEU1 expression under iron deprivation. In contrast, the iron-replete condition showed significant derepression of LEU1 expression in the cir1 mutant. These findings suggest that the Hap proteins repress LEU1 expression upon iron deprivation, whereas Cir1 does so in the iron-replete condition. Previously, we suggested that the TCA cycle and electron transport chain are negatively influenced by the Hap proteins. Examples include the Fe–S cluster enzymes aconitase and succinate dehydrogenase in the TCA cycle, and the heme-containing enzyme complexes I–IV in the electron transport chain. Transcript levels of genes encoding these enzymes were significantly increased in the mutant lacking HAP3 and HAPX (Jung et al., 2010). Therefore, the Hap proteins may be the key regulator that reduces consumption of iron upon iron deprivation by inhibiting expression of the genes encoding iron-containing enzymes; as we showed in this study, the Fe-S cluster enzyme Leu1 is one such enzyme. Our conclusion is also supported by the existence of the putative ‘CCAAT’ element, the Hap protein-binding site, in the promoter region of LEU1. Moreover, Cir1 contributes to regulation of LEU1 expression, but, in contrast to the Hap proteins, its negative influence only occurs in cells grown in the iron-replete condition. Negative influences of Cir1 were also observed on the expression of putative low-affinity iron uptake systems encoded by Cft2 and Cfo2, and the laccase-encoding gene LAC1 in C. neoformans (Jung et al., 2006). Hyper-activation of these genes may be harmful as a result of hyper-accumulation of intracellular metals, and over-production of Fe-S proteins including Leu1 under the iron-replete condition appeared to be prevented by Cir1 in C. neoformans.
In S. cerevisiae, Leu1 and Aco1 are abundant 4Fe-4S proteins that are often used as markers to monitor Fe-S cluster biosynthesis and distribution in the cytoplasm and mitochondria (Gardner, 2002; Kispal et al., 1999). The influence of Leu1 on the regulation of mitochondrial Fe-S cluster synthesis and distribution has been well characterized. For instance, up-regulation of Leu1 is associated with down-regulation of the mitochondrial 4Fe-4S protein Aco1, and absence of Leu1 resulted in up- regulation of proteins required for mitochondrial Fe-S cluster synthesis. These findings suggest that Leu1 plays a key role in the mitochondrial-cytoplasmic Fe-S cluster balance and influences mitochondrial Fe-S cluster synthesis (Bedekovics et al., 2011). We observed that the C. neoformans mutant lacking LEU1 displayed up-regulation of the mitochondrial Fe-S proteins Aco1 and Nfu1, the latter of which is the homolog of Nfu1 in S. cerevisiae that is involved in mitochondrial Fe-S cluster synthesis. These findings suggest that Leu1 influences mitochondrial Fe-S cluster synthesis and plays a role in mitochondrial-cytoplasmic Fe-S cluster balance in C. neoformans, as it does in S. cerevisiae. In contrast to the result for the mitochondrial Fe-S cluster proteins, the leu1 mutant displayed similar levels of the cytosolic Fe-S cluster protein Fra2 compared to the wild type. This finding suggests that although the mitochondrial-cytoplasmic Fe-S cluster balance was disrupted, cytoplasmic Fe-S cluster proteins were only marginally influenced by deletion of LEU1. Altered expression of Aco1 and Nfu1 in the leu1 mutant was mainly observed in the cells grown in the low-iron condition, as the mutant cultured in high-iron medium showed wild-type levels of the proteins. These observations imply that leucine biosynthesis plays a stronger role when iron availability in the environment is reduced and suggest a close relationship between amino acid biosynthesis and iron metabolism.
Deficiency in Fe-S cluster synthesis and homeostasis in mitochondria in the leu1 mutant was supported by our observations of phenotypes related to mitochondrial dysfunction such as increased sensitivity of the mutant cells to cell wall and membrane-disrupting agents. It has been shown that mitochondrial dysfunction alters membrane lipid homeostasis, ergosterol synthesis and cell wall integrity, and causes increased sensitivity to the azole antifungal drug miconazole, the cell wall and membrane-disrupting agent SDS, and hydrogen peroxide in C. glabrata, C. albicans and S. cerevisiae (Shingu-Vazquez and Traven, 2011). Moreover, our previous study showed that mitochondrial dysfunction increased sensitivity to azole antifungal drugs and cell wall and membrane-disrupting agent in C. neoformans (Jung et al., 2009). Mitochondria have also been shown to influence pathogenesis and are considered an effective target for antifungal drugs (Kim et al., 2014; Shingu-Vazquez and Traven, 2011). Therefore, considering that deletion of LEU1 results in dysfunctional mitochondria, the Leu1 protein may be an attractive novel target for antifungal treatment.
