Abstract
The high-affinity reductive iron uptake system that includes a ferroxidase (Cfo1) and an iron permease (Cft1) is critical for the pathogenesis of Cryptococcus neoformans. In addition, a mutant lacking CFO1 or CFT1 not only has reduced iron uptake but also displays a markedly increased susceptibility to azole antifungal drugs. Altered antifungal susceptibility of the mutants was of particular interest because the iron uptake system has been proposed as an alternative target for antifungal treatment. In this study, we used transcriptome analysis to begin exploring the molecular mechanisms of altered antifungal susceptibility in a cfo1 mutant. The wild-type strain and the cfo1 mutant were cultured with or without the azole antifungal drug fluconazole and their transcriptomes were compared following sequencing with Illumina Genome Analyzer IIx (GAIIx) technology. As expected, treatment of both strains with fluconazole caused elevated expression of genes in the ergosterol biosynthetic pathway that includes the target enzyme Erg11. Additionally, genes differentially expressed in the cfo1 mutant were involved in iron uptake and homeostasis, mitochondrial functions and respiration. The cfo1 mutant also displayed phenotypes consistent with these changes including a reduced ratio of NAD+/NADH and down-regulation of Fe-S cluster synthesis. Moreover, combination treatment of the wild-type strain with fluconazole and the respiration inhibitor diphenyleneiodonium dramatically increased susceptibility to fluconazole. This result supports the hypothesis that down-regulation of genes required for respiration contributed to the altered fluconazole susceptibility of the cfo1 mutant. Overall, our data suggest that iron uptake and homeostasis play a key role in antifungal susceptibility and could be used as novel targets for combination treatment of cryptococcosis. Indeed, we found that iron chelation in combination with fluconazole treatment synergistically inhibited the growth of C. neoformans.
Keywords: C. neoformans, iron uptake, ferroxidase, CFO1, antifungal drugs, fluconazole
1. Introduction
Cryptococcus neoformans is a basidiomycete fungal pathogen of humans that frequently causes life-threatening fungal meningitis in immunocompromised individuals. Cryptococcal meningitis associated with AIDS is fatal in the absence of antifungal therapy and the mortality rate can be as high as 40% in sub-Saharan Africa (Hakim et al., 2000). Non-HIV-associated cryptococcosis has also become an important issue, especially among patients with organ transplantation or long-term anticancer therapy (Bicanic and Harrison, 2004). A limited number of antifungal drugs are available to treat cryptococcosis with the polyene drug amphotericin B and azole drugs such as fluconazole being the most widely used. Amphotericin B interacts directly with sterols in the fungal membrane and this results in the production of pores that alter permeability and cause cytoplasmic leakage leading to cell death (Ghannoum and Rice, 1999; Lupetti et al., 2002). Although amphotericin B is considered the most effective antifungal drug, severe side effects such as nephrotoxicity limit its clinical use. To reduce side effects, liposomal forms of amphotericin B have been developed. Moreover, combination therapy with amphotericin B and 5-flucytosine has been established to treat cryptococcal meningitis (Bennett et al., 1979).
Besides amphotericin B, azole antifungals are also widely used in the treatment of cryptococcal infections. Fluconazole, in particular, has been the agent of choice for the long term management of cryptococcosis because of its clinical efficacy and safety (Zonios and Bennett, 2008). Azole antifungal drugs such as fluconazole are generally considered fungistatic and act by inhibiting the ergosterol biosynthetic enzyme lanosterol 14 -demethylase (Erg11), which belongs to the hemoprotein cytochrome P450 protein family. Inhibition of Erg11 interferes with the conversion of lanosterol to ergosterol, a membrane component, and this leads to accumulation of 14- -methylsterols that cause membrane disorganization, cytoplasmic leakage and growth arrest (Ghannoum and Rice, 1999). Although they are effective, long-term use of azole antifungal drugs often leads to the emergence of resistance that causes treatment failure and recurrence of disease. The mechanisms of resistance to azole antifungal drugs have been extensively studied in another well-known pathogenic yeast, Candida albicans. In C. albicans, several mechanisms result in the emergence of resistance: 1) reduced intracellular accumulation of azoles, which is often associated with the up-regulation of genes encoding efflux pumps; 2) increased levels of the azole cellular target by altered regulation; 3) alteration in sterol synthesis, which is usually achieved by a deficiency in the sterol 5,6–desaturase encoded by ERG3 and; 4) decreased affinity of azoles to their target due to mutations in ERG11 (Lupetti et al., 2002). Moreover, a role of Hsp90 in evolution of azole resistance has been demonstrated (Cowen et al., 2006; Cowen and Lindquist, 2005).
Azole resistant strains have been reported for C. neoformans, although few studies have examined the mechanisms of azole resistance. One example is a recent study that employed comparative genome hybridization to reveal that the formation of disomic chromosomes carrying ERG11 or a gene for an efflux pump caused increased resistance (Sionov et al., 2010). Furthermore, another recent study reported that mutations in the ERG11 gene were associated with azole resistance (Sionov et al., 2011). The emergence of resistance can be countered by combination antifungal therapy and this approach has been successfully applied to cryptococcosis through the use of amphotericin B in combination with 5-flucytocine (Wirk and Wingard, 2008). Combinations of drugs that target new functions also hold promise for expanding antifungal therapeutic options. For example, the combination of Hsp90 inhibition and micafungin, a member of the echinocandin class of inhibitors of fungal 1,3-β-D-glucan synthase, has shown enhanced fungicidal effect (Singh et al., 2009).
Interference with iron acquisition by pathogens may yield new opportunities for combination therapy. For C. neoformans, the high-affinity reductive iron uptake pathway comprised of the iron permease Cft1 and the ferroxidase Cfo1 plays an important role in virulence. Mutants deficient in the pathway are not able to utilize iron from transferrin and display significant attenuation of virulence in a mouse model of cryptococcosis (Jung et al., 2009; Jung et al., 2008). Moreover, our previous data suggested that the high-affinity reductive iron uptake pathway influences susceptibility to azole antifungal drugs in C. neoformans (Jung et al., 2009). For example, a mutant lacking CFT1 shows increased susceptibility to miconazole, and a mutant lacking CFO1 displays increased susceptibility to fluconazole (Jung et al., 2009; Jung et al., 2008). These results support the possibility of a novel combination therapy that uses currently available azole drugs and inhibition of the high-affinity reductive iron uptake pathway. This idea prompted our current study of the mechanism of altered azole susceptibility for the cfo1 mutant. Our approach was to examine the transcriptome of the cfo1 mutant grown in the presence or absence of fluconazole using deep RNA sequencing (RNA-Seq) and to compare it with the transcriptome of the wild-type strain. We found that the expression of genes encoding iron-dependent enzymes including those required for ergosterol biosynthesis and respiration were significantly changed by deletion of CFO1. Moreover, our data suggested that reduced expression of genes required for mitochondrial functions such as respiration and Fe-S cluster synthesis made important contributions to the increased fluconazole susceptibility of the cfo1 mutant. We also found a consistent pattern of regulation in which these genes were transcriptionally regulated by the iron-regulatory proteins Hap3 and HapX; mutations in these regulators also influenced azole susceptibility in C. neoformans.
2. Materials and methods
2.1 Strains and growth conditions
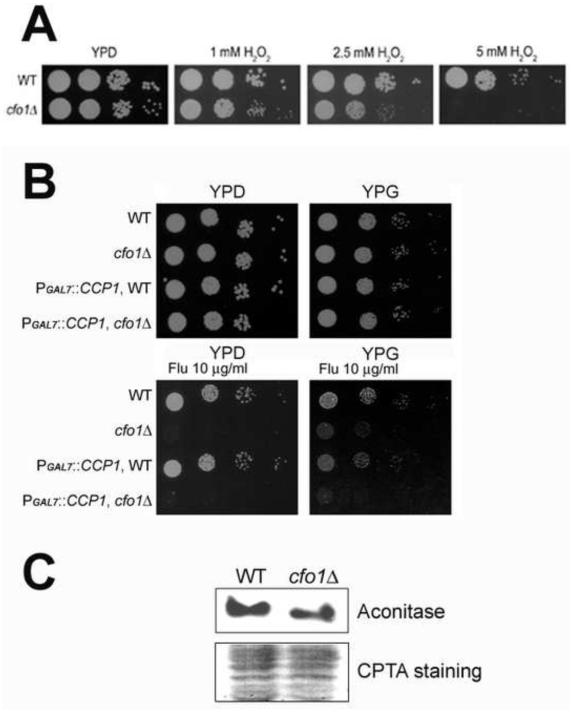
Strains used in this study were Cryptococcus neoformans var. grubii H99 and a cfo1 mutant of the same strain. The mutant was constructed previously and complementation demonstrated that the phenotypes of the strain were due to the deletion of CFO1 (Jung et al., 2009). Cells were routinely grown in YPD medium (1% yeast extract, 2% bacto-peptone and 2% glucose) or YNB medium (yeast nitrogen base, Difco) with 2.0% glucose. YPG medium (1% yeast extract, 2% bacto-peptone and 2% galactose) was used to induce overexpression of genes via the GAL7 promoter.
