Abstract
Although the opportunistic bacterial pathogen Enterococcus faecium is a leading source of nosocomial infections, it appears to lack many of the overt virulence factors produced by other bacterial pathogens, and the underlying mechanism of pathogenesis is not clear. Using E. faecium-mediated killing of the nematode worm Caenorhabditis elegans as an indicator of toxicity, we determined that E. faecium produces hydrogen peroxide at levels that cause cellular damage. We identified E. faecium transposon insertion mutants with altered C. elegans killing activity, and these mutants were altered in hydrogen peroxide production. Mutation of an NADH oxidase-encoding gene eliminated nearly all NADH oxidase activity and reduced hydrogen peroxide production. Mutation of an NADH peroxidase-encoding gene resulted in the enhanced accumulation of hydrogen peroxide. E. faecium is able to produce hydrogen peroxide by using glycerol-3-phosphate oxidase, and addition of glycerol to the culture medium enhanced the killing of C. elegans. Conversely, addition of glucose, which leads to the down-regulation of glycerol metabolism, prevented both C. elegans killing and hydrogen peroxide production. Lastly, detoxification of hydrogen peroxide either by exogenously added catalase or by a C. elegans transgenic strain overproducing catalase prevented E. faecium-mediated killing. These results suggest that hydrogen peroxide produced by E. faecium has cytotoxic effects and highlight the utility of C. elegans pathogenicity models for identifying bacterial virulence factors.
Enterococci are gram-positive bacteria that usually reside in the gastrointestinal tract as commensal organisms, but they are also capable of causing severe infections (18). Enterococci are the third leading source of nosocomial infections, causing endocarditis, peritonitis, bacteremia, and urinary tract infections. Two enterococcal species are responsible for almost all of these infections. According to a 1997 survey, the majority (85 to 90%) of enterococcal infections are caused by Enterococcus faecalis, and the remaining infections are due to Enterococcus faecium (35). However, in certain settings, the frequency of infections due to E. faecium has been increasing in recent years (50). The acquisition of antibiotic resistance is believed to be the major contributing factor for the elevated incidence of E. faecium infections. Approximately one-half of E. faecium clinical isolates are now resistant to vancomycin, while only a small fraction of E. faecalis clinical isolates are vancomycin resistant (50).
The mechanism underlying E. faecium pathogenesis is obscure, due in part to the fact that few E. faecium virulence-related factors have been identified. The espfm and hylEfm genes, encoding a surface protein and hyaluronidase, respectively, are more likely to be present in pathogenic E. faecium strains than in strains isolated from healthy individuals (11, 45, 61). The espfm gene appears to be located on a pathogenicity island containing genes implicated in virulence, antibiotic resistance, and transcriptional regulation (29). Other E. faecium factors predicted to contribute to pathogenesis are the Acm and SagA proteins, which bind to extracellular matrix components (34, 58).
One of the limitations of studying E. faecium infections is the lack of a simple animal model. Our laboratory has pioneered the use of simple model hosts to identify microbial virulence factors. We have used the plant Arabidopsis thaliana, the wax moth caterpillar Galleria mellonella, and the nematode Caenorhabditis elegans as simple model hosts to study infections caused by E. faecalis, Staphylococcus aureus, Pseudomonas aeruginosa, Salmonella enterica serovar Typhimurium, and Cryptococcus neoformans (1, 17, 26, 31, 33, 44, 51, 57). Importantly, the majority of virulence factors required for infection of the simple model hosts are also required for infection in mouse infection models, indicating that the pathogenic mechanisms used in mammalian infections are also used in the simple model hosts (43).
The C. elegans model system has been used to study two different types of pathogenic mechanisms. C. elegans can be killed by pathogens either through an infection-like process or by diffusible toxins. To assay either mechanism, C. elegans, which feeds on bacteria, is transferred from its normal food source, lawns of an auxotrophic strain of Escherichia coli, to lawns of the pathogen. In the infection model system, the nematodes ingest the bacteria and then die after 2 to 5 days, depending on the pathogen. In contrast, killing by diffusible toxins occurs more quickly and does not require direct bacterial contact. P. aeruginosa strain PAO1 produces cyanide that paralyzes and kills C. elegans (9, 16). P. aeruginosa strain PA14 produces different toxins, including phenazines, when it is grown on high-osmolarity medium, which quickly kill but do not paralyze C. elegans (31).
Previously, our laboratory showed that E. faecalis kills C. elegans by an infectious process but that E. faecium does not (17). Here, however, we show that E. faecium is capable of killing C. elegans by a diffusible toxin after the bacteria are grown under anaerobic conditions. By identifying mutants altered in nematode killing, we found that E. faecium releases substantial amounts of the reactive oxygen species hydrogen peroxide and that hydrogen peroxide is responsible for nematode killing. Other bacterial pathogens, including Streptococcus pneumoniae and Streptococcus pyogenes, also produce hydrogen peroxide that kills C. elegans (5, 27, 53). Hydrogen peroxide produced by S. pneumoniae has been shown to contribute to virulence by damaging host tissue (6, 10, 22). An additional activity of hydrogen peroxide is the growth inhibition of competing bacterial species (20, 39). We suggest that the hydrogen peroxide produced by E. faecium causes damage to surrounding cells, and this may play a role in colonization or infection.
MATERIALS AND METHODS
Bacterial and nematode strains.
Enterococcal strains were grown on brain heart infusion (BHI) media (Difco Becton Dickinson, Sparks, Md.) containing kanamycin (75 μg/ml) or nalidixic acid (10 μg/ml) at 37°C. Liquid cultures were typically grown without shaking. We used E. faecium strains GE-1 (= ATCC 51558) (12), E007 (17), E0158 (61), E0238 (61), E0318 (61), E0734 (61), SE34 (= TX1330) (59), and DO (= TX0016 = ATCC BAA-472) (2) and E. faecalis strains OG1RF (32) and VS583. The vanHB genes were deleted in strain V583 (49) to produce the vancomycin-sensitive strain VS583.
C. elegans strains were maintained by using standard practices (56). Wild-type Bristol strain N2 (7) was used for all experiments unless otherwise noted. We also used strains daf-2(e1370), daf-16(mgDf47) (37), and daf-2(e1370) daf-16(mgDf47).
Nematode killing assay.
Bacterial cultures were grown in BHI medium at 37°C and diluted to an optical density at 600 nm (OD600) of 0.1 to 0.2 in fresh medium, and then 10-μl portions were spread onto BHI agar containing kanamycin (75 μg/ml) or nalidixic acid (10 μg/ml) in 35-mm petri plates. The plates were incubated overnight at 37°C. GasPak Plus envelopes and containers (Becton Dickinson) were used to generate an anaerobic environment. Before the nematodes were transferred to the anaerobically grown lawns of E. faecium, the plates were cooled to room temperature in ambient air for 30 to 60 min. Nematodes at the L4 developmental stage were resuspended and washed two times in M9 buffer. Nematodes were then transferred to the bacterial lawns in 5- to 10-μl drops and incubated at 25°C. At the times indicated below, nematodes were scored for survival. Nematodes that did not respond to touch with a platinum wire pick were considered to be dead. Assays for each bacterial strain and condition were done at least twice, and each experiment was done in duplicate or triplicate.
In order to estimate the numbers of viable bacteria on the assay plates, the bacterial lawns were resuspended in 50 mM sodium phosphate (pH 7), diluted, plated, and counted to determine the number of CFU. Approximately 6.9 × 109 ± 2.3 × 109 CFU were recovered from an E. faecium lawn grown anaerobically on a 35-mm petri plate. For certain experiments, either catalase (catalogue no. C-1345; Sigma, St. Louis, Mo.) was mixed into unsolidified BHI agar at a concentration of 1,000 U/ml or 1,000 U of catalase in 100 μl was spread onto solidified BHI agar. Superoxide dismutase (catalogue no. S-2515; Sigma) was spread onto solidified BHI agar in 100 μl containing 1,000 U.
Screening for Tn917 mutants.
Tn917 contained in the temperature-sensitive plasmid pTV1OK (19) was transformed into E. faecium SE34 by electroporation as described previously (15). Four independent overnight cultures were grown at 25°C in BHI medium containing 2 mg of kanamycin per ml and 0.05 μg of erythromycin per ml, plated onto BHI medium containing 2 μg of erythromycin per ml, and incubated at 47°C to select for transposon integrants. Colonies were picked by using a Q-bot (Genetix, Boston, Mass.) into 384-well plates, grown overnight in BHI medium containing 50 μg of erythromycin per ml and 15% glycerol, and frozen at −80°C. BHI medium containing 50 μg of erythromycin per ml was inoculated with the frozen strains and incubated at 37°C. Portions (20 μl) of the saturated cultures were spread onto 1 ml of BHI agar in duplicate 24-well plates and grown at 37°C. For each set of duplicate plates one plate was grown in a GasPak container overnight, and the other was grown in ambient air. The plates were cooled to room temperature, and the anaerobically grown plate was exposed to air. A mixture of starved L1-L2 stage and well-fed L4 stage nematodes was suspended in M9 buffer and pipetted onto the lawns of E. faecium. After 2 h, the anaerobically grown plates were examined for wells in which worms remained mobile. After 6 h, the aerobically grown plates were examined for wells in which worms became immobile.
A total of 3,120 erythromycin-resistant isolates were tested by both procedures, and 4 of these strains were found to have a reproducibly altered effect on nematode mobility. Mutants 2F22, 3C23, and 11M12, identified on the anaerobically grown plates, exhibited diminished killing activity, whereas mutant 2C4, identified on the aerobically grown plates, exhibited enhanced killing activity.
The transposon insertion sites of the E. faecium mutants were identified by using two rounds of arbitrary primed PCR followed by DNA sequencing (38). Two mutants (2F22 and 3C23) that were derived from the same liquid culture, which was grown under nonselective conditions, were found to have identical transposon insertion sites. These mutants are most likely siblings, and only the 2F22 mutant was characterized further. The genes surrounding the transposon insertion site of the 11M12 mutant were not found in the currently available genomic sequence of E. faecium strain DO/TX0016 (http://www.hgsc.bcm.tmc.edu/microbial/Efaecium/), and these genes were PCR amplified and sequenced.
Hydrogen peroxide measurement.
To measure hydrogen peroxide accumulation upon aeration of saturated cultures grown under anaerobic conditions, 100-ml BHI medium cultures of the relevant E. faecium strains in 500-ml flasks were grown at 37°C without agitation in GasPak containers. Cultures were removed from the GasPak containers and aerated on an orbital shaker at 300 rpm. To measure bacterial growth and hydrogen peroxide accumulation in aerobic cultures, overnight cultures were grown without shaking, diluted into 100 ml of prewarmed BHI medium in 500-ml flasks, and aerated at 37°C by shaking at 300 rpm. Samples were filtered through 0.22-μm-pore-size filters (Costar Spin-x; Costar, Corning, N.Y.), and the hydrogen peroxide levels of the bacterium-free culture eluates were measured with a 907-015 Correlate assay kit by using a 1:10 dilution with buffer (Assay Designs, Ann Arbor, Mich.) or with an Amplex Red kit by using a 1:100 dilution (Molecular Probes, Eugene, Oreg.).
Northern blotting.
Overnight saturated cultures were diluted 1:100 in prewarmed BHI medium and either shaken at 300 rpm or incubated without agitation in GasPak containers. Logarithmic-phase cultures were harvested at an OD600 of 0.2 to 0.3, and saturated cultures were harvested at an OD600 of ≥1.2. Cultures were treated with RNAprotect and were purified with RNeasy kits (QIAGEN, Valencia, Calif.). Five micrograms of RNA of each sample was run on 1.2% agarose-formaldehyde gels and transferred to positively charged nylon membranes. RNA blot hybridizations were carried out as previously described (3) by using 400-base probes labeled with [32P]dCTP by random priming. To analyze the effect of heavy metal exposure, logarithmic-phase aerobic cultures were treated with heavy metals at a concentration of 1 mM for 10 min at 37°C before cells were harvested. One molar stock solutions of cadmium chloride, cobalt(II) chloride, sodium dichromate, chromium trioxide, copper(II) chloride, iron(II) chloride, iron(III) chloride, magnesium chloride, manganese(II) chloride, nickel(II) sulfate, silver nitrate, and zinc chloride were used.
NADH oxidase assay.
Aerobic cultures were harvested at an OD600 of 0.2 to 0.3. Cells were washed with ice-cold buffer containing 50 mM potassium phosphate (pH 7.0) and 0.5 mM EDTA. Cells were disrupted by using glass beads (catalogue no. G-1145; Sigma) and a Mini Bead Beater (Biospec Products, Bartlesville, Okla.) and were microcentrifuged for 15 min at 4°C at 16,000 × g. The protein concentration was determined by the Pierce protein assay (catalogue no. 23200; Pierce, Rockford, Ill.) by using bovine serum albumin as a standard. Typically, 10 μg of extract was added to a 1-ml reaction mixture containing 150 μM NADH (catalogue no. N-8129; Sigma) in the buffer described above, and the absorbance at 340 nm was monitored for 2 to 3 min. For lysates from the 2F22 mutant, 100 μg of protein was added, and the reaction was monitored for 10 min. An extinction coefficient of 6.2 mM−1cm−1 was used to calculate NADH concentrations. One unit was defined as 1 μmol of NADH oxidized per min at 25°C.
Nucleotide sequence accession number.
The sequence of the genes surrounding the transposon insertion site of the 11M12 mutant has been deposited in the GenBank database under accession number AY527733.
RESULTS
E. faecium-mediated killing of C. elegans.
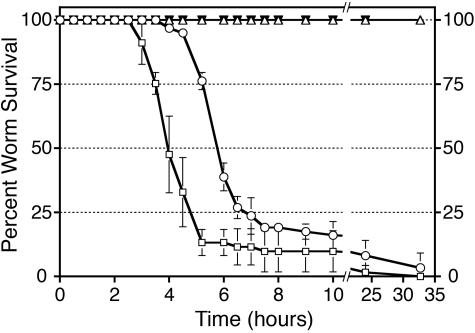
Our laboratory previously reported that several strains of E. faecium grown aerobically on BHI medium do not have any apparent deleterious effect on the nematode C. elegans when the bacteria are provided to the worms as the sole source of food (17). However, when grown under anaerobic conditions on BHI medium and then exposed to aerobic conditions, a variety of different E. faecium strains appeared to produce a toxin that rapidly killed C. elegans (Fig. 1 and data not shown). Depending on the strain tested, the initial effect of the toxicity was observed in as little as 45 min as a decrease in nematode mobility. After 1 to 2 h of exposure to anaerobically grown E. faecium, the nematodes lost muscle tone and became immobile. Fifty percent of the nematodes died within 4 to 6 h after the initial exposure, and nearly all of the nematodes were dead after 24 h (Fig. 1). Eight assorted E. faecium strains (strains DO, E007, E0158, E0238, E0318, E0734, GE-1, and SE34) killed C. elegans with similar kinetics when they were grown under anaerobic conditions. Similar to previously reported results, no killing activity was observed when the eight strains were grown aerobically (Fig. 1 and data not shown). In contrast to E. faecium, E. faecalis (strain OG1RF or VS583), killed C. elegans relatively slowly over a 5-day period, irrespective of whether it was grown aerobically or anaerobically (Fig. 1, Table 1, and data not shown).
FIG. 1.
C. elegans survival when it was fed enterococci. E. faecium and E. faecalis were grown on solid BHI medium under aerobic (solid symbols) or anaerobic (open symbols) conditions. L4-stage C. elegans was transferred onto the lawns of enterococci, incubated at 25°C, and scored for survival. Symbols: ○, E. faecium strain SE34 grown anaerobically; □, E. faecium strain E007 grown anaerobically; ▵, E. faecalis strain VS583 grown anaerobically; ▾, E. faecium strain SE34 grown aerobically.
TABLE 1.
Worm survival on lawns of Enterococcus
| Bacterial strain | Growth conditiona | % Survival of nematodes | Mobilityb |
|---|---|---|---|
| E007 | Anaerobic | 0.0 ± 0.0 | − |
| VS583 | Anaerobic | 100 ± 0.0 | +++ |
| SE34 | Anaerobic | 0.0 ± 0.0 | − |
| 2C4 | Anaerobic | 0.0 ± 0.0 | − |
| 2F22 | Anaerobic | 86.6 ± 5.4 | −/+ |
| 11M12 | Anaerobic | 100 ± 0.0 | +++ |
| SE34 | Aerobic | 100 ± 0.0 | +++ |
| 2C4 | Aerobic | 23.1 ± 3.7 | −/+ |
Worm survival was scored after 15 h of incubation for anaerobic growth and after 18 h of incubation for aerobic growth.
Average worm mobility was scored as follows: +++, normal movement; ++, slowed movement; +, movement restricted to the head and tail; −, no movement.
Identification of E. faecium mutants.
In other C. elegans pathogenicity models developed in our laboratory and in other laboratories, relatively rapid killing of C. elegans is indicative of the production of a low-molecular-weight toxin (9, 16, 31). To identify the putative E. faecium fast-killing toxin, E. faecium was mutagenized with transposon Tn917 as described in Materials and Methods, and 3,120 transposon-mutagenized clones were screened for mutants that exhibited aberrant C. elegans killing activity.
In order to identify mutants with diminished killing activity, mutant clones were grown anaerobically, returned to air, and then incubated with wild-type C. elegans. This screening procedure led to identification of two mutants (2F22 and 11M12) with diminished killing activity. Fifteen hours of exposure to anaerobically grown parental strain SE34 killed all of the nematodes. In contrast, 87 and 100% of the nematodes survived exposure to mutants 2F22 and 11M12, respectively (Table 1). Some toxicity was retained in mutant 2F22 since it was able to impair nematode mobility, whereas no inhibition of nematode mobility was observed with mutant 11M12.
To identify E. faecium mutants with enhanced killing activity, we screened for mutants that acquired the ability to kill nematodes when they were grown aerobically. One such mutant, 2C4, was identified. Under these assay conditions, mutant 2C4 killed 77% of the nematodes within 18 h, whereas no nematodes were killed by the aerobically grown parental strain SE34 (Table 1). The differences in C. elegans killing between the mutants and the parental strain could not be attributed to differences in the number of viable CFU on the assay plates (as described in Materials and Methods, the numbers of CFU of the mutant strains recovered from the assay plates were similar to the numbers of CFU of the parental strain).
Tn917 is inserted just upstream of the npr gene encoding an NADH peroxidase in the hypervirulent mutant 2C4.
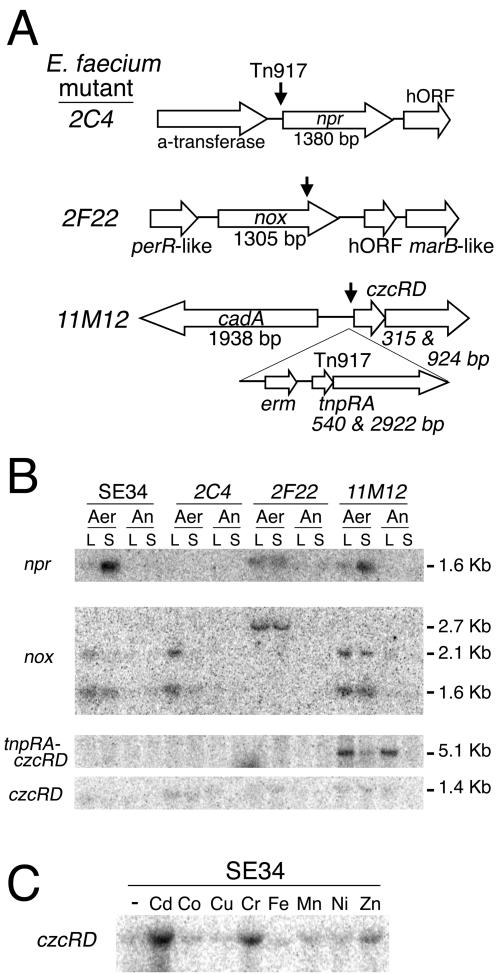
Molecular analysis of the enhanced-killing mutant 2C4 showed that Tn917 had integrated into the E. faecium chromosome 16 bp upstream of the start codon of the npr gene, which encodes an NADH peroxidase (NPX) (Fig. 2A). The E. faecium NADH peroxidase exhibits 46% amino acid identity with the E. faecalis 10C1 NADH peroxidase (46) and is homologous over the entire length of the protein. In the parental strain SE34, npr is transcribed as a 1.6-kb monocistronic transcript that is expressed at low levels during aerobic growth and is highly induced during stationary-phase growth. However, the npr transcript was not detectable in the 2C4 mutant (Fig. 2B).
FIG. 2.
(A) DNA map of transposon Tn917 insertion sites in E. faecium mutants 2C4, 2F22, and 11M12. The vertical arrows indicate Tn917 insertion sites. (B) Northern blot of E. faecium strains SE34, 2C4, 2F22, and 11M12 hybridized against the npr, nox, and czcRD transcripts. Cultures were grown aerobically (Aer) or anaerobically (An) and harvested from logarithmic-phase (L) or saturated (S) cultures. (C) Northern blot of SE34 treated with various heavy metals hybridized against the czcRD transcript. Aerobic log-phase cultures were treated with cadmium, cobalt, copper, chromium, iron(II), manganese, nickel, or zinc at a concentration of 1 mM for 10 min. The first lane (−) contained a mock-treated culture.
E. faecium is a facultative anaerobe that uses oxidases that reduce molecular oxygen to form hydrogen peroxide (23). E. faecium is catalase negative, and the major mechanism to scavenge hydrogen peroxide is NADH peroxidase. We hypothesized that the absence of NADH peroxidase allows increased amounts of hydrogen peroxide to accumulate in 2C4 cultures and that hydrogen peroxide might be the cause of E. faecium-mediated rapid killing of C. elegans.
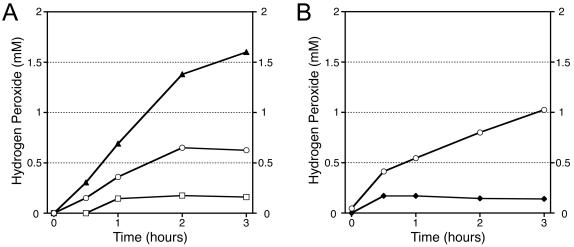
In aerobic liquid cultures, both parental strain SE34 and mutant 2C4 produced hydrogen peroxide as the cultures entered the stationary phase (see below). To simulate the amount of hydrogen peroxide putatively produced by E. faecium in the worm-killing assays, bacteria were grown to saturation in liquid cultures under anaerobic conditions and then aerated in the presence of atmospheric oxygen. No hydrogen peroxide was detectable in anaerobic cultures, but upon aeration, hydrogen peroxide accumulated at a rate of approximately 0.24 mM per h per 109 CFU/ml in the case of wild-type strain SE34 (Fig. 3A). Under the same experimental conditions, the 2C4 mutant culture accumulated hydrogen peroxide at a rate that was approximately two times the rate of the parental strain. The culture densities of SE34 and 2C4 were similar and remained constant during the 3 h of aeration.
