Abstract
Legionella pneumophila, the causative agent of Legionnaires' disease, expresses a type IVB secretion apparatus that translocates bacterial proteins into amoeba and macrophage hosts. When stationary-phase cultures are used to infect hosts, the type IVB apparatus encoded by the icm/dot genes is required for entry, delay of phagosome-lysosome fusion, and intracellular multiplication within host cells. Null mutants with mutations in icm/dot genes are defective in these phenotypes. Here a new model is described in which hosts are infected with stationary-phase cultures that have been incubated overnight in pH 6.5 buffer. This model is called Ers treatment because it enhances the resistance to acid, hydrogen peroxide, and antibiotic stress beyond that of stationary-phase cultures. Following Ers treatment entry into amoeba and macrophage hosts does not require dotA, which is essential for Legionella virulence phenotypes when hosts are infected with stationary-phase cultures, dotB, icmF, icmV, or icmX. Defective host entry is also suppressed for null mutants with mutations in the KatA and KatB catalase-peroxidase enzymes, which are required for proper intracellular growth in amoeba and macrophage hosts. Ers treatment-induced suppression of defective entry is not associated with increased bacterial adhesion to host cells or with morphological changes in the bacterial envelope but is dependent on protein expression during Ers treatment. By using proteomic analysis, Ers treatment was shown to induce a protein predicted to contain eight tetratricopeptide repeats, a motif previously implicated in enhanced entry of L. pneumophila. Characterization of Ers treatment-dependent changes in expression is proposed as an avenue for identifying icm/dot-independent factors that function in the entry of Legionella into amoeba and macrophage hosts.
Legionella pneumophila, the causative agent of Legionnaires' disease, is a facultative intracellular pathogen found in aquatic environments as planktonic cells, in biofilms, and as an intracellular parasite of amoebae (14, 61, 62). When internalized by alveolar macrophages (54), L. pneumophila resides in specialized phagosomes that are delayed in fusion with lysosomes and are permissive to bacterial replication (78, 81). Formation of the specialized phagosomes is attributed to proteins secreted into the host (16, 23, 48, 53) by a type IVB secretion apparatus encoded by the icm/dot loci (intracellular multiplication/defective organelle trafficking) (63, 69, 73, 80). Null mutants with mutations in icm/dot genes are defective in entry into macrophage and amoeba hosts, intracellular multiplication, delay of phagosome-lysosome fusion, and host cell killing (11, 39, 63, 66, 70, 80). Involvement of non-icm/dot factors in Legionella virulence is implicated by mutations outside the icm/dot loci associated with defective intracellular multiplication and host cell killing. Such mutations implicate type II secretion, iron sequestration, pilin synthesis, hydrogen peroxide decomposition, amino acid biosynthesis, and sugar transport (5, 17, 80). Delayed fusion of Legionella-containing phagosomes with lysosomes requires type IVB secretion. However, type IVB secretion is not required for the delayed acquisition of certain lysosomal markers, which occurs in phagosomes containing mutants with mutations in dotA and dotB, which are considered essential for type IVB secretion (43, 79). These data implicate the presence of icm/dot-independent virulence factors, which are poorly defined.
The importance of environmental stress in the transition of L. pneumophila from environmental bacterium to intracellular pathogen is supported by several lines of evidence. Nutrient, thermal, and osmotic stresses are imposed in the environmental reservoirs from which Legionnaires' disease is spread, including water cooling towers, hot water tanks, and evaporative condensers of air conditioning systems. Increased resistance to antibiotics, to biocides, and to acid, H2O2, osmotic, and thermal stresses following growth within amoebae (19, 20, 62) suggests that stress is imposed by environmental amoeba hosts (2, 29, 32, 38, 74, 80) belonging to the genera Acanthamoeba and Hartmannella (14, 62). Stress genes are induced when Legionella is grown in amoebae or in macrophage hosts (1, 2, 59; M. Miyaki, T. Fukui, Y. Imai, and H. A. Shuman, Abstr. 103th Gen. Meet. Am. Soc. Microbiol., abstr. B-013, 2003). Participants in the oxidative stress response, the KatA and KatB catalase-peroxidase enzymes, are required for optimal intracellular multiplication in macrophages (5). Finally, the stress of growth to the stationary phase endows L. pneumophila with a full panel of virulence traits compared to rapidly dividing exponential cultures, which are essentially avirulent (15).
We report here a new model for studying the effect of stress on entry of L. pneumophila into amoebae and macrophages: overnight incubation of stationary-phase cultures in a phosphate-saline buffer. The model is called Ers treatment (enhanced resistance to stress) because it enhances resistance of stationary cultures to acid, H2O2, and antibiotic stresses. Ers treatment suppresses the defective entry into amoeba and macrophage hosts of null mutants with mutations in dotA and dotB, other icm/dot genes, and katA and katB genes. Since the icm/dot genes are not required for host entry of Ers-treated Legionella, icm/dot-independent entry factors are implicated in the Ers treatment model. Thus, analysis of Ers treatment-induced gene expression may be an avenue for identifying icm/dot-independent factors involved in the entry of Legionella into eukaryotic hosts.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All L. pneumophila mutants were derived from serotype 1 wild-type strain JR32 (66, 83) (Table 1). Legionella strains were revived from frozen storage on charcoal-N-(2-acetamido)-2-aminoethanesulfonic acid (ACES)-buffered pH 6.9 yeast extract agar plates and then cultured in ACES-buffered pH 6.9 yeast extract (AYE) broth (27, 40). Exponential- and stationary-phase cultures were grown to optical densities at 600 nm of 0.5 to 0.9 and >2.5, respectively (15). When required, chloramphenicol, hygromycin sulfate, and gentamicin sulfate were present at concentrations of 5, 100, and 10 μg/ml, respectively. Ers-treated cultures were prepared by resuspension of stationary-phase cultures in Acanthamoeba castellanii buffer (AC buffer) (50), followed by incubation for 16 to 19 h. AC buffer contained 4 mM magnesium sulfate, 0.4 mM calcium chloride, 3.4 mM sodium citrate, 0.051 mM ferrous ammonium sulfate, 2.5 mM sodium monohydrogen phosphate, and 2.5 mM potassium dihydrogen phosphate, and the pH was adjusted to 6.5 with HCl. For recombinant construction we used Escherichia coli strain DH5α cultured in Luria-Bertani medium (75) with chloramphenicol, hygromycin sulfate, and gentamicin sulfate at concentrations of 25, 150, and 5 μg/ml, respectively. All cultures were incubated at 37°C with aeration. Bacterial viability was determined by the LIVE/DEAD BacLight technique (Molecular Probes, Eugene, Oreg.).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or features | Reference |
|---|---|---|
| Strains | ||
| JR32 | Salt-sensitive isolate of AM511 (Philadelphia-1 Smr r−m+) | 66 |
| katA (PB126) | JR32 katA::Gmr | 6 |
| katB (PB117) | JR32 katB::Ω Cmr | 7 |
| icmS (GS3001) | JR32 icmS3001::Km | 68 |
| icmP (GS3002) | JR32 icmP3002::Km | 68 |
| icmX (LELA921) | JR32 icmX921::Tn903dIIlacZ | 66 |
| icmF (LELA1275) | JR32 icmF1275::Tn903dIIlacZ | 66 |
| icmV (LELA1747) | JR32 icmV1747::Tn903dIIlacZ | 66 |
| dotB (LELA2883) | JR32 dotB2883::Tn903dIIlacZ | 66 |
| dotA (LELA3118) | JR32 dotA3118::Tn903dIIlacZ | 66 |
| icmS complemented | GS3001/pGS-Lc-37-14 | 71 |
| icmP complemented | GS3002/pGS-Lc-34-14 | 71 |
| icmF complemented | LELA1275/pGS-Lc-55-14 | 71 |
| katA complemented | PB126/pMMB207αB-Km-14::katA | 6 |
| Plasmids | ||
| pMV206.hyg | BspHI to SmaI hygromycin resistance cassette from p16R1 in pMV206 with Kmr resistance cassette removed | 33 |
| pJN105 | pBBR1MCS-5 with araC and PBAD regions before the polylinker, Gmr | 55 |
| pJN105-hygro-GFP | pJN105 with hygromycin resistance and GFP | This study |
| pGS-Lc-37-14 | icmTS in pMMB207αB-Km-14 | 71 |
| pMMB207 | RSF1010 derivative, IncQ, oriT, lacIq, Ptac, Cmr | 52 |
| pGS-Lc-34-14 | icmPO in pMMB207αB-Km-14 | 71 |
| pGS-Lc-55-14 | icmF and tphA in pMMB207αB-Km-14 | 71 |
| pMMB207αB-Km-14 | pMMB207, lacZα, MCS, with mobA::Kmr | 70 |
| pMMB207αB-Km-14::katA | katA in BamHI site of pMMB207αB-Km-14 opposite the direction of Ptac | 6 |
| pGS-GFP-04 | gfp in pMMB207 | 39 |
Amoeba and macrophage lines and culture conditions.
A. castellanii ATCC 30234 was cultured at 28°C in peptone yeast extract glucose (PYG) medium, as described previously (5, 39, 71). The HL60 monocyte cell line was maintained in RPMI medium supplemented with 2 mM glutamine, 10% heat-inactivated fetal bovine serum, and penicillin-streptomycin (5,000 U/ml) at 37°C in a humidified atmosphere containing 5% CO2 (39, 71).
Growth of L. pneumophila in A. castellanii.