2.2 Drug susceptibility
To estimate antifungal drug susceptibility, the wild-type strain and the cfo1 mutant were grown in 3 ml of YPD medium at 30°C overnight with shaking. After incubation, 10-fold dilutions of cells (starting at 104 cells) were spotted onto YPD agar medium plates containing various azole antifungal drugs as indicated in the text. To investigate fluconazole susceptibility of the strains that contain the GAL7 overexpression promoter, cells were grown in 3 ml of YPD medium overnight, washed twice with PBS (pH 7.4), and resuspended in PBS. Cell suspensions were spotted on YPD or YPG medium with or without fluconazole. The plates were incubated at 30°C for 2 days and photographed. Minimal inhibitory concentrations (MIC) were determined according to Clinical and Laboratory Standards (CLS) M27-A3, and minimal fungicidal concentrations (MFC) were determined as described elsewhere (Clinical and Laboratory Standards, 2008; Sanglard et al., 2003).
2.3 Quantitative real-time PCR
Primers for real-time PCR were designed using Primer Express software 3.0 (Applied Biosystems) and are listed in Table S1. Total RNA was extracted with the RiboPure™-Yeast Kit (Ambion) and cDNA was synthesized using the RevertAid™ First Strand cDNA synthesis Kit (Fermentas). Relative gene expression was quantified using a 7500 system (Applied Biosystems) based on the 2-LlLlC method. TEF2 (CNAG_00044, translation elongation factor 2) was used as a reference.
2.4 Construction of ERG11 or CCP1 overexpression strains
The native promoter of ERG11 (CNAG_00040) or CCP1 (CNAG_01138) was replaced with the GAL7 promoter by homologous recombination. The 5’-flanking region containing the native promoter sequences of the target gene, and the 3’-flanking region containing the ATG start codon and the coding sequences were amplified by PCR with the wild-type genomic DNA as a template and primers Gal7-1/Gal7-2 and Gal7-3/Gal7-4 (Table S1). The DNA fragment that contains the NAT (nourseothricin acetyltransferase) resistance gene and the GAL7 promoter were amplified by PCR using primers Sed5-F and Sed5-R and genomic DNA of the C. neoformans PGAL7::SED5 strain (J. C., unpublished data). The 5’-flanking region, the 3’-flanking region and the NAT- PGAL7 region were fused with overlapping PCR. The resulting fragments were biolistically transformed into the wild-type strain or the cfo1 mutant as described (Toffaletti et al., 1993).
2.5 RNA extraction, sequencing and data analysis
Overnight cultures of the wild-type strain and the cfo1 mutant were diluted to 1.0 × 106 cells in 50 ml of YPD medium with or without 10 g/ml of fluconazole (dissolved in dimethyl sulfoxide (DMSO)). DMSO was added to YPD medium without fluconazole as a drug-free control. Cultures were incubated with shaking at 30°C for 3 hours and harvested for RNA extraction. Total RNA was extracted by the RiboPureTM-Yeast Kit (Ambion) following the manufacturer's instructions. Contaminating DNA was eliminated by RNase-free DNase (Invitrogen), and the quantity and quality of the total RNA were evaluated using RNA electropherograms (Agilent 2100 Bioanalyzer) and the RNA integrity number (RIN) (Schroeder et al., 2006). Total RNA (10 g) from each sample was used as starting material to prepare sequencing libraries with the Illumina mRNA-Seq sample preparation kit (Illumina) following the manufacturer's instructions. One lane per sample was used for sequencing with the Illumina Genome Analyzer IIx to generate non-directional, single-ended 36 base pair reads. Quality-filtered reads were mapped to the reference genome sequence (http://www.broadinstitute.org/annotation/genome/cryptococcus_neoformans/MultiHome.html) using CLC Genomics Workbench 4.0 (CLC bio). The relative transcript abundance was computed by counting the reads per kilobase of exon model per million mapped sequence reads (RPKM) (Mortazavi et al., 2008). Processed data was deposited in Gene Express Omnibus (GEO) with the accession number GSE37875. GO (Gene Ontology) analysis was performed by assigning GO categories to each gene according to the Gene ontology consortium database (Ashburner et al., 2000). Each gene was compared to the GO database (http://www.geneontology.org/GO.downloads.database.shtml) using BLASTP searches and the best hit with an e-value under 10−5 and hit coverage over 50% was adopted.
2.6 ICP-AES (Inductively Coupled Plasma-Atomic Emission Spectroscopy)
Total cellular iron was determined by ICP-AES. The wild-type strain and the cfo1 mutant were grown in 50 ml of YPD medium at 30°C overnight. Cells were washed twice with low-iron water and resuspended in 200 ml of defined low-iron YNB medium followed by incubation at 30°C for 2 days (Jung et al., 2009). Cells were washed three times with low-iron water and lyophilized. A total of 0.15 g of cell biomass was digested with 3 ml of H2O2 and 5 ml of HNO3 using a microwave digestion system (START D). ICP-AES analysis was performed using the OPTIMA 5300 DV (PerkinElmer) system. The scaling and normalization process were based on total cell mass.
2.7 Aconitase assay (zymography)
The activity of aconitase was determined by zymography (Shi et al., 2009). The wild-type strain and the cfo1 mutant were grown in YPD medium at 30°C for 12 hours and harvested by centrifugation at 13,000 rpm for 5 min. Cell pellets were lysed with 1% triton/citrate lysis buffer containing 36.6 mM KCl, 22.9 mM Tris-HCl pH 7.5, 1% Triton, 1.0 mM DTT, 1 mM PMSF, 2.0 mM sodium citrate, 0.6 mM MnCl2 and 1% protease inhibitor cocktail (Sigma). The zymography gel consisted of an 8% separating gel containing 132 mM Tris-base, 3.6 mM citrate and 132 mM borate, and 4% stacking gel containing 67 mM Tris base, 3.6 mM citrate and 67 mM borate. The sample mixture contained 20 g of protein lysate, 25 mM Tris-HCl pH 8.0, 10% glycerol and 0.025% bromophenol blue, and the running buffer contained 25 mM Tris-HCl pH 8.3, 192 mM glycine and 3.6 mM citrate. Cell extracts were analyzed by native polyacrylamide gel electrophoresis at 160V at 4°C for 2 hours. Aconitase activities were visualized by photographing the gel, which was placed in staining buffer containing 100 mM Tris-Cl pH 8.0, 1 mM NADP, 2.5 mM cis-aconitic acid, 5 mM MgCl2, 1.2 mM MTT, 0.3 mM phenazin methosulfate and 5 U/ml isocitrate dehydrogenase, and was incubated in the dark at 37°C for 10 minutes. The nitrocellulose membrane containing the same protein samples was stained with copper phthalocyanine 3,4′,4′′,4′′′-tetrasulfonic acid tetrasodium salt (CPTS) and staining intensity was used for normalization.
2.8 NAD+/NADH assay
To determine the NAD+/NADH ratio in the wild-type strain or the cfo1 mutant, cells were grown in YPD medium at 30°C overnight and washed twice with cold PBS. Cell pellets were homogenized, and the supernatant was used for the assay. The NAD+/NADH ratio was calculated by the EnzyChromTM NAD+/NADH Assay Kit (BioAssay Systems) at 565 nm following the manufacturer’s instructions.
3. Results
3.1 Deletion of CFO1 increases susceptibility and reduces tolerance to azole antifungal drugs.
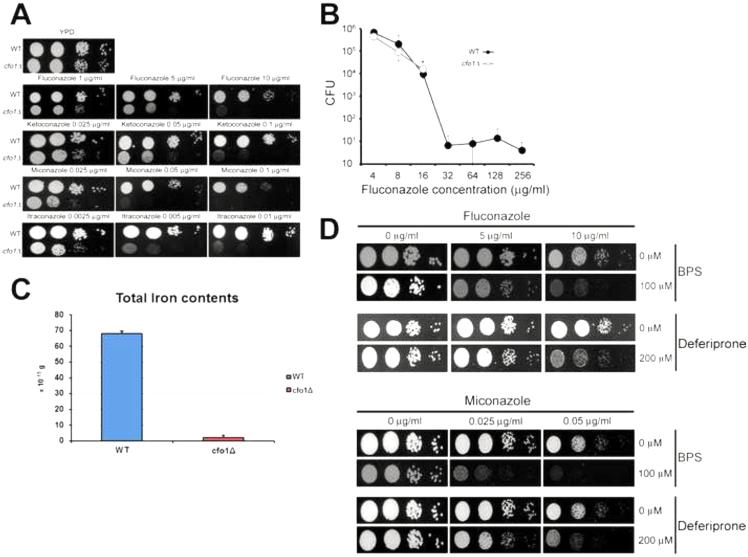
We previously reported that deletion of the CFO1 gene encoding the ferroxidase component of the high-affinity reductive iron uptake system increased the susceptibility of C. neoformans to the antifungal drug fluconazole. We hypothesized that a possible underlying mechanism could be low intracellular iron and a concomitant deficiency in heme synthesis resulting in reduced activity of heme-containing enzymes for ergosterol biosynthesis (e.g., Erg11) (Jung et al., 2009). To begin to test this hypothesis and explain the enhanced susceptibility phenotype, we initiated a more detailed examination of the cfo1 mutant. Our previous study had only tested the susceptibility of the cfo1 mutant to fluconazole, and we therefore first investigated whether the phenotype was common to other azole antifungal drugs including ketoconazole, miconazole and itraconazole. Figure 1A shows that the growth of the cfo1 mutant was significantly reduced compared to the wild-type strain in media containing any of the azole drugs. In general, these results confirmed and extended our previous findings regarding loss of CFO1 and increased susceptibility to fluconazole. We also determined the minimal inhibitory concentration (MIC) of fluconazole and found that the wild-type strain and the cfo1 mutant exhibited the MICs of 16 g/ml and 8 g/ml, respectively. It was surprising that the mutant displayed only a marginal MIC difference compared to the wild-type strain and this led us to speculate that deletion of CFO1 might influence drug tolerance to fluconazole. Tolerance of a drug is defined as the ability to survive at drug concentrations above the MIC and can be quantified as the ratio of a minimal fungicidal concentration to the MIC (MFC/MIC) (Sanglard et al., 2003). We determined tolerance of the wild-type strain and the cfo1 mutant, and found that they were >128 and 4, respectively (Table 1). These data suggested that fluconazole tolerance was affected by deletion of CFO1. For further confirmation of altered fluconazole tolerance in the cfo1 mutant, the viability of the cells was analyzed in the presence of various concentrations of fluconazole. The results showed that fluconazole, which is generally considered to be fungistatic, exhibited a fungicidal effect on the cfo1 mutant (Figure 1B).
Figure 1. Deletion of CFO1 increased susceptibility to azole antifungal drugs.
(A) The growth of the cfo1 mutant in media containing various azole antifungal drugs was monitored. Ten-fold serial dilutions of cells (starting at 104 5 cells) were spotted onto YPD plates with or without the antifungal drug at the concentrations indicated. Plates were incubated at 30°C for 2 days. (B) Tolerance to fluconazole was assayed with time kill curves in YPD liquid media with various concentrations of fluconazole, as indicated. No viable cells were observed for the cfo1 mutant cultured in the presence of fluconazole above 16 μg/ml while a small number of the wild-type cells grew in the presence of higher concentrations of the drug. Data are the averages of three separate measurements with the standard deviations. (C) Total cellular iron levels were determined using ICP analysis. Values indicate weight per cell and are averages from three independent experiments with standard deviations. (D) Combination treatment of an iron chelator and fluconazole or miconazole. The growth of the wildtype strain was tested in media containing azole drug with or without the iron chelator indicated. Tenfold serial dilutions of cells (starting at 104 15 cells) were spotted onto plates with subsequent incubation at 30°C for 2 days.
Table 1.
Minimal inhibitory concentration and tolerance to fluconazole
| MFC ( g/ml) | MIC ( g/ml) | MFC/MIC | |
|---|---|---|---|
| WT | > 2048 | 16 | > 128 |
| cfo1Δ | 32 | 8 | 4 |
The Cfo1 protein plays an essential role in the high-affinity iron uptake pathway. Therefore, as mentioned above, we hypothesized that the deletion of the CFO1 reduced the intracellular iron concentration. To test this directly, the total cellular iron concentration of the strains was measured by ICP-AES. Indeed, we found that the cfo1 mutant displayed reduced iron levels compared to the wild-type strain with 2.0×10−15±0.017 g and 68×10−15±0.186 g per cell, respectively (Figure 1C). These data suggested that any perturbation of intracellular iron levels might alter azole susceptibility for C. neoformans. We therefore investigated the possibility of a novel combination treatment using iron chelation together with an azole antifungal drug to inhibit growth. In this case, we tested the iron chelators bathophenanthroline disulfonate (BPS) or deferiprone in combination with fluconazole, or with miconazole, and found that these combinations showed a synergistic effect to the wild-type strain as predicted (Figure 1D). Overall, these results were consistent with our hypothesis and indicated that a more detailed investigation of the impact of the cfo1 perturbation on the cell was warranted.
3.2 Deletion of CFO1 causes large changes in the transcriptome in the presence and absence of fluconazole.
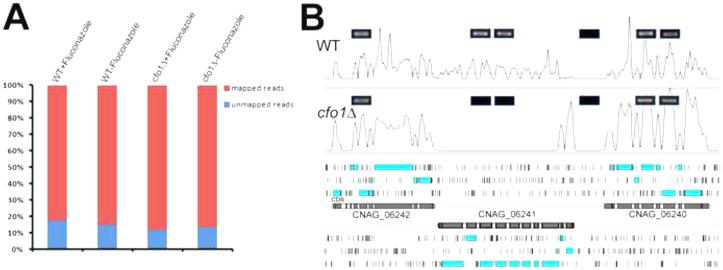
C. neoformans is known to demonstrate a transcriptional response for the ERG and other genes in response to fluconazole (Florio et al., 2011). We therefore reasoned that the abundance of transcripts for genes involved in antifungal drug susceptibility may be altered by deletion of CFO1. To test this idea, we sequenced the transcriptomes of the wild-type strain and the cfo1 mutant grown in the presence or absence of fluconazole. We focused our attention on fluconazole because it is the most commonly prescribed azole for treating cryptococcosis and because of the known transcriptional response (Florio et al., 2011). Four different cDNA libraries were synthesized using mRNA purified from the wild-type strain or the cfo1 mutant in the presence or absence of fluconazole and sequenced for cross-comparison (see Materials and Methods). In total, 138,663,841 sequence reads were obtained from four cDNA libraries. Of these, 118,962,848 (85.79%) sequence reads were mapped against the genome of C. neoformans var. grubii H99 (Figure 2A). The current annotation of the genome of the wild-type strain, C. neoformans var. grubii H99, has 6,980 predicted coding sequences including 13 mitochondrial transcripts. Of these, we detected expression of 6,951 coding sequences (Table S2). The high-reliability of our data sets was evident from the low read-counts in the intergenic regions and the high read-counts in the coding regions; these features were confirmed by RT-PCR for six selected regions including the CFO1 locus (Figure 2B).
Figure 2. Sequence read mapping statistics and representative transcript signals at the CFO1 locus.
(A) Each bar represents the total number of sequence reads obtained. The reads that mapped to the reference genome (Cryptococcus neoformans var. grubii H99) are shown in red and those that failed to map to the genome are shown in blue. (B) Representative RNA sequencing data from the wild-type strain (WT) or the cfo1 mutant (cfo1Δ) grown in YPD, show the expression at the CFO1 locus of each strain. Boxes with blue borders represent the RT-PCR results confirming the existence of transcript signals, and no RT-PCR signal was detected from the region of the CFO1 locus (CNAG_06241) in the cfo1 mutant (thus illustrating the reliability of the RNA-Seq data). Light blue boxes in the lower panel represent predicted open reading frames and black vertical bars represent stop codons in the reference genome.
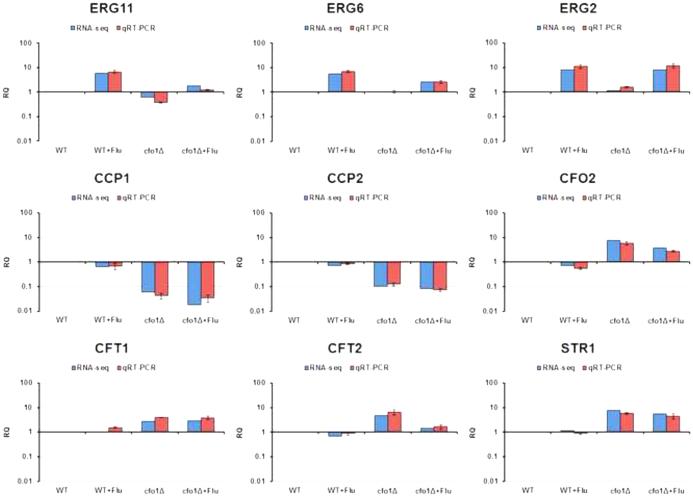
Comparisons of the transcriptomes of the wild-type strain and the cfo1 mutant in the presence or absence of fluconazole revealed that subsets of genes were affected by fluconazole treatment and/or deletion of CFO1. Fluconazole treatment triggered more than 2-fold differential expression of 315 genes in the wild-type strain (Table 2). Of these 315 genes, 138 were up-regulated and 177 were down-regulated. The number of differentially expressed genes was significantly increased in the cfo1 mutant when its transcriptome was compared with that of the wild-type strain upon fluconazole treatment. The transcript abundance of total 1,359 genes was altered, which included 838 up-regulated and 521 down-regulated genes. This result indicated that the absence of CFO1 has a dramatic influence on the transcriptome of C. neoformans, with the implication that altered expression of key functions caused the increased susceptibility of the mutant to fluconazole. Differential expression of a selected subset of genes was confirmed by quantitative real-time PCR (Q-RT-PCR) (Figure 3). These included genes for ergosterol biosynthetic enzymes (ERG2, ERG 6 and ERG11), for mitochondrial functions (CCP1 and CCP2) and for iron uptake (CFO2, CFT1, CFT2 and STR1). As described below, these genes were among the differential set that defined a signature for the influence of the cfo1 defect on iron homeostasis, ergosterol biosynthesis, mitochondrial functions, respiration, and the TCA cycle. Notably, we identified a similar signature in our previous analysis of the Hap3 and HapX transcription factors (Jung et al., 2010). We therefore focused on functions from this signature pattern to test interactions with fluconazole, and included a comparison with data from our study with Hap3 and HapX.
Table 2.
The number of differentially expressed genes caused by fluconazole treatment and/or by deletion of CFO1
|
cfo1Δ vs. WT w/o fluconazole |
cfo1Δ vs. WT with fluconazole |
WT with fluconazole vs. w/o fluconazole |
cfo1Δ with fluconazole vs. w/o fluconazole |
|
|---|---|---|---|---|
| Up-regulation | 388 | 838 | 138 | 270 |
| Down- regulation |
544 | 521 | 177 | 155 |
Figure 3. Confirmation of RNA-Seq data by quantitative RT-PCR in the wild-type strain and the cfo1 mutant.
Nine genes were selected and their transcripts in the wild-type strain (WT) or the cfo1 mutant (cfo1Δ) were quantified by Q-RT-PCR. The Q-RT-PCR results are shown in parallel with the RNA-Seq data, and both analyses revealed identical patterns. The data were averages from three independent experiments with three biological replicates, and they were normalized with TEF2 as an internal control (Livak and Schmittgen, 2001). The data are presented as relative expression in comparison to the wild-type value set at 1. The bars represent standard deviations.
3.3 Deletion of CFO1 influenced expression of the genes associated with ergosterol synthesis.
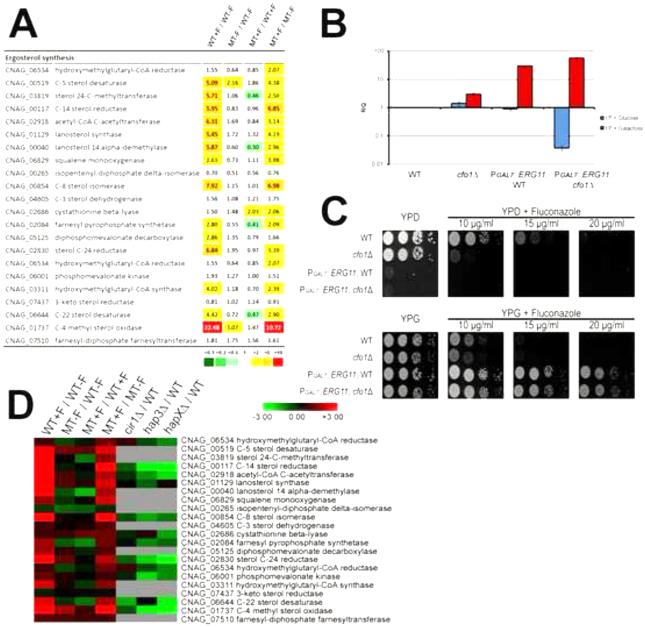
Genes involved in ergosterol synthesis were among the differentially expressed genes in the cfo1 mutant. We paid particular attention to these genes because ergosterol synthesis is a primary target of azole antifungal drugs. Figure 4A shows differentially expressed genes that are associated with ergosterol synthesis (the ERG genes hereafter) in the transcriptome data sets. Overall, transcriptional levels of the ERG genes were significantly elevated in the wild-type cells treated with fluconazole compared to the untreated cells, a result consistent with compensation for fluconazole inhibition of ergosterol synthesis. The gene, CNAG_01737, encoding an ortholog of the S. cerevisiae C-4 methyl sterol oxidase (Erg25) provides a dramatic example because its expression was increased ~22-fold upon fluconazole treatment. A similar increase in transcript abundance for the ERG genes was observed in the cfo1 mutant treated with fluconazole compared to the untreated mutant. For example, the transcript level of the ERG11 homolog was increased 2.96-fold in this condition, while it was elevated 5.87-fold in the wild-type strain upon fluconazole treatment. In contrast, the abundance of many of the ERG gene transcripts was not dramatically increased in the cfo1 mutant upon fluconazole treatment compared to the wild-type strain. However, we noted a different pattern for the genes CNAG_03819, CNAG_00040, CNAG_02084 and CNAG_06644, which are orthologs of S. cerevisiae ERG6, ERG11, ERG20 and ERG5, respectively. The transcript levels for these genes were reduced 2-3 fold in the mutant versus wild type in the presence of fluconazole. In general, these results suggested that ergosterol synthesis was inhibited by fluconazole in both the wild-type strain and the cfo1 mutant, and that expression of relatively few ERG genes were influenced by deletion of CFO1. We therefore hypothesized that the reduced expression of some of the ERG genes in the cfo1 mutant might have caused the increased susceptibility. To test this, we overexpressed the ERG11 homolog in the cfo1 mutant using the inducible GAL7 promoter (PGAL7) and investigated whether this influenced the susceptibility phenotype. As shown in Figure 4B, Q-RT-PCR analysis demonstrated that ERG11 expression was controlled by the GAL7 promoter in both the wild-type strain and the cfo1 mutant, and that the expression patterns of PGAL7::ERG11 in both strains were similar. We did note a much lower level of the GAL7 promoter-controlled ERG11 transcript on glucose in the cfo1 mutant relative to the wild-type strain. This result may indicate different post-transcriptional regulation of PGAL7::ERG11 between the wild-type strain and the cfo1 mutant. Also, we observed no growth of the wild-type strain or the cfo1 mutant containing PGAL7::ERG11 under repressing conditions indicating the gene is essential in C. neoformans (Figure 4C). Under inducing conditions on galactose, both the wild-type strain and the cfo1 mutant containing PGAL7::ERG11 were viable. As expected, overexpression of ERG11 reduced susceptibility of the cfo1 mutant and the wild-type strain to fluconazole. Both strains grew under overexpression conditions in the presence of fluconazole up to 20 g/ml, although the cfo1 mutant continued to display slightly greater susceptibility. Therefore, reduced expression of ERG11 appears to contribute to the susceptibility of the cfo1 mutant, but it may not be the sole cause.