FIG. 3.
(A) Hydrogen peroxide accumulation upon aeration of saturated cultures grown under anaerobic conditions. (B) Hydrogen peroxide accumulation upon aeration of saturated cultures grown under semiaerobic conditions. Symbols: ○, parental strain SE34; ▴, mutant 2C4; □, mutant 2F22; ♦, mutant 11M12.
Hydrogen peroxide produced by E. faecium is sufficient to kill C. elegans.
Several lines of evidence are consistent with the hypothesis that hydrogen peroxide is the lethal toxin produced by E. faecium. First, nematodes were killed by hydrogen peroxide in a bacterium-free liquid assay at a rate similar to the rate of killing by lawns of E. faecium (Table 2). After 15 h of incubation with concentrations of hydrogen peroxide greater than 0.2 mM in M9 buffer, nematode mobility was decreased. Incubation with 0.3 to 0.4 mM hydrogen peroxide killed 50% of the nematodes, and 98% of the nematodes were killed by 1 to 2 mM hydrogen peroxide (Table 2).
TABLE 2.
Worm survival after exposure to hydrogen peroxidea
| H2O2 concn (mM) | % Survival | Mobility |
|---|---|---|
| 0 | 100 ± 0.0 | +++ |
| 0.1 | 100 ± 0.0 | +++ |
| 0.2 | 100 ± 0.0 | +/++ |
| 0.3 | 70.5 ± 23.7 | −/+ |
| 0.4 | 30.3 ± 13.6 | − |
| 0.5 | 4.8 ± 1.1 | − |
| 1 | 1.3 ± 1.9 | − |
| 2 | 2.0 ± 1.4 | − |
| 5 | 0.0 ± 0.0 | − |
Worm survival was scored after 15 h of incubation. Survival was assayed in M9 buffer containing hydrogen peroxide.
Second, addition of exogenous catalase to the solid BHI medium completely rescued nematode killing by E. faecium. In addition, incorporation of catalase allowed the nematodes to remain mobile (Table 3). In contrast to catalase treatment, addition of superoxide dismutase to the medium did not have any effect on nematode killing (data not shown).
TABLE 3.
Effect of catalase on worm survivala
| Bacteria grown anaerobically | Untreated
|
Treated with catalase
|
||
|---|---|---|---|---|
| % Survival | Mobility | % Survival | Mobility | |
| SE34 | 6.3 | − | 100 ± 0.0 | +++ |
| 2C4 | 0 | − | 100 ± 0.0 | ++/+++ |
| 2F22 | 83.3 | − | 100 ± 0.0 | +++ |
| 11M12 | 100 | +++ | 100 ± 0.0 | +++ |
| E007 | 0 | − | 100 ± 0.0 | +++ |
| VS583 | 100 | +++ | 100 ± 0.0 | +++ |
Worm survival was scored after 15 h of incubation.
Third, C. elegans daf-2 mutants, which are resistant to a variety of oxidative stresses, exhibit enhanced longevity, and constitutively overexpress catalase (60), were more resistant to killing by anaerobically grown E. faecium (Table 4). daf-2 encodes an insulin-like receptor that negatively regulates the fork head transcription factor DAF-16 (37). Thus, daf-16 mutations suppress the long-lived and stress-resistant phenotypes of daf-2 mutations. As expected, a daf-2 daf-16 double mutant was as susceptible to E. faecium-mediated killing as wild-type C. elegans (Table 4).
TABLE 4.
Survival of worms mutated in daf-2 or daf-16a
| C. elegans strain | % Survival | Mobility |
|---|---|---|
| N2 | 0.0 ± 0.0 | − |
| daf-2 | 100 ± 0.0 | + |
| daf-16 | 2.0 ± 2.8 | − |
| daf-2 daf-16 | 1.0 ± 1.4 | − |
Worm survival was scored after 24 h of incubation with anaerobically grown SE34.
E. faecium mutants defective in nematode killing.
Based on the observation that E. faecium produces hydrogen peroxide, which kills C. elegans, we predicted that the E. faecium mutants identified in our screening analysis that are defective in nematode killing would produce less hydrogen peroxide. We confirmed this prediction for mutants 2F22 and 11M12 by measuring the hydrogen peroxide produced after aeration of stationary-phase cultures. When cultures of 2F22 and SE34 were grown under anaerobic conditions and then aerated for 3 h, the 2F22 culture accumulated 75% less hydrogen peroxide than the SE34 culture accumulated even though the culture densities were equivalent (Fig. 3A). Although cultures of 11M12 grown anaerobically to saturation had a lower number of viable CFU per milliliter than SE34 cultures had for unexplained reasons (data not shown), both 11M12 and SE34 grew to the same density in semiaerobic cultures (grown without agitation in the presence of atmospheric air). When semiaerobic saturated cultures of 11M12 and SE34 were aerated for 3 h, the 11M12 mutant, like 2F22, accumulated 75% less hydrogen peroxide than SE34 accumulated (Fig. 3B).
In mutant 2F22, Tn917 integrated into the nox gene that apparently encodes the major NADH oxidase of E. faecium (Fig. 2A and see below). NADH oxidases use molecular oxygen as an electron acceptor to regenerate NAD+ from NADH. There are two categories of NADH oxidases; one type produces water, whereas the second type produces hydrogen peroxide. In the parental SE34 strain under aerobic growth conditions, there are two nox gene transcripts, a 1.6-kb monocistronic transcript and a 2.1-kb transcript that also contains the adjacent 300-bp hypothetical open reading frame with an unknown function (Fig. 2B). In the 2F22 mutant, these two nox transcripts are replaced by a single 2.7-kb transcript (Fig. 2B). As measured by a spectroscopic assay, the total NADH oxidase activity was decreased 98% in the 2F22 mutant. The parental strain had an NADH oxidation activity of 0.882 ± 0.051 U per mg of total soluble protein, whereas the 2F22 mutant had an activity of only 0.019 ± 0.003 U per mg of protein. The partially sequenced E. faecium strain DO contains five other putative NADH oxidase genes, which may account for the remaining small amount of NADH oxidase activity in extracts of 2F22.
Interestingly, even though 2F22 produces significantly less H2O2 than the wild type produces, the nox gene mutated in 2F22 appears to encode the water-forming type of NADH oxidase since no hydrogen peroxide was detectable in the NOX assay when wild-type extracts were used. Moreover, mutant 11M12, which is also defective in hydrogen peroxide production, has the same NADH oxidase activity as the parental strain. The regeneration of NADH by NADH oxidases allows bacteria to use alternative mixed acid fermentation pathways when they are grown in the presence of molecular oxygen (21, 30). Mutation of nox constrains bacteria toward anaerobic metabolism, which in turn is predicted to lead to reduced activity of oxidases that produce hydrogen peroxide. Although this explains the phenotype of the 2F22 mutant, we have not ruled out the possibility that the decreased hydrogen peroxide production of the 2F22 mutant is due to a polar effect on downstream genes. Unfortunately, we were unable to transform plasmids into mutant 2F22 because the strain lysed upon centrifugation while the cells were being prepared for electroporation. Consequently, we were unable to express nox+ in mutant 2F22, and we were not able to confirm that the nox mutation is responsible for the phenotype of the 2F22 mutant.