The acapsular phenotype of the leu1 mutant may also be influenced by mitochondrial dysfunction resulting in reduced cell wall and membrane integrity. The cell wall is particularly important for the formation of cell-associated capsule because it is the site of polysaccharide attachment. Increased sensitivity to oxidative stress and the host body temperature of 37°C were also caused by deficiency in mitochondrial function in the leu1 mutant. These phenotypic characteristics likely also contributed to the attenuated virulence of the mutant. In contrast, it is uncertain whether leucine auxotrophy contributed significantly to the attenuated virulence. Early studies showed that adult human plasma contains 122 - 160 −mol/L leucine, and 10.9 −mol/L leucine was found, on average, in the cerebrospinal fluid of humans with various health statuses (Armstrong and Stave, 1973; Dickinson and Hamilton, 1966). Taken together, our results suggest that the leucine biosynthesis pathway and Leu1 in particular are required for mitochondrial function, iron homeostasis and full virulence in C. neoformans.
Supplementary Material
Highlights.
▶ The current study characterized the role of Leu1 in iron homeostasis and the virulence of Cryptococcus neoformans.
▶ Leu1 influenced mitochondrial iron metabolism and sensitivity to oxidative stress and cell wall/membrane disrupting agents.
▶ Leu1 function was required for capsule formation and virulence in C. neoformans.
Acknowledgements
This study was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT, and Future Planning NRF-2013R1A1A1A05007037 (WJ), and the National Institute of Allergy and Infectious Diseases (RO1 AI053721) (JWK).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adour L, et al. Sequential use of ammonium and leucine as nitrogen sources during growth of Geotrichum candidum on a glucose based medium. Electronic Journal of Biotechnology. 2010:13. [Google Scholar]
- Armstrong MD, Stave U. A study of plasma free amino acid levels. II. Normal values for children and adults. Metabolism. 1973;22:561–9. doi: 10.1016/0026-0495(73)90069-3. [DOI] [PubMed] [Google Scholar]
- Batova M, et al. Functional characterization of the CgPGS1 gene reveals a link between mitochondrial phospholipid homeostasis and drug resistance in Candida glabrata. Curr Genet. 2008;53:313–22. doi: 10.1007/s00294-008-0187-9. [DOI] [PubMed] [Google Scholar]
- Beauchamp C, Fridovich I. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Analytical biochemistry. 1971;44:276–287. doi: 10.1016/0003-2697(71)90370-8. [DOI] [PubMed] [Google Scholar]
- Bedekovics T, et al. Leucine biosynthesis regulates cytoplasmic iron-sulfur enzyme biogenesis in an Atm1p-independent manner. J Biol Chem. 2011;286:40878–88. doi: 10.1074/jbc.M111.270082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical biochemistry. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brun S, et al. Biological consequences of petite mutations in Candida glabrata. J Antimicrob Chemother. 2005;56:307–14. doi: 10.1093/jac/dki200. [DOI] [PubMed] [Google Scholar]
- Dagley MJ, et al. Cell wall integrity is linked to mitochondria and phospholipid homeostasis in Candida albicans through the activity of the post-transcriptional regulator Ccr4-Pop2. Mol Microbiol. 2011;79:968–89. doi: 10.1111/j.1365-2958.2010.07503.x. [DOI] [PubMed] [Google Scholar]
- Davidson JF, Schiestl RH. Mitochondrial respiratory electron carriers are involved in oxidative stress during heat stress in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:8483–9. doi: 10.1128/MCB.21.24.8483-8489.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson JF, et al. Oxidative stress is involved in heat-induced cell death in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1996;93:5116–21. doi: 10.1073/pnas.93.10.5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson JC, Hamilton PB. The free amino acids of human spinal fluid determined by ion exchange chromatography. J Neurochem. 1966;13:1179–85. doi: 10.1111/j.1471-4159.1966.tb04275.x. [DOI] [PubMed] [Google Scholar]
- Foury F, Cazzalini O. Deletion of the yeast homologue of the human gene associated with Friedreich's ataxia elicits iron accumulation in mitochondria. FEBS Lett. 1997;411:373–7. doi: 10.1016/s0014-5793(97)00734-5. [DOI] [PubMed] [Google Scholar]
- Friden P, Schimmel P. LEU3 of Saccharomyces cerevisiae activates multiple genes for branched-chain amino acid biosynthesis by binding to a common decanucleotide core sequence. Mol Cell Biol. 1988;8:2690–7. doi: 10.1128/mcb.8.7.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gakh O, et al. Assembly of the iron-binding protein frataxin in Saccharomyces cerevisiae responds to dynamic changes in mitochondrial iron influx and stress level. J Biol Chem. 2008;283:31500–10. doi: 10.1074/jbc.M805415200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner PR. Aconitase: sensitive target and measure of superoxide. Methods Enzymol. 2002;349:9–23. doi: 10.1016/s0076-6879(02)49317-2. [DOI] [PubMed] [Google Scholar]
- Hakim JG, et al. Impact of HIV infection on meningitis in Harare, Zimbabwe: a prospective study of 406 predominantly adult patients. AIDS. 2000;14:1401–7. doi: 10.1097/00002030-200007070-00013. [DOI] [PubMed] [Google Scholar]
- Hausmann A, et al. Cellular and mitochondrial remodeling upon defects in iron-sulfur protein biogenesis. J Biol Chem. 2008;283:8318–30. doi: 10.1074/jbc.M705570200. [DOI] [PubMed] [Google Scholar]
- Hofman-Bang J. Nitrogen catabolite repression in Saccharomyces cerevisiae. Mol Biotechnol. 1999;12:35–73. doi: 10.1385/MB:12:1:35. [DOI] [PubMed] [Google Scholar]
- Hu G, Kronstad JW. A putative P-type ATPase, Apt1, is involved in stress tolerance and virulence in Cryptococcus neoformans. Eukaryotic cell. 2010;9:74–83. doi: 10.1128/EC.00289-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihrig J, et al. Iron regulation through the back door: iron-dependent metabolite levels contribute to transcriptional adaptation to iron deprivation in Saccharomyces cerevisiae. Eukaryot Cell. 2010;9:460–71. doi: 10.1128/EC.00213-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, et al. Role of ferroxidases in iron uptake and virulence of Cryptococcus neoformans. Eukaryot Cell. 2009;8:1511–20. doi: 10.1128/EC.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, et al. HapX positively and negatively regulates the transcriptional response to iron deprivation in Cryptococcus neoformans. PLoS Pathog. 2010;6:e1001209. doi: 10.1371/journal.ppat.1001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, et al. Iron source preference and regulation of iron uptake in Cryptococcus neoformans. PLoS Pathog. 2008;4:e45. doi: 10.1371/journal.ppat.0040045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, et al. Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans. PLoS Biol. 2006;4:e410. doi: 10.1371/journal.pbio.0040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaptain S, et al. A Regulated Rna-Binding Protein Also Possesses Aconitase Activity. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:10109–10113. doi: 10.1073/pnas.88.22.10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, et al. A defect in iron uptake enhances the susceptibility of Cryptococcus neoformans to azole antifungal drugs. Fungal Genet Biol. 2012;49:955–66. doi: 10.1016/j.fgb.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, et al. A vanillin derivative causes mitochondrial dysfunction and triggers oxidative stress in Cryptococcus neoformans. PLoS One. 2014;9:e89122. doi: 10.1371/journal.pone.0089122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury JM. Cryptococcus neoformans Ilv2p confers resistance to sulfometuron methyl and is required for survival at 37°C and in vivo. Microbiology. 2004;150:1547–1558. doi: 10.1099/mic.0.26928-0. [DOI] [PubMed] [Google Scholar]
- Kingsbury JM, McCusker JH. Cytocidal amino acid starvation of Saccharomyces cerevisiae and Candida albicans acetolactate synthase (ilv2Δ) mutants is influenced by the carbon source and rapamycin. Microbiology. 2010;156:929–39. doi: 10.1099/mic.0.034348-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury JM, et al. Cryptococcus neoformans Ilv2p confers resistance to sulfometuron methyl and is required for survival at 37 degrees C and in vivo. Microbiology. 2004;150:1547–58. doi: 10.1099/mic.0.26928-0. [DOI] [PubMed] [Google Scholar]
- Kispal G, et al. The ABC transporter Atm1p is required for mitochondrial iron homeostasis. FEBS Lett. 1997;418:346–50. doi: 10.1016/s0014-5793(97)01414-2. [DOI] [PubMed] [Google Scholar]
- Kispal G, et al. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 1999;18:3981–9. doi: 10.1093/emboj/18.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmetzsch L, et al. The GATA-type transcriptional activator Gat1 regulates nitrogen uptake and metabolism in the human pathogen Cryptococcus neoformans. Fungal Genet Biol. 2011;48:192–9. doi: 10.1016/j.fgb.2010.07.011. [DOI] [PubMed] [Google Scholar]
