Abstract
Background
Methods for parentage control in cattle have changed since their initial implementation in the late 1950’s from blood group typing to more current single nucleotide polymorphism determination. In the early 1990’s, 12 microsatellites were selected by the International Society for Animal Genetics based on their informativeness and robustness in a variety of different cattle breeds. Since then this panel is used as standard in cattle herd book breeding and its application is accompanied by recurrent international comparison tests ensuring permanent validity for the most common commercial dairy and beef cattle breeds for example Holstein Friesian, Simmental, Angus, and Hereford. Although, nearly every parentage can be resolved using these microsatellites, cases with very close relatives became an emerging resolution problem during recent years. This is mainly due to an increase of monomorphism and a trend to the fixation of alleles, although no direct selection against their variability was applied. Thus other effects must be presumed resulting in a loss of polymorphism information content, heterozygosity, and exclusion probabilities.
Results
To determine changes of allele frequencies and exclusion probabilities, we analyzed the development of these parameters for the 12 microsatellites from 2004 to 2014. One hundred sixty eight thousand recorded Holstein Friesian cattle genotypes were evaluated. During this period certain alleles of nine microsatellites increased significantly (t-values >5). When calculating the exclusion probabilities for 11 microsatellites, reduction was determined for the three situations, i.e. one parent is wrongly identified (p = 0.01), both parents are wrongly identified (p = 0.005), and the genotype of one parent is missing (p = 0.048). With the addition of BM1818 to the marker set in 2009, this development was corrected leading to significant increases in exclusion probabilities. Although, the exclusion probabilities for the three family situations using the 12 microsatellites are >99 %, the clarification of 142 relationships in 40,000 situations where one parent is missing will still be impossible.
Twenty-five sires were identified that are responsible for the most significant microsatellite allele increases in the population. The corresponding alleles are mainly associated with milk protein and fat yield, body weight at birth and weaning, as well as somatic cell score, milk fat percentage, and longissimus muscle area.
Conclusions
Our data show that most of the microsatellites used for parentage control in cattle show directional changes in allele frequencies consistent with the history of artificial selection in the German Holstein population.
Keywords: Parentage control, Cattle, Holstein Friesian, Microsatellite, Single nucleotide polymorphism, Allele frequency, Exclusion probability, Hitchhiking
Background
Parentage control and traceability is an important issue in animal production and usually obligatory for animals used in breeding programs [1–6]. For routine parentage diagnosis, analyses should fulfill a variety of technical requirements, for instance easy handling, robustness, reproducibility, standardization, possibility of automation, short processing time, and reasonable costs. However, the most important prerequisite for markers used in parentage control is the ability to discriminate between even very close relatives [3]. Therefore, during the last seven decades methods for parentage control in cattle have changed considerably. In the early 1940’s and late 1950’s cattle blood groups were identified and shown to be useful in parentage control [7–10]. However, due to intensive inbreeding in the Holstein Friesian population and limited variability, blood groups became increasingly uninformative over time. Hence, approximately 40 years later, with the rapid development of molecular biological techniques and genome data, blood group typing was replaced by the use of highly polymorphic DNA markers, so-called mini- and microsatellites. The use of minisatellites in DNA fingerprinting and identification of individuals was first described in humans [11, 12], rapidly also entering the area of domestic animal identification and pedigree analysis [13]. Initial steps in using mini- and microsatellites in cattle identification and parentage control were done only a few years later [14, 15] and further actions were taken to establish a robust and internationally comparable panel of markers [16–20]. In international comparison tests under the direction of the International Society for Animal Genetics (ISAG) a panel of at least 12 microsatellite markers (short tandem repeats-STR) was established for parentage control in cattle. The 12 markers comprised BM1814, BM1818, BM2113, ETH3, ETH10, ETH225, INRA023, SPS115, TGLA53, TGLA122, TGLA126, TGLA227 [21–27]. Since the mid 1990’s this panel is used worldwide for parentage control and after approximately 15 years this is now in the process of being replaced again by the use of 100 and/or 200 single nucleotide polymorphisms (SNP). These SNPs are a sub-set taken from markers used for genomic selection or genome wide association analysis [28–30].
However, microsatellites are still the gold standard for parentage control in most breeding programs of beef as well as dairy cattle, based on the ease of testing, testing availability and million of results in the breeding databases. In this context it is important to review continuously the genetic variability and exclusion probabilities of the applied microsatellites [17, 19]. Ideally, microsatellites should be neutral DNA markers maintaining their characteristics relatively constant. Neutral DNA markers are solely subjected to stochastic processes such as mutation and genetic drift [31]. However, several of the microsatellites in the ISAG parentage control panel are under artificial selection and therefore actually not completely neutral. ETH10 on bovine chromosome 5 for example seems to be associated with growth and carcass traits in Angus, Brangus, and other cattle breeds [32, 33]. The ETH10 locus was also associated with coat colour in a Charolais x Holstein resource population and arachnomelia in Brown Swiss cattle [34, 35]. BM1818 was shown to be associated with somatic cell score (SCS) and specific alleles of this microsatellite are either favourable or unfavourable for mastitis resistance [36]. In another study, significant differences in allelic frequencies for BM1824, ETH10, INRA023, SPS115 and TGLA53 alleles were described in lines of Japanese Black cattle depending on selection of sires for intramascular fat [37]. It must therefore be assumed that due to selection for specific traits the variability and exclusion probabilities will decrease. As a consequence, the microsatellite panel will become increasingly uninformative especially in situations where very close relatives have to be tested. This seems in particular foreseeable in livestock with an active and well established breeding program, where certain sires can become predominant, if their breeding value is exceptional. To prove this hypothesis, we have evaluated the development in allele distribution of the internationally used STR markers in the German Holstein population over the last decade.
Results
Microsatellite genotypes of the German Holstein Friesian population (GHF) were analyzed, generated in the frame of routine parentage control using the standardized microsatellite panel recommended by ISAG. From 2004 to 2008, no data were available for BM1818, which was added to the panel only in 2009. Table 1 shows the number and lengths of alleles (standardized according to animal No. 13 of the ISAG cattle comparison test 2005) detected for each microsatellite marker in GHF together with the respective repeat numbers that have been determined by sequencing elsewhere [38]. For microsatellites BM1818, BM2113, ETH3 and TGLA227 several alleles that have been described previously were not detected in the GHF. On the other hand, a larger number of markers i.e. BM1824, ETH10, ETH225, INRA023, SPS115, TGLA122, TGLA126, TGLA227 and TGLA53, showed alleles that have not yet been described.
Table 1.
Microsatellites alleles (bp) detected in the Holstein Friesian population
| Microsatellite allele (bp)a | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Repeat number | 1b | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| 9 | 71 | |||||||||||
| 10 | 174 d | 192 | ||||||||||
| 11 | 194 | 75 | ||||||||||
| 12 | 178 | 121 | 196 | 105 | 77 | |||||||
| 13 | 256 c | 180 | 123 | 198 | 107 | 79 | ||||||
| 14 | 258 | 182 | 125 | 209 | 200 | 137 | 109 | 81 | ||||
| 15 | 260 | 184 | 127 | 103 | 211 | 202 | 139 | 111 | 83 | |||
| 16 | 262 | 186 | 129 | 213 | 204 | 141 | 113 | 85 | ||||
| 17 | 264 | 188 | 131 | 215 | 206 | 240 | 143 | 115 | 87 | 150 | ||
| 18 | 266 | 190 | 133 | 109 | 217 | 208 | 145 | 117 | 89 | 152 | ||
| 19 | 268 | 135 | 219 | 140 | 210 | 244 | 147 | 119 | 91 | 154 | ||
| 20 | 270 | 137 | 221 | 142 | 212 | 246 | 149 | 121 | 93 | 156 | ||
| 21 | 272 | 139 | 115 | 223 | 144 | 214 | 248 | 151 | 123 | 95 | 158 | |
| 22 | 141 | 117 | 225 | 146 | 216 | 250 | 153 | 125 | 97 | 160 | ||
| 23 | 143 | 119 | 148 | 218 | 252 | 155 | 99 | 162 | ||||
| 24 | 121 | 150 | 220 | 254 | 157 | 101 | 164 | |||||
| 25 | 280 | 123 | 152 | 222 | 256 | 159 | 103 | 166 | ||||
| 26 | 125 | 154 | 258 | 161 | 168 | |||||||
| 27 | 127 | 156 | 260 | 163 | 170 | |||||||
| 28 | 129 | 158 | 262 | 165 | 172 | |||||||
| 29 | 131 | 160 | 167 | 174 | ||||||||
| 30 | 169 | 176 | ||||||||||
| 31 | 171 | 178 | ||||||||||
| 32 | 173 | 180 | ||||||||||
| 33 | 175 | 182 | ||||||||||
| 34 | 177 | 184 | ||||||||||
| 35 | 179 | 186 | ||||||||||
| 36 | 181 | 188 | ||||||||||
| 37 | 183 | 190 | ||||||||||
| 38 | 185 | 192 | ||||||||||
| 39 | ||||||||||||
| 40 | 196 | |||||||||||
aAllele sizes are standardized according animal No. 13 of the ISAG cattle comparison test 2005. b1: BM1818, 2: BM1824, 3: BM2113, 4: ETH3, 5: ETH10, 6: ETH225, 7: INRA023, 8: SPS115, 9: TGLA122, 10: TGLA126, 11: TGLA227, 12: TGLA53. cAlleles that have been described, but are not present in the HF population are shown in bold. dAlleles that have not been described, but are present in the HF population are italicized
Allele frequencies for all markers that were calculated for each year and alleles with significant changes in frequencies during the 11 years are shown in Figs. 1, 2, 3 and 4. Those alleles showing a significant trend during the observation period are summarized in Table 2. All microsatellites showed either increases or decreases of specific alleles during the analyzed period. Only BM2113, showed no significant frequency increase of a single allele. For all other markers at least one allele increased significantly over the evaluated 11 years period.
Fig. 1.
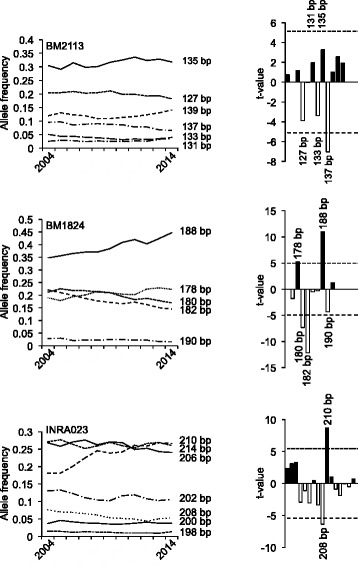
Development of allele frequencies of microsatellite markers BM2113, BM1824, and INRA023 from 2004 to 2014. The left side of the figure illustrates the development of the major alleles of the respective microsatellite markers from 2004 to 2014. The significance of the increases or decreases in allele frequencies was calculated as described above and is shown on the right side. Sizes of alleles with t-values >5 are indicated. Black bars correspond to alleles with increased, open bars correspond to alleles with decreased allele frequencies
Fig. 2.
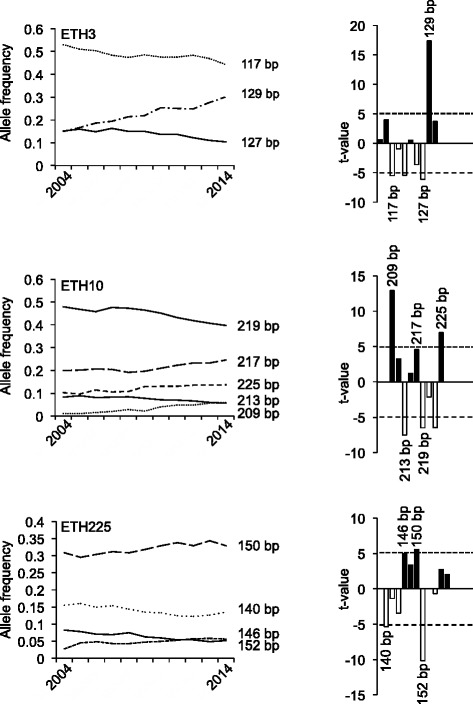
Development of allele frequencies of microsatellite markers ETH3, ETH10, and ETH225 from 2004 to 2014. The left side of the figure illustrates the development of the major alleles of the respective microsatellite markers from 2004 to 2014. The significance of the increases or decreases in allele frequencies was calculated as described above and is shown on the right side. Sizes of alleles with t-values >5 are indicated. Black bars correspond to alleles with increased, open bars correspond to alleles with decreased allele frequencies
Fig. 3.
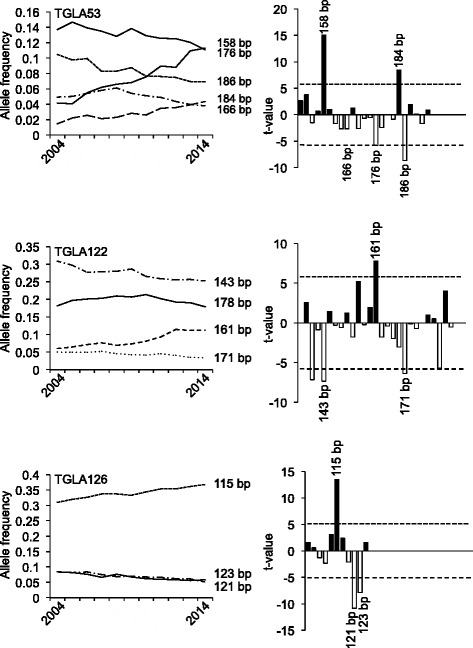
Development of allele frequencies of microsatellite markers TGLA53, TGLA122, and TGLA126 from 2004 to 2014. The left side of the figure illustrates the development of the major alleles of the respective microsatellite markers from 2004 to 2014. The significance of the increases or decreases in allele frequencies was calculated as described above and is shown on the right side. Sizes of alleles with t-values >5 are indicated. Black bars correspond to alleles with increased, open bars correspond to alleles with decreased allele frequencies
Fig. 4.
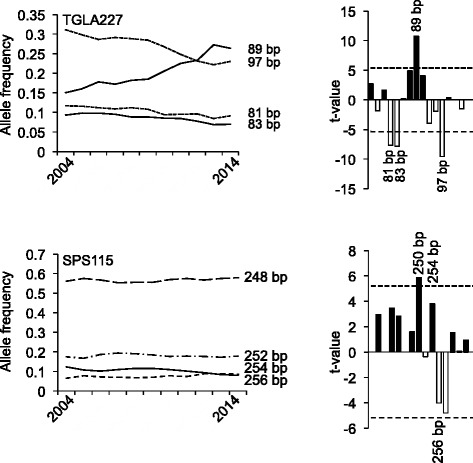
Development of allele frequencies of microsatellite markers TGLA227 and SPS115 from 2004 to 2014. The left side of the figure illustrates the development of the major alleles of the respective microsatellite markers from 2004 to 2014. The significance of the increases or decreases in allele frequencies was calculated as described above and is shown on the right side. Sizes of alleles with t-values >5 are indicated. Black bars correspond to alleles with increased, open bars correspond to alleles with decreased allele frequencies
Table 2.
Development of microsatellite allele frequencies from 2004 to 2014
| STRa | Repeat number | Allele (bp) | f(2014) b | t-valuec | t-valued |
|---|---|---|---|---|---|
| BM1824 | 17 | 188 | 0.446 | 10.99 | 8.73 |
| 12 | 178 | 0.223 | 5.25 | 3.85 | |
| ETH3 | 28 | 129 | 0.301 | 17.38 | 15.05 |
| ETH10 | 14 | 209 | 0.059 | 12.9 | 11.62 |
| 22 | 225 | 0.138 | 6.97 | 5.62 | |
| ETH225 | 24 | 150 | 0.329 | 5.57 | 3.68 |
| INRA023 | 19 | 210 | 0.267 | 8.67 | 7.23 |
| SPS115 | 22 | 250 | 0.002 | 5.86 | 5.28 |
| TGLA53 | 21 | 158 | 0.113 | 15.07 | 13.31 |
| 34 | 184 | 0.044 | 8.42 | 3.47 | |
| TGLA122 | 26 | 161 | 0.112 | 7.79 | 6.72 |
| TGLA126 | 17 | 115 | 0.681 | 13.45 | 9.65 |
| TGLA227 | 18 | 89 | 0.264 | 10.75 | 9.23 |
| BM1824 | 13 | 180 | 0.169 | −7.34 | −5.58 |
| 14 | 182 | 0.145 | −12.17 | −9.65 | |
| BM2113 | 20 | 137 | 0.066 | −7.04 | −5.38 |
| ETH3 | 22 | 117 | 0.443 | −5.04 | −3.95 |
| 24 | 121 | 0.009 | −5.43 | −4.29 | |
| 27 | 127 | 0.104 | −6.16 | −4.89 | |
| ETH10 | 16 | 213 | 0.056 | −7.53 | −5.96 |
| 19 | 219 | 0.397 | −6.50 | −4.94 | |
| 21 | 223 | 0.047 | −6.45 | −5.07 | |
| ETH225 | 19 | 140 | 0.135 | −5.36 | −3.93 |
| 25 | 152 | 0.052 | −10.18 | −8.16 | |
| INRA023 | 18 | 208 | 0.053 | −6.37 | −5.04 |
| TGLA53 | 30 | 176 | 0.111 | −5.84 | −4.10 |
| 35 | 186 | 0.069 | −8.65 | −3.56 | |
| TGLA122 | 15 | 139 | 0.001 | −7.16 | −6.14 |
| 17 | 143 | 0.252 | −7.35 | −5.23 | |
| 31 | 171 | 0.034 | −6.37 | −4.92 | |
| TGLA126 | 20 | 121 | 0.051 | −10.84 | −8.57 |
| 21 | 123 | 0.058 | −7.85 | −6.14 | |
| TGLA227 | 14 | 81 | 0.092 | −7.68 | −5.78 |
| 15 | 83 | 0.069 | −7.83 | −6.06 | |
| 22 | 97 | 0.231 | −9.57 | −7.62 |
a STR short tandem repeat, b f (2014) Allele frequency at the end of 2014. c0-hypothesis: slope = 0; dvs expected maximal random change
Those detected significant linear trends were compared to the theoretic values that could occur by random genetic drifts. For this, the expected development of allele frequencies in a random population over 10 years with an effective population size of 103 [39] was assessed and compared to the observed trends.
For all STR alleles that show a highly significant trend vs. a zero-slope, the significance was still <0.05 when compared to the maximum expected random slope (Table 2).
To analyze, whether the changes in allele frequencies had an influence on the informativeness of the marker panel, we calculated the exclusion probabilities (EP) as previously described [40]. In Fig. 5 the development of the EPs are shown. In three situations the EPs displayed a similar course over the years with a reduction in exclusion probabilities when using only the initially recommended 11 microsatellites. With the addition of BM1818 in 2009 the maker panel reached an acceptable level of EP again.
Fig. 5.
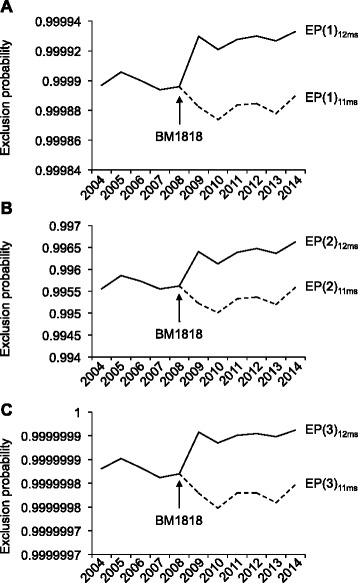
Development of the exclusion probabilities using 11 or 12 microsatellite markers from 2004 to 2014. Exclusion probabilities were calculated for the three family situations [40], (a) two parents and one offspring is given; exclude a parent, (b) one parent and one offspring is given; exclude their relationship, (c) two parents and one offspring are given; exclude both parents. The addition of marker BM1818 in 2009 is indicated with an arrow
In an approach to define the founder(s) of the detected changes in STR frequencies, we searched our database for all sires harboring alleles with the highest positive t-value, which are BM1824 (188 bp), ETH3 (129 bp), ETH10 (209 bp), INRA023 (210 bp), TGLA53 (158 bp), TGLA126 (115 bp) and TGLA227 (89 bp), and identified a total of 193 sires. The search criteria were then refined in a following step by the addition of the second most significantly increased alleles, i.e. ETH10 (225 bp), ETH225 (146 bp), SPS115 (250 bp), TGLA53 (184 bp), TGLA122 (161 bp) and BM1818 (268 bp). In addition alleles were included that showed an increasing tendency, i.e. BM2113 (135 bp), SPS115 (240 bp) and TGLA122 (183 bp). With these two refinements, the number of sires was reduced to 25 individuals.
Discussion
For almost two decades, microsatellites have been used in parentage control in cattle breeding under the assumption that these markers remain informative even under intensive inbreeding and selection, as being trait neutral. Currently even though still relatively rare, in recent years problems arose in parentage control whenever ancestries of closely related animals had to be determined. Hence, although microsatellites used for parentage control should be neutral DNA markers, the data presented here clearly show that this is not the case. This is supported by earlier analyses describing that ETH10, BM1818, BM1824, INRA023, SPS115 and TGLA53 are somehow associated with different economical important traits [32–37].
The data presented here also indicate that ETH3, ETH225, TGLA122, TGLA126 and TGLA227 are most likely influenced either by selective breeding or by hitchhiking effect. Although BM2113 did not show frequency increase of any single allele, the significant reduction of the allele with 137 bp in GHF (Fig. 1) might indicate an unfavourable effect of this allele in breeding. This can also be assumed for significantly reduced alleles of the other microsatellites in the population shown in Table 2.
To analyze whether the increase or decrease in allele frequencies was due to the fact that most of the microsatellite markers are associated with economical important traits, we looked for QTLs at the chromosomal locations of the microsatellites. At least 40 different QTLs have been described flanking the microsatellite chromosomal positions and the most frequent traits included milk protein yield, milk fat yield, somatic cell score, milk fat percentage, body weight at birth and body weight at weaning [41]. From this it can be hypothesized that the allele frequency changes over the last several years are–at least in part–a consequence of selection for these traits.
According to the German Holstein Friesian Association, 1.61 million HF cows were registered in 2012 in Germany [42]. If an estimated effective population size of 103 is assumed [39], the number of males (Nm) can be calculated to be 25.75 [43]. As this number seems to be rather low at a first sight, we wanted to see, whether a similar number would be obtained, when searching the GHF population for sires transmitting the most significantly increased alleles over the last decade (Figs. 1, 2, 3 and 4). Interestingly, this number agrees perfectly to the estimated number of males calculated from the effective population size. With the reverse search using the allele frequencies several famous HF sires were identified, e.g. Goldwin, Shottle, Hayden, Atwood, and Laudan.
Finally, we wanted to proof that the changes in allele frequencies are not due to random genetic drift. Therefore, we compared the expected development of allele frequencies in a random with the observed development in the GHF. The slopes calculated for both scenarios (genetic drift or hitchhiking) are significantly different and hence the process influencing the changes in allele frequencies is not only due to genetic drift.
Conclusions
In summary, we were able to show that the microsatellite markers recommended for parentage control in cattle are influenced by selective breeding and are therefore adaptive DNA markers. Nearly all of the microsatellites are located in QTL regions or are associated with genes influencing the breeding value. Consequently, during the last 10 years several alleles significantly increased or decreased in frequency resulting in a reduced overall informativeness and exclusion power of the marker panel. This problem can only be solved by the inclusion of additional markers to the panel. Similar recommendations can be given for the foreseeable exclusive use of SNPs in the near future. The evaluation and applicability of SNPs in parentage control has been shown in several studies [3, 30, 44–46]. However, it is also clear that the currently recommended minimal number of SNPs might not be sufficient to eliminate false-negative results [28, 47].
Methods
Ethical statement
Data are based on rotuine diagnostic parentage control performed with written owner consent. Collection of blood samples was conducted exclusively by local veterinarians. Blood sampling by veterinarians with state examination is in accordance with the German Animal Welfare Act (§6 Abs. 1 Satz 2 TierSchG). Therefore no formal ethical approval was required, since no other samples were collected for this study.
DNA samples and genotyping
A total of 168,000 Holstein Friesian cattle were genotyped. DNA from blood samples was extracted using a salting out procedure [48] or the MagNA Pure LC DNA Isolation Kit I (Roche Diagnostics). For the isolation of DNA from tissue/hair samples the DNeasy Blood and Tissue Kit (Qiagen) or the MagNA Pure LC DNA Isolation Kit II (Roche Diagnostics) was used according to the manufacturer’s protocols.
For genotyping the StockMarks® for Cattle Genotyping Kit (Life Technologies™) or after cessation of that a laboratory developed multiplex method was used and allele sizes were adjusted to the reference animal No. 13 from the ISAG cattle comparison test 2005. Reactions were separated on an ABI PRISM® 3130xl Genetic Analyzer (Life Technologies™) according to the manufacturers’ protocols. DNA profiles were recorded with Data Collection v3.1.1 and evaluted using GeneMapper v4.1 (Life Technologies™). From the database, the allelic frequencies were calculated on a yearly basis over the period from 2004 to 2014.
Statistical analysis
All statistical analyses were done with Microsoft® Excel® for Mac version 14.4.8 (150116). Exclusion probabilities were calculated as described previously [40]. The variance in allele frequency after t generations (Vt) was calculated as described [49]. The effective population size (Ne) of the German Holstein population was set to 103 in accordance to estimations based on linkage disequilibrium data published earlier [39].
For each of the STRs, a linear trend was assessed for any occurring allele using the least-square linear regression model. For statistical purposes, the usual definition of the regression t-value was used (slope/SEslope). Such a trend was considered significant if the calculated t-value (0-hypothesis as slope = 0) exceeded the corresponding p-value, considering the degrees of freedom (9) and after Bonferroni correction for the individual number of alleles for the STR. The calculation was performed against the maximum theoretical slope (in the same direction as the observed slope) based on Vt, using the observed standard error of the slope as denominator ([slopeobserved − slopeexpected]/SEslope(obs)).
Acknowledgements
The authors are thankful to Melanie Scharfenstein, Susann Loos, and Louisa Jüttner for excellent technical assistance. This work was supported by a grant of the Erxleben Research & Innovation Council to BB (ERIC-BR1959-2014-01).
Abbreviations
- EP
exclusion probability
- GHF
German Holstein Friesian
- HF
Holstein Friesian
- ISAG
International Society for Animal Genetics
- QTL
quantitative trait locus
- SCS
somatic cell score
- SNP
single nucleotide polymorphism
- STR
short tandem repeat
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
ES performed the statistical analyses. BB wrote the manuscript. Both authors read and approved the final version of the manuscript.
Contributor Information
Bertram Brenig, Phone: +49-551-3933383, Email: bbrenig@gwdg.de.
Ekkehard Schütz, Email: eschuet@gwdg.de.
References
- 1.Golden BL, Garrick DJ, Benyshek LL. Milestones in beef cattle genetic evaluation. J Anim Sci. 2009;87(14 Suppl):E3–10. doi: 10.2527/jas.2008-1430. [DOI] [PubMed] [Google Scholar]
- 2.Carolino I, Sousa CO, Ferreira S, Carolino N, Silva FS, Gama LT. Implementation of a parentage control system in Portuguese beef-cattle with a panel of microsatellite markers. Genet Mol Biol. 2009;32(2):306–11. doi: 10.1590/S1415-47572009005000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernandez ME, Goszczynski DE, Liron JP, Villegas-Castagnasso EE, Carino MH, Ripoli MV, et al. Comparison of the effectiveness of microsatellites and SNP panels for genetic identification, traceability and assessment of parentage in an inbred Angus herd. Genet Mol Biol. 2013;36(2):185–91. doi: 10.1590/S1415-47572013000200008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orru L, Napolitano F, Catillo G, Moioli B. Meat molecular traceability: how to choose the best set of microsatellites? Meat Sci. 2006;72(2):312–7. doi: 10.1016/j.meatsci.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 5.Vazquez JF, Perez T, Urena F, Gudin E, Albornoz J, Dominguez A. Practical application of DNA fingerprinting to trace beef. J Food Prot. 2004;67(5):972–9. doi: 10.4315/0362-028x-67.5.972. [DOI] [PubMed] [Google Scholar]
- 6.Dimauro C, Cellesi M, Steri R, Gaspa G, Sorbolini S, Stella A, et al. Use of the canonical discriminant analysis to select SNP markers for bovine breed assignment and traceability purposes. Anim Genet. 2013;44(4):377–82. doi: 10.1111/age.12021. [DOI] [PubMed] [Google Scholar]
- 7.Ferguson LC. The blood groups of cattle. J Am Vet Med Assoc. 1947;111(849):466–9. [PubMed] [Google Scholar]
- 8.Ferguson LC. Heritabe antigens in the erythrocytes of cattle. J Immun Balt. 1941;40:213–42. [Google Scholar]
- 9.Irwin MR, editor. Blood grouping and its utilization in animal breeding. VII International Congress of Animal Husbandry. Madrid: Executive Commission of the Congress; 1956. [Google Scholar]
- 10.Stormont C, editor. On the application of blood groups in animal breeding. X International Congress of Genetics. Montreal, Canada: University of Toronto Press; 1958. [Google Scholar]
- 11.Jeffreys AJ, Wilson V, Thein SL. Individual-specific ‘fingerprints’ of human DNA. Nature. 1985;316(6023):76–9. doi: 10.1038/316076a0. [DOI] [PubMed] [Google Scholar]
- 12.Jeffreys AJ, Wilson V, Thein SL. Hypervariable ‘minisatellite’ regions in human DNA. Nature. 1985;314(6006):67–73. doi: 10.1038/314067a0. [DOI] [PubMed] [Google Scholar]
- 13.Jeffreys AJ, Morton DB. DNA fingerprints of dogs and cats. Anim Genet. 1987;18(1):1–15. doi: 10.1111/j.1365-2052.1987.tb00739.x. [DOI] [PubMed] [Google Scholar]
- 14.Trommelen GJ, Den Daas JH, Vijg J, Uitterlinden AG. DNA profiling of cattle using micro- and minisatellite core probes. Anim Genet. 1993;24(4):235–41. doi: 10.1111/j.1365-2052.1993.tb00305.x. [DOI] [PubMed] [Google Scholar]
- 15.Trommelen GJ, Den Daas NH, Vijg J, Uitterlinden AG. Identity and paternity testing of cattle: application of a deoxyribonucleic acid profiling protocol. J Dairy Sci. 1993;76(5):1403–11. doi: 10.3168/jds.S0022-0302(93)77471-8. [DOI] [PubMed] [Google Scholar]
- 16.Glowatzki-Mullis ML, Gaillard C, Wigger G, Fries R. Microsatellite-based parentage control in cattle. Anim Genet. 1995;26(1):7–12. doi: 10.1111/j.1365-2052.1995.tb02612.x. [DOI] [PubMed] [Google Scholar]
- 17.Heyen DW, Beever JE, Da Y, Evert RE, Green C, Bates SR, et al. Exclusion probabilities of 22 bovine microsatellite markers in fluorescent multiplexes for semiautomated parentage testing. Anim Genet. 1997;28(1):21–7. doi: 10.1111/j.1365-2052.1997.t01-1-00057.x. [DOI] [PubMed] [Google Scholar]
- 18.Kemp SJ, Hishida O, Wambugu J, Rink A, Longeri ML, Ma RZ, et al. A panel of polymorphic bovine, ovine and caprine microsatellite markers. Anim Genet. 1995;26(5):299–306. doi: 10.1111/j.1365-2052.1995.tb02663.x. [DOI] [PubMed] [Google Scholar]
- 19.Peelman LJ, Mortiaux F, Van Zeveren A, Dansercoer A, Mommens G, Coopman F, et al. Evaluation of the genetic variability of 23 bovine microsatellite markers in four Belgian cattle breeds. Anim Genet. 1998;29(3):161–7. doi: 10.1111/j.1365-2052.1998.00280.x. [DOI] [PubMed] [Google Scholar]
- 20.Ma RZ, Russ I, Park C, Heyen DW, Beever JE, Green CA, et al. Isolation and characterization of 45 polymorphic microsatellites from the bovine genome. Anim Genet. 1996;27(1):43–7. doi: 10.1111/j.1365-2052.1996.tb01175.x. [DOI] [PubMed] [Google Scholar]
- 21.Barendse W, Armitage SM, Kossarek LM, Shalom A, Kirkpatrick BW, Ryan AM, et al. A genetic linkage map of the bovine genome. Nat Genet. 1994;6(3):227–35. doi: 10.1038/ng0394-227. [DOI] [PubMed] [Google Scholar]
- 22.Bishop MD, Kappes SM, Keele JW, Stone RT, Sunden SL, Hawkins GA, et al. A genetic linkage map for cattle. Genetics. 1994;136(2):619–39. doi: 10.1093/genetics/136.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kappes SM, Keele JW, Stone RT, McGraw RA, Sonstegard TS, Smith TP, et al. A second-generation linkage map of the bovine genome. Genome Res. 1997;7(3):235–49. doi: 10.1101/gr.7.3.235. [DOI] [PubMed] [Google Scholar]
- 24.Moore SS, Byrne K, Berger KT, Barendse W, McCarthy F, Womack JE, et al. Characterization of 65 bovine microsatellites. Mamm Genome. 1994;5(2):84–90. doi: 10.1007/BF00292333. [DOI] [PubMed] [Google Scholar]
- 25.Steffen P, Eggen A, Dietz AB, Womack JE, Stranzinger G, Fries R. Isolation and mapping of polymorphic microsatellites in cattle. Anim Genet. 1993;24(2):121–4. doi: 10.1111/j.1365-2052.1993.tb00252.x. [DOI] [PubMed] [Google Scholar]
- 26.Toldo SS, Fries R, Steffen P, Neibergs HL, Barendse W, Womack JE, et al. Physically mapped, cosmid-derived microsatellite markers as anchor loci on bovine chromosomes. Mamm Genome. 1993;4(12):720–7. doi: 10.1007/BF00357796. [DOI] [PubMed] [Google Scholar]
- 27.Vaiman D, Mercier D, Moazami-Goudarzi K, Eggen A, Ciampolini R, Lepingle A, et al. A set of 99 cattle microsatellites: characterization, synteny mapping, and polymorphism. Mamm Genome. 1994;5(5):288–97. doi: 10.1007/BF00389543. [DOI] [PubMed] [Google Scholar]
- 28.Schutz E, Brenig B. Analytical and statistical consideration on the use of the ISAG-ICAR-SNP bovine panel for parentage control, using the Illumina BeadChip technology: example on the German Holstein population. Genet Sel Evol. 2015;47(1):3. doi: 10.1186/s12711-014-0085-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stock KF, Reents R. Genomic selection: status in different species and challenges for breeding. Reprod Domest Anim. 2013;48(Suppl 1):2–10. doi: 10.1111/rda.12201. [DOI] [PubMed] [Google Scholar]
- 30.Werner FA, Durstewitz G, Habermann FA, Thaller G, Kramer W, Kollers S, et al. Detection and characterization of SNPs useful for identity control and parentage testing in major European dairy breeds. Anim Genet. 2004;35(1):44–9. doi: 10.1046/j.1365-2052.2003.01071.x. [DOI] [PubMed] [Google Scholar]
- 31.Mariani S, Bekkevold D. The nuclear genome: neutral and adaptive markers in fisheries. Stock identification methods: applications in fishery science. 2. Oxford, UK: Elsevier Inc.; 2014. [Google Scholar]
- 32.DeAtley KL, Rincon G, Farber CR, Medrano JF, Luna-Nevarez P, Enns RM, et al. Genetic analyses involving microsatellite ETH10 genotypes on bovine chromosome 5 and performance trait measures in Angus- and Brahman-influenced cattle. J Anim Sci. 2011;89(7):2031–41. doi: 10.2527/jas.2010-3293. [DOI] [PubMed] [Google Scholar]
- 33.Meirelles SL, Gouveia GV, Gasparin G, Alencar MM, Gouveia JJ, Regitano LC. Candidate gene region for control of rib eye area in Canchim beef cattle. Genet Mol Res. 2011;10(2):1220–6. doi: 10.4238/vol10-2gmr1175. [DOI] [PubMed] [Google Scholar]
- 34.Drogemuller C, Rossi M, Gentile A, Testoni S, Jorg H, Stranzinger G, et al. Arachnomelia in Brown Swiss cattle maps to chromosome 5. Mamm Genome. 2009;20(1):53–9. doi: 10.1007/s00335-008-9157-2. [DOI] [PubMed] [Google Scholar]
- 35.Gutierrez-Gil B, Wiener P, Williams JL. Genetic effects on coat colour in cattle: dilution of eumelanin and phaeomelanin pigments in an F2-Backcross Charolais x Holstein population. BMC Genet. 2007;8:56. doi: 10.1186/1471-2156-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chu MX, Zhou GL, Jin HG, Shi WH, Cao FC, Fang L, et al. Study on relationships between seven microsatellite loci and somatic cell score in Beijing Holstein cows. Yi Chuan Xue Bao. 2005;32(5):471–5. [PubMed] [Google Scholar]
- 37.Smith SB, Zembayashi M, Lunt DK, Sanders JO, Gilbert CD. Carcass traits and microsatellite distributions in offspring of sires from three geographical regions of Japan. J Anim Sci. 2001;79(12):3041–51. doi: 10.2527/2001.79123041x. [DOI] [PubMed] [Google Scholar]
- 38.Butler JM, Reeder DJ. Cattle (bovine) STRs. 2015. [Google Scholar]
- 39.Qanbari S, Pimentel EC, Tetens J, Thaller G, Lichtner P, Sharifi AR, et al. The pattern of linkage disequilibrium in German Holstein cattle. Anim Genet. 2010;41(4):346–56. doi: 10.1111/j.1365-2052.2009.02011.x. [DOI] [PubMed] [Google Scholar]
- 40.Jamieson A, Taylor SC. Comparisons of three probability formulae for parentage exclusion. Anim Genet. 1997;28(6):397–400. doi: 10.1111/j.1365-2052.1997.00186.x. [DOI] [PubMed] [Google Scholar]
- 41.Hu ZL, Park CA, Wu XL, Reecy JM. Animal QTLdb: an improved database tool for livestock animal QTL/association data dissemination in the post-genome era. Nucleic Acids Res. 2013;41(Database issue):D871–9. doi: 10.1093/nar/gks1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DHV . Facts and figures 2012. 2015. [Google Scholar]
- 43.Caballero A. Developments in the prediction of effective population size. Heredity. 1994;73(Pt 6):657–79. doi: 10.1038/hdy.1994.174. [DOI] [PubMed] [Google Scholar]
- 44.Hayes BJ. Efficient parentage assignment and pedigree reconstruction with dense single nucleotide polymorphism data. J Dairy Sci. 2011;94(4):2114–7. doi: 10.3168/jds.2010-3896. [DOI] [PubMed] [Google Scholar]
- 45.Seroussi E, Glick G, Shirak A, Ezra E, Zeron Y, Ron M, et al. Maternity validation using sire-only BovineSNP50 BeadChip data. Anim Genet. 2013;44(6):754–7. doi: 10.1111/age.12062. [DOI] [PubMed] [Google Scholar]
- 46.Strucken EM, Gudex B, Ferdosi MH, Lee HK, Song KD, Gibson JP, et al. Performance of different SNP panels for parentage testing in two East Asian cattle breeds. Anim Genet. 2014;45(4):572–5. doi: 10.1111/age.12154. [DOI] [PubMed] [Google Scholar]
- 47.Strucken EM, Lee SH, Lee HK, Song KD, Gibson JP, Gondro C. How many markers are enough? Factors influencing parentage testing in different livestock populations. J Anim Breed Genet. 2015 doi: 10.1111/jbg.12179. [DOI] [PubMed] [Google Scholar]
- 48.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barton NH, Briggs DEG, Eisen JA, Goldstein DB, Patel NH. Random drift of allele frequencies. Evolution. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2007. p. 415. [Google Scholar]