For growth of L. pneumophila in A. castellanii (39, 71), adherent amoebae were fed with fresh PYG medium the previous day in a 75-cm2 tissue culture flask. The PYG medium was removed from the flask, 25 ml of AC buffer was added, and stationary-phase wild-type or katA null mutant L. pneumophila cells were added at a multiplicity of infection (MOI) of 3 to 5. After incubation for 1 h at 37°C, the amoeba monolayer was washed twice and incubated for 48 h at 37°C with 25 ml of fresh AC buffer. The AC buffer, containing Legionella released following intracellular multiplication and lysis of the amoeba hosts, was removed. Fresh AC buffer was added, and adherent amoebae containing intracellular bacteria were dislodged (71) and recovered by centrifugation. The amoeba pellet was resuspended in AC buffer and RPMI medium containing glutamine and normal human serum for stress and HL60 entry experiments, respectively, and amoebae were lysed by repeated aspiration through a 30-gauge needle (20, 50). Amoeba debris was removed by centrifugation (1 min at 150 × g) (20), and the supernatant containing amoeba-grown L. pneumophila was used immediately for stress or entry experiments.
Plasmids.
Transformation of L. pneumophila with plasmids (Table 1) expressing hygromycin resistance was accomplished by natural competence (51, 77) by using a protocol described previously (51). All other plasmids were transformed by electroporation by using a previously described protocol (18), modified as follows. Cultures of L. pneumophila in AYE broth (optical density at 600 nm, 0.4 to 1.0) were centrifuged and resuspended in cold 10% glycerol three times, the last time at a ratio of 2 ml of 10% glycerol per liter of culture. Electroporation was performed in 0.2-cm cuvettes with a Gene Pulser (Bio-Rad, Hercules, Calif.) set at 25 μF and 2.5 kV, and this was followed by 4 to 5 h of outgrowth in AYE broth at 37°C before plating. To perform immunofluorescence entry assays with complemented L. pneumophila mutants, it was necessary to construct a green fluorescent protein (GFP)-expressing plasmid compatible with the IncQ plasmids conventionally used for complementation. The pBBR1MCS-5 derivative of the Bordetella bronchiseptica BHR plasmid has a pBBR/BHR replicon (45) that is maintained in L. pneumophila (47, 48) and is compatible with IncP, IncW, IncQ, and IncRi incompatibility groups (45). PCR primers with BglII sites at their 5′ ends were used to amplify the region outside the gentamicin cassette in pJN105, a derivative of pBBR1MCS-5 with araC and PBAD regions before the polylinker (55). The PCR fragment was digested with BglII, blunted with the Klenow fragment, and ligated to the blunted DdeI-DdeI hygromycin resistance fragment from pMV206.hygro (33) to form pJN105-hygro. The Ptac-GFP fragment of pGS-GFP04 (39) was amplified by PCR by using primers with NheI sites at their 5′ ends. After digestion with NheI, this fragment was ligated with NheI-digested pJN105-hygro to form pJN105-hygro-GFP. To our knowledge, hygromycin has not previously been reported to be a selectable marker in L. pneumophila.
Acid and hydrogen peroxide stress treatments.
L. pneumophila cultures (exponential, stationary phase, or Ers treated stationary phase) were washed, resuspended in AC buffer, and diluted to the desired cell concentration by assuming that an optical density at 600 nm of 0.3 corresponded to 5 × 108 CFU/ml (39). For acid treatment, 0.5 ml of a bacterial suspension was rapidly mixed with 0.5 ml of 40 mM citric acid in AC buffer to obtain a final pH of 3.0. For H2O2 treatment, H2O2 was added to 1 ml of a bacterial suspension to a final concentration of 1 mM. After incubation at 37°C with aeration for the desired time, aliquots were diluted into AC buffer and then plated on CYE agar for colony counting. Acid and H2O2 stress experiments with wild-type strain JR32 and the katA mutant were performed three or more times. Acid stress experiments with icmF, icmP, and icmS mutants were performed two or more times.
Immunofluorescence assay for entry of Legionella into A. castellanii and HL60-derived macrophages.
For the immunofluorescence assay for entry of Legionella into A. castellanii (39), adherent A. castellanii cells were resuspended in PYG medium to a concentration of 1 × 107 cells/ml, and then 0.5 ml was added to wells of a 24-well tissue culture plate containing glass coverslips (diameter, 12 mm). After incubation for 1 h at 28°C in a humidified atmosphere, the PYG medium was removed, and the amoeba monolayer was washed three times with AC buffer. To each well 0.5 ml of AC buffer and an L. pneumophila culture in AC buffer were added to obtain an MOI of 3 to 5. The tissue culture plate was centrifuged at room temperature (700 × g, 10 min) to synchronize the infection and then incubated for 30 min at 28°C in a humidified atmosphere. Subsequent washing, formaldehyde fixation, and incubation with rhodamine-conjugated rabbit anti-L. pneumophila serotype 1 antibody (m-Tech, Atlanta, Ga.) were performed as described previously (39). Coverslips were inverted and mounted on microscopy slides by using Fluoromount G mounting medium (Southern Biotechnology Associates, Birmingham, Ala.).
HL60 cells were resuspended in RPMI-glutamine-PennStrep medium without fetal bovine serum to a concentration of 3 × 106 cells/ml, and 0.5 ml was added to wells of a 24-well tissue culture plate containing glass coverslips. After incubation for 1 h at 37°C in a humidified 5% CO2 atmosphere, the medium was removed, and the monolayer was incubated for 20 to 24 h in RPMI-glutamine-PennStrep medium with 10% fetal bovine serum containing 20 nM phorbol 12-myristate 13-acetate to promote differentiation into macrophages. The monolayer was then washed three times with medium without PennStrep or phorbol 12-myristate 13-acetate. Then to each well 0.5 ml of RPMI-glutamine medium containing 10% normal human AB serum (Gemini Bioproducts, Woodland, Calif.) was added, followed by L. pneumophila in the same medium at an MOI of 100. The tissue culture plate was centrifuged for 10 min at room temperature and then incubated for 30 min at 37°C. Subsequent washing, formaldehyde fixation, incubation with rhodamine-conjugated anti-L. pneumophila antibody, and mounting were performed as described above for the fluorescence-based A. castellanii entry experiments (39).
The L. pneumophila strains used for the immunofluorescence assay contained one of two plasmids expressing GFP. For strains harboring plasmid pGS-GFP-04 (39), the Legionella medium contained chloramphenicol and 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). For strains harboring plasmid pJN105-hygro-GFP, the medium contained hygromycin and 0.5 mM IPTG. Whenever tested, entry with the pGS-GFP-04 (IncQ) GFP-expressing plasmid and entry with the pJN105-hygro-GFP (pBBR/BHR) GFP-expressing plasmid were identical. Each amoeba or macrophage entry experiment was performed in duplicate. In addition, for amoeba entry, duplicate experiments were performed three or more times with the JR32, katA, icmF, icmP, icmS, and dotA strains. Duplicate macrophage entry experiments were performed two or more times with the JR32, katA, icmF, icmP, icmX, and dotA strains.
Fluorescence microscopy.
For fluorescence microscopy (39), microscope slides with immunofluorescent samples were observed at a magnification of ×90 by using HiQ band-pass GFP and CY3/rhodamine filters (Chroma Technology Corp., Brattleboro, Vt.) with an Olympus 1X70 fluorescence microscope. Only L. pneumophila cells that are external or adherent to the host cell surface are accessible to rhodamine-conjugated anti-serotype 1 antibody; these cells are green with the GFP filter and red with the rhodamine filter. Legionella cells within a host cell are not accessible to the antibody. Internalized bacteria were defined as the bacteria within the confines of a host cell envelope that were green with the GFP filter and not visible with the rhodamine filter. Microscope slides could be stored in the dark at 4°C for 2 weeks prior to observation with the fluorescence microscope without quantitative changes in the phagocytic index (number of bacteria internalized per 50 host cells).
Gentamicin protection assay of entry into HL60-derived macrophages.
For the gentamicin protection assay of entry into HL60-derived macrophages (39), HL60 monocytes (1.5 × 106 cells/well) were plated, differentiated, washed, and incubated with L. pneumophila cultures at an MOI 100 as described above for the immunofluorescence method, except that no glass coverslips were added to the 24-well tissue culture plates. Following 10 min of synchronizing centrifugation and an additional 30 min of incubation at 37°C, the HL60 monolayers were washed twice with phosphate-buffered saline and then incubated for 40 min at 37°C with RPMI-glutamine medium with 100 μg of gentamicin per ml to kill external, adherent Legionella. The monolayers were washed twice with phosphate-buffered saline and lysed for 30 min with 0.5 ml of water to release internalized bacteria. Aliquots of lysates were plated on CYE agar for colony counting.
Negative-stain electron microscopy.
Cultures of the L. pneumophila dotA and icmS null mutants in AYE broth or after Ers treatment were transferred to Formvar- and carbon-coated grids and negatively stained with 1% phosphotungstic acid or 1% ammonium molybdate. The grids were blotted dry and immediately observed with a JEOL 1200 EX transmission electron microscope at 80 kV. Images for different strains and culture conditions were compared by measuring cell width and length, by observing features of the cell periphery from stain pooled along the cell edges, and by observing the speckling over the cell body from stain pooled in involutions on the cell surface.
Two-dimensional gel electrophoresis.
Cell extracts were prepared from broth-grown stationary-phase cultures and Ers-treated stationary-phase cultures by resuspending bacteria in 25 mM potassium phosphate (pH 7.1) containing 1 mM EDTA and 1× Complete EDTA-free protease inhibitor cocktail (Boehringer, Indianapolis, Ind.), followed by sonication (76) and centrifugation at 15,000 × g for 40 min. Supernatant proteins were precipitated by phenol-ethyl ether extraction (67) and then dissolved at a concentration of ∼8 mg ml−1 in isoelectric focusing buffer containing 9 M urea, 2% Triton X-100, 2% dithioerythritol, 0.5% IPG buffer, and pH 4-7L Immobiline strips (Amersham Biosciences, Piscataway, N.J.). The pH 4-7L Immobiline strips (13 cm) were hydrated with a solution containing ∼2 mg of protein for 14 h at 20°C and subjected to isoelectric focusing in an IPGphor apparatus (Amersham Biosciences), typically for a total of 40,000 V · h. After isoelectric focusing, each strip was equilibrated with 10 ml of 50 mM Tris HCl (pH 8.8)-6 M urea-30% glycerol-2% sodium dodecyl sulfate (SDS)-2% dithioerythritol for 40 min at room temperature and then incubated for an additional 15 min after addition of iodoacetamide to a final concentration of 2.5%. The strips were then placed in 0.5% agarose on top of 10% acrylamide-SDS gels. After electrophoresis, the gels were stained with 0.05% Coomassie brilliant blue in 7% acetic acid-50% methanol, destained in 7% acetic acid-50% methanol, and then soaked overnight in water and stored at 4°C until they were used for protein identification.
Protein identification by mass spectrometry.
Spots of interest that were cut from acrylamide gels following two-dimensional electrophoresis were washed once with 25% acetonitrile in 25 mM Tris HCl (pH 8.5) and twice with 50% acetonitrile in 25 mM Tris HCl (pH 8.5) and then dried under a vacuum and stored at −20°C. For trypsin digestion, each gel spot was incubated overnight at 32°C in 8 μl of sequencing grade modified trypsin (0.01 mg ml−1 in 25 mM Tris HCl [pH 8.5]; Promega, Madison Wis.) and 15 μl of water. The peptides in the digestion buffer plus those extracted from the gel with 2% trifluoroacetic acid in 50% acetonitrile were dried under a vacuum and then dissolved in a 4-hydroxy-α-cyanocinnamic acid matrix solution (10 mg ml−1 in 0.1% trifluoroacetic acid-50% acetonitrile) and applied to a matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometer sample plate. The masses of tryptic peptides in digestion mixtures were determined by MALDI-TOF mass spectrometry with a Voyager-DE mass spectrometer (PerSeptive Biosystems, Inc. Farmingham, Mass.). Proteins in gel spots were identified by matching experimentally determined masses with masses predicted from open reading frames (ORFs) in the L. pneumophila strain JR32 genome (http://genome3.cpmc.columbia.edu/∼legion/index.html) by using the Peptide Search Program (http://www.narrador.embl-heidelberg.de/GroupPages/PageLink/peptidesearchpage.html). Unambiguous matches were established by considering the number of peptides matched, the percentage of the ORF covered, the matched uncleaved peptides, and the agreement between the experimental and predicted masses and the isoelectric points for the S-acetamidomethylated polypeptide.
RESULTS
Intra-amoebic growth enhances resistance of wild-type L. pneumophila to acid and H2O2 stresses.
Intracellular multiplication of L. pneumophila in environmental amoebae is integral to the spread of Legionnaires' disease (3, 8, 74, 80). We examined if intracellular multiplication in A. castellanii—known to enhance entry of L. pneumophila into macrophage and amoeba hosts (19, 20)—enhanced the resistance of Legionella stationary-phase cultures used to infect amoebae to H2O2 and acid stresses. Since Legionella is likely to encounter reactive oxygen species following phagocytosis (5, 35, 41, 46) and to encounter acid stress when Legionella-containing phagosomes ultimately fuse with lysosomes (78), these stresses are physiologically relevant.
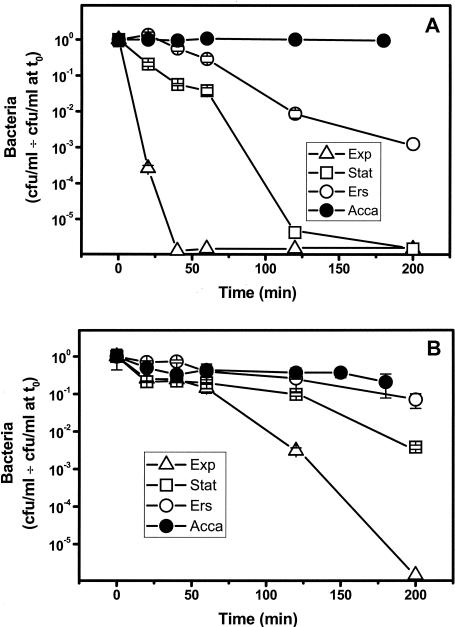
Amoeba-grown Legionella maintained an approximately constant titer after challenge by pH 3 (Fig. 1A) or 1 mM H2O2 (Fig. 1B). In contrast, the titers of broth-grown stationary-phase cultures decreased by more than 5 orders of magnitude and by 2 orders of magnitude, respectively. Exponential-phase cultures were considerably less resistant to acid and H2O2 stresses than stationary-phase cultures (Fig. 1). This cross-resistance of stationary-phase cultures to stress, initially observed in E. coli (42), is also found in L. pneumophila (4, 36). Wild-type L. pneumophila strain AA100 showed increases in resistance to acid, H2O2, osmotic, and thermal stresses in bacteria isolated after intracellular multiplication in Hartmannella vermiformis compared with the resistance of exponential-phase broth cultures (2). Our demonstration of increased stress resistance following intra-amoebic growth is in qualitative agreement with the results of H. vermiformis studies (2). However, a quantitative comparison was not possible due to differences in the stress resistance of the stationary-phase and exponential-phase cultures to which the amoeba-grown Legionella cells were compared.
FIG. 1.
Effect of growth conditions on stress resistance of wild-type L. pneumophila. Exponential-phase (Exp), stationary-phase (Stat), or Ers-treated stationary-phase (Ers) cultures or bacteria recovered from A. castellanii following intracellular multiplication (Acca) were challenged with pH 3 (A) or 1 mM H2O2 (B) and titrated after different times (mean ± standard deviation). The points plotted on the x axis are the values when there were no CFU.
Identification of enhanced resistance to stress (Ers treatment).
Intracellular multiplication of L. pneumophila in the amoeba Acanthamoeba polyphagia enhances resistance to antibiotic and biocide stresses (9, 10). In the previous studies, amoeba-grown bacteria were isolated from the medium 3 days after infection. Legionella replicates intracellularly, lyses the amoeba host, and begins to be released into the infection medium within a few hours after infection (5, 36, 71). Thus, the amoeba-grown bacteria used for antibiotic and biocide studies were exposed for as long as 3 days to the infection medium, a neutral-pH phosphate-saline buffer that does not support L. pneumophila replication. It is possible that incubation in the infection medium—without prior exposure to amoebae—contributes to phenotypic changes attributed to intra-amoebic growth. In our experiments (Fig. 1) and in the experiments with H. vermiformis (2), intra-amoebic L. pneumophila cells were recovered by shear or osmotic lysis of the amoebae, and any bacteria released into the infection medium by bacterial lysis of amoebae were removed and not used for stress studies.
We tested if resistance to acid and H2O2 stresses is enhanced by incubation of Legionella in the infection medium for A. castellanii, AC buffer (5, 19, 20, 50, 71), a phosphate-saline buffer similar to A. polyphagia infection medium (2). Incubation in AC buffer in the absence of amoebae significantly increased the resistance of stationary-phase L. pneumophila cultures to pH 3 and H2O2 (Fig. 1). We called this incubation in AC buffer Ers treatment (enhanced resistance to stress) to indicate its effect on resistance of stationary-phase cultures to acid and H2O2.
Ers treatment enhances stress resistance of Legionella katA and icm/dot mutants.
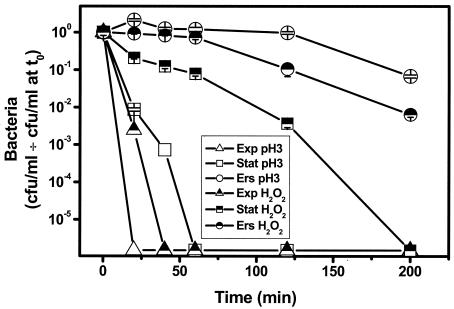
Mutants with defective virulence phenotypes were not examined in prior studies of the effect of in vitro and intra-amoebic growth on stress resistance (2, 9, 10). Therefore, we examined the effects of these growth conditions on an L. pneumophila periplasmic catalase-peroxidase mutant with a Gmr element inserted in the katA gene. This mutant is defective in intracellular multiplication in primary macrophages, macrophage lines, and A. castellanii (5, 6). Stationary-phase cultures of the katA oxidative stress response mutant were considerably more sensitive than wild-type stationary cultures to pH 3 and H2O2 stresses (compare Fig. 2 with Fig. 1). However, Ers treatment restored the level of resistance of the katA mutant to that of wild-type stationary-phase cultures for both acid and H2O2 stresses (compare Fig. 2 with Fig. 1).
FIG. 2.
Effect of growth conditions on the stress resistance of the katA null mutant. Exponential-phase (Exp), stationary-phase (Stat), or Ers-treated stationary-phase (Ers) cultures of the katA::Gmr null mutant or bacteria recovered from A. castellanii following intracellular multiplication (Acca) were challenged with pH 3 or 1 mM H2O2 and titrated after different times (mean ± standard deviation). The points on the x axis are the values when there were no CFU.
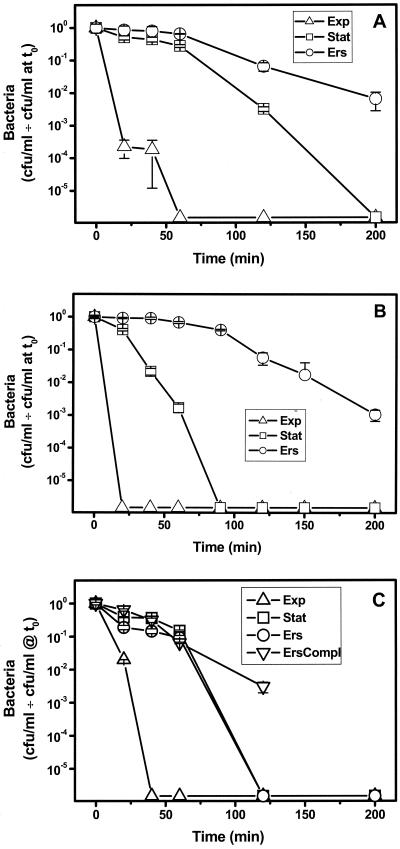
Next, the effect of growth conditions on acid resistance was examined in icm/dot mutants defective in intracellular multiplication in HL60-derived macrophages and A. castellanii (icmF, icmP, and icmS Tn insertion mutants) (71). (Fig. 3) Ers treatment increased the level of resistance to that of Ers-treated wild-type Legionella for the icmF and icmP mutants, while the icmS mutant was not responsive to Ers treatment. Restoration of the response to Ers treatment in the complemented icmS mutant shows that loss of the response is related to inactivation of icmS. These results suggest that icmS is essential for the stress response to Ers treatment but that neither katA, icmF, nor icmP is required. Since for all mutants there was increased resistance of stationary-phase cultures compared to exponential-phase cultures, neither katA, icmF, icmP, nor icmS is required for the cross-resistance stress phenotype (Fig. 2 and 3).
FIG. 3.
Effect of growth conditions on acid resistance of icm null mutants. Acid challenge experiments were performed with icmF (A), icmP (B), and icmS (C) null mutants. Exponential-phase (Exp), stationary-phase (Stat), or Ers-treated stationary-phase (Ers) cultures were challenged with pH 3 and titrated after different times (mean ± standard deviation). Ers compl, Ers-treated stationary-phase culture of icmS mutant containing an icmS+ complementing plasmid. The points on the x axis are the values when there were no CFU.
Ers treatment suppresses the defective entry of katA, katB, and dot/icm mutants into amoebae.
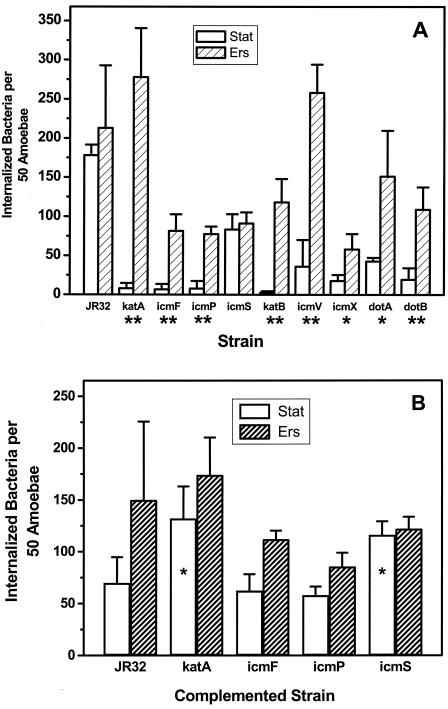
The entry of amoeba-grown wild-type L. pneumophila into A. castellanii is increased 10-fold compared to the entry of stationary-phase bacteria (20). The amoeba-grown bacteria used for this study were isolated from the AC buffer infection medium following lysis of amoeba hosts by Legionella. Since both intra-amoebic growth and Ers treatment enhanced the acid and H2O2 resistance of wild-type Legionella (Fig. 1), we examined if Ers treatment enhanced entry into A. castellanii. No significant change in entry was seen following Ers treatment of wild-type Legionella (Fig. 4A).
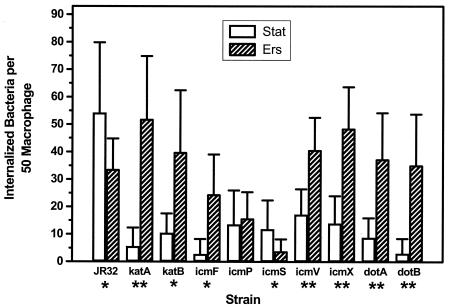
FIG. 4.
Ers treatment reverses the defect in entry of katA, katB, and icm/dot mutants into A. castellanii. A. castellanii was infected at an MOI of 3 to 5 with wild-type L. pneumophila strain JR32 or a mutant strain grown to the stationary phase (Stat) or after Ers treatment of stationary-phase cultures (Ers). Internalized bacteria were quantified by the fluorescence microscopy method (mean ± standard deviation). (A) Strains containing a GFP-expressing plasmid. (B) Strains containing the pJN105-hygro-GFP plasmid and a second plasmid, pMMB207αB-Km14 for wild-type strain JR32 or a complementing plasmid for mutant strains. P values were determined by one-sided t tests by comparing entry of stationary-phase and Ers-treated cultures of the same strain (A) or by comparing a mutant stationary-phase or Ers-treated culture with a similar wild-type culture (B) (two asterisks, P < 0.005; one asterisk, P < 0.02; no asterisk, P > 0.05).
We next examined if Ers treatment enhanced entry of katA and icm mutants, mutants that exhibited defective intracellular multiplication in A. castellanii. Consistent with previous results obtained by using fluorescence microscopy (39), icmF, icmP, and icmS mutants were reduced in entry when stationary-phase cultures of Legionella were used to infect A. castellanii (Fig. 4A). Defective entry for the katA mutant was not examined previously. Remarkably, entry of the katA, icmF, and icmP mutants was increased 10- to 30-fold when Ers-treated stationary-phase cultures were used to infect A. castellanii (Fig. 4A). Ers treatment restored entry of the katA mutant to wild-type values, and entry of icmF and entry of icmP were restored to 40% of the wild-type values. These effects parallel the effects of Ers treatment on stress resistance in that the katA, icmF, and icmP mutants (Fig. 2 and 3A and B) were responsive, while the icmS mutant was not affected by Ers treatment (Fig. 3C).
When the icm mutants were complemented with wild-type alleles on plasmids, entry of stationary-phase cultures was no longer defective compared to entry of wild-type strain JR32 containing the empty complementing vector (Fig. 4B). These findings are consistent with the results of previous studies that demonstrated complementation of the entry defect in stationary-phase cultures (39). Similarly, the entry of Ers-treated complemented mutants was not significantly different from the entry of the wild type containing the empty vector (Fig. 4B). The complementation data demonstrated that suppression of defective entry by Ers treatment was associated with loss of function of the katA, icmF, and icmP genes and was unlikely to be associated with spontaneous mutations outside the inactivated genes.
To ascertain if Ers treatment suppresses defective entry of other Legionella mutants, we examined a katB null mutant, whose intracellular multiplication phenotype was like that of the katA mutant (5, 6), and null mutants with mutation in four other icm/dot genes. One of these was a dotA mutant, with a Tn insertion in the dotA gene, required for many aspects of Legionella virulence when stationary-phase cultures are used for infection, from entry through cytotoxicity to the host cell (11, 43, 44, 63, 64). All five mutants exhibited statistically significant increases in entry following Ers treatment, which ranged from 3- to 4-fold for the icmX and dotA mutants to 40-fold for the katB mutant (Fig. 4A). Moreover, Ers treatment restored the levels of entry to >50% of the wild-type value for the katB, icmV, dotA, and dotB mutants and to 30% of the wild-type value for the icmX mutant.
These data show that the defective entry of katA, katB, icmF, icmP, icmV, icmX, dotA, and dotB mutants into A. castellanii is conditional and dependent on culture conditions. The entry deficit was evident in stationary-phase cultures, as described previously (39), but was suppressed by Ers treatment of stationary-phase cultures. To our knowledge, suppression of an entry defect by varying the culture conditions of a Legionella icm/dot or other virulence mutant has not been reported previously.
Ers treatment suppresses the defective entry of katA, katB, and icm/dot mutants into HL60-derived macrophages.
Many L. pneumophila icm/dot mutants are defective in entry (39) and in intracellular multiplication (71) in both macrophages and amoeba hosts. We therefore examined if Ers treatment, which suppressed the defective entry of stationary-phase cultures of katA/B and icm/dot mutants into amoebae, similarly suppressed the defective entry into HL60-derived macrophages. Consistent with previous work (39), stationary-phase cultures of icm/dot mutants exhibited decreased entry into HL60-derived macrophages (Fig. 5). Ers treatment did indeed suppress the defective entry of katA, katB, icmF, icmV, icmX, dotA, and dotB null mutants into macrophages. Statistically significant two- to ninefold increases were seen, and for all mutants entry into macrophages was restored to wild-type levels. Entry of the icmP mutant into macrophages was not significantly changed, and entry of the icmS mutant was decreased.
FIG. 5.
Ers treatment reverses the defect in entry of katA, katB, and icm/dot mutants into HL60-derived macrophages. The entry of wild-type strain JR32 or a mutant strain into macrophages derived from HL60 monocytes (MOI, 100) was determined as described for entry into A. castellanii in the legend of Fig. 4. P values were determined by one-sided t tests by comparing entry of stationary-phase (Stat) and Ers-treated (Ers) cultures of the same strain (one asterisk, P < 0.05; two asterisks, P < 0.005; no asterisk, P > 0.05).
Intra-amoebic growth suppresses the defective entry of the katA virulence mutant into HL60-derived macrophages.
Only wild-type L. pneumophila cells were used in previous studies that demonstrated enhanced entry into macrophage and amoeba hosts following intra-amoebic growth (19, 20). To examine the effect of intra-amoebic growth on a Legionella mutant with defective intracellular multiplication, we used the katA mutant, which replicates in A. castellanii at a reduced rate (5) and was defective in amoeba and macrophage entry (Fig. 4A and 5). The dotA mutant and most other icm/dot Tn insertion mutants were unsuitable for this experiment because they are nonreplicative in A. castellanii (71). This is likely why only wild-type L. pneumophila was examined in previous studies of phenotypic changes following intra-amoebic growth (9, 10, 19, 20, 32).
A. castellanii was infected with wild-type or katA Legionella, and intracellular bacteria were isolated 2 days later by shear lysis of the amoeba host, as was done for isolation of amoeba-grown Legionella for the acid and H2O2 stress experiments (Fig. 1). Entry into HL60-derived macrophages was quantified by examining the gentamicin protection, the method used previously to show enhanced entry of amoeba-grown wild-type Legionella (19, 20). The 8.1-fold increased entry of wild-type L. pneumophila which we found is in good agreement with the previously described 10-fold increased entry of amoeba-grown wild type into cultured macrophage lines (20). Entry of the katA mutant was increased 12-fold, in good agreement with the 9.3-fold increase determined by GFP fluorescence after Ers treatment of this mutant (Fig. 5). Thus, intra-amoebic growth, like Ers treatment, can suppress the defective entry of the Legionella katA null mutant.
Ers treatment increases antibiotic resistance of wild-type and dotA mutant L. pneumophila.
Increased resistance to biocides used in water purification and to antibiotics used to treat Legionella infections, including erythromycin (65), is shown by Legionella grown in the amoeba A. polyphagia (9, 10). Since both intra-amoebic growth and Ers treatment increased resistance to acid and H2O2 stresses (Fig. 1) and suppressed defective entry of the katA mutant, we examined if Ers treatment increased antibiotic resistance.
The resistance of wild-type Legionella to gentamicin and erythromycin was increased 400- and 2-fold, respectively, following Ers treatment of stationary-phase cultures (Table 2). The resistance of the dotA Tn insertion mutant to gentamicin, erythromycin, and rifampin was increased 20-, 5-, and 95-fold, respectively, by Ers treatment. Previous studies reported that intra-amoebic growth increased resistance to erythromycin and rifampin 100- and 1,000-fold, respectively, for wild-type Legionella (10), but icm/dot mutants were not examined. In the previous studies the workers used 10- to 20-fold-lower drug concentrations and resuspended Legionella in fresh broth during the 6 to 24 h of antibiotic challenge. In our experiments stationary-phase or Ers-treated cultures were exposed to antibiotics for 40 min in AC buffer, which does not support Legionella growth. Thus, the Ers-induced increase in antibiotic resistance is qualitatively similar to that observed for intra-amoebically grown Legionella (10), but quantitative comparisons are not possible due to differences in experimental design.
TABLE 2.
Effect of Ers treatment on the antibiotic resistance of stationary-phase Legionellaa
| Antibiotic | Concn (μg/ml) | JR32
|
dotA mutant
|
||||
|---|---|---|---|---|---|---|---|
| % Survival
|
Ers/Stat | % Survival
|
Ers/Stat | ||||
| Stat | Ers | Stat | Ers | ||||
| Gentamicin | 100 | 1.75 × 10−3 ± 1.2 × 10−3 | 0.73 ± 0.41 | 420 | 0.041 ± 0.0042b | 0.87 ± 0.06 | 21 |
| Erythromycin | 100 | 19 ± 2 | 46 ± 16 | 2.4 | 2.5 ± 0.9 | 12 ± 6 | 4.8 |
| Rifampin | 100 | 54 ± 0.5 | 69 ± 16 | 1.3 | 0.74 ± 0.04 | 70 ± 11 | 95 |
Bacteria in the stationary phase (Stat) or following Ers treatment after the stationary phase (Ers) were resuspended in phosphate-buffered saline at a concentration of 1 × 108 to 5 × 109 CFU/ml, incubated at 37°C for 40 min with the concentration of antibiotic indicated, and then titrated on CYE plates. The titer obtained compared to titer before antibiotic treatment is expressed as the percent survival; the values are means ± standard deviations.
The higher level of survival of the stationary-phase dotA mutant than of stationary-phase strain JR32 is attributable to the kanamycin resistance element in dotA::Tn903, which conveys a low level of gentamicin resistance.
In the studies that demonstrated increased antibiotic resistance following intra-amoebic growth, L. pneumophila cells were isolated from the infection medium 3 days after infection of A. polyphagia (10). As noted above, Legionella begans to be released into the medium within a few hours after infection of amoebae. The 16- to 19-h duration of Ers treatment, in which Legionella was exposed to a phosphate-saline buffer similar to the A. polyphagia infection medium (10), is within the 3-day time frame of exposure to infection medium in the A. polyphagia studies. Therefore, exposure to the infection medium following Legionella lysis of amoeba hosts may have contributed to the increased antibiotic resistance previously attributed to intra-amoebic growth (10).
Mechanistic aspects of Ers suppression of mutant phenotypes in katA, katB, and icm/dot Legionella mutants
Ers treatment suppressed defective entry associated with null mutations in a diverse group of genes encoding structural and chaperone components of the Icm/Dot type IVB secretion apparatus (22, 63, 69, 74, 80) and catalase-peroxidase enzymes. To investigate the mechanism(s) by which suppression was accomplished, we first considered the possibility that Ers treatment induces multiple icm/dot genes, which then compensate for inactivation of individual icm/dot genes. This mechanism is unlikely because several icm::lacZ translational fusions show no significant change in expression during incubation of stationary-phase cultures in AC buffer (Gil Segal, Tel Aviv University, 2004, personal communication). In addition, induction of icm/dot genes is an unlikely mechanism for suppression of entry defects in mutants with mutations in KatA and KatB catalase-peroxidase enzymes that are not components of the Icm/Dot type IVB secretion machinery. Second, we considered the possibility that Ers treatment enhances adhesion of Legionella to host cells, thus enhancing entry. This possibility is not supported by the results obtained by the fluorescence microscopy method that distinguished internalized bacteria from external bacteria. Specifically, Ers treatment does not increase association of external L. pneumophila with amoebae or macrophage envelopes. Third, we considered the possibility that Ers treatment increases piliation, which has recently been implicated in entry by isolation of a gene with homology to the genes encoding pilin subunits of type IV secretion systems (60). This possibility is not supported by the results of electron microscopy with phosphotungstate and ammonium molybdate negative staining. No difference in piliation was seen between stationary-phase and Ers-treated cultures of the dotA mutant, which responded to Ers treatment, or the icmS mutant, which did not respond to Ers treatment (data not shown). Fourth, we considered the possibility that Ers treatment changes the bacterial surface morphology, eliciting changes responsible for enhanced entry. This possibility is not supported by electron microscopy data for stationary-phase and Ers-treated cultures of dotA and icmS mutants. In both mutants and in both culture conditions, the cell envelope is smooth and unruffled, and the speckling patterns that are reflective of involutions in the cell surface are comparable. The cell widths are identical for the two mutants and are not changed by the culture conditions, and the cell length is shorter in Ers-treated cultures, which is consistent with starvation, utilization of energy stores, and decreased entry into a replicative state (data not shown). The dramatic differences in cell morphology seen in amoeba-grown Legionella by light and electron microscopy (20, 32) are not evident in Ers-treated cultures. Fifth, we considered the possibility that Ers treatment decreases viability and that enhanced entry can be attributed to increased phagocytosis of dead bacteria. This possibility is not consistent with the viability measurements. The levels of viability of broth-grown stationary-phase cultures of wild-type strain JR32 and the katA and dotA mutants are 95% ± 5%, 92% ± 3%, and 96% ± 4%, respectively. The levels of viability of Ers-treated broth cultures are 94% ± 5%, 95% ± 2%, and 94% ± 2%, respectively. Thus, Ers treatment does not significantly decrease the viability of broth-grown stationary-phase cultures, and high levels of viability are evident in both stationary-phase and Ers-treated cultures.
Protein expression differences between stationary-phase L. pneumophila and intra-amoebically grown bacteria are evident as determined by SDS-polyacrylamide gel electrophoresis (20). To establish if protein expression is required for Ers suppression of defective entry, entry into A. castellanii was determined for stationary-phase cultures and for stationary-phase cultures exposed to Ers treatment in the absence or in the presence chloramphenicol (Fig. 6). Chloramphenicol inhibition of protein synthesis abolished Ers treatment-enhanced entry of katA and icmF mutants. Entry of wild-type Legionella into amoebae was not significantly altered by Ers treatment in the absence or presence of chloramphenicol. These results demonstrated that protein synthesis occurs during Ers treatment and is required for Ers suppression of the entry defect in katA and icmF mutants and presumably is required for suppression of other mutants.
FIG. 6.
Chloramphenicol inhibits Ers-enhanced entry into A. castellanii for icmF and katA mutants. Entry into A. castellanii was determined for wild-type strain JR32 and icmF and katA mutant strains as described in the legend to Fig. 4 with stationary-phase cultures (Stat), Ers-treated stationary-phase cultures (Ers), and Ers-treated stationary-phase cultures in the presence of chloramphenicol (ErsCm and Ers + Cm).
Proteomic analysis of Ers treatment-induced changes in expression.
To directly examine expression changes implicated in the chloramphenicol inhibition experiments, stationary-phase and Ers-treated L. pneumophila cultures were compared by using two-dimensional gel electrophoresis, MALDI-TOF analysis of tryptic digests, and data mining of the L. pneumophila genome. This approach has been used with other bacterial species to assess changes in protein expression in different culture conditions (12, 34, 56, 82). Two-dimensional gels were prepared by using proteins in sonic extracts of katA, dotA, dotB, icmF, icmS, and icmP mutant strains.
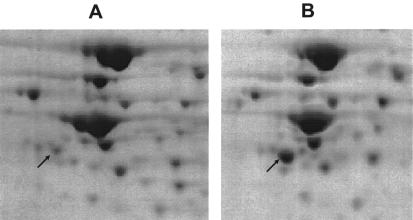
Intensity differences between stationary-phase and Ers-treated cultures were consistently seen in about 15 spots. One protein whose level was consistently decreased after Ers treatment is the Legionella homologue of cell division protein FtsZ, ORF lpg 2609 in the L. pneumophila genome (James Russo, Columbia Genome Center, personal communication). A decrease in the FtsZ level is consistent with the decreased replication expected in cultures exposed to Ers treatment, in which stationary-phase bacteria are incubated in a nutrient-deficient buffer. The decreased expression of FtsZ after Ers treatment is consistent with electron microscopy changes in cell length that are suggestive of decreased replication after Ers treatment. All strains whose defective entry into macrophages was suppressed by Ers treatment (katA, dotA, dotB, and icmF mutants) (Fig. 5) showed a large increase in the intensity of a spot in Ers-treated cultures compared to stationary-phase cultures (Fig. 7). In strains whose defective entry into macrophages was not suppressed by Ers treatment (icmP and icmS mutants) (Fig. 5), the intensity of the spot was not increased by Ers treatment. The protein in this spot, ORF lpg2222 in the L. pneumophila genome (Russo, personal communication), is predicted (49; http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) to containeight consecutive tetratricopeptide repeats (TPRs) (13) and is highly homologous (E value, <e−25) to known TPR proteins. The TPR motif is a degenerate repeat consisting of ∼34 amino acids that functions in protein-protein interactions, often in multiprotein complexes. This motif is found in yeast transcription factors, eukaryotic chaperones involved in protein import to peroxisomes, and mitochondria (13) and has recently been implicated in chaperones and regulators of bacterial type III secretion systems (57).
FIG. 7.
Two-dimensional gel electrophoresis analysis of L. pneumophila katA mutant. Isoelectric focusing (horizontal) and SDS-polyacrylamide gel electrophoresis (vertical) were performed with sonic extracts from stationary-phase (A) and Ers-treated stationary-phase (B) cultures. The regions shown are ∼20% of each gel surrounding ORF lpg2222 (arrows) (calculated molecular weight, 41,400; calculated pI 5.35). The amount of protein loaded in panel A was greater than the amount loaded in panel B.
DISCUSSION
Environmental stress plays a major role in the transition of L. pneumophila from a planktonic pond resident to an intracellular pathogen of alveolar macrophages. In the laboratory, growth to the stationary phase is widely used to model this stress-induced transition. When stationary-phase cultures are used as the infectious inocula, Legionella homologues of enteric regulators of stress, starvation, and/or stationary-phase responses play a role in expression of intracellular multiplication and other Legionella virulence phenotypes. Among these regulators are L. pneumophila homologues of RpoS (4, 36, 84), an alternative sigma factor, GacA (LetA) (31, 37), a global activator, and CsrA (28, 51), an RNA-binding protein in the carbon starvation response.
A major focus in studying the pathways connecting stress with virulence phenotypes is the icm/dot loci, which encode a type IVB secretion apparatus proposed to translocate Legionella proteins into host cells. The type IVB apparatus and its translocated proteins are involved in entry, evasion or delay of fusion between lysosomes and Legionella-containing phagosomes, association of the phagosome with the endoplasmic reticulum, intracellular multiplication, cytotoxicity, and the release of replicated bacteria from host cells (63, 69, 73, 80). However, Legionella RpoS and GacA have no significant effect on the expression of the icm/dot transcriptional units (31, 84). CpxR, a regulator of the enteric response to envelope stress (58), activates expression of IcmR (30), which influences formation of pores or channels in host cells by IcmQ (26). The effect of CsrA on icm/dot gene expression has not been reported. Since these stress regulators have a limited effect or no effect on expression of the icm/dot genes, they appear to control non-icm/dot virulence genes outside the icm/dot loci.
The presence of icm/dot-independent virulence genes is indicated by data showing that certain attributes of the Legionella-containing phagosome do not require a functional Icm/Dot apparatus. DotA, an essential component of the Icm/Dot secretion apparatus (11, 43, 44, 63, 64), and DotB (72) are not required for the delayed acquisition of cathepsin D or Texas Red ovalbumin lysosomal markers by Legionella-containing phagosomes (43). These findings indicate that icm/dot-independent genes are present. Icm/Dot-independent genes are non-icm/dot genes that play a role in Legionella virulence phenotypes in the absence of fully functional icm/dot genes. The identities of such icm/dot-independent genes and methods to screen for them are poorly defined.
In this paper we propose Ers treatment as an avenue for identifying icm/dot-independent Legionella virulence factors. In Ers treatment, broth-grown stationary-phase cultures are incubated overnight in AC buffer, a pH 6.5 phosphate-saline buffer which does not support replication of L. pneumophila (39, 50, 71). When stationary-phase cultures of icm/dot mutants are used for infection, defective entry into amoeba and macrophage hosts is observed (39). When the same icm/dot mutants are subjected to Ers treatment, entry into both amoeba and macrophage hosts is significantly increased. Entry is restored to wild-type or nearly wild-type values for dotA, dotB, icmF, icmV, and icmX Tn insertion mutants. Ers treatment also significantly increases and restores to wild-type values the entry of null mutants with mutations in the katA- and katB-encoded catalase-peroxidase enzymes, which are similarly defective in entry when stationary-phase cultures are used for infection. To our knowledge, Ers treatment is the first instance of suppressing a virulence defect in an L. pneumophila mutant by a change in culture conditions. For a null mutant with a mutation in the Legionella general stress gene, gspA, intra-amoebic growth suppresses sensitivity to oxidative, osmotic, acid, and thermal stresses (2). However, the gspA mutant is not defective in intracellular multiplication and cytotoxicity.
Ers suppression of defective entry indicates that Ers treatment-induced changes in gene expression can restore entry into amoeba and macrophage hosts to wild-type values in the absence of the icm/dot or the catalase-peroxidase gene that is inactivated. Therefore, Ers treatment is a screening procedure for icm/dot-independent factors that act in Legionella entry into eukaryotic hosts, an early step in Legionella pathogenicity. A remarkable feature of Ers treatment is its pleiotropic nature. Ers treatment suppresses defective entry into both amoebae and HL60-derived macrophages. It restores entry for mutants with mutations in catalase-peroxidase enzymes (periplasmic KatA and cytosolic KatB) (6) and membrane-associated (DotA, IcmV), cytosolic (DotB), and predicted periplasmic (IcmX) components of the type IVB secretion apparatus (69). Thus, identifying the molecular players in Ers treatment suppression and their mechanism of action may lead to new insights into L. pneumophila pathogenicity.
Since protein synthesis is required for Ers suppression, analysis of expression differences between stationary-phase and Ers-treated cultures is a means of investigating the mechanism of Ers suppression and identifying icm/dot-independent virulence factors. We developed a proteomic approach for identifying Ers-induced expression changes and observed Ers-dependent induction of ORF lpg2222 in the L. pneumophila genome. The ORF lpg2222 protein is predicted to contain TPRs. This protein-protein interaction motif is also present in EnhC (ORF lpg2639), previously associated with enhanced entry in a screening of mutagenized L. pneumophila strain AA100 for enhanced entry into macrophage and epithelial cell hosts (21). Enhanced entry of the enhC mutant is attributed to elevated enhC expression. A comparison of enhC mutant and wild-type stationary-phase cultures revealed no difference in pili or flagella, similar to the results obtained when we compared stationary-phase and Ers-treated L. pneumophila. It is not known if enhC is upregulated by Ers treatment, if elevated expression of enhC suppresses the defective entry of dotA, dotB, and other icm/dot mutants, or if expression of enhC or ORF lpg2222 is influenced by Legionella stress regulators.
Association of two TPR-containing proteins with entry into macrophage hosts raises the possibility that the TPR motif is important for protein-protein interactions in this step in Legionella infection. EnhC contains 13 TPR motif regions (49;http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). ORFlpg 2222 contains eight TPR motifs, one 35 residues long and seven 36 residues long. Initially found in yeast cell division cycle proteins, TPR motifs participate in protein-protein interactions of transcription factors and cochaperones and in the import of proteins into the matrix of peroxisomes and mitochondria (13). Recently, TPR motifs have been predicted in known chaperones and regulators of the bacterial type III secretion apparatus (57). However, no TPRs are predicted (49; http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) in VirE1 and IcmR, which are known chaperones in the type IVB secretion systems of Agrobacterium tumefaciens (24) and L. pneumophila (22, 25). Knowledge of the protein partners with which the TPR motif proteins EnhC and ORF lpg2222 interact—whether they interact with Icm/Dot proteins, with one another, or with novel proteins—is important information for determining their roles in the entry of L. pneumophila into host cells. For ORF lpg2222, this information may also provide insight into the icm/dot-independent factors that act in suppression of defective entry of icm/dot and katA/B Legionella mutants into amoeba and macrophage hosts by Ers treatment.
Acknowledgments
This research was supported by NSF grant MCB-980992 to H.M.S. and by REU supplements that supported the summer research of H.A.C. and A.P.-W.
We thank Howard Shuman, Department of Microbiology and Immunology, Columbia University College of Physicians and Surgeons, for icm/dot mutant strains and advice on the immunofluorescence entry method, and we thank colleagues at Albert Einstein College of Medicine, including the staff of the Laboratory for Macromolecular Analysis and Proteomics (LMAP) for instruction on the use of LMAP isoelectric focusing and LMAP MALDI-TOF equipment, Frank Macaluso of the Analytical Ultrastructure Laboratory for electron microscopy, Magdia De Jesus for analyses of the electron micrographs, and Anne Bresnick for use of her fluorescence microscope.
Editor: D. L. Burns
REFERENCES
- 1.Abu Kwaik, Y., B. I. Eisenstein, and N. C. Engleberg. 1993. Phenotypic modulation by Legionella pneumophila upon infection of macrophages. Infect. Immun. 61:1320-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abu Kwaik, Y., L. Y. Gao, O. S. Harb, and B. J. Stone. 1997. Transcriptional regulation of the macrophage-induced gene (gspA) of Legionella pneumophila and phenotypic characterization of a null mutant. Mol. Microbiol. 24:629-642. [DOI] [PubMed] [Google Scholar]
- 3.Abu Kwaik, Y., L. Y. Gao, B. J. Stone, C. Venkataraman, and O. S. Harb. 1998. Invasion of protozoa by Legionella pneumophila and its role in bacterial ecology and pathogenesis. Appl. Environ. Microbiol. 64:3127-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachman, M. A., and M. S. Swanson. 2001. RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. Mol. Microbiol. 40:1201-1214. [DOI] [PubMed] [Google Scholar]
- 5.Bandyopadhyay, P., B. Byrne, Y. Chan, M. S. Swanson, and H. M. Steinman. 2003. The Legionella pneumophila catalase-peroxidases are required for proper trafficking and growth in primary macrophages. Infect. Immun. 71:4526-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bandyopadhyay, P., and H. M. Steinman. 2000. Catalase-peroxidases of Legionella pneumophila: cloning of the katA gene and studies of KatA function. J. Bacteriol. 182:6679-6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bandyopadhyay, P., and H. M. Steinman. 1998. Legionella pneumophila catalase peroxidases: cloning of the katB gene and studies of KatB function. J. Bacteriol. 180:5369-5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barker, J., and M. R. Brown. 1995. Speculations on the influence of infecting phenotype on virulence and antibiotic susceptibility of Legionella pneumophila. J. Antimicrob. Chemother. 36:7-21. [DOI] [PubMed] [Google Scholar]
- 9.Barker, J., M. R. Brown, P. J. Collier, I. Farrell, and P. Gilbert. 1992. Relationship between Legionella pneumophila and Acanthamoeba polyphaga: physiological status and susceptibility to chemical inactivation. Appl. Environ. Microbiol. 58:2420-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barker, J., H. Scaife, and M. R. Brown. 1995. Intraphagocytic growth induces an antibiotic-resistant phenotype of Legionella pneumophila. Antimicrob. Agents Chemother. 39:2684-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berger, K. H., and R. R. Isberg. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7:7-19. [DOI] [PubMed] [Google Scholar]
- 12.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 13.Blatch, G. L., and M. Lassle. 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21:932-939. [DOI] [PubMed] [Google Scholar]
- 14.Brown, M. R., and J. Barker. 1999. Unexplored reservoirs of pathogenic bacteria: protozoa and biofilms. Trends Microbiol. 7:46-50. [DOI] [PubMed] [Google Scholar]
- 15.Byrne, B., and M. S. Swanson. 1998. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect. Immun. 66:3029-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, J., K. S. de Felipe, M. Clarke, H. Lu, O. R. Anderson, G. Segal, and H. A. Shuman. 2004. Legionella effectors that promote nonlytic release from protozoa. Science 303:1358-1361. [DOI] [PubMed] [Google Scholar]
- 17.Cianciotto, N. P. 2001. Pathogenicity of Legionella pneumophila. Int. J. Med. Microbiol. 291:331-343. [DOI] [PubMed] [Google Scholar]
- 18.Cianciotto, N. P., and B. S. Fields. 1992. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc. Natl. Acad. Sci. USA 89:5188-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cirillo, J. D., S. L. Cirillo, L. Yan, L. E. Bermudez, S. Falkow, and L. S. Tompkins. 1999. Intracellular growth in Acanthamoeba castellanii affects monocyte entry mechanisms and enhances virulence of Legionella pneumophila. Infect. Immun. 67:4427-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cirillo, J. D., S. Falkow, and L. S. Tompkins. 1994. Growth of Legionella pneumophila in Acanthamoeba castellanii enhances invasion. Infect. Immun. 62:3254-3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cirillo, S. L., J. Lum, and J. D. Cirillo. 2000. Identification of novel loci involved in entry by Legionella pneumophila. Microbiology 146:1345-1359. [DOI] [PubMed] [Google Scholar]
- 22.Coers, J., J. C. Kagan, M. Matthews, H. Nagai, D. M. Zuckman, and C. R. Roy. 2000. Identification of Icm protein complexes that play distinct roles in the biogenesis of an organelle permissive for Legionella pneumophila intracellular growth. Mol. Microbiol. 38:719-736. [DOI] [PubMed] [Google Scholar]
- 23.Conover, G. M., I. Derre, J. P. Vogel, and R. R. Isberg. 2003. The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol. Microbiol. 48:305-321. [DOI] [PubMed] [Google Scholar]
- 24.Deng, W., L. Chen, W. T. Peng, X. Liang, S. Sekiguchi, M. P. Gordon, L. Comai, and E. W. Nester. 1999. VirE1 is a specific molecular chaperone for the exported single-stranded-DNA-binding protein VirE2 in Agrobacterium. Mol. Microbiol. 31:1795-1807. [DOI] [PubMed] [Google Scholar]
- 25.Dumenil, G., and R. R. Isberg. 2001. The Legionella pneumophila IcmR protein exhibits chaperone activity for IcmQ by preventing its participation in high-molecular-weight complexes. Mol. Microbiol. 40:1113-1127. [DOI] [PubMed] [Google Scholar]
- 26.Dumenil, G., T. P. Montminy, M. Tang, and R. R. Isberg. 2004. IcmR-regulated membrane insertion and efflux by the Legionella pneumophila IcmQ protein. J. Biol. Chem. 279:4686-4695. [DOI] [PubMed] [Google Scholar]
- 27.Feeley, J. C., R. J. Gibson, G. W. Gorman, N. C. Langford, J. K. Rasheed, D. C. Mackel, and W. B. Baine. 1979. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J. Clin. Microbiol. 10:437-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fettes, P. S., V. Forsbach-Birk, D. Lynch, and R. Marre. 2001. Overexpresssion of a Legionella pneumophila homologue of the E. coli regulator csrA affects cell size, flagellation, and pigmentation. Int. J. Med. Microbiol. 291:353-360. [DOI] [PubMed] [Google Scholar]
- 29.Fields, B. S. 1996. The molecular ecology of legionellae. Trends Microbiol. 4:286-290. [DOI] [PubMed] [Google Scholar]
- 30.Gal-Mor, O., and G. Segal. 2003. Identification of CpxR as a positive regulator of icm and dot virulence genes of Legionella pneumophila. J. Bacteriol. 185:4908-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gal-Mor, O., and G. Segal. 2003. The Legionella pneumophila GacA homolog (LetA) is involved in the regulation of icm virulence genes and is required for intracellular multiplication in Acanthamoeba castellanii. Microb. Pathog. 34:187-194. [DOI] [PubMed] [Google Scholar]
- 32.Garduno, R. A., E. Garduno, M. Hiltz, and P. S. Hoffman. 2002. Intracellular growth of Legionella pneumophila gives rise to a differentiated form dissimilar to stationary-phase forms. Infect. Immun. 70:6273-6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.George, K. M., Y. Yuan, D. R. Sherman, and C. E. Barry III. 1995. The biosynthesis of cyclopropanated mycolic acids in Mycobacterium tuberculosis. Identification and functional analysis of CMAS-2. J. Biol. Chem. 270:27292-27298. [DOI] [PubMed] [Google Scholar]
- 34.Gupta, S., S. B. Pandit, N. Srinivasan, and D. Chatterji. 2002. Proteomics analysis of carbon-starved Mycobacterium smegmatis: induction of Dps-like protein. Protein Eng. 15:503-512. [DOI] [PubMed] [Google Scholar]
- 35.Halablab, M. A., M. Bazin, L. Richards, and J. Pacy. 1990. Ultra-structure and localisation of formazan formed by human neutrophils and amoebae phagocytosing virulent and avirulent Legionella pneumophila. FEMS Microbiol. Immunol. 2:295-301. [DOI] [PubMed] [Google Scholar]
- 36.Hales, L. M., and H. A. Shuman. 1999. The Legionella pneumophila rpoS gene is required for growth within Acanthamoeba castellanii. J. Bacteriol. 181:4879-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hammer, B. K., E. S. Tateda, and M. S. Swanson. 2002. A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol. Microbiol. 44:107-118. [DOI] [PubMed] [Google Scholar]
- 38.Harb, O. S., L. Y. Gao, and Y. Abu Kwaik. 2000. From protozoa to mammalian cells: a new paradigm in the life cycle of intracellular bacterial pathogens. Environ. Microbiol. 2:251-265. [DOI] [PubMed] [Google Scholar]
- 39.Hilbi, H., G. Segal, and H. A. Shuman. 2001. Icm/Dot-dependent upregulation of phagocytosis by Legionella pneumophila. Mol. Microbiol. 42:603-617. [DOI] [PubMed] [Google Scholar]
- 40.Horwitz, M. A., and S. C. Silverstein. 1983. Intracellular multiplication of Legionnaires' disease bacteria (Legionella pneumophila) in human monocytes is reversibly inhibited by erythromycin and rifampin. J. Clin. Investig. 71:15-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jacobs, R. F., R. M. Locksley, C. B. Wilson, J. E. Haas, and S. J. Klebanoff. 1984. Interaction of primate alveolar macrophages and Legionella pneumophila. J. Clin. Investig. 73:1515-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jenkins, D. E., J. E. Schultz, and A. Matin. 1988. Starvation-induced cross-protection against heat or H2O2 challenge in Escherichia coli. J. Bacteriol. 170:3910-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joshi, A. D., S. Sturgill-Koszycki, and M. S. Swanson. 2001. Evidence that Dot-dependent and -independent factors isolate the Legionella pneumophila phagosome from the endocytic network in mouse macrophages. Cell. Microbiol. 3:99-114. [DOI] [PubMed] [Google Scholar]
- 44.Kirby, J. E., J. P. Vogel, H. L. Andrews, and R. R. Isberg. 1998. Evidence for pore-forming ability by Legionella pneumophila. Mol. Microbiol. 27:323-336. [DOI] [PubMed] [Google Scholar]
- 45.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 46.Kura, F., K. Suzuki, H. Watanabe, Y. Akamatsu, and F. Amano. 1994. Difference in Legionella pneumophila growth permissiveness between J774.1 murine macrophage-like JA-4 cells and lipopolysaccharide (LPS)-resistant mutant cells, LPS1916, after stimulation with LPS. Infect. Immun. 62:5419-5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liles, M. R., P. H. Edelstein, and N. P. Cianciotto. 1999. The prepilin peptidase is required for protein secretion by and the virulence of the intracellular pathogen Legionella pneumophila. Mol. Microbiol. 31:959-970. [DOI] [PubMed] [Google Scholar]
- 48.Luo, Z. Q., and R. R. Isberg. 2004. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl. Acad. Sci. USA 101:841-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marchler-Bauer, A., J. B. Anderson, C. DeWeese-Scott, N. D. Fedorova, L. Y. Geer, S. He, D. I. Hurwitz, J. D. Jackson, A. R. Jacobs, C. J. Lanczycki, C. A. Liebert, C. Liu, T. Madej, G. H. Marchler, R. Mazumder, A. N. Nikolskaya, A. R. Panchenko, B. S. Rao, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, S. Vasudevan, Y. Wang, R. A. Yamashita, J. J. Yin, and S. H. Bryant. 2003. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31:383-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moffat, J. F., and L. S. Tompkins. 1992. A quantitative model of intracellular growth of Legionella pneumophila in Acanthamoeba castellanii. Infect. Immun. 60:296-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Molofsky, A. B., and M. S. Swanson. 2003. Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol. Microbiol. 50:445-461. [DOI] [PubMed] [Google Scholar]
- 52.Morales, V. M., A. Backman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 53.Nagai, H., J. C. Kagan, X. Zhu, R. A. Kahn, and C. R. Roy. 2002. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295:679-682. [DOI] [PubMed] [Google Scholar]
- 54.Nash, T. W., D. M. Libby, and M. A. Horwitz. 1984. Interaction between the Legionnaires' disease bacterium (Legionella pneumophila) and human alveolar macrophages. Influence of antibody, lymphokines, and hydrocortisone. J. Clin. Investig. 74:771-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Newman, J. R., and C. Fuqua. 1999. Broad-host-range expression vectors that carry the l-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227:197-203. [DOI] [PubMed] [Google Scholar]
- 56.Novotna, J., J. Vohradsky, P. Berndt, H. Gramajo, H. Langen, X. M. Li, W. Minas, L. Orsaria, D. Roeder, and C. J. Thompson. 2003. Proteomic studies of diauxic lag in the differentiating prokaryote Streptomyces coelicolor reveal a regulatory network of stress-induced proteins and central metabolic enzymes. Mol. Microbiol. 48:1289-1303. [DOI] [PubMed] [Google Scholar]
- 57.Pallen, M. J., M. S. Francis, and K. Futterer. 2003. Tetratricopeptide-like repeats in type-III-secretion chaperones and regulators. FEMS Microbiol. Lett. 223:53-60. [DOI] [PubMed] [Google Scholar]
- 58.Raivio, T. L., and T. J. Silhavy. 1999. The σE and Cpx regulatory pathways: overlapping but distinct envelope stress responses. Curr. Opin. Microbiol. 2:159-165. [DOI] [PubMed] [Google Scholar]
- 59.Rankin, S., Z. Li, and R. R. Isberg. 2002. Macrophage-induced genes of Legionella pneumophila: protection from reactive intermediates and solute imbalance during intracellular growth. Infect. Immun. 70:3637-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ridenour, D. A., S. L. Cirillo, S. Feng, M. M. Samrakandi, and J. D. Cirillo. 2003. Identification of a gene that affects the efficiency of host cell infection by Legionella pneumophila in a temperature-dependent fashion. Infect. Immun. 71:6256-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rogers, J., and C. W. Keevil. 1992. Immunogold and fluorescein immunolabeling of Legionella pneumophila within an aquatic biofilm visualized by using episcopic differential interference contrast microscopy. Appl. Environ. Microbiol. 58:2326-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rowbotham, T. J. 1986. Current views on the relationships between amoebae, legionellae and man. Isr. J. Med. Sci. 22:678-689. [PubMed] [Google Scholar]
- 63.Roy, C. R. 2002. The Dot/Lcm transporter of Legionella pneumophila: a bacterial conductor of vesicle trafficking that orchestrates the establishment of a replicative organelle in eukaryotic hosts. Int. J. Med. Microbiol. 291:463-467. [DOI] [PubMed] [Google Scholar]
- 64.Roy, C. R., K. H. Berger, and R. R. Isberg. 1998. Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake. Mol. Microbiol. 28:663-674. [DOI] [PubMed] [Google Scholar]
- 65.Ryan, K. J. 2004. Legionella, p. 415-420. In K. J. Ryan and C. G. Ray (ed.), Sherris medical microbiology. An introduction to infectious diseases, 4th ed. McGraw-Hill, New York, N.Y.
- 66.Sadosky, A. B., L. A. Wiater, and H. A. Shuman. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 61:5361-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sauve, D. M., D. T. Ho, and M. Roberge. 1995. Concentration of dilute protein for gel electrophoresis. Anal. Biochem. 226:382-383. [DOI] [PubMed] [Google Scholar]
- 68.Segal, G., and H. A. Shuman. 1997. Characterization of a new region required for macrophage killing by Legionella pneumophila. Infect. Immun. 65:5057-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Segal, G., and H. A. Shuman. 1998. How is the intracellular fate of the Legionella pneumophila phagosome determined? Trends Microbiol. 6:253-255. [DOI] [PubMed] [Google Scholar]
- 70.Segal, G., and H. A. Shuman. 1998. Intracellular multiplication and human macrophage killing by Legionella pneumophila are inhibited by conjugal components of IncQ plasmid RSF1010. Mol. Microbiol. 30:197-208. [DOI] [PubMed] [Google Scholar]
- 71.Segal, G., and H. A. Shuman. 1999. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect. Immun. 67:2117-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sexton, J. A., J. S. Pinkner, R. Roth, J. E. Heuser, S. J. Hultgren, and J. P. Vogel. 2004. The Legionella pneumophila PilT homologue DotB exhibits ATPase activity that is critical for intracellular growth. J. Bacteriol. 186:1658-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sexton, J. A., and J. P. Vogel. 2002. Type IVB secretion by intracellular pathogens. Traffic 3:178-185. [DOI] [PubMed] [Google Scholar]
- 74.Shuman, H. A., M. Purcell, G. Segal, L. Hales, and L. A. Wiater. 1998. Intracellular multiplication of Legionella pneumophila: human pathogen or accidental tourist? Curr. Top. Microbiol. Immunol. 225:99-112. [DOI] [PubMed] [Google Scholar]
- 75.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 76.Steinman, H. M. 1985. Bacteriocuprein superoxide dismutases in pseudomonads. J. Bacteriol. 162:1255-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stone, B. J., and Y. A. Kwaik. 1999. Natural competence for DNA transformation by Legionella pneumophila and its association with expression of type IV pili. J. Bacteriol. 181:1395-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sturgill-Koszycki, S., and M. S. Swanson. 2000. Legionella pneumophila replication vacuoles mature into acidic, endocytic organelles. J. Exp. Med. 192:1261-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Swanson, M. S., and E. Fernandez-Moreia. 2002. A microbial strategy to multiply in macrophages: the pregnant pause. Traffic 3:170-177. [DOI] [PubMed] [Google Scholar]
- 80.Swanson, M. S., and B. K. Hammer. 2000. Legionella pneumophila pathogenesis: a fateful journey from amoebae to macrophages. Annu. Rev. Microbiol. 54:567-613. [DOI] [PubMed] [Google Scholar]
- 81.Swanson, M. S., and R. R. Isberg. 1996. Identification of Legionella pneumophila mutants that have aberrant intracellular fates. Infect. Immun. 64:2585-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tremoulet, F., O. Duche, A. Namane, B. Martinie, and J. C. Labadie. 2002. A proteomic study of Escherichia coli O157:H7 NCTC 12900 cultivated in biofilm or in planktonic growth mode. FEMS Microbiol. Lett. 215:7-14. [DOI] [PubMed] [Google Scholar]
- 83.Wiater, L. A., A. B. Sadosky, and H. A. Shuman. 1994. Mutagenesis of Legionella pneumophila using Tn903dIIlacZ: identification of a growth-phase-regulated pigmentation gene. Mol. Microbiol. 11:641-653. [DOI] [PubMed] [Google Scholar]
- 84.Zusman, T., O. Gal-Mor, and G. Segal. 2002. Characterization of a Legionella pneumophila relA insertion mutant and roles of RelA and RpoS in virulence gene expression. J. Bacteriol. 184:67-75. [DOI] [PMC free article] [PubMed] [Google Scholar]