Abstract
Culturomics coupled with taxonogenomics is currently used to isolate and characterize new bacteria. Here we describe the features and complete genome sequence of Gabonia massiliensis strain GM3, an anaerobic Gram negative, non-spore-forming and catalase-positive bacillus isolated from a stool specimen of a healthy Gabonese male youth. Belonging to a new genus called Gabonia, it exhibits a genome of 4 261 752 bp including 37.9% GC content and 3,288 predicted genes.
Keywords: Culturomics, Gabonia massiliensis gen. nov. et sp. nov., genome, taxonogenomics, gut microbiota
Introduction
Culturomics was recently introduced in our laboratory (URMITE, Marseille, France) as an alternative method to expand the human gut repertoire [1], thanks to the multiplication of culture conditions with a rapid identification method by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF) [1]. Consequently, new genera and species of bacteria are found thanks to this technique [2], [3], [4], [5], [6].
The characterization of new bacteria was previously based on a combination of phylogenetic and genotypic characteristics, including 16S rRNA sequence similarity, GC percentage and DNA-DNA hybridization [7], [8], [9], [10]. Unfortunately, significant limits such as their threshold values, which were not applicable to all species or genera, did not promote their utilization [11]. The advent of high-throughput sequencing techniques made many bacterial genome sequences available in public databases [12]. Our laboratory recently developed a taxonogenomic approach based on a systematic comparison of phenotypic (especially the MALDI-TOF spectrum) characteristics and genomic characteristics with the phylogenetically closest bacteria found in the databases [2], [3], [4], [5], [6]. Here we present an analysis of the characteristics that allowed us to describe Gabonia massiliensis strain GM3, a bacterium isolated from a stool specimen from a healthy Gabonese male youth and classified into the Porphyromonadaceae family, created in 2011 by Krieg [13].
We describe the classification, biochemical features and complete genomic sequencing and annotation of G. massiliensis gen. nov., sp. nov. strain GM3 (= CSUR P1909 = DSM 100571).
Material and Methods
Ethics and sample collection
The stool sample was collected in Lébamba (Gabon) in January 2015 after approval from the National Ethics Committee of Gabon (registration 0023/2013/SG/CNE) and IFR48 (Marseille, France) (registration 09-022) and after receipt of the patient's signed consent. The specimen was taken from a healthy 16-year-old Gabonese male youth (body mass index, 19.03 kg/m2). He was a member of the Nzebi tribe, which is one of the biggest tribes in Gabon. The stool sample was stored at −80°C until it was sent to URMITE (Marseille, France).
Strain isolation
In April 2015, the stool sample was diluted in phosphate-buffered saline (Life Technologies, Carlsbad, CA, USA). Obtained inoculum was preincubated anaerobically at 37°C in an anaerobic blood culture bottle (bioMérieux, Marcy l’Étoile, France) enriched with blood and rumen [14]. After the preincubation stage, 100 μL was incubated for 24 to 48 hours on 5% sheep's blood–enriched Columbia agar (bioMérieux) at 37°C under anaerobic conditions using an anaerobic atmosphere generator system (GENbag anaer; bioMérieux). The bacteria colonies obtained were isolated on 5% sheep's blood–enriched Columbia agar solid medium (bioMérieux) and identified by MALDI-TOF and 16S rRNA sequencing.
MALDI-TOF and 16S rRNA identifications
MALDI-TOF identification consisted of picking one isolated bacterial colony with a pipette tip from a culture agar plate and spreading it as a thin film on an MTP 384 MALDI-TOF target plate (Bruker Daltonics, Leipzig, Germany) [2], [3], [4], [5], [6]. Twelve deposits from 12 distinct isolated colonies were performed for strain GM3. Then 2 μL of matrix solution (saturated solution of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile and 2.5% trifluoroacetic acid) was overlaid on each smear and allowed to dry for 5 minutes. Measurements and MALDI-TOF analysis were conducted as previously described [2], [3], [4], [5], [6]. Identification depended on the score obtained; a score of ≥2 with a validly published species enabled identification at the species level; a score of ≥1.7 but <2 enabled identification at the genus level; and a score of <1.7 did not enable any identification [2], [3], [4], [5], [6]. If the colonies corresponded to a new bacterium, confirmation of the identification had to occur with a 16S rRNA PCR coupled with sequencing. That was performed using GeneAmp PCR System 2720 thermal cyclers (Applied Biosystems, Bedford, MA, USA) and ABI Prism 3130xl Genetic Analyzer capillary sequencer (Applied Biosystems), respectively [15]. If the confirmation was positive, the spectrum of the new bacterium was entered into the Bruker database. The 16S rRNA nucleotide sequence was corrected using Chromas Pro 1.34 software (Technelysium, Tewantin, Australia), and the BLASTn searches were performed by the National Center for Biotechnology Information tool (http://blast.ncbi.nlm.nih.gov.gate1.inist.fr/Blast.cgi). For phylogeny, sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained using the neighbour-joining method within MEGA software. Numbers at the nodes are percentages of bootstrap values obtained by repeating the analysis 1000 times to generate a majority consensus tree. The 16S rRNA for G. massiliensis strain GM3 was deposited in GenBank under accession number LN849789.
Growth conditions
Strain GM3 was cultivated on 5% sheep's blood–enriched Colombia agar (bioMérieux) in anaerobic conditions at different temperatures (28, 37, 45 and 55°C) to assess its range of growth temperatures. The GENbag anaer and GENbag microaer systems (bioMérieux) were respectively used to assess the ability of the bacterium to grow anaerobically or in microaerophilic conditions at 37°C, using the same medium culture. For halophilia and pH testing, four NaCl concentrations (0, 5, 15 and 45%) and three different pHs (5, 7 and 8.5) were tested.
Biochemical, sporulation and motility assays
API Gallery systems were performed with API ZYM, API 20 A and API 50CH (bioMérieux) for biochemical assays. Oxidase (Becton Dickinson, Le Pont de Claix, France) and catalase assays (bioMérieux) were done separately. Sporulation was tested by performing thermal shock on bacterial colonies (diluted in phosphate-buffered saline) at 80°C for 10 minutes. The motility of strain GM3 was tested by observing its fresh colony between blades and slats using a DM1000 photonic microscope (Leica Microsystems, Nanterre, France) with a 40× objective lens.
Antibiotic susceptibility
The antibiotic susceptibility of G. massiliensis was tested using SirScan Discs of amoxicillin, doxycycline, rifampicin, nitrofurantoin, vancomycin, amoxicillin/clavulanic acid, clindamycin, gentamicin, imipenem, erythromycin, metronidazole, trimethoprim/sulfamethoxazole, amikacin, ciprofloxacin and tobramycin antibiotics (i2a, Montpellier, France).
Microscopy
G. massiliensis strain GM3 was observed after negative coloration using a Tecnai G20 (FEI, Limeil-Brevannes, France) transmission electron microscope at an operating voltage of 60 kV. Gram coloration was performed using a Color Gram 2 Kit (bioMérieux). This coloration was observed by using the DM1000 photonic microscope (Leica Microsystems) with a 100× oil-immersion objective lens.
Genome sequencing and assembly
Using the mate-pair strategy, the genomic DNA (gDNA) of G. massiliensis GM3 was sequenced on the MiSeq sequencer (Illumina, San Diego, CA, USA) [2], [3], [4], [5], [6]. The gDNA was barcoded in order to be mixed with 11 other projects with the Nextera Mate-Pair sample prep kit (Illumina). The mate-pair library was prepared with 1 μg of gDNA using the Nextera Mate-Pair Illumina guide, and the gDNA sample was simultaneously fragmented and tagged with a mate-pair junction adapter. The pattern of fragmentation was validated on an Agilent 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA, USA) with a DNA 7500 labchip. The DNA fragments ranged in size from 1 to 10 kb, with an optimal size of 4.08 kb. No size selection was performed, and only 464 ng of tagmented fragments were circularized [2], [3], [4], [5], [6]. The circularized DNA was mechanically sheared to small fragments with an optimal size of 569 bp in microtubes on the Covaris S2 device (Covaris, Woburn, MA, USA). The library profile was visualized on a High Sensitivity Bioanalyzer LabChip (Agilent Technologies), and the final library concentration was measured at 24.4 nmol/L. The libraries were normalized at 2 nM and pooled. After a denaturation step and dilution at 15 pM, the pool of libraries was loaded onto the reagent cartridge and then onto the instrument along with the flow cell. An automated cluster generation and sequencing run were performed in a single 39-hour run on 2 × 251 bp. A total of 10.1 Gb of information was obtained from a 1189K/mm2 cluster density with a cluster passing quality control filters of 99.1% (22 579 000 clusters). The reads obtained were trimmed and then assembled using the CLC genomics WB4 software [2], [3], [4], [5], [6].
Genome annotation and comparison
Using Prodigal (http://prodigal.ornl.gov/) with default parameters, we predicted open reading frames (ORFs). However, if they spanned a sequencing gap region, the predicted ORFs were excluded. The predicted bacterial protein sequences were searched against the GenBank [16] and Clusters of Orthologous Groups (COGs) databases using BLASTP. Both tRNAs and rRNAs were predicted using the tRNAScan-SE [17] and RNAmmer [18] tools, respectively. Signal peptides and the number of transmembrane helices were predicted using SignalP [19] and TMHMM [20], respectively. Mobile genetic elements were predicted using PHAST [21] and RAST [22]. ORFans were identified when their BLASTP E value was lower than 1e-03 for an alignment length greater than 80 amino acids. If alignment lengths were smaller than 80 amino acids, we used an E value of 1e-05. We used such parameter thresholds in previous works to define ORFans. Artemis [23] and a DNAPlotter [24] were used for data management and visualization of genomic features, respectively. Finally, we used the Mauve alignment tool (version 2.3.1) for multiple genomic sequence alignments [25].
The mean level of nucleotide sequence similarity at the genome level between G. massiliensis and other bacteria was estimated using the average genomic identity of orthologous gene sequences (AGIOS) homemade software [6]. Overall, this software is combined with other software: Proteinortho [26] (to detect orthologous proteins between genomes compared two by two, and then retrieves the corresponding genes) and the Needleman-Wunsch global alignment algorithm (to determine the mean percentage of nucleotide sequence identity among orthologous ORFs).
Annotation and comparison analyses were performed using the multiagent software system DAGOBAH [27] that includes Figenix [28].