In mutant 11M12, Tn917 is integrated in a locus involved in heavy metal regulation and is integrated between the czcRD operon and the cadA gene. Tn917 is integrated approximately 50 bp upstream of the start codon of czcR and approximately 300 bp upstream of the start codon of cadA on the opposite DNA strand (Fig. 3A). cadA encodes a putative P-type cation efflux transmembrane pump that exhibits 58% identity with a predicted cadmium transporter from E. faecalis (GenBank accession no. AAO80575) (4) and 84% identity with the predicted cation transporter CopA from Staphylococcus epidermidis (GenBank accession no. AAO03659). cadA transcription is induced by cadmium, chromium, and zinc but is not altered by cobalt, copper, iron, manganese, or nickel (data not shown). cadA transcript levels are not altered in the 11M12 mutant (data not shown), making it unlikely that the phenotype of 11M12 is a consequence of aberrant cadA transcription.
CzcR belongs to the ArsR/SmtB family of metal-sensing transcriptional repressors, while CzcD belongs to the Czc (Co/Zn/Cd) family of heavy metal exporters (52). The two-gene operon organization of the czcRD genes is similar to the organization of the czrAB genes of S. aureus, and the protein sequences exhibit 43 and 27% identity, respectively (28). In response to the presence of specific heavy metal ions, ArsR-type proteins derepress transcription of heavy metal efflux transporters and stress response proteins (52). In the wild-type strain, the 1.4-kb czcRD operon is expressed at very low levels under normal growth conditions but is strongly induced in the presence of cadmium or chromium. Mild induction is seen upon exposure to zinc, whereas no induction is detectable upon exposure to cobalt, copper, iron, magnesium, manganese, nickel, or silver (Fig. 3C and data not shown). In mutant 11M12, the czcRD operon is aberrantly expressed as a fusion to the Tn917 tnpRA operon, resulting in a 5.1-kb transcript (Fig. 2B). Toxicity from chromium and cadmium has been linked to oxidative stress (55), and the decreased hydrogen peroxide phenotype of 11M12 suggests a link between the regulation of genes involved in heavy metal detoxification and hydrogen peroxide production. Specifically, we hypothesize that the elevated levels of the transcriptional repressor CzcR in mutant 11M12 may result in the repression of oxidases, resulting in lowered hydrogen peroxide production. In an attempt to reproduce the phenotype of the 11M12 mutant, the czcR or czcRD genes were overexpressed from plasmids in SE34. Even though highly expressed transcripts were detected by Northern blotting, the strains overexpressing czcR or czcRD did not produce smaller amounts of hydrogen peroxide (data not shown). Thus, we cannot definitively conclude that the diminished hydrogen peroxide production phenotype of 11M12 is a consequence of the insertion of Tn917 upstream of the czcRD operon or that another unlinked mutation is responsible.
Hydrogen peroxide production is modulated by glycerol and glucose.
Enterococci synthesize glycerol-3-phosphate oxidase, which uses molecular oxygen as a reductant to form dihydroxyacetone and hydrogen peroxide (13). Addition of glycerol to liquid cultures of E. faecium that were incubated without agitation resulted in increased accumulation of hydrogen peroxide. After overnight culture, we determined that strain SE34 accumulated 232 ± 37.5 μM hydrogen peroxide in the absence of glycerol and 636 ± 48.5 μM hydrogen peroxide in the presence of 0.2% glycerol. We therefore tested whether nematode killing was enhanced when glycerol was added to the E. faecium solid growth medium. We found that addition of 0.2% glycerol to the medium enabled aerobically grown E. faecium to kill nematodes (Table 5).
TABLE 5.
Effects of glycerol and glucose on worm survival
| Bacterial strain | Glycerol expta
|
Glucose exptb
|
||||||
|---|---|---|---|---|---|---|---|---|
| Untreated
|
Treated with 0.2% glycerol
|
Untreated
|
Treated with 1% glucose
|
|||||
| % Survival | Mobility | % Survival | Mobility | % Survival | Mobility | % Survival | Mobility | |
| SE34 | 100 ± 0.0 | +++ | 0.0 ± 0.0 | − | 5.5 ± 3.5 | − | 99.8 ± 0.4 | +++ |
| 2C4 | 34.0 ± 4.6 | − | 2.8 ± 2.0 | − | 2.0 ± 1.4 | − | 99.5 ± 0.6 | +++ |
| 2F22 | 100 ± 0.0 | +++ | 43.3 ± 18.5 | +/− | 78.1 ± 2.3 | − | 100 ± 0.0 | +++ |
| 11M12 | 97.1 ± 0.7 | +++ | 100 ± 0.0 | +++ | 99.0 ± 1.4 | +++ | 100 ± 0.0 | +++ |
| E007 | 97.4 ± 1.9 | +++ | 2.9 ± 0.4 | +/− | 0.5 ± 0.7 | − | 100 ± 0.0 | +++ |
| VS583 | 95.5 ± 0.4 | +++ | 100 ± 0.0 | +++ | 98.1 ± 2.7 | +++ | 99.5 ± 0.7 | +++ |
Worm survival was scored after 24 h of incubation with aerobically grown bacteria.
Worm survival was scored after 15 h of incubation with anaerobically grown bacteria.
The glycerol metabolic pathway is down-regulated by glycolytic intermediates, which inhibit glycerol kinase and glycerol-3-phosphate oxidase (8, 13, 23). Therefore, glucose addition should lead to a decrease in hydrogen peroxide production by reducing glycerol-3-phosphate oxidase activity and by possibly inhibiting other oxidases through catabolite repression. In liquid aerobic cultures, addition of 1% glucose to BHI medium completely eliminated hydrogen peroxide accumulation (data not shown) and nematode killing (Table 5) by all of the E. faecium strains tested.
Hydrogen peroxide produced in aerobic liquid cultures.
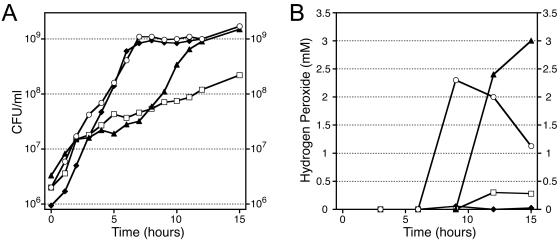
We showed that E. faecium produces hydrogen peroxide after anaerobic cultures are exposed to aerobic conditions. We also determined, however, that growth under anaerobic conditions is not a prerequisite for hydrogen peroxide production in liquid cultures. When the parental strain SE34 was grown in a highly agitated and aerated culture, 1 to 2 mM hydrogen peroxide accumulated as the culture entered the stationary phase (Fig. 4). For the mutant strains, differences in hydrogen peroxide production did not correlate with changes in aerobic growth rates. Mutant 11M12 grew at the same rate as the wild type, but the 11M12 culture did not accumulate hydrogen peroxide (Fig. 4). Mutant 2F22 grew at a lower rate and entered the stationary phase at a density that was 90% less than that of the wild type. Because the amount of hydrogen peroxide that accumulated in aerobic cultures of 2F22 was also decreased by 90%, each viable cell of 2F22 was producing an amount of hydrogen peroxide that was equivalent to the amount produced by a wild-type cell under these culture conditions. The initial growth rate of the 2C4 mutant was similar to that of the wild type upon dilution of a semiaerobic saturated culture into fresh media (see Materials and Methods). Then the growth rate of the 2C4 mutant temporarily decreased, thereby delaying entry of the culture into the stationary phase (Fig. 4A). In the stationary phase, the 2C4 mutant accumulated up to 3 mM hydrogen peroxide (Fig. 4B).
FIG. 4.
(A) Growth of E. faecium mutants in aerobic cultures. (B) Hydrogen peroxide accumulation in aerobic cultures of the E. faecium mutants. Symbols: ○, parental strain SE34; ▴, mutant 2C4; □, mutant 2F22; ♦, mutant 11M12.
We determined that the amount of hydrogen peroxide generated by wild-type E. faecium in an aerobic culture was sufficient to kill C. elegans. Bacterium-free supernatants taken from aerobic cultures were able to kill C. elegans, while supernatants treated with catalase were not able to kill C. elegans (data not shown).
DISCUSSION
In this study, we identified conditions in which E. faecium kills C. elegans, and in this report we present evidence that the killing is due to the production by E. faecium of hydrogen peroxide. First, we identified E. faecium mutants that exhibited either decreased or increased killing of C. elegans. The ability of the mutants to kill C. elegans was proportional to the amount of hydrogen peroxide produced. Second, we found that the amount of hydrogen peroxide produced by E. faecium could be modulated by glucose or glycerol, which in turn affected C. elegans killing. Third, hydrogen peroxide killed C. elegans at the same concentrations that were produced by E. faecium. Finally, nematode killing was prevented by addition of catalase to cultures of E. faecium that killed the worms.
When our laboratory originally developed the Enterococcus-C. elegans model system, we observed that C. elegans could use E. faecium as a food source and have a life span that is only slightly shorter than that of nematodes grown on the normal food source (E. coli on NGM medium). Because E. faecium is able to cause severe and potentially fatal infections, we hypothesized that the original E. faecium-C. elegans system lacked elements required for E. faecium pathogenicity. We tried growing E. faecium under a variety of different conditions, and we found that E. faecium acquires C. elegans killing activity after the bacteria are grown on solid BHI medium under anaerobic conditions. Initially, we hypothesized that E. faecium was producing a toxin during the anaerobic growth phase, but in actuality, the toxin hydrogen peroxide was not produced until the lawns of E. faecium were exposed to oxygen.
Hydrogen peroxide is scavenged by NADH peroxidase, which is expressed under aerobic conditions due to transcriptional regulation by OxyR (47). Under anaerobic conditions, npr is transcribed at very low levels in E. faecium. We speculate that during the C. elegans killing assay with anaerobically grown E. faecium, the abrupt exposure to atmospheric oxygen allows E. faecium to produce and accumulate large amounts of hydrogen peroxide because the low levels of NADH peroxidase in the cells cannot adequately scavenge hydrogen peroxide. On the other hand, anaerobic growth of E. faecium is not required for hydrogen peroxide production in liquid cultures. We have found that E. faecium produces high levels of hydrogen peroxide as aerobic cultures enter the stationary phase. Previous studies that only examined logarithmically growing cultures may have overlooked the production of hydrogen peroxide by E. faecium (39).
In contrast to E. faecium, E. faecalis does not kill C. elegans by a fast, toxin-mediated mechanism, and E. faecalis does not produce detectable amounts of hydrogen peroxide under our assay conditions. One difference between the two enterococcal species is that E. faecium is catalase negative and E. faecalis is catalase positive when the organisms are grown on media such as BHI medium containing hematin (14, 42). Another difference between E. faecium and E. faecalis is that E. faecalis is capable of respiration. Although E. faecalis possesses glycerol-3-phosphate oxidase and can produce hydrogen peroxide when glycerol is its sole carbon source (41), it appears that E. faecalis prefers to use respiration to meet its energetic needs. While E. faecalis did not produce detectable levels of hydrogen peroxide (≤25 μM) under the conditions which we used in this study, E. faecalis produces extracellular superoxide when it is grown in the presence of molecular oxygen and in the absence of hematin and fumurate (25). In rat intestines colonized by E. faecalis, there is sufficient molecular oxygen to allow the production of superoxide, and Hucyke et al. have postulated that E. faecalis is a potential source of oxidative stress for intestinal cells which may contribute to carcinogenesis (24, 25). Our data suggest that E. faecium may be another source of oxidative stress for intestinal cells.
E. faecium is a facultative anaerobe whose metabolism relies exclusively on glycolysis and fermentation (23). In anaerobic environments, lactate fermentation is used to regenerate NADH. In aerobic environments, NADH is regenerated by water-forming NADH oxidases, thereby allowing the use of mixed acid fermentation. Another type of NADH oxidase catalyzes the two-electron reduction of molecular oxygen, resulting in hydrogen peroxide formation (21, 40). The hydrogen peroxide-forming NADH oxidase works in concert with the AhpC protein to scavenge alky hydroperoxides (21, 40). In this study, we found an E. faecium nox mutant, 2F22, that produces less hydrogen peroxide. Although our results indicate that a nox gene is mutated in 2F22, the gene appears to encode a water-forming NADH oxidase, and we believe that the decreased hydrogen peroxide production in this mutant is a secondary phenotype due to alterations in energy metabolism. First, the amino acid sequence of the protein encoded by the mutated nox gene is more similar to the sequences of previously studied water-forming NADH oxidases than to sequences of hydrogen peroxide-forming NADH oxidases (48). Second, the growth defect phenotypes of mutant 2F22 are similar to the phenotypes of streptococcal strains mutated in the water-forming NADH oxidase (21, 62). Third, we were not able to detect hydrogen peroxide in the NADH oxidase assays of extracts of wild-type cells.
The release of hydrogen peroxide by bacteria has been shown to inhibit or kill other competing bacteria or host cells. This may be beneficial for both the host and the bacteria, as in the case of Lactobacillus species colonizing the vaginal tract, where hydrogen peroxide synthesized by Lactobacillus inhibits the growth of pathogenic species such as S. aureus, Neisseria gonorrhoeae, and Gardnerella vaginalis (20, 36, 54). Alternatively, hydrogen peroxide production can be detrimental to the host, as in the case of S. pneumoniae, in which hydrogen peroxide inhibits the growth of Haemonphilus influenzae, thereby allowing S. pneumoniae to colonize the respiratory tract without competition from other pathogens (39). The release of hydrogen peroxide by S. pneumoniae can also damage host cells and has been shown to contribute to the induction of apoptosis of neuronal cells in a meningitis infection model (6, 10, 22). Interestingly, similar to our results, Bolm et al. have shown that many streptococcal species, including S. pneumoniae and S. pyogenes, kill C. elegans by producing hydrogen peroxide (5, 27).
It appears that hydrogen peroxide generation is a common trait among catalase-negative streptococci and the related enterococci, and it seems likely that the hydrogen peroxide released by E. faecium damages nearby host cells. However, the relatively low levels of hydrogen peroxide released from E. faecium may be difficult to detect in vivo. Similarly, it may be difficult to determine whether hydrogen peroxide production plays an important role in E. faecium pathogenesis. To date, controlled genetic manipulation of pathogenic E. faecium strains has not been achieved, and no E. faecium mutants have been found to be attenuated in a mammalian infection model. The difficulties in studying E. faecium prompted the development of a C. elegans pathogenesis model as a system to discover potential virulence factors. Use of this simple model led us to the finding that E. faecium produces hydrogen peroxide at levels that are high enough to kill C. elegans. Additional studies are necessary to determine the significance of hydrogen peroxide in E. faecium pathogenesis in mammalian hosts.
Acknowledgments
We thank G. M. Eliopoulos, M. S. Gilmore, B. E. Murray, and R. J. Willems for generous gifts of strains and advice. We thank Johannes Huebner, Markus Hufnagel, and Stephanie Koch for their help.
This work was supported by a grant from Aventis SA to F.M.A. and S.B.C., by NIH grant GM48707, and by postdoctoral fellowships from the Howard Hughes Medical Institute (to E.M.) and the American Cancer Society (grant PF-02-130-01-MBC to T.I.M.).
Editor: J. T. Barbieri
REFERENCES
- 1.Aballay, A., P. Yorgey, and F. M. Ausubel. 2000. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr. Biol. 10:1539-1542. [DOI] [PubMed] [Google Scholar]
- 2.Arduino, R. C., K. Jacques-Palaz, B. E. Murray, and R. M. Rakita. 1994. Resistance of Enterococcus faecium to neutrophil-mediated phagocytosis. Infect. Immun. 62:5587-5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1996. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 4.Axelsen, K. B., and M. G. Palmgren. 1998. Evolution of substrate specificities in the P-type ATPase superfamily. J. Mol. Evol. 46:84-101. [DOI] [PubMed] [Google Scholar]
- 5.Bolm, M., W. T. Jansen, R. Schnabel, and G. S. Chhatwal. 2004. Hydrogen peroxide-mediated killing of Caenorhabditis elegans: a common feature of different streptococcal species. Infect. Immun. 72:1192-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun, J. S., J. E. Sublett, D. Freyer, T. J. Mitchell, J. L. Cleveland, E. I. Tuomanen, and J. R. Weber. 2002. Pneumococcal pneumolysin and H2O2 mediate brain cell apoptosis during meningitis. J. Clin. Investig. 109:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics 77:71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charrier, V., E. Buckley, D. Parsonage, A. Galinier, E. Darbon, M. Jaquinod, E. Forest, J. Deutscher, and A. Claiborne. 1997. Cloning and sequencing of two enterococcal glpK genes and regulation of the encoded glycerol kinases by phosphoenolpyruvate-dependent, phosphotransferase system-catalyzed phosphorylation of a single histidyl residue. J. Biol. Chem. 272:14166-14174. [DOI] [PubMed] [Google Scholar]
- 9.Darby, C., C. L. Cosma, J. H. Thomas, and C. Manoil. 1999. Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:15202-15207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duane, P. G., J. B. Rubins, H. R. Weisel, and E. N. Janoff. 1993. Identification of hydrogen peroxide as a Streptococcus pneumoniae toxin for rat alveolar epithelial cells. Infect. Immun. 61:4392-4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eaton, T. J., and M. J. Gasson. 2002. A variant enterococcal surface protein Esp(fm) in Enterococcus faecium; distribution among food, commensal, medical, and environmental isolates. FEMS Microbiol. Lett. 216:269-275. [DOI] [PubMed] [Google Scholar]
- 12.Eliopoulos, G. M., C. Wennersten, S. Zighelboim-Daum, E. Reiszner, D. Goldmann, and R. C. Moellering, Jr. 1988. High-level resistance to gentamicin in clinical isolates of Streptococcus (Enterococcus) faecium. Antimicrob. Agents Chemother. 32:1528-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esders, T. W., and C. A. Michrina. 1979. Purification and properties of l-alpha-glycerophosphate oxidase from Streptococcus faecium ATCC 12755. J. Biol. Chem. 254:2710-2715. [PubMed] [Google Scholar]
- 14.Frankenberg, L., M. Brugna, and L. Hederstedt. 2002. Enterococcus faecalis heme-dependent catalase. J. Bacteriol. 184:6351-6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friesenegger, A., S. Fiedler, L. A. Devriese, and R. Wirth. 1991. Genetic transformation of various species of Enterococcus by electroporation. FEMS Microbiol. Lett. 63:323-327. [DOI] [PubMed] [Google Scholar]
- 16.Gallagher, L. A., and C. Manoil. 2001. Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J. Bacteriol. 183:6207-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garsin, D. A., C. D. Sifri, E. Mylonakis, X. Qin, K. V. Singh, B. E. Murray, S. B. Calderwood, and F. M. Ausubel. 2001. A simple model host for identifying Gram-positive virulence factors. Proc. Natl. Acad. Sci. USA 98:10892-10897. Epub 2001 Sep 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilmore, M. S., P. S. Coburn, S. R. Nallapareddy, and B. E. Murray. 2002. Enterococcal virulence, p. 301-354. In M. S. Gilmore, D. B. Clewell, P. Courvalin, G. M. Dunny, B. E. Murray, and L. B. Rice (ed.), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, D.C.
- 19.Gutierrez, J. A., P. J. Crowley, D. P. Brown, J. D. Hillman, P. Youngman, and A. S. Bleiweis. 1996. Insertional mutagenesis and recovery of interrupted genes of Streptococcus mutans by using transposon Tn917: preliminary characterization of mutants displaying acid sensitivity and nutritional requirements. J. Bacteriol. 178:4166-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hawes, S. E., S. L. Hillier, J. Benedetti, C. E. Stevens, L. A. Koutsky, P. Wølner-Hanssen, and K. K. Holmes. 1996. Hydrogen peroxide-producing lactobacilli and acquisition of vaginal infections. J. Infect. Dis. 174:1058-1063. [DOI] [PubMed] [Google Scholar]
- 21.Higuchi, M., Y. Yamamoto, L. B. Poole, M. Shimada, Y. Sato, N. Takahashi, and Y. Kamio. 1999. Functions of two types of NADH oxidases in energy metabolism and oxidative stress of Streptococcus mutans. J. Bacteriol. 181:5940-5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirst, R. A., K. S. Sikand, A. Rutman, T. J. Mitchell, P. W. Andrew, and C. O'Callaghan. 2000. Relative roles of pneumolysin and hydrogen peroxide from Streptococcus pneumoniae in inhibition of ependymal ciliary beat frequency. Infect. Immun. 68:1557-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huycke, M. M. 2002. Physiology of enterococci, p. 133-175. In M. S. Gilmore, D. B. Clewell, P. Courvalin, G. M. Dunny, B. E. Murray, and L. B. Rice (ed.), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, D.C.
- 24.Huycke, M. M., V. Abrams, and D. R. Moore. 2002. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis 23:529-536. [DOI] [PubMed] [Google Scholar]
- 25.Huycke, M. M., D. Moore, W. Joyce, P. Wise, L. Shepard, Y. Kotake, and M. S. Gilmore. 2001. Extracellular superoxide production by Enterococcus faecalis requires demethylmenaquinone and is attenuated by functional terminal quinol oxidases. Mol. Microbiol. 42:729-740. [DOI] [PubMed] [Google Scholar]
- 26.Jander, G., L. G. Rahme, and F. M. Ausubel. 2000. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J. Bacteriol. 182:3843-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jansen, W. T., M. Bolm, R. Balling, G. S. Chhatwal, and R. Schnabel. 2002. Hydrogen peroxide-mediated killing of Caenorhabditis elegans by Streptococcus pyogenes. Infect. Immun. 70:5202-5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuroda, M., H. Hayashi, and T. Ohta. 1999. Chromosome-determined zinc-responsible operon czr in Staphylococcus aureus strain 912. Microbiol. Immunol. 43:115-125. [DOI] [PubMed] [Google Scholar]
- 29.Leavis, H., J. Top, N. Shankar, K. Borgen, M. Bonten, J. van Embden, and R. J. Willems. 2004. A novel putative enterococcal pathogenicity island linked to the esp virulence gene of Enterococcus faecium and associated with epidemicity. J. Bacteriol. 186:672-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez de Felipe, F., M. Kleerebezem, W. M. de Vos, and J. Hugenholtz. 1998. Cofactor engineering: a novel approach to metabolic engineering in Lactococcus lactis by controlled expression of NADH oxidase. J. Bacteriol. 180:3804-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahajan-Miklos, S., M. W. Tan, L. G. Rahme, and F. M. Ausubel. 1999. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96:47-56. [DOI] [PubMed] [Google Scholar]
- 32.Murray, B. E., K. V. Singh, R. P. Ross, J. D. Heath, G. M. Dunny, and G. M. Weinstock. 1993. Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function. J. Bacteriol. 175:5216-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mylonakis, E., F. M. Ausubel, J. R. Perfect, J. Heitman, and S. B. Calderwood. 2002. Killing of Caenorhabditis elegans by Cryptococcus neoformans as a model of yeast pathogenesis. Proc. Natl. Acad. Sci. USA 99:15675-15680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nallapareddy, S. R., G. M. Weinstock, and B. E. Murray. 2003. Clinical isolates of Enterococcus faecium exhibit strain-specific collagen binding mediated by Acm, a new member of the MSCRAMM family. Mol. Microbiol. 47:1733-1747. [DOI] [PubMed] [Google Scholar]
- 35.National Nosocomial Infections Surveillance System. 1997. National Nosocomial Infections Surveillance (NNIS) report, data summary from October 1986-April 1997, issued May 1997. A report from the NNIS System. Am. J. Infect. Control 25:477-487. [PubMed] [Google Scholar]
- 36.Ocaña, V. S., A. A. de Ruiz Holgado, and M. E. Nader-Macías. 1999. Growth inhibition of Staphylococcus aureus by H2O2-producing Lactobacillus paracasei subsp. paracasei isolated from the human vagina. FEMS Immunol. Med. Microbiol. 23:87-92. [DOI] [PubMed] [Google Scholar]
- 37.Ogg, S., S. Paradis, S. Gottlieb, G. I. Patterson, L. Lee, H. A. Tissenbaum, and G. Ruvkun. 1997. The fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans Nature 389:994-999. [DOI] [PubMed] [Google Scholar]
- 38.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to study of biofilms. Methods Enzymol. 310:91-109. [DOI] [PubMed] [Google Scholar]
- 39.Pericone, C. D., K. Overweg, P. W. Hermans, and J. N. Weiser. 2000. Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect. Immun. 68:3990-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poole, L. B., M. Higuchi, M. Shimada, M. L. Calzi, and Y. Kamio. 2000. Streptococcus mutans H2O2-forming NADH oxidase is an alkyl hydroperoxide reductase protein. Free Radic. Biol. Med. 28:108-120. [DOI] [PubMed] [Google Scholar]
- 41.Pugh, S. Y., and C. J. Knowles. 1982. Growth of Streptococcus faecalis var. zymogenes on glycerol: the effect of aerobic and anaerobic growth in the presence and absence of haematin on enzyme synthesis. J. Gen. Microbiol. 128:1009-1017. [DOI] [PubMed] [Google Scholar]
- 42.Pugh, S. Y., and C. J. Knowles. 1983. Synthesis of catalase by “Streptococcus faecalis subsp. zymogenes.” Arch. Microbiol. 136:60-63. [DOI] [PubMed] [Google Scholar]
- 43.Rahme, L. G., F. M. Ausubel, H. Cao, E. Drenkard, B. C. Goumnerov, G. W. Lau, S. Mahajan-Miklos, J. Plotnikova, M. W. Tan, J. Tsongalis, C. L. Walendziewicz, and R. G. Tompkins. 2000. Plants and animals share functionally common bacterial virulence factors. Proc. Natl. Acad. Sci. USA 97:8815-8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902. [DOI] [PubMed] [Google Scholar]
- 45.Rice, L. B., L. Carias, S. Rudin, C. Vael, H. Goossens, C. Konstabel, I. Klare, S. R. Nallapareddy, W. Huang, and B. E. Murray. 2003. A potential virulence gene, hylEfm, predominates in Enterococcus faecium of clinical origin. J. Infect. Dis. 187:508-512. [DOI] [PubMed] [Google Scholar]
- 46.Ross, R. P., and A. Claiborne. 1991. Cloning, sequence and overexpression of NADH peroxidase from Streptococcus faecalis 10C1. Structural relationship with the flavoprotein disulfide reductases. J. Mol. Biol. 221:857-871. [DOI] [PubMed] [Google Scholar]
- 47.Ross, R. P., and A. Claiborne. 1997. Evidence for regulation of the NADH peroxidase gene (npr) from Enterococcus faecalis by OxyR. FEMS Microbiol. Lett. 151:177-183. [DOI] [PubMed] [Google Scholar]
- 48.Ross, R. P., and A. Claiborne. 1992. Molecular cloning and analysis of the gene encoding the NADH oxidase from Streptococcus faecalis 10C1. Comparison with NADH peroxidase and the flavoprotein disulfide reductases. J. Mol. Biol. 227:658-671. [DOI] [PubMed] [Google Scholar]
- 49.Sahm, D. F., J. Kissinger, M. S. Gilmore, P. R. Murray, R. Mulder, J. Solliday, and B. Clarke. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33:1588-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sahm, D. F., M. K. Marsilio, and G. Piazza. 1999. Antimicrobial resistance in key bloodstream bacterial isolates: electronic surveillance with the Surveillance Network Database—USA. Clin. Infect. Dis. 29:259-263. [DOI] [PubMed] [Google Scholar]
- 51.Sifri, C. D., J. Begun, F. M. Ausubel, and S. B. Calderwood. 2003. Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis. Infect. Immun. 71:2208-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silver, S., and M. Walderhaug. 1992. Gene regulation of plasmid- and chromosome-determined inorganic ion transport in bacteria. Microbiol. Rev. 56:195-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spellerberg, B., D. R. Cundell, J. Sandros, B. J. Pearce, I. Idänpään-Heikkilä, C. Rosenow, and H. R. Masure. 1996. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol. Microbiol. 19:803-813. [DOI] [PubMed] [Google Scholar]
- 54.St. Amant, D. C., I. E. Valentin-Bon, and A. E. Jerse. 2002. Inhibition of Neisseria gonorrhoeae by Lactobacillus species that are commonly isolated from the female genital tract. Infect. Immun. 70:7169-7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stohs, S. J., and D. Bagchi. 1995. Oxidative mechanisms in the toxicity of metal ions. Free Radic. Biol. Med. 18:321-336. [DOI] [PubMed] [Google Scholar]
- 56.Sulston, J., and J. Hodgkin 1988. Methods, p. 587-606. In W. B. Wood (ed.), The nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 57.Tan, M. W., S. Mahajan-Miklos, and F. M. Ausubel. 1999. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. USA 96:715-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Teng, F., M. Kawalec, G. M. Weinstock, W. Hryniewicz, and B. E. Murray. 2003. An Enterococcus faecium secreted antigen, SagA, exhibits broad-spectrum binding to extracellular matrix proteins and appears essential for E. faecium growth. Infect. Immun. 71:5033-5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Teng, F., B. E. Murray, and G. M. Weinstock. 1998. Conjugal transfer of plasmid DNA from Escherichia coli to enterococci: a method to make insertion mutations. Plasmid 39:182-186. [DOI] [PubMed] [Google Scholar]
- 60.Vanfleteren, J. R., and A. De Vreese. 1995. The gerontogenes age-1 and daf-2 determine metabolic rate potential in aging Caenorhabditis elegans. FASEB J. 9:1355-1361. [DOI] [PubMed] [Google Scholar]
- 61.Willems, R. J., W. Homan, J. Top, M. van Santen-Verheuvel, D. Tribe, X. Manzioros, C. Gaillard, C. M. Vandenbroucke-Grauls, E. M. Mascini, E. van Kregten, J. D. van Embden, and M. J. Bonten. 2001. Variant esp gene as a marker of a distinct genetic lineage of vancomycin-resistant Enterococcus faecium spreading in hospitals. Lancet 357:853-855. [DOI] [PubMed] [Google Scholar]
- 62.Yu, J., A. P. Bryant, A. Marra, M. A. Lonetto, K. A. Ingraham, A. F. Chalker, D. J. Holmes, D. Holden, M. Rosenberg, and D. McDevitt. 2001. Characterization of the Streptococcus pneumoniae NADH oxidase that is required for infection. Microbiology 147:431-438. [DOI] [PubMed] [Google Scholar]