Figure 4. Differentially expressed genes involved in ergosterol synthesis.
(A) Differentially expressed ERG genes are shown. The values in each column indicate the ratio of expression level as follows: WT+F/WT−F: the wild-type strain with fluconazole versus (vs.) wild-type strain without fluconazole; MT−F/WT−F: the cfo1 mutant without fluconazole vs. the wild-type strain without fluconazole; MT+F/WT+F: the cfo1 mutant with fluconazole versus vs. the wild-type strain with fluconazole; MT+F/MT−F: the cfo1 mutant with fluconazole versus vs. the cfo1 mutant without fluconazole. (B) Overexpression of ERG11 under the control of the GAL7 promoter by galactose was confirmed by Q-RT-PCR. The data were averages from three independent experiments with three biological replicates, normalized with TEF2 as an internal control, and were presented as relative expression in comparison to the wild-type (grown in YP media with glucose) value set at 1. The bars represent standard deviations. (C) The growth of the strains in media containing either glucose (YPD, repression condition) or galactose (YPG, induction condition) as a carbon source and in the presence of different concentrations of fluconazole was monitored. Ten-fold serial dilutions of cells (starting at104 cells) were spotted onto plates, incubated at 30°C for 2 days, and photographed. (D) Changes in transcript abundance of the ERG genes obtained from the current study were compared with previously collected transcriptome data using the cir1, hap3 and hapX mutants (Jung et al., 2010), and the results were represented by the heat-map. Grey indicates excluded values due to no statistical significance.
Cir1 and the Hap protein complex including Hap3, Hap5 and HapX are major iron regulatory proteins in C. neoformans, and their transcriptional regulatory networks were recently revealed by extensive transcriptome analysis using microarrays (Jung et al., 2010). The same analysis revealed that some of the ERG genes (e.g., ERG25 (CNAG_00117) and ERG24 (CNAG_01737)) were down-regulated in the mutant lacking HAP3 or HAPX suggesting positive regulation. We took advantage of the previous microarray data to gain a more detailed understanding of transcriptional regulation of the ERG genes in the wild-type strain and the cfo1 mutant. Specifically, we compared the differentially expressed ERG genes in the current study with the genes in the mutants lacking CIR1, HAP3 or HAPX (Figure 4D). This comparison further highlighted the similarities in the expression patterns of the wild-type strain and the cfo1 mutant, as mentioned above, and illustrated that the up-regulated ERG genes in the wild-type strain and the cfo1 mutant upon fluconazole treatment were significantly down-regulated in the hap3 or hapX mutant. Taken together, our results indicated that expression of the ERG genes was influenced by fluconazole, but not significantly by deletion of CFO1, and that Hap3 and/or HapX positively regulated the ERG genes. This reciprocal pattern of regulation is examined further below.
3.4 Loss of CFO1 alters the expression of mitochondrial functions and the response to oxidative stress.
The transcriptome data showed that many genes encoding mitochondrial proteins were particularly enriched within the set of differentially expressed genes in the cfo1 mutant. For example, 43 genes were down-regulated more than 10-fold in the cfo1 mutant compared to the wild-type strain in the presence of fluconazole and at least 30 of these were predicted to encode mitochondrial proteins based on GO terms and a manual analysis of their localization with TargetP (Emanuelsson et al., 2000) (Table S3). These proteins were predicted to function in processes such as respiration, redox homeostasis, antioxidant defense, and Fe-S cluster synthesis and assembly. To examine these functions in more detail, we focused on a cytochrome c peroxidase (CCP1, CNAG_01138), which was previously characterized as a mitochondrial antioxidant in C. neoformans (Giles et al., 2005). Considering that an additional mechanism of growth inhibition by azole antifungal drugs is the generation of Reactive Oxygen Species (ROS), we hypothesized that reduced expression of CCP1 in the cfo1 mutant led to reduce oxidative stress defense, which in turn caused increased fluconazole susceptibility (Kobayashi et al., 2002). Consistent with this idea, we found that the cfo1 mutant showed increased susceptibility to hydrogen peroxide (Figure 5A). We therefore tested whether CCP1 contributed to the increased susceptibility of the cfo1 mutant to fluconazole by examining strains with repression or overexpression of the gene with the GAL7 promoter (Figure 5B). However, we found that repression of CCP1 in the wild-type background or in the cfo1 mutant did not show altered fluconazole susceptibility. Moreover, overexpression of the gene failed to influence the fluconazole susceptibility of the cfo1 mutant, thus suggesting that CCP1 alone was not directly involved (Figure 5B). We did note that regulation of CCP1 influenced susceptibility to hydrogen peroxide (Figure S1).
Figure 5. Altered mitochondrial functions in the cfo1 mutant.
(A) Susceptibility of the strains to hydrogen peroxide (H2O2) was tested. Ten-fold serial dilutions of cells (starting at 104 26 cells) were spotted onto plates and incubated at 30°C for 2 days. (B) The growth of the strains in media containing either glucose (YPD, repressing condition) or galactose (YPG, induction condition) as a carbon source and in the presence of fluconazole was monitored. Ten-fold serial dilutions of cells (starting at 104 cells) were spotted onto plates, incubated at 30°C for 2 days and photographed. (C) Activity of aconitase in the strains was compared using zymography. Each lane was loaded with 20μg of total protein from the wild-type strain or the cfo1 mutant using 1 native gel conditions, and aconitase activity was measured (upper panel). The nitrocellulose membrane containing the same protein samples was stained with CPTS and a partial image is shown to indicate that the same amount of each sample was loaded (lower panel). A total of three independent experiments were performed and the results were nearly identical. A representative image is displayed.
A number of mitochondrial proteins are involved in respiration, especially the tricarboxylic acid (TCA) cycle and the electron transport chain, and many of them require Fe-S clusters. Two of the down-regulated genes in the cfo1 mutant encoded the predicted mitochondrial proteins homoaconitate hydratase (CNAG_02565) and the NifU-like protein c (CNAG_03395), which are associated with Fe-S cluster synthesis and homeostasis. To confirm an influence of the cfo1 mutation on mitochondrial functions, we employed zymography to investigate the activity of the aconitase enzyme that contains an Fe-S cluster and that functions in the TCA cycle. The analysis of aconitase activity has been used previously to evaluate Fe-S cluster synthesis (Wallace et al., 2004). The zymography results showed that aconitase activity was significantly reduced in the cfo1 mutants with 58.01±14.23% of the activity found in the wild-type strain (Figure 5C). This result suggested that loss of Cfo1 had an impact on mitochondrial functions such as Fe-S cluster synthesis, as predicted by the transcriptome analysis.
3.5 A change in the expression of respiration functions may increase the fluconazole susceptibility of the cfo1 mutant.
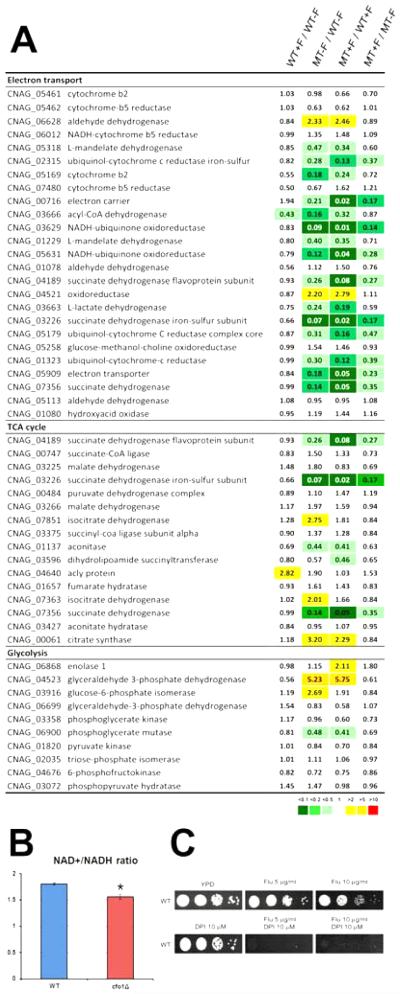
As mentioned above, the expression of a number of mitochondrial proteins was significantly altered in the cfo1 mutant, and many of these were involved in aspects of cellular respiration such as the TCA cycle and the electron-transport chain (Figure 6A). In contrast, most of the genes involved in glycolysis, which occurs in the cytosol, were unchanged. For the TCA cycle, four and five genes were down-regulated more than two-fold in the cfo1 mutant compared to the wild-type strain in the absence or presence of fluconazole, respectively. In addition, 17 of the 25 genes annotated in the C. neoformans genome as required for the electron-transport chain (with some degree of redundancy) were differentially expressed in the cfo1 mutant. Strikingly, the expression of seven and 11 of the genes was down-regulated more than 5-fold in the cfo1 mutant compared to the wild-type strain in the absence or presence of fluconazole, respectively. These results indicated that deletion of CFO1 significantly influenced expression of genes involved in respiration, a result consistent with the iron-dependent nature of this process.
Figure 6. Differentially expressed genes involved in cellular respiration and intracellular redox state in the cfo1 mutant.
(A) The values in each column indicate the ratio of expression level as follows: WT+F/WT−F: the wild-type strain with fluconazole versus (vs.) wild-type strain without fluconazole; MT−F/WT−F: the cfo1 mutant without fluconazole vs. the wild-type strain without fluconazole; MT+F/WT+F: the cfo1 mutant with fluconazole versus vs. the wild-type strain with fluconazole; MT+F/MT−F: the cfo1 mutant with fluconazole versus vs. the cfo1 mutant without fluconazole. (B) The cfo1 mutant has a reduced ratio of NAD+ /NADH compared to the wild-type strain. *p = 0.021. (C) Susceptibility of the wild-type strain (WT) to fluconazole in combination with DPI was investigated. The growth of the strains in media containing fluconazole with or without DPIwas monitored. Ten-fold serial dilutions of cells (starting at 104 cells) were spotted onto plates, incubated at 30°C for 2 days and photographed.
To functionally test the influence of the cfo1 mutation, the NAD+/NADH ratio was evaluated as a measure of the intracellular redox state and metabolic activity. Several studies have shown that an increase in the NAD+/NADH ratio correlated with higher respiratory activity in S. cerevisiae (Barros et al., 2004; Lin et al., 2004; Reverter-Branchat et al., 2007). We therefore measured the NAD+/NADH ratio to evaluate respiration and found that the cfo1 mutant had a reduced ratio of NAD+/NADH compared to the wild-type strain (Figure 6B). This result is consistent with the observed down-regulation of the genes required for the electron transport due to deletion of CFO1. To investigate whether reduced expression of the genes for respiration, especially in the electron transport chain, caused increased fluconazole susceptibility, we treated the wild-type strain with fluconazole in the presence of diphenyleneiodonium (DPI). This compound binds and inhibits the substrate side of the Fe-S clusters of mitochondrial NADH-ubiquinone oxidoreductase (Complex I) in the electron transport chain (Majander et al., 1994). DPI treatment dramatically increased the susceptibility of the wild-type strain to fluconazole in support of the idea that decreased expression of electron transport chain functions could contribute to the altered fluconazole susceptibility of the cfo1 mutant (Figure 6C). Overall, these data indicate that broader cellular changes, beyond ergosterol biosynthesis and including decreased respiration, result from loss of Cfo1 and influence azole susceptibility.
3.6 Cfo1 influenced expression of the genes governed by Cir1 and Hap proteins.
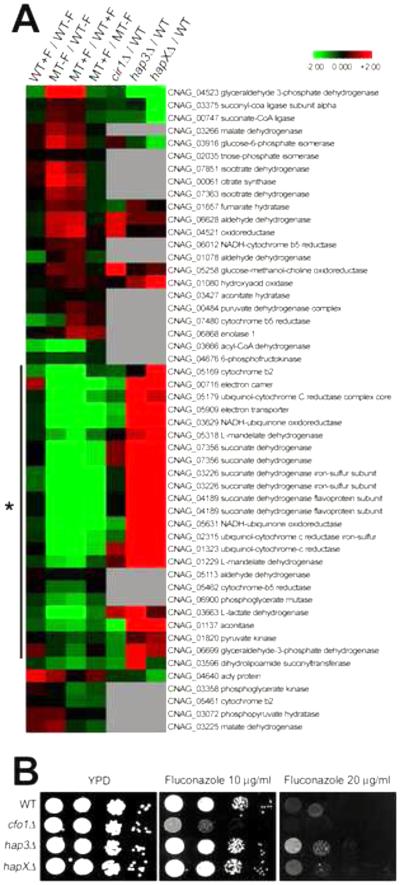
We extended our analysis to further define the mechanisms underlying the altered transcriptional regulation in the cfo1 mutant with regard to metabolic changes. As with the ERG genes above, the differentially expressed genes in the cfo1 mutant were clustered with differentially expressed genes in the cir1, hap3 or hapX mutants to identify potential patterns of co-regulation with these iron regulators. This analysis identified sets of co-regulated genes. Two of the sets were particularly enriched for genes for iron homeostasis and transport in one case, and for carbon catabolism in the other case. The genes required for iron homeostasis and transport were generally up-regulated in the cfo1 mutant suggesting that their expression might compensate for the reduced intracellular iron levels in the mutant (data not shown). A connection with the iron regulatory network is likely, given that the expression of the same set of the genes was significantly up-regulated in the cir1 mutant lacking the major iron regulator in C. neoformans (Jung et al., 2006).
Other co-regulated genes were related to carbon catabolism, especially those encoding enzymes for the TCA cycle and components of the electron transport chain; these genes were down-regulated in the cfo1 mutant. Interestingly, the same genes were dramatically up-regulated in the hap3 or the hapX mutants, while no significant change was observed in the cir1 mutant (Figure 7A). These results suggested that Hap3 or/and HapX negatively regulated expression of the genes in the TCA cycle and the electron transport chain, and that the expression of these genes was also influenced by Cfo1. We therefore hypothesized that the hap3 and/or hapX mutants would display reduced antifungal susceptibility to fluconazole based on the reciprocal expression patterns of the genes in the TCA cycle and the electron transport chain relative to the cfo1 mutant. We found that the hap3 mutant, and to a lesser extent the hapX mutant, displayed decreased fluconazole susceptibility compared to the wild-type strain (Figure 7B). This result is consistent with the idea that the decreased respiration, especially due to defects in the TCA cycle and the electron transport chain, is an important contributor to the increased fluconazole susceptibility of the cfo1 mutant. However, the less dramatic change of fluconazole susceptibility for the hapX mutant implies additional regulatory mechanisms, as expected by the distinct global regulatory influence of HapX (Jung et al., 2010).
Figure 7. Differentially expressed genes in the cfo1 mutant and regulation by the Cir1 and Hap iron-regulatory proteins.
(A) Transcriptome data from the current study were compared with previously collected transcriptome data using the cir1, hap3 and hapX mutants (Jung et al., 2010), and results were represented by the heat-map. The genes related to carbon catabolism (the TCA cycle and the electron transport chain) were indicated by a vertical line with an asterisk. Grey indicates excluded values due to no statistical significance. (B) The hap3 mutant and the hapX mutant to a lesser extent showed decreased susceptibility to fluconazole. The growth of the strains in media containing fluconazole was monitored. Ten-fold serial dilutions of cells (starting at 104 cells) were spotted onto plates and incubated at 30°C for 2 days.
4. Discussion
In this study, we examined phenotypic and gene expression changes in the cfo1 mutant lacking the ferroxidase for the high-affinity iron uptake system in C. neoformans to obtain a more comprehensive view of the reasons underlying increased fluconazole susceptibility in the mutant. Initially, we found that fluconazole tolerance was significantly reduced by deletion of CFO1 and that the normally fungistatic activity of fluconazole became fungicidal upon loss of Cfo1. Similar results have been obtained with azoles and specific mutants in other fungal pathogens. For example, Sanglard et al. showed that a C. albicans mutant lacking the gene encoding the calcineurin A subunit lost viability in the presence of fluconazole due to its reduced tolerance to the drug (Sanglard et al., 2003). Based on these findings, they proposed combination treatment with fluconazole and the immunosuppressive drug cyclosporin A that inhibits calcineurin activity in C. albicans (Marchetti et al., 2000a; Marchetti et al., 2000b; Sanglard et al., 2003). Similarly, our study also suggested a potential synergistic effect of fluconazole and miconazole in combination with an iron chelating agent to treat cryptococcosis, and we demonstrated this possibility with the chelators BPS and deferiprone. Deferiprone is particularly attractive because this chelator has been developed to treat iron overload disorders and it has been shown to be effective in a mouse model of mucormycosis (Hoffbrand et al., 2003; Ibrahim et al., 2006). The use of synthetic iron chelators in combination with azole antifungal drugs has also been proposed to treat C. albicans and Aspergillus fumigatus infections, although the underlying mechanisms for efficacy have not been examined (Prasad et al., 2006; Zarember et al., 2009).
To gain a more detailed understanding of the changes in the cfo1 mutant that might influence azole susceptibility, we performed a transcriptome analysis and identified the expression patterns resulting from interactions between fluconazole and the mutation. Our observed changes in the transcriptome of the wild-type strain in response to fluconazole were similar to the recently published study of Florio et al. as well as to previous studies in C. albicans and S. cerevisiae (Bammert and Fostel, 2000; Florio et al., 2011; Lepak et al., 2006; Rogers and Barker, 2002). Changes in the expression of the ERG genes were particularly interesting. It is known that iron deficiency in S. cerevisiae caused heme depletion and in turn reduced ergosterol synthesis. Two of the enzymes encoded by ERG11 and ERG25 are indeed heme-dependent enzymes and were influenced by heme depletion (Shakoury-Elizeh et al., 2010). We focused on the Erg11 homolog because the enzyme is a direct target of fluconazole; decreased activity of Erg11 causes increased susceptibility to the azole drugs and increased expression of Erg11 results in resistance (Du et al., 2004; Heilmann et al., 2010; Sionov et al., 2010). In this context, we tested whether reduced expression of ERG11 caused by deletion of CFO1 contributed to increased susceptibility to fluconazole. ERG11 was overexpressed in the cfo1 mutant and reduced fluconazole susceptibility of the cfo1 mutants was observed. As expected, reduced susceptibility was also seen for the wild-type strain and it was therefore not clear whether ERG11 was the sole cause of the phenotype of the cfo1 mutant.
The results of our transcriptome analysis revealed that deletion of CFO1 influenced a number of other metabolic processes besides ergosterol biosynthesis in C. neoformans. These included processes involved in respiration, especially those localized in mitochondria, such as the TCA cycle and the electron transport chain. Specifically, the TCA cycle and the electron transport chain are negatively impacted by iron deprivation because these processes depend on specific iron-containing enzymes. For example, two of the TCA cycle enzymes, aconitase and succinate dehydrogenase, contain Fe-S clusters and complex I–IV in the electron transport chain contain Fe-S clusters and heme prosthetic groups. We hypothesized that the TCA cycle function and decreased respiration were important contributors to the increased fluconazole susceptibility of the cfo1 mutants. We were able to demonstrate reduced aconitase activity as a result of loss of Cfo1. However, it was difficult to genetically test this idea because many of the genes in the respiratory pathway belong to a multiple gene families and/or encode subunits for protein complexes. For example, the genes encoding NADH ubiquinone oxidoreductases, which form a complex I in the electron transport chain, possesses at least 31 genes encoding subunits of the complex (unpublished data). Instead, we turned to a pharmacological approach and showed that wild-type cells treated with an inhibitor of the electron transport chain had increased susceptibility to fluconazole. We also found that loss of Cfo1 results in reduced intracellular iron levels and this likely causes reduced respiration, as reported for iron deficiency in S. cerevisiae (Shakoury-Elizeh et al., 2010).
The patterns of gene expression that we observed for fluconazole treatment and loss of Cfo1 were reminiscent of the regulatory impact of the hap3 and hapX mutations in our previous analysis of these genes (Jung et al., 2010). Specifically, our transcriptional profiling with the hap3 and hapX mutants revealed that the proteins negatively influenced the expression of the genes required for respiration, such as the TCA cycle and electron transport (Jung et al., 2010). We were interested in the Hap proteins in C. neoformans because mutations in the HAP genes reduced the ability of the fungus to use heme as an iron source. Our present comparisons here of the transcriptome data revealed that genes required for respiration and the TCA cycle were down-regulated in the cfo1 mutant and largely up-regulated in the hap3 or hapX mutants. Importantly, deletion of HAP3 and HAPX (to a lesser extent) resulted in increased resistance to fluconazole, thus providing a strong correlation between gene expression and susceptibility. These results suggested that down-regulation of the genes involved in iron dependent metabolic processes in the cfo1 mutant are transcriptionally regulated, and further confirmed that Hap3 and/or HapX play regulatory roles for the metabolic shift upon iron availability in C. neoformans.
The role of the Hap proteins is interesting in the broader context of metabolic remodeling during iron deficiency in fungi from iron-dependent to iron independent metabolic pathways (Philpott et al., 2012). In S. cerevisiae in particular, many genes involved in iron-dependent metabolic pathways are down-regulated in response to iron deficiency, and this process is governed by two distinct mechanisms, transcriptional regulation in association with metabolites that are synthesized in an iron-dependent manner and post-transcriptional regulation involving mRNA degradation (Ihrig et al., 2010). The Hap1 and Hap4 transcription factors, in combination with Hap2/Hap3/Hap5 CCAAT-box binding complex provide transcriptional regulation of a number of genes involved in respiration. Post-transcriptional regulation is governed by Cth1 and Cth2 (Ihrig et al., 2010; Puig et al., 2005). Furthermore, a study using A. fumigatus showed that the CCAAT-box binding protein HapX represses genes involved in iron-dependent metabolic pathways such as TCA cycle, amino acid metabolism, Fe-S cluster and heme biosynthesis (Schrettl et al., 2010). We speculate that the Hap3 and HapX proteins in C. neoformans are also involved in metabolic remodeling of iron utilization functions and that this regulation contributes to the altered sensitivity to fluconazole.
5. Conclusions
A C. neoformans mutant lacking the ferroxidase Cfo1 displayed decreased intracellular iron levels and increased susceptibility to several azole drugs. A combination of transcriptional profiling, mutant analysis, overexpression studies and the use of inhibitors demonstrated that the altered fluconazole susceptibility of the cfo1 mutant was caused by decreased respiration in combination with other mitochondrial functions such as redox homeostasis and Fe-S cluster synthesis. Furthermore, we identified a signature pattern of gene expression that demonstrated the interconnected influence of Hap3/HapX regulation, iron deficiency due to loss of Cfo1 and treatment with azole drugs. We suggest that a potent synergistic effect of azole drugs in combination with iron chelating agents could provide a novel therapy for cryptococcosis.
Supplementary Material
Highlights.
▶ Altered antifungal susceptibility of a mutant lacking CFO1 was investigated.
▶ Iron uptake plays a key role in antifungal susceptibility of C. neoformans.
▶ Iron uptake pathway could be used as a novel target for combination therapy.