G. massiliensis strain GM3 was compared with Bacteroides salanitronis strain DSM 18170, Alistipes shahii strain WAL 8301, Alistipes finegoldii strain DSM 17242, Barnesiella viscericola strain DSM 18177, Ornithobacterium rhinotracheale strain ORT-UMN 88, Bacteroides fragilis strain YCH46, Bacteroides dorei strain HS1 and Helicobacter pylori strain 26695.
Results
MALDI-TOF and phylogenic analysis
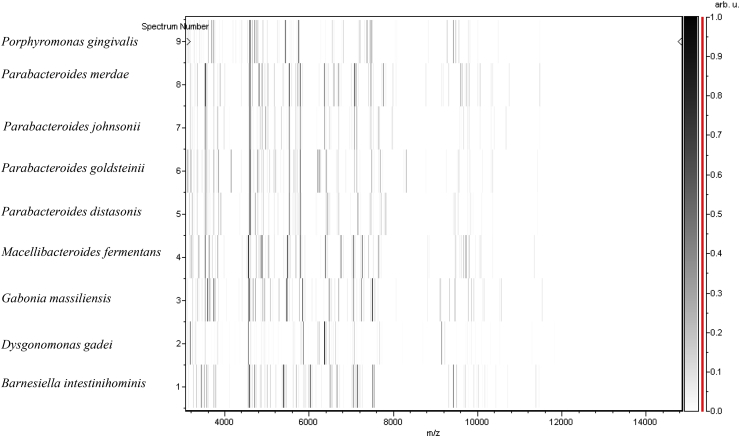
The spectrum resulting from the 12 clean G. massiliensis spots did not identify bacteria because there was no spectrum match with those of Bruker database. Using 16S rRNA sequence, the phylogenic analysis revealed that G. massiliensis gen. nov., sp. nov., strain GM3, exhibited 91% identification with Barnesiella viscericola (AB267809.1) (Fig. 1). The similarity of 16S rRNA in these two bacteria led to classification of this new bacterium into the Porphyromonadaceae family, created in 2011 by Krieg (Table 1) [13]. However, this 16S rRNA nucleotide similarity to Barnesiella viscericola was lower than the threshold of 95% recommended by Stackebrandt and Ebers [8] to delineate a new genus. Consequently, G. massiliensis gen. nov., sp. nov., strain GM3, was classified as new genus called Gabonia. The spectrum resulting from G. massiliensis was thus added into the Bruker database (Fig. 2), and a gel view was performed in order to see the spectra differences between other close bacteria (Fig. 3). Then the 16S rRNA sequence of G. massiliensis was deposited in GenBank under accession number LN849789.
Fig. 1.
Phylogenetic tree highlighting position of Gabonia massiliensis strain GM3 compared to other most phylogenetically close bacteria. GenBank accession numbers are indicated in parentheses. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained using neighbour-joining method within MEGA software. Numbers at nodes are percentages of bootstrap values obtained by repeating analysis 1000 times to generate majority consensus tree. Helicobacter pylori strain 26695 (NR_074393) was used as outgroup. Scale bar = 5% nucleotide sequence divergence.
Table 1.
Classification and general features of Gabonia massiliensis strain GM3
| Property | Term |
|---|---|
| Current classification | Domain: Bacteria |
| Phylum: Bacteroidetes | |
| Class: Bacteroidia | |
| Order: Bacteroidales | |
| Family: Porphyromonadaceae | |
| Genus: Gabonia | |
| Species: Gabonia massiliensis | |
| Type strain: GM3 | |
| Gram stain | Negative |
| Cell shape | Rod |
| Motility | Not motile |
| Sporulation | Nonsporulating |
| Temperature range | Mesophilic |
| Optimum temperature | 37°C |
Fig. 2.
MALDI-TOF reference spectrum of Gabonia massiliensis strain GM3. Spectra from 12 individual colonies were compared and reference spectrum was generated. MALDI-TOF, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry.
Fig. 3.
Gel view comparing Gabonia massiliensis strain GM3 with other members of Porphyromonadaceae family. Gel view displays raw spectra of loaded spectrum files arranged in pseudo-gel like look. x-axis records m/z value, left y-axis running spectrum number originating from subsequent spectra loading. Peak intensity is expressed by greyscale scheme code. Color bar and right y-axis indicate relation between color peak is displayed with and peak intensity in arbitrary units. Displayed species are indicated at left.
Phenotypic and biochemical characterization
G. massiliensis gen. nov., sp. nov., strain GM3 (= CSUR P1909 = DSM 100571), is a Gram-negative bacillus (Fig. 4). Strain GM3 is a non motile, non-spore-forming, oxidase-negative but catalase-positive bacterium. Growth was observed from 28 to 45°C, with optimal growth at 37°C, and colonies were obtained after 48 hours of culture. G. massiliensis grew in salinity conditions of 0 and at a pH of 7. It exhibited translucent colonies with a diameter of 1.5 mm on 5% sheep's blood–enriched Colombia agar. Strain GM3 is a preferentially anaerobic bacillus but was able to grow in microaerophilic atmosphere and unable to grow in aerobic conditions. Individual cells had a mean diameter of 0.5 μm and a length of 1.25 μm when observed with electron microscopy (Fig. 5).
Fig. 4.

Gram staining of Gabonia massiliensis strain GM3.
Fig. 5.

Transmission electron microscopy of Gabonia massiliensis strain GM3 using Tecnai G20 (FEI) at operating voltage of 60 kV. Scale bar = 500 nm.
Using API 20A, we observed positive reactions with d-glucose, d-lactose, d-maltose, d-cellobiose, d-mannose, d-raffinose, d-rhamnose and d-trehalose but not with d-mannitol, d-sucrose, salicin, d-xylose, l-arabinose, glycerol, d-melezitose and d-sorbitol. Negative reactions were recorded for indole formation, urease and gelatin using the same gallery. Using API 50CH, we concluded that G. massiliensis is able to ferment inositol, methyl-αd-mannopyranoside, methyl-αd-glucopyranoside, N-acetylglucosamine, esculin ferric citrate, d-cellobiose, d-maltose, d-lactose, d-sucrose, inulin, starch, d-arabitol and potassium-5-ketogluconate. Results from the API ZYM Gallery showed enzymatic activities of alkaline phosphatase, esterase (C4), esterase lipase (C8), acid phosphatase, naphthol-AS-BI-phosphohydrolase, α- and β-galactosidase and N-acetyl-β-glucosaminidase. Antibiotic susceptibility testing revealed that G. massiliensis strain GM3 is sensitive to nitrofurantoin, doxycycline, rifampicin, amoxicillin/clavulanic acid, and imipenem. However, it remained resistant against amoxicillin, vancomycin, clindamycin, gentamicin, erythromycin/metronidazole, tobramycin, trimethoprim/sulfamethoxazole, amikacin and ciprofloxacin. The main phenotypic and biochemical characteristics of G. massiliensis compared to the closest bacteria are summarized in Table 2.
Table 2.
Differential characteristics of G. massiliensis strain GM3 (data from this study) with Barnesiella viscericola strain C46T [31], Barnesiella intestinihominis strain YIT 11860 [32], Coprobacter fastidiosus strain NSB1 [33] and Porphyromonas gingivicanis strain JCM 15907 [34]
| Property | G. massiliensis | B. viscericola | B. intestinihominis | C. fastidiosus | P. gingivicanis |
|---|---|---|---|---|---|
| Cell diameter (μm) | 0.5 | 0.8–1.6 | 0.4–1.0 | 0.2–0.3 | NA |
| Oxygen requirement | − | − | − | − | − |
| Gram stain | − | − | − | − | − |
| Motility | − | − | − | − | − |
| Endospore formation | − | − | NA | − | − |
| Indole | − | − | − | − | NA |
| Production of: | |||||
| Alkaline phosphatase | + | + | + | + | NA |
| Catalase | + | − | − | + | NA |
| Oxidase | − | NA | − | NA | NA |
| Urease | − | − | − | − | NA |
| β-Galactosidase | + | + | + | + | NA |
| N-acetyl-glucosaminidase | + | + | + | + | NA |
| Acid from: | |||||
| l-Arabinose | − | − | − | − | NA |
| Ribose | − | − | − | NA | NA |
| Mannose | + | + | + | + | NA |
| Mannitol | − | − | − | − | NA |
| Sucrose | + | + | − | − | NA |
| d-Glucose | + | + | + | + | NA |
| d-Fructose | − | − | − | − | NA |
| d-Maltose | + | + | + | + | NA |
| d-Lactose | + | − | + | + | NA |
| G+C content (%) | 37.9 | 51.7 | 43.9 | 38.3 | 42.7 |
| Habitat | Human gut | Chicken gut | Human gut | Human gut | Beagles |
NA, not applicable.
Genome properties
The genome was 4,261,752 bp long with 37.9% GC content. It was composed of 19 scaffolds (composed of 66 contigs) (Fig. 6). Of the 3,288 predicted genes, 3,227 were protein-coding genes and 61 were RNA genes (two genes were 5S rRNA, two genes were 16S rRNA, two genes were 23S rRNA and 55 genes were tRNA genes). A total of 1,807 genes (56.00%) were assigned as putative function (by COGs or by NR blast), and 113 genes (3.50%) were identified as ORFans (Table 3). The remaining genes were annotated as encoding hypothetical proteins (1,229 genes, 38.08%). Nucleotide content and gene count levels of the genome are summarized in Table 3, while the distribution of genes into COGs functional categories is presented in Table 4. The genome sequence has been deposited in GenBank under accession number CYPV00000000.
Fig. 6.
Graphical circular map of genome. From outside to center: contigs (red/grey), Clusters of Orthologous Groups (COGs) database category of genes on forward strand (three circles), genes on forward (blue circle) and reverse strands (red circle), COGs category on reverse strand (three circles), GC content.
Table 3.
Nucleotide content and gene count levels of chromosome
| Attribute | Genome (total) |
|
|---|---|---|
| Value | % of total | |
| Size (bp) | 4,261,752 | 100 |
| G+C content (bp) | 1,611,220 | 37.9 |
| Coding sequence size (bp) | 3,718,423 | 87.3 |
| Extrachromosomal elements | 0 | 0 |
| Total genes | 3,288 | 100 |
| RNA genes | 61 | 1.9 |
| Protein-coding genes | 3,227 | 98.1 |
| Genes with function prediction | 1,807 | 55.9 |
| Genes assigned to COGs | 1,380 | 42.8 |
| Protein with peptid signal | 1,024 | 31.7 |
| Genes with transmembrane helices | 627 | 19.4 |
| No of antibiotic resistant genes | 0 | 0 |
| Genes associated with PKS or NRPS | 7 | 0.21 |
| No. of genes with Pfam-A domains | 2,907 | 88.41 |
| No. of CRISPRs | 1 | 0.03 |
COGs, Clusters of Orthologous Groups database; CRISPR, clustered regularly interspaced short palindromic repeat.
Table 4.
Number of genes associated with 25 general COGs functional categories
| Code | Value | % of totala | Description |
|---|---|---|---|
| J | 138 | 4.3 | Translation |
| A | 0 | 0 | RNA processing and modification |
| K | 81 | 2.5 | Transcription |
| L | 87 | 2.7 | Replication, recombination and repair |
| B | 0 | 0 | Chromatin structure and dynamics |
| D | 13 | 0.4 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0 | Nuclear structure |
| V | 39 | 1.2 | Defense mechanisms |
| T | 26 | 0.8 | Signal transduction mechanisms |
| M | 119 | 3.7 | Cell wall/membrane biogenesis |
| N | 0 | 0 | Cell motility |
| Z | 0 | 0 | Cytoskeleton |
| W | 0 | 0 | Extracellular structures |
| U | 16 | 0.50 | Intracellular trafficking and secretion |
| O | 52 | 1.6 | Posttranslational modification, protein turnover, chaperones |
| C | 110 | 3.4 | Energy production and conversion |
| G | 149 | 4.6 | Carbohydrate transport and metabolism |
| E | 130 | 4.0 | Amino acid transport and metabolism |
| F | 57 | 1.8 | Nucleotide transport and metabolism |
| H | 76 | 2.4 | Coenzyme transport and metabolism |
| I | 50 | 1.5 | Lipid transport and metabolism |
| P | 73 | 2.3 | Inorganic ion transport and metabolism |
| Q | 19 | 0.6 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 206 | 6.4 | General function prediction only |
| S | 78 | 2.4 | Function unknown |
| — | 1847 | 57.2 | Not in COGs |
Total is based on either size of genome (bp) or total number of protein coding genes in annotated genome. COGs, Clusters of Orthologous Groups database.
Genome comparison
The draft genome of Gabonia massiliensis strain GM3 was larger in size (4.26 Mb) than Bacteroides salanitronis strain DSM 18170, Alistipes shahii strain WAL 8301, Alistipes finegoldii strain DSM 17242, Barnesiella viscericola strain DSM 18177 and Ornithobacterium rhinotracheale strain ORT-UMN 88 (4.24, 3.76, 3.73, 3.08 and 2.4 Mb, respectively) but smaller than Bacteroides fragilis strain YCH46 and Bacteroides dorei strain HS1 (5.28 and 5.24 Mb). The G+C content (37.9%) of Gabonia massiliensis strain GM3 is larger than that of Ornithobacterium rhinotracheale strain ORT-UMN 88 (37.2%) but smaller than those of Alistipes shahii strain WAL 8301, Alistipes finegoldii strain DSM 17242, Barnesiella viscericola strain DSM 18177, Bacteroides fragilis strain YCH46 and Bacteroides dorei strain HS1 (57.2, 56.6, 51.7, 46.6, 43.3 and 41.6, respectively). The gene content (3228) of Gabonia massiliensis strain GM3 is larger compared to those of Ornithobacterium rhinotracheale strain ORT-UMN 88, Barnesiella viscericola strain DSM 18177, Alistipes shahii strain WAL 8301 and Alistipes finegoldii strain DSM 17242 (2,289, 2,557, 3,090 and 3,226) but smaller than that of Bacteroides fragilis strain YCH46, Bacteroides dorei strain HS1 and Bacteroides salanitronis strain DSM 18170 (4,670, 4,024 and 3,543). Similarly, the number of protein-coding genes (3,227) of Gabonia massiliensis strain GM3 were higher than those of Ornithobacterium rhinotracheale strain ORT-UMN 88, Barnesiella viscericola strain DSM 18177, Alistipes shahii strain WAL 8301 and Alistipes finegoldii strain DSM 17242 (2,210, 2,464, 2,996 and 3,108) but lower than those of Bacteroides fragilis strain YCH46, Bacteroides dorei strain HS1 and Bacteroides salanitronis strain DSM 18170 (4,578, 4,024 and 3,543). However, the distribution of genes into COGs categories was similar in all genomes compared (Fig. 7). In addition, Gabonia massiliensis strain GM3 shared 3,231, 3,132, 4,269, 4,373, 3,686, 2,499, 1,579 and 2,312 orthologous genes with Alistipes finegoldii strain DSM 17242, Alistipes shahii strain WAL 8301, Bacteroides dorei strain HS1, Bacteroides fragilis strain YCH46, Bacteroides salanitronis strain DSM 18170, Barnesiella viscericola strain DSM 18177, Helicobacter pylori strain 26695 and Ornithobacterium rhinotracheale strain ORT-UMN 88 (Table 5). Although Gabonia massiliensis is a draft genome, making the comparison complex, we propose a summary of the comparison of genome content of Gabonia massiliensis and other bacteria in Table 6.
Fig. 7.
Distribution of functional classes of predicted genes according to clusters of orthologous groups of proteins.
Table 5.
Numbers of orthologous protein-coding genes shared between genomes (upper right)a
| Alistipes finegoldii | Alistipes shahii | Bacteroides dorei | Bacteroides fragilis | Bacteroides salanitronis | Barnesiella viscericola | Gabonia massiliensis | Helicobacter pylori | Ornithobacterium rhinotracheale | |
|---|---|---|---|---|---|---|---|---|---|
| Alistipes finegoldii | 3,231 | 1,091 | 1,060 | 1,072 | 931 | 916 | 953 | 269 | 633 |
| Alistipes shahii | 83.25 | 3,132 | 1,011 | 1,040 | 911 | 881 | 936 | 256 | 620 |
| Bacteroides dorei | 59.43 | 58.96 | 4,269 | 1,682 | 1,476 | 1,248 | 1,454 | 309 | 734 |
| Bacteroides fragilis | 61.02 | 61.06 | 71.11 | 4,373 | 1,448 | 1,258 | 1,454 | 307 | 754 |
| Bacteroides salanitronis | 62.34 | 62.53 | 71.23 | 69.52 | 3,686 | 1,166 | 1,277 | 287 | 687 |
| Barnesiella viscericola | 64.82 | 65.16 | 63.85 | 65.37 | 66.25 | 2,499 | 1,238 | 288 | 673 |
| Gabonia massiliensis | 56.41 | 55.84 | 65.64 | 65.48 | 63.62 | 64.46 | 3,349 | 302 | 699 |
| Helicobacter pylori | 52.40 | 52.06 | 56.20 | 56.07 | 55.61 | 53.74 | 56.77 | 1,579 | 262 |
| Ornithobacterium rhinotracheale | 56.44 | 56.41 | 61.12 | 61.46 | 60.98 | 59.38 | 61.07 | 57.86 | 2,312 |
Average percentage similarity of nucleotides corresponding to orthologous protein shared between genomes (lower left) and numbers of proteins per genome (bold).
Table 6.
Genome comparison between Gabonia massiliensis and closely related species
| Specimen no. | Organism name | INSDC | Size (Mb) | G+C% | Protein coding genes | Total genes |
|---|---|---|---|---|---|---|
| 1 | Gabonia massiliensis | 4.26 | 37.9 | 3227 | 3288 | |
| 2 | Barnesiella viscericola DSM 18177 | CP007034.1 | 3.08 | 51.7 | 2464 | 2557 |
| 3 | Bacteroides fragilis YCH46 | AP006841.1 | 5.28 | 43.3 | 4578 | 4670 |
| 4 | Ornithobacterium rhinotracheale ORT-UMN 88 | CP006828.1 | 2.4 | 37.2 | 2210 | 2289 |
| 5 | Alistipes finegoldii DSM 17242 | CP003274.1 | 3.73 | 56.6 | 3108 | 3226 |
| 6 | Bacteroides salanitronis DSM 18170 | CP002530.1 | 4.24 | 46.6 | 3543 | 3681 |
| 7 | Bacteroides dorei HS1 | CP008741.1 | 5.24 | 41.6 | 4024 | 4186 |
| 8 | Alistipes shahii WAL 8301 | FP929032.1 | 3.76 | 57.2 | 2996 | 3090 |
| 9 | Helicobacter pylori 26695 | AE000511.1 | 1.67 | 38.9 | 1445 | 1555 |
Among species with standing in nomenclature, AGIOS values ranged from 83.25 between Alistipes shahii strain WAL 8301 and Alistipes finegoldii strain DSM 17242 to 56.41 between Gabonia massiliensis strain GM3 and Alistipes finegoldii strain DSM 17242 (Table 5).
To evaluate genomic similarity among the studied strains, we determined two parameters, AGIOS (Table 5) [6], which was designed to be independent from DNA-DNA hybridization (DDH), and digital DDH, which exhibits a high correlation with DDH [29], [30] (Table 7).
Table 7.
Pairwise comparison of Gabonia massiliensis with eight other speciesa
| Alistipes finegoldii | Alistipes shahii | Bacteroides dorei | Bacteroides fragilis | Bacteroides salanitronis | Barnesiella viscericola | Gabonia massiliensis | Ornithobacterium rhinotracheale | Helicobacter pylori | |
|---|---|---|---|---|---|---|---|---|---|
| Alistipes finegoldii | 100% ± 00 | 30.5% ± 2.99 | 39.1% ± 2.54 | 35% ± 2.54 | 30.1% ± 2.54 | 25.6% ± 2.54 | 24.7% ± 2.52 | 32.9% ± 2.53 | 15.5% ± 2.52 |
| Alistipes shahii | 100% ± 00 | 28.8% ± 2.53 | 45.4% ± 2.54 | 31.4% ± 2.54 | 26.7% ± 2.54 | 25.1% ± 2.52 | 45.5% ± 2.53 | 15.3% ± 2.52 | |
| Bacteroides dorei | 100% ± 00 | 26.9% ± 2.59 | 22.3% ± 2.58 | 23% ± 2.55 | 23.2% ± 2.54 | 27.1% ± 2.53 | 15.6% ± 2.52 | ||
| Bacteroides fragilis | 100% ± 00 | 28.4% ± 2.56 | 34.8% ± 2.56 | 24.1% ± 2.54 | 41% ± 2.54 | 15.4% ± 2.52 | |||
| Bacteroides salanitronis | 100% ± 00 | 30.6% ± 2.58 | 22.9% ± 2.54 | 39.1% ± 2.55 | 15.2% ± 2.52 | ||||
| Barnesiella viscericola | 100% ± 00 | 20.9% ± 2.54 | 38% ± 2.55 | 15.1% ± 2.53 | |||||
| Gabonia massiliensis | 100% ± 00 | 23.9% ± 2.52 | 15.3% ± 2.52 | ||||||
| Ornithobacterium rhinotracheale | 100% ± 00 | 17.9% ± 2.52 | |||||||
| Helicobacter pylori | 100% ± 00 |
GGDC, Genome-to-Genome Distance Calculator; DDH, DNA-DNA hybridization.
Data were generated using GGDC, formula 2 (DDH estimates based on identities/HSP length). Confidence intervals indicate inherent uncertainty in estimating DDH values from intergenomic distances based on models derived from empirical test data sets (which are always limited in size). These results are in accordance with the 16S rRNA (Fig. 1) and phylogenomic analyses as well as GGDC results.
Conclusion
On the basis of phenotypic, phylogenetic and genomic analyses, we formally propose the creation of Gabonia massiliensis gen. nov., sp. nov. that contains the strain GM3. This bacterium was isolated from a stool sample from a healthy Gabonese male youth in Marseille, France.
Taxonomic and nomenclatural proposals
Description of Gabonia gen. nov.
Gabonia (Ga.bo.nia. N.L. gen. n. Gabonia, the Latin name of Gabon, the sub-Saharan African country where the stool specimen was collected).
This is a Gram-negative, non-spore-forming and non motile bacillus. G. massiliensis is preferentially anaerobic. It is oxidase and indole negative but catalase positive. It showed positive activity for alkaline phosphatase, esterase (C4), esterase lipase (C8), acid phosphatase, naphthol-AS-BI-phosphohydrolase, α- and β-galactosidase and N-acetyl-β-glucosaminidase. Its habitat is the human gut. Its type species is Gabonia massiliensis strain GM3.
Description of Gabonia massiliensis gen. nov., sp. nov.
Gabonia massiliensis (ma.si.li.en'sis. L. masc. adj. massiliensis, of Massilia, the Latin name of Marseille, where G. massiliensis was isolated).
G. massiliensis exhibited translucent colonies with a 1.5 mm diameter. Individual cells exhibited a diameter of 0.5 μm and a length of 1.25 μm. G. massiliensis is preferentially anaerobic and grows at an optimal temperature of 37°C. It is a Gram-negative, non-spore-forming and nonmotile bacillus. It is oxidase negative but catalase positive. G. massiliensis showed positive reactions for alkaline phosphatase, esterase (C4), esterase lipase (C8), acid phosphatase, naphthol-AS-BI-phosphohydrolase, α- and β-galactosidase and N-acetyl-β-glucosaminidase. It was sensitive for nitrofurantoin, doxycycline, rifampicin, amoxicillin/clavulanic acid and imipenem. G. massiliensis type strain GM3 (= CSUR P1909 = DSM 100571) was isolated from a stool sample of a healthy Gabonese male youth. This strain exhibited a GC content of 37.9%. Its 16S rRNA sequence was deposited in GenBank under accession number LN849789, and the whole genome shotgun sequence has been deposited in GenBank under accession number CYPV00000000.
Acknowledgements
The authors thank the Xegen Company (www.xegen.fr) for automating the genomic annotation process and C. Andrieu for administrative assistance. Funded by the Méditerranée Infection Foundation.
Conflict of Interest
None declared.