- Kobayashi D, et al. Endogenous reactive oxygen species is an important mediator of miconazole antifungal effect. Antimicrob Agents Chemother. 2002;46:3113–7. doi: 10.1128/AAC.46.10.3113-3117.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlhaw GB. Leucine biosynthesis in fungi: entering metabolism through the back door. Microbiol Mol Biol Rev. 2003;67:1–15. doi: 10.1128/MMBR.67.1.1-15.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad J, et al. Beyond the big three: systematic analysis of virulence factors in Cryptococcus neoformans. Cell Host Microbe. 2008;4:308–10. doi: 10.1016/j.chom.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Kronstad JW, et al. Expanding fungal pathogenesis: Cryptococcus breaks out of the opportunistic box. Nat Rev Microbiol. 2011;9:193–203. doi: 10.1038/nrmicro2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IR, et al. Nitrogen metabolite repression of metabolism and virulence in the human fungal pathogen Cryptococcus neoformans. Genetics. 2011;188:309–23. doi: 10.1534/genetics.111.128538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IR, et al. Characterization of an Nmr homolog that modulates GATA factor-mediated nitrogen metabolite repression in Cryptococcus neoformans. PLoS One. 2012;7:e32585. doi: 10.1371/journal.pone.0032585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluf GA. Genetic regulation of nitrogen metabolism in the fungi. Microbiol Mol Biol Rev. 1997;61:17–32. doi: 10.1128/mmbr.61.1.17-32.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCourt JA, Duggleby RG. Acetohydroxyacid synthase and its role in the biosynthetic pathway for branched-chain amino acids. Amino Acids. 2006;31:173–210. doi: 10.1007/s00726-005-0297-3. [DOI] [PubMed] [Google Scholar]
- Nazi I, et al. Role of homoserine transacetylase as a new target for antifungal agents. Antimicrob Agents Chemother. 2007;51:1731–6. doi: 10.1128/AAC.01400-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver JD, et al. The Aspergillus fumigatus dihydroxyacid dehydratase Ilv3A/IlvC is required for full virulence. PLoS One. 2012;7:e43559. doi: 10.1371/journal.pone.0043559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BJ, et al. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–30. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- Pascon RC, et al. Cryptococcus neoformans methionine synthase: expression analysis and requirement for virulence. Microbiology. 2004;150:3013–23. doi: 10.1099/mic.0.27235-0. [DOI] [PubMed] [Google Scholar]
- Puig S, et al. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell. 2005;120:99–110. doi: 10.1016/j.cell.2004.11.032. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. CSHL press; 2001. [Google Scholar]
- Schilke B, et al. Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1999;96:10206–11. doi: 10.1073/pnas.96.18.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingu-Vazquez M, Traven A. Mitochondria and fungal pathogenesis: drug tolerance, virulence, and potential for antifungal therapy. Eukaryot Cell. 2011;10:1376–83. doi: 10.1128/EC.05184-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze JY, et al. In vitro transcriptional activation by a metabolic intermediate: activation by Leu3 depends on alpha-isopropylmalate. Science. 1992;258:1143–5. doi: 10.1126/science.1439822. [DOI] [PubMed] [Google Scholar]
- Thomas E, et al. Mitochondria influence CDR1 efflux pump activity, Hog1-mediated oxidative stress pathway, iron homeostasis, and ergosterol levels in Candida albicans. Antimicrob Agents Chemother. 2013;57:5580–99. doi: 10.1128/AAC.00889-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toffaletti DL, et al. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. Journal of Bacteriology. 1993;175:1405–1411. doi: 10.1128/jb.175.5.1405-1411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisman R, et al. Regulation of leucine uptake by tor1+ in Schizosaccharomyces pombe is sensitive to rapamycin. Genetics. 2005;169:539–50. doi: 10.1534/genetics.104.034983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, et al. Molecular and genetic analysis of the Cryptococcus neoformans MET3 gene and a met3 mutant. Microbiology. 2002;148:2617–25. doi: 10.1099/00221287-148-8-2617. [DOI] [PubMed] [Google Scholar]
- Zhou K, et al. Structure of yeast regulatory gene LEU3 and evidence that LEU3 itself is under general amino acid control. Nucleic Acids Res. 1987;15:5261–73. doi: 10.1093/nar/15.13.5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou KM, et al. Yeast regulatory protein LEU3: a structure-function analysis. Nucleic Acids Res. 1990;18:291–8. doi: 10.1093/nar/18.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.