Acknowledgements
We thank Kyunghwan Han for help with CFU analysis, and Byung Hee Kim and Kyoung Kyu Kang for help with ICP analysis. This work was supported by awards from the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology 2012-0004062 (W.H.J.) and 2011-0029843 (W.H.J, Y.L), and the National Institutes of Health R01 AI053721 (J.W.K.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bammert GF, Fostel JM. Genome-wide expression patterns in Saccharomyces cerevisiae: comparison of drug treatments and genetic alterations affecting biosynthesis of ergosterol. Antimicrob Agents Chemother. 2000;44:1255–65. doi: 10.1128/aac.44.5.1255-1265.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros MH, et al. Higher respiratory activity decreases mitochondrial reactive oxygen release and increases life span in Saccharomyces cerevisiae. J Biol Chem. 2004;279:49883–8. doi: 10.1074/jbc.M408918200. [DOI] [PubMed] [Google Scholar]
- Bennett JE, et al. A comparison of amphotericin B alone and combined with flucytosine in the treatment of cryptoccal meningitis. N Engl J Med. 1979;301:126–31. doi: 10.1056/NEJM197907193010303. [DOI] [PubMed] [Google Scholar]
- Bicanic T, Harrison TS. Cryptococcal meningitis. Br Med Bull. 2004;72:99–118. doi: 10.1093/bmb/ldh043. [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI) Reference method for broth dilution antifungal susceptibility testing of yeasts, vol. 28, no. 14. Approved standard M27-A3. National Committee for Clinical and Laboratory Standards; Villanova, Pa: 2008. [Google Scholar]
- Cowen LE, et al. Genetic architecture of Hsp90-dependent drug resistance. Eukaryot Cell. 2006;5:2184–8. doi: 10.1128/EC.00274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowen LE, Lindquist S. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science. 2005;309:2185–9. doi: 10.1126/science.1118370. [DOI] [PubMed] [Google Scholar]
- Du W, et al. Shuttle vectors for Candida albicans: control of plasmid copy number and elevated expression of cloned genes. Curr Genet. 2004;45:390–8. doi: 10.1007/s00294-004-0499-3. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, et al. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–16. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Florio AR, et al. Genome-wide expression profiling of the response to short-term exposure to fluconazole in Cryptococcus neoformans serotype A. BMC Microbiol. 2011;11:97. doi: 10.1186/1471-2180-11-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghannoum MA, Rice LB. Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin Microbiol Rev. 1999;12:501–17. doi: 10.1128/cmr.12.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles SS, et al. Cytochrome c peroxidase contributes to the antioxidant defense of Cryptococcus neoformans. Fungal Genet Biol. 2005;42:20–9. doi: 10.1016/j.fgb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Hakim JG, et al. Impact of HIV infection on meningitis in Harare, Zimbabwe: a prospective study of 406 predominantly adult patients. AIDS. 2000;14:1401–7. doi: 10.1097/00002030-200007070-00013. [DOI] [PubMed] [Google Scholar]
- Heilmann CJ, et al. An A643T mutation in the transcription factor Upc2p causes constitutive ERG11 upregulation and increased fluconazole resistance in Candida albicans. Antimicrob Agents Chemother. 2010;54:353–9. doi: 10.1128/AAC.01102-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffbrand AV, et al. Role of deferiprone in chelation therapy for transfusional iron overload. Blood. 2003;102:17–24. doi: 10.1182/blood-2002-06-1867. [DOI] [PubMed] [Google Scholar]
- Ibrahim AS, et al. Deferiprone iron chelation as a novel therapy for experimental mucormycosis. J Antimicrob Chemother. 2006;58:1070–3. doi: 10.1093/jac/dkl350. [DOI] [PubMed] [Google Scholar]
- Ihrig J, et al. Iron regulation through the back door: iron-dependent metabolite levels contribute to transcriptional adaptation to iron deprivation in Saccharomyces cerevisiae. Eukaryot Cell. 2010;9:460–71. doi: 10.1128/EC.00213-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, et al. Role of ferroxidases in iron uptake and virulence of Cryptococcus neoformans. Eukaryot Cell. 2009;8:1511–20. doi: 10.1128/EC.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, et al. HapX positively and negatively regulates the transcriptional response to iron deprivation in Cryptococcus neoformans. PLoS Pathog. 2010;6:e1001209. doi: 10.1371/journal.ppat.1001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, et al. Iron source preference and regulation of iron uptake in Cryptococcus neoformans. PLoS Pathog. 2008;4:e45. doi: 10.1371/journal.ppat.0040045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, et al. Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans. PLoS Biol. 2006;4:e410. doi: 10.1371/journal.pbio.0040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi D, et al. Endogenous reactive oxygen species is an important mediator of miconazole antifungal effect. Antimicrob Agents Chemother. 2002;46:3113–7. doi: 10.1128/AAC.46.10.3113-3117.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepak A, et al. Time course of microbiologic outcome and gene expression in Candida albicans during and following in vitro and in vivo exposure to fluconazole. Antimicrob Agents Chemother. 2006;50:1311–9. doi: 10.1128/AAC.50.4.1311-1319.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, et al. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18:12–6. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lupetti A, et al. Molecular basis of resistance to azole antifungals. Trends Mol Med. 2002;8:76–81. doi: 10.1016/s1471-4914(02)02280-3. [DOI] [PubMed] [Google Scholar]
- Majander A, et al. Diphenyleneiodonium inhibits reduction of iron-sulfur clusters in the mitochondrial NADH-ubiquinone oxidoreductase (Complex I) J Biol Chem. 1994;269:21037–42. [PubMed] [Google Scholar]
- Marchetti O, et al. Fluconazole plus cyclosporine: a fungicidal combination effective against experimental endocarditis due to Candida albicans. Antimicrob Agents Chemother. 2000a;44:2932–8. doi: 10.1128/aac.44.11.2932-2938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti O, et al. Potent synergism of the combination of fluconazole and cyclosporine in Candida albicans. Antimicrob Agents Chemother. 2000b;44:2373–81. doi: 10.1128/aac.44.9.2373-2381.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Philpott CC, et al. Metabolic remodeling in iron-deficient fungi. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbamcr.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad T, et al. Unexpected link between iron and drug resistance of Candida spp.: iron depletion enhances membrane fluidity and drug diffusion, leading to drug-susceptible cells. Antimicrob Agents Chemother. 2006;50:3597–606. doi: 10.1128/AAC.00653-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig S, et al. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell. 2005;120:99–110. doi: 10.1016/j.cell.2004.11.032. [DOI] [PubMed] [Google Scholar]
- Reverter-Branchat G, et al. Chronological and replicative life-span extension in Saccharomyces cerevisiae by increased dosage of alcohol dehydrogenase 1. Microbiology. 2007;153:3667–76. doi: 10.1099/mic.0.2007/009340-0. [DOI] [PubMed] [Google Scholar]
- Rogers PD, Barker KS. Evaluation of differential gene expression in fluconazole-susceptible and -resistant isolates of Candida albicans by cDNA microarray analysis. Antimicrob Agents Chemother. 2002;46:3412–7. doi: 10.1128/AAC.46.11.3412-3417.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanglard D, et al. Calcineurin A of Candida albicans: involvement in antifungal tolerance, cell morphogenesis and virulence. Mol Microbiol. 2003;48:959–76. doi: 10.1046/j.1365-2958.2003.03495.x. [DOI] [PubMed] [Google Scholar]
- Schrettl M, et al. HapX-mediated adaption to iron starvation is crucial for virulence of Aspergillus fumigatus. PLoS Pathog. 2010;6:e1001124. doi: 10.1371/journal.ppat.1001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder A, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakoury-Elizeh M, et al. Metabolic response to iron deficiency in Saccharomyces cerevisiae. J Biol Chem. 2010;285:14823–33. doi: 10.1074/jbc.M109.091710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, et al. Human ISD11 is essential for both iron-sulfur cluster assembly and maintenance of normal cellular iron homeostasis. Hum Mol Genet. 2009;18:3014–25. doi: 10.1093/hmg/ddp239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SD, et al. Hsp90 governs echinocandin resistance in the pathogenic yeast Candida albicans via calcineurin. PLoS Pathog. 2009;5:e1000532. doi: 10.1371/journal.ppat.1000532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sionov E, et al. Identification of a Cryptococcus neoformans Cytochrome P450 Lanosterol 14alpha-Demethylase (Erg11) Residue Critical for Differential Susceptibility between Fluconazole/Voriconazole and Itraconazole/Posaconazole. Antimicrob Agents Chemother. 2011 doi: 10.1128/AAC.05502-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sionov E, et al. Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathog. 2010;6:e1000848. doi: 10.1371/journal.ppat.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toffaletti DL, et al. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J Bacteriol. 1993;175:1405–11. doi: 10.1128/jb.175.5.1405-1411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace MA, et al. Superoxide inhibits 4Fe-4S cluster enzymes involved in amino acid biosynthesis. Cross-compartment protection by CuZn-superoxide dismutase. J Biol Chem. 2004;279:32055–62. doi: 10.1074/jbc.M403590200. [DOI] [PubMed] [Google Scholar]
- Wirk B, Wingard JR. Combination antifungal therapy: from bench to bedside. Curr Infect Dis Rep. 2008;10:466–72. doi: 10.1007/s11908-008-0076-x. [DOI] [PubMed] [Google Scholar]
- Zarember KA, et al. Antifungal activities of natural and synthetic iron chelators alone and in combination with azole and polyene antibiotics against Aspergillus fumigatus. Antimicrob Agents Chemother. 2009;53:2654–6. doi: 10.1128/AAC.01547-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonios DI, Bennett JE. Update on azole antifungals. Semin Respir Crit Care Med. 2008;29:198–210. doi: 10.1055/s-2008-1063858. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.