References
- 1.Lagier J.C., Armougom F., Million M. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 2.Lagier J.C., El Karkouri K., Nguyen T.T., Armougom F., Raoult D., Fournier P.E. Non-contiguous-finished genome sequence and description of Anaerococcus senegalensis sp. nov. Stand Genomic Sci. 2012;6:116–125. doi: 10.4056/sigs.2415480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lagier J.C., Armougom F., Mishra A.K., Nguyen T.T., Raoult D., Fournier P.E. Non-contiguous-finished genome sequence and description of Alistipes timonensis sp. nov. Stand Genomic Sci. 2012;6:315–324. doi: 10.4056/sigs.2685971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mishra A.K., Lagier J.C., Robert C., Raoult D., Fournier P.E. Non-contiguous-finished genome sequence and description of Clostridium senegalense sp. nov. Stand Genomic Sci. 2012;6:386–395. doi: 10.4056/sigs.2766062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mishra A.K., Lagier J.C., Robert C., Raoult D., Fournier P.E. Non-contiguous-finished genome sequence and description of Peptinophilus timonensis sp. nov. Stand Genomic Sci. 2012;7:1–11. doi: 10.4056/sigs.2956294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mishra A.K., Lagier J.C., Robert C., Raoult D., Fournier P.E. Non-contiguous-finished genome sequence and description of Peptinophilus senegalensis sp. nov. Stand Genomic Sci. 2013;7:370–381. doi: 10.4056/sigs.3366764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tindall B.J., Rosselló-Móra R., Busse H.J., Ludwig W., Kämpfer P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol. 2010;60:249–266. doi: 10.1099/ijs.0.016949-0. [DOI] [PubMed] [Google Scholar]
- 8.Stackebrandt E., Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152–155. [Google Scholar]
- 9.Wayne L.G., Brenner D.J., Colwell P.R. Report of the ad hoc committee on reconciliation of approaches to bacterial systematic. Int J Syst Bacteriol. 1987;37:463–464. [Google Scholar]
- 10.Rossello-Mora R. DNA-DNA reassociation methods applied to microbial taxonomy and their critical evaluation. In: Stackebrandt E., editor. Molecular identification, systematics, and population structure of prokaryotes. Springer; Berlin: 2006. pp. 23–50. [Google Scholar]
- 11.Welker M., Moore E.R. Applications of whole-cell matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry in systematic microbiology. Syst Appl Microbiol. 2011;34:2–11. doi: 10.1016/j.syapm.2010.11.013. [DOI] [PubMed] [Google Scholar]
- 12.Ramasamy D., Mishra A.K., Lagier J.C. A polyphasic strategy incorporating genomic data for the taxonomic description of new bacterial species. Int J Syst Evol Microbiol. 2014;64:384–391. doi: 10.1099/ijs.0.057091-0. [DOI] [PubMed] [Google Scholar]
- 13.Krieg N.R. “Family IV. Porphyromonadaceae fam. nov.”. In: Krieg N.R., Staley J.T., Brown D.R., et al, editors. Bergey's manual of systematic bacteriology. 2nd ed. Springer-Verlag; New York: 2011. 4:61. [Google Scholar]
- 14.Lagier J.C., Hugon P., Khelaifia S., Fournier P.E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nkamga V.D., Huynh H.T., Aboudharam G., Ruimy R., Drancourt M. Diversity of human-associated Methanobrevibacter smithii isolates revealed by multispacer sequence typing. Curr Microbiol. 2015;70:810–815. doi: 10.1007/s00284-015-0787-9. [DOI] [PubMed] [Google Scholar]
- 16.Benson D.A., Karsch-Mizrachi I., Clark K., Lipman D.J., Ostell J., Sayers E.W. GenBank. Nucl Acids Res. 2012;40:D48–D53. doi: 10.1093/nar/gkr1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowe T.M., Eddy S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucl Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lagesen K., Hallin P., Rodland E.A., Staerfeldt H.H., Rognes T., Ussery D.W. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucl Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bendtsen J.D., Nielsen H., von Heijne G., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 20.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 21.Zhou Y., Liang Y., Lynch K.H., Dennis J.J., Wishart D.S. PHAST: a fast phage search tool. Nucl Acids Res. 2011;39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aziz R.K., Bartels D., Best A.A. The RAST Server: rapid annotations using subsystems tchnology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rutherford K., Parkhill J., Crook J. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 24.Carver T., Thomson N., Bleasby A., Berriman M., Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darling A.C., Mau B., Blattner F.R., Perna N.T. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lechner M., Findeiss S., Steiner L., Marz M., Stadler P.F., Prohaska S.J. Proteinortho: detection of (co)orthologs in large-scale analysis. BMC Bioinformatics. 2011;12:124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gouret P, Paganini J, Dainat J et al. Integration of evolutionary biology concepts for functional annotation and automation of complex research in evolution: the multi-agent software system DAGOBAH. New York: Springer; p. 71–87.
- 28.Gouret P., Vitiello V., Balandraud N., Gilles A., Pontarotti P., Danchin E.G. FIGENIX: intelligent automation of genomic annotation: expertise integration in a new software platform. BMC Bioinformatics. 2005;6:198. doi: 10.1186/1471-2105-6-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Auch A.F., Von Jan M., Klenk H.P., Göker M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci. 2010;2:117–134. doi: 10.4056/sigs.531120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meier-Kolthoff J.P., Auch A.F., Klenk H.P., Göker M. Genome sequence–based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakamoto M., Lan P.T., Benno Y. Barnesiella viscericola gen. nov., sp. nov., a novel member of the family Porphyromonadaceae isolated from chicken caecum. Int J Syst Evol Microbiol. 2007;57:342–346. doi: 10.1099/ijs.0.64709-0. [DOI] [PubMed] [Google Scholar]
- 32.Morotomi M., Nagai F., Sakon H., Tanaka R. Dialister succinatiphilus sp. nov. and Barnesiella intestinihominis sp. nov., isolated from human faeces. Int J Syst Evol Microbiol. 2008;58:2716–2720. doi: 10.1099/ijs.0.2008/000810-0. [DOI] [PubMed] [Google Scholar]
- 33.Shkoporov A.N., Khokhlova E.V., Chaplin AV Coprobacter fastidiosus gen. nov., sp. nov., a novel member of the family Porphyromonadaceae isolated from infant faeces. Int J Syst Evol Microbiol. 2013;63:4181–4188. doi: 10.1099/ijs.0.052126-0. [DOI] [PubMed] [Google Scholar]
- 34.Hirasawa M., Takada K. Porphyromonas gingivicanis sp. nov. and Porphyromonas crevioricanis sp. nov., isolated from beagles. Int J Syst Bacteriol. 1994;44:637–640. doi: 10.1099/00207713-44-4-637. [DOI] [PubMed] [Google Scholar]