Abstract
Whereas the cellular basis of the hematopoietic stem cell (HSC) niche in the bone marrow has been characterized, the nature of the fetal liver (FL) niche is not yet elucidated. We show that Nestin+NG2+ pericytes associate with portal vessels, forming a niche promoting HSC expansion. Nestin+NG2+ cells and HSCs scale during development with the fractal branching patterns of portal vessels, tributaries of the umbilical vein. After closure of the umbilical inlet at birth, portal vessels undergo a transition from Neuropilin-1+Ephrin-B2+ artery to EphB4+ vein phenotype, associated with a loss of periportal Nestin+NG2+ cells and emigration of HSCs away from portal vessels. These data support a model in which HSCs are titrated against a periportal vascular niche with a fractal-like organization enabled by placental circulation.
Hematopoietic stem cells (HSCs) are generated in the mouse fetus around embryonic day 10.5 (E10.5) from hemogenic endothelium of the dorsal aorta (1, 2), then migrate to the placenta via the umbilical arteries (3) and return to the fetus via the umbilical vein (4). The umbilical vein delivers oxygenated blood to the fetus via the portal sinus whose branches give rise to the portal vessels in the fetal liver (FL). In this organ, HSCs undergo marked expansion (5). The predictable growth curve of HSCs during development suggests hitherto unknown determinants set the numbers of these cells.
Although FL HSCs are highly proliferative, a hallmark of adult bone marrow (BM) HSCs is their cell cycle quiescence (6). Perivascular cells expressing Nestin (7), CXCL12 (8) and the leptin receptor (9) contribute to HSC maintenance. Nestin+NG2+ arteriolar pericytes (10) as well as megakaryocytes (11, 12) maintain quiescent HSCs in the BM. Much less is known about the FL niche promoting HSC proliferation. FL-derived stromal cell lines support HSC expansion in vitro (13, 14). However, a HSC niche in the liver has not been demonstrated in vivo.
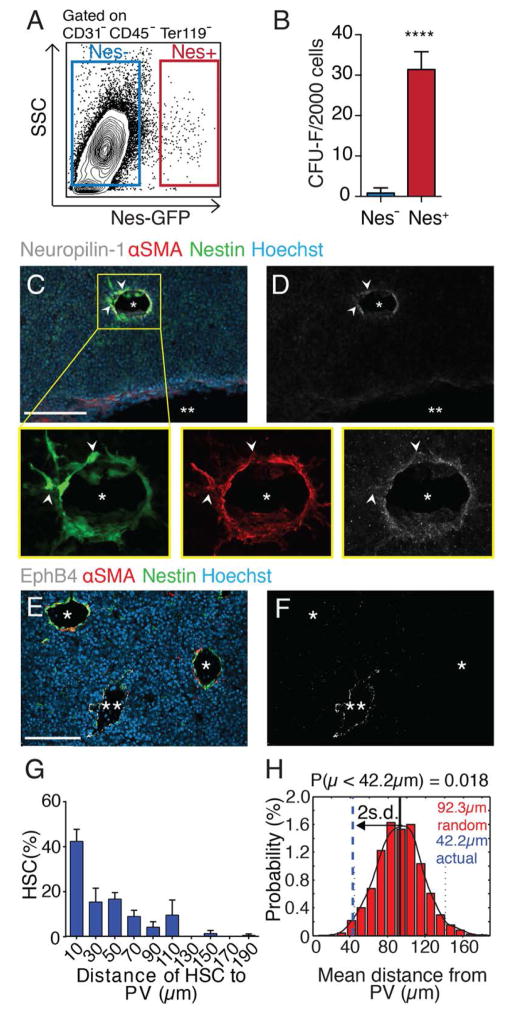
Within the E14.5 FL of Nestin-GFP transgenic mice, endothelial cells and a rare population of stromal cells (Fig. 1A, 0.045% ± 0.007% of total nucleated cells), are marked by GFP. These stromal cells, hereafter termed Nestin+ cells, are highly enriched in colony-forming unit-fibroblast activity (CFU-F) (Fig. 1B), and expressed mesenchymal lineage markers and DLK1, but neither the biliary marker EpCAM (15) nor hepatic genes (fig. S1, A and B). FL CFU-F colonies derived from Nestin+ had trilineage mesenchymal lineage capacity when cultured in defined conditions (fig. S1, C to F). Nestin+ cells expressed α-smooth muscle actin (αSMA) (fig. S1G), the pericyte marker NG2 (fig. S2, C and D) and colocalized with αSMA staining on portal vessels expressing the endothelial arterial markers Ephrin-B2 and Neuropilin-1, but not the venular marker EphB4 (fig. S1, H to J, and Fig. 1, C to F). Thus, Nestin+ cells are pericytes abutting Efnb2 and Neuropilin-1-expressing endothelia on portal vessels. Lastly, Nestin+ cells were enriched for HSC niche and expansion factors (fig. S1, K and L), raising the potential for regulating FL HSCs.
Fig. 1. Peri-arterial Nestin+ stromal cells associate with HSCs in the fetal liver.
(A) FACS analysis of Nestin-GFP stroma from E14.5 FL. (B) CFU-F from sorted FL cells, n = 4. (C to F) Immunofluorescence analyses of Nestin-GFP E14.5 FL cryosections stained for Neuropilin-1 (white; C, D) or EphB4 (white; E, F). Colocalization of αSMA+ and Nestin+ pericytes around Neuropilin-1+ portal vessels (*) but not EphB4+ veins (**). Scale bar, 100 μm. (G) Distance distribution between CD150+CD48−CD41−Lineage− HSCs and Nestin+ cells from whole-mount stained Nestin-GFP FL (n = 105 from 12 E14.5 FLs), binned into 20 μm intervals. (H) Probability distribution of mean distances from simulations of randomly positioned HSCs in relation to Nestin+ cells. Actual mean distance shown in relation to mean of simulations ± 2 s.d. ****P < 10−4.
To evaluate the spatial relationships between HSCs and Nestin+ cells, we stained CD150+CD48−CD41−Lineage− HSCs in whole-mount FLs and evaluated the significance of the associations by computational modeling (10). A large HSC fraction (>40%) was located within 20 μm from Nestin+ cells on portal vessels (Fig. 1G). We then simulated nonpreferential HSC placement on images of whole-mount prepared FLs to define the distribution of randomly localized HSC to portal vessels. The observed HSC mean distance to Nestin+ cells (42.2 μm) was statistically different from that of randomly placed HSCs (92.3 μm, P = 0.018) (Fig. 1H). The close physical associations between HSCs and Nestin+ periportal cells suggest that the portal vasculature may harbor a HSC niche.
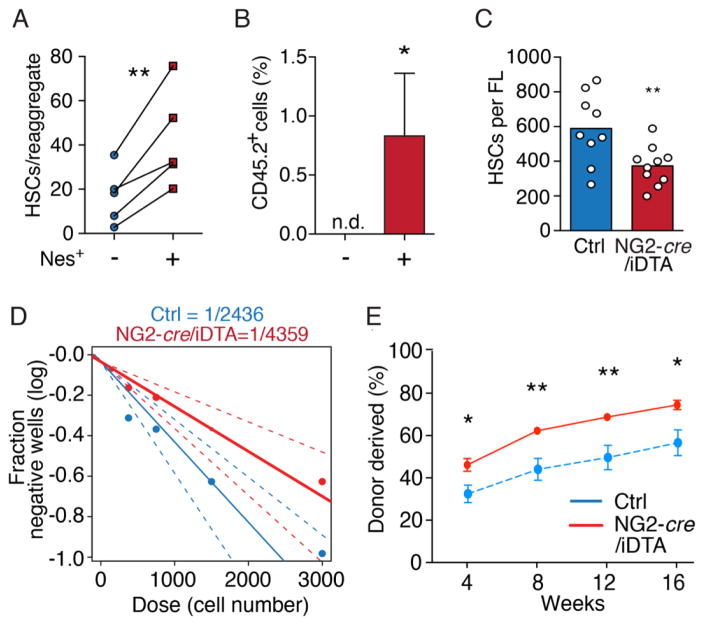
We adapted the reaggregate organ culture assay in which selected populations are pelleted and cultured on the surface of a porous membrane (16). A FL cell mixture containing hematopoietic progenitor cells (Lineage−CD45+), CD31+ endothelial cells, and hepatic parenchymal/stromal cells (CD45−Lineage−) were separated by cell sorting and were reaggregated with or without Nestin+ cells (ratio ~235/1) and cultured in the absence of exogenous cytokines or serum (fig. S2A). Significantly higher numbers of phenotypic HSCs (17) were maintained in the reaggregates containing Nestin+ cells (Fig. 2A and fig. S2B), and these reaggregates contained detectable long-term repopulating FL HSC activity after transplantation whereas the controls did not (Fig. 2B). These results suggest that factors produced by the FL Nestin+ cells were sufficient to maintain HSCs in culture.
Fig. 2. Nestin+ perivascular cells drive HSC expansion in vivo.
(A) FACS quantification of CD150+Sca1+CD45+CD48−Lin− HSCs in reaggregates with or without Nestin+ cells. (B) Donor engraftment 16 weeks after transplantation of reaggregates together with congenic competitor cells. n.d. = not detectable. n = 6–7 per group. (C) CD150+CD48−Sca1+CD11b+CD45+CD41−Lineage−DAPI− HSC numbers in control and NG2-depleted littermates. (D) Limiting dilution analyses of LTC-ICs in Lineage− cells from NG2-depleted (red) and control (blue) littermate FLs. (E) Competitive repopulation assays of NG2-depleted and control FL. n = 6/7 mice per control/depleted littermates. *P < 0.05, **P < 0.01.
By contrast to Nestin-GFP which also labels FL endothelial cells, NG2 expression was specific to Nestin+ pericytes (~98% overlap) (fig. S2, C to G) and αSMA+ portal vessels, as determined by NG2-cre transgenic mice (18) crossed with inducible TdTomato transgenic mice (fig. S2, H and I). We then crossed male NG2-cre mice with female Cre-inducible Diphtheria toxin A (iDTA) mice to deplete NG2+ cells by selective expression of DTA following NG2-cre-mediated excision of a floxed-STOP cassette (19). Embryos were staged in utero by crown-rump length (CRL) using ultrasound imaging (fig. S2J). NG2-cre/iDTA fetuses did not exhibit gross developmental defects compared to littermates (fig. S3A), and had similar CRL (fig. S3B). Although NG2-cre activity was detected in mural cells surrounding the midline dorsal aorta at E11 (fig. S3C), we found no difference in HSCs and LSKs in E12–12.5 FLs of NG2-cre/iDTA transgenic mice (fig. S3, D to G), suggesting that HSC specification and homing to FL was not significantly altered. At E14.5, however, HSCs were significantly reduced (by ~45%) (Fig. 2C and fig. S3H) in FLs from NG2-cre/iDTA fetuses compared to control littermates, whereas progenitors were unaffected (fig. S3, I and J). Limiting dilution analyses also revealed a 45% reduction of long-term culture-initiating colonies (LTC-IC) (20) in NG2-cre;iDTA FLs compared to control littermates (Fig. 2D). These data suggest that Nestin+NG2+ cells provide a niche in the FL.
HSCs from NG2-cre/iDTA mice FLs exhibited reductions (~43%) in BrdU uptake compared to littermate controls, suggesting that Nestin+NG2+ cells are required for HSC proliferation (fig. S3K). Consistent with previous reports suggesting a reduced capacity of S/G2/M-phase HSCs to engraft irradiated recipient mice (20, 21), we observed an increase in HSC engraftment in NG2-cre/iDTA FLs compared to control littermates (Fig. 2E and fig. S4A). Indeed, engraftment-competent G0/G1-phase HSCs were enriched in NG2-cre/iDTA mice compared to littermate controls, whereas the S/G2/M-phase HSCs (DNA>2n) were reduced (fig. S4, B to F). Together, these data indicate that Nestin+ cells are required to drive fetal HSC expansion.
Since arteriole-associated Nestin+NG2+ cells promote HSC quiescence in adult BM (10) and analogous cells appear to exert opposite functions in the FL, we next sequenced cDNA libraries of Nestin+NG2+ cells sorted from BM and FL stroma from Nestin-GFP;NG2-DsRed double-transgenic mice (fig. S5). Most (95.6%) BM and FL Nestin+NG2+ cell transcripts were statistically indistinguishable, but FL transcripts accounted for the majority of differentially expressed (DE) genes (fig. S5, A and B) suggesting that these cells are highly similar. Indeed, BM and FL Nestin+NG2+ cells had similar expression of many HSC niche and pericyte genes (fig. S5, F and H). Accordingly, reaggregation of E14.5 FL cell mixtures with similar numbers of adult BM Nestinbright cells (10) or FL Nestin+ cells revealed a similar capacity to support HSCs (fig. S4G). In addition, Scf, Angptl2, and Igf2 expression in Nestin+ cells were similar at E12, E13, and E14.5, suggesting that Nestin+ cells do not expand HSCs by increasing expression of these factors (fig. S4, H and I). FL DE genes were analyzed with Enrichr, gene-set enrichment analysis tool (22); genes involved in mitosis, metabolism and growth, as well as knock-out phenotypes including abnormal blood vessel development, blood circulation and cell proliferation were significantly enriched (fig. S5, C to E). ChIP-seq enrichment analyses (23) applied to identify upstream regulatory factors highly connected to FL DE genes pointed to key drivers of cell cycle progression (fig. S6, A to C). Accordingly, FL Nestin+ cells differentially expressed cell cycle genes including Ki67 and were actively proliferating (figs. S5I and S1M).
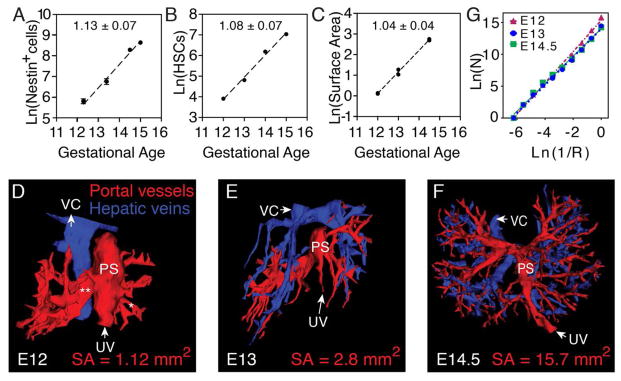
The enrichment of genes promoting cell cycle progression and vascular development supports the hypothesis—as implied by the niche concept—that the expansion of Nestin+NG2+ cell and portal vessel are synchronized with HSC expansion. The absolute numbers of Nestin+ cells increased logarithmically over E12 to E14.5 (Fig. 3A). When compared to numbers of HSCs (Fig. 3B) (5), the rates of expansion were indistinguishable (Fig. 3, A and B) (P = 0.69, ANCOVA). We next mapped the vasculature by tracing vessels in consecutive cryosections of Nestin-GFP+ FLs from E12 to E14.5 (fig. S7, A to C) to generate complete 3D surface maps (Fig. 3, D to F). Portal vessels receiving blood from the umbilical vein (UV) were lined with Nestin+ cells (fig. S7, A to C and Fig. 3, D to F), in contrast to αSMA-negative hepatic veins that lacked Nestin+ cell lining (fig. S7C). At E13 and E14.5, portal vessel branching became progressively more intricate (Fig. 3, E and F) and the surface area of these structures increased logarithmically. Nestin+ cell numbers and portal vessels’ surface area scaled over time with indistinguishable slopes (P = 0.47, ANCOVA) (Fig. 3, A and C), suggesting that the HSC niche expansion is also related to portal vessel surface area.
Fig. 3. Fractal geometry underlies HSC expansion.
(A) Nestin+ cells expand from E12–E15 according to a power law (R2 = 0.95). (B) HSC numbers relative to gestational age plotted from quantification of Ema et al. (5). (C) Portal vessel surface area from E12–E14.5 scales according to similar power law (R2 = 0.99; n = 6). Slopes indicated in panels. (D to F) 3D reconstructions of afferent PVs (red) and efferent veins (blue) in E12 (D), E13 (E), and E14.5 (F) FLs. PS, portal sinus; VC, vena cava; SA, surface area. (G) Calculation of the scale-invariant fractal dimension of FL afferent vessels using box-counting.
That Nestin+ cell and portal vessel expansion fit a line when plotted on logarithmic ordinates suggests that they follow a fractal-like organization. Fractals are geometries exhibiting self-similarity; the observed structures share characteristics across spatial levels (24). To characterize the multi-scale behavior of portal vessels, we applied a 3D box-counting algorithm. This method consists of overlaying the object with a series of 3D grids of exponentially decreasing block sizes (R) and counting the number of boxes intersecting the object (N). By analyzing the slope of N plotted against R−1 on a bi-logarithmic scale, the dimensionality of the object was determined. Box-counting analysis revealed that portal vessels were self-similar over three decades of scales. Furthermore, the dimensionality was relatively constant as portal vessels branched over E12 to E14.5 (2.15–2.35) (Fig. 3G). These findings suggest that the expansion of HSCs and Nestin+ cells during fetal development is governed by fractal-like geometries of the portal vessel niche.
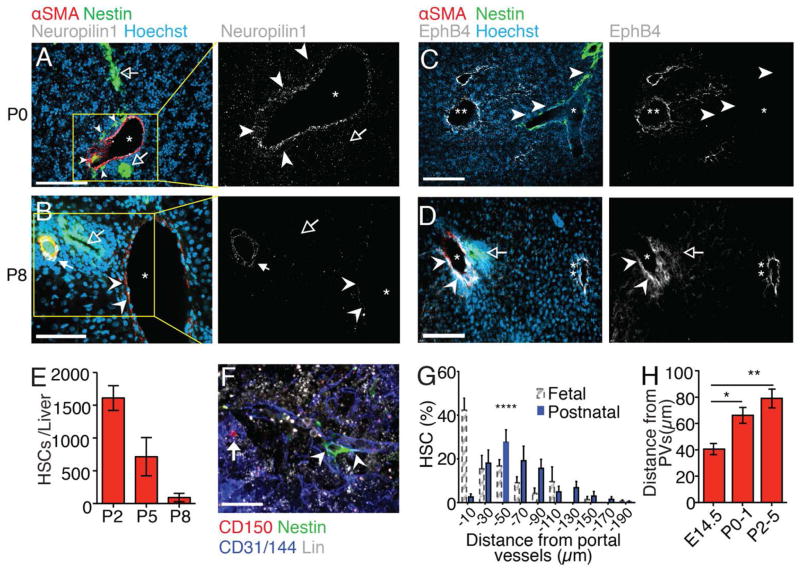
After birth, ligation of the umbilical inlet leads to dramatic hemodynamic changes in portal vessel flow (fig. S8, A to E). Whereas at P0 portal vessels expressed the arterial markers Neuropilin-1 and Ephrin-B2, and were accompanied by Nestin+ cells (Fig. 4A and fig. S8F), their expression levels were markedly reduced in P8 portal vessels which were, by then, devoid of Nestin+ cells (Fig. 4B and fig. S8G), in part due to Nestin+ cell apoptosis as detected by TUNEL staining (fig. S8, H and I). At this time, portal vessels expressed EphB4 (Fig. 4, C and D) suggesting that they transited into a vein phenotype.
Fig. 4. Ligation of the umbilical vein inlet terminates the portal vessel HSC niche.
(A to D) Immunofluorescence analyses of postnatal day 0 (P0; A and C) and P8 (B and D) liver cryosections stained for αSMA (red), Nestin-GFP (green), nuclear dye Hoechst (blue), and Neuropilin-1 or EphB4 (white). *portal vessels; **hepatic veins. Arrowheads: Nestin+ cells (P0) or outline of portal vessel; Solid arrow: hepatic artery, open arrows: bile ducts. (E) FACS analysis showing decreasing numbers of CD150+CD48−Sca1+CD11b+CD45+CD41−Lineage−DAPI− HSCs between P2 and P8. (F) Representative whole-mount immunofluorescence imaging of a CD150+CD48−Lin− HSC (arrow) in P3 fetal liver. Arrowheads: portal vessel. (G) Distance distribution of HSCs from portal vessels (n = 77 from 20 livers). FL HSC measurements from Fig. 1G (dashed-line bars) are shown for comparison. (H) HSC distances from portal vessels in fetal and postnatal livers showing increasing mean distances with age. Scale bars: A–D, 100 μm, F, 20 μm. *P < 0.05, **P < 0.01, ****P < 0.0001.
These remarkable changes were associated with a marked reduction in liver HSC content at P8 (Fig. 4E). Perinatal HSCs were also rapidly established in the neonatal spleen (fig. S8J) and might also contribute to sustaining blood production until the BM becomes fully functional (25, 26). Using whole-mount imaging, we found that in contrast to the fetal liver, few HSCs (<3%) were located within 20 μm of postnatal portal vessels (P0–P5) (Fig. 4, F and G), and mean HSC distances to portal vessels increased significantly (Fig. 4H). These results thus further underscore the critical role for the arterial portal vessels in forming a FL niche.
Our data support the concept that HSCs are titrated against a branching portal vessel network. Fractal geometries of vessel branching optimize the delivery of blood (27), with each division serving a smaller compartment within the organ. The availability of niche cells to sustain proliferating HSCs may thus be tied to the innate growth of the portal vascular tree. Our results also provide a biological explanation for the rapid loss of HSCs in the postnatal liver where dramatic postnatal changes in portal vessels lead to a loss of niche cells and the migration of HSC away from the portal niche. As HSCs emerge from the largest artery (aorta), expand around arterial portal vessels of the fetal liver, and are later maintained quiescent in the adult marrow near small arterioles (11), the arterial vasculature may provide an adaptive niche, serving hematopoiesis at multiple stages of mammalian life.
Supplementary Material
Acknowledgments
We are grateful to the National Institutes of Health (NIH) for the support: J.A.K. was supported from Integrated Training in Pharmacological Science program (NIGMS T32 063754) and NHLBI Ruth L. Kirschstein NRSA predoctoral M.D./Ph.D. fellowship (F30 943257); A. Me. is supported by NHLBI Ruth L. Kirschstein NRSA postdoctoral fellowship F32 HL123224; R01 grants HL097700, DK056638, HL069438 to P.S.F., CA164468 and DA033788 to A.B. S.P. is a New York Stem Cell Foundation–Druckenmiller Fellow. A. Ma. is supported by U54HL127624, U54CA189201 and R01GM098316. We thank A. L. Kolodkin for providing anti-Nrp1 antibody and L. Silberstein for RNA-sequencing recommendations. We thank O. Uche and L. Tesfa for technical assistance with sorting and K. O’Connell for technical assistance with Vevo Ultrasound imaging technology. We are also grateful to the New York State Department of Health (NYSTEM Program) for shared facility (C029154) and research support (N13G-262). This work was funded by NIH grants R01 DK056638, R01 HL116340, R01 HL069438 and NYSTEM grants (C029154 and C029570). J.A.K. designed and analyzed experiments in this study. Y.K. performed transplantations. J.A.K. and Y.Ku. performed the in vivo experiments. J.A.K., Y.Ku. and A.Me. performed whole-mount experiments; J.A.K., A.A., P.C., A.Me, A.Bi. performed immunostaining. J.A.K. and S.P. performed the LTC-IC experiments; A.Me. and S.P. performed differentiation assays;. J.A.K., F.N. and A.Me. performed expression analyses; J.A.K. performed the RNA-sequencing experiments; Y.Ko., A.Ma., and J.A.K. analyzed the RNA-seq data; J.A.K. and A.B. performed computational modelling and statistical analysis of the data; J.A.K. and P.S.F. wrote the manuscript.
Footnotes
www.sciencemag.org/cgi/content/full/science.aad0084/DC1
Materials and Methods
The authors declare no competing financial interests.
REFERENCES AND NOTES
- 1.Medvinsky A, Dzierzak E. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell. 1996;86:897–906. doi: 10.1016/S0092-8674(00)80165-8. [DOI] [PubMed] [Google Scholar]
- 2.Boisset JC, van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak E, Robin C. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464:116–120. doi: 10.1038/nature08764. [DOI] [PubMed] [Google Scholar]
- 3.de Bruijn MF, Speck NA, Peeters MC, Dzierzak E. Definitive hematopoietic stem cells first develop within the major arterial regions of the mouse embryo. EMBO J. 2000;19:2465–2474. doi: 10.1093/emboj/19.11.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gekas C, Dieterlen-Lièvre F, Orkin SH, Mikkola HKA. The placenta is a niche for hematopoietic stem cells. Dev Cell. 2005;8:365–375. doi: 10.1016/j.devcel.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 5.Ema H, Nakauchi H. Expansion of hematopoietic stem cells in the developing liver of a mouse embryo. Blood. 2000;95:2284–2288. [PubMed] [Google Scholar]
- 6.Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, Jaworski M, Offner S, Dunant CF, Eshkind L, Bockamp E, Lió P, Macdonald HR, Trumpp A. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 7.Méndez-Ferrer S, Michurina TV, Ferraro F, Mazloom AR, Macarthur BD, Lira SA, Scadden DT, Ma’ayan A, Enikolopov GN, Frenette PS. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenbaum A, Hsu YM, Day RB, Schuettpelz LG, Christopher MJ, Borgerding JN, Nagasawa T, Link DC. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 2013;495:227–230. doi: 10.1038/nature11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding L, Saunders TL, Enikolopov G, Morrison SJ. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481:457–462. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kunisaki Y, Bruns I, Scheiermann C, Ahmed J, Pinho S, Zhang D, Mizoguchi T, Wei Q, Lucas D, Ito K, Mar JC, Bergman A, Frenette PS. Arteriolar niches maintain haematopoietic stem cell quiescence. Nature. 2013;502:637–643. doi: 10.1038/nature12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruns I, Lucas D, Pinho S, Ahmed J, Lambert MP, Kunisaki Y, Scheiermann C, Schiff L, Poncz M, Bergman A, Frenette PS. Megakaryocytes regulate hematopoietic stem cell quiescence through CXCL4 secretion. Nat Med. 2014;20:1315–1320. doi: 10.1038/nm.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao M, Perry JM, Marshall H, Venkatraman A, Qian P, He XC, Ahamed J, Li L. Megakaryocytes maintain homeostatic quiescence and promote post-injury regeneration of hematopoietic stem cells. Nat Med. 2014;20:1321–1326. doi: 10.1038/nm.3706. [DOI] [PubMed] [Google Scholar]
- 13.Moore KA, Pytowski B, Witte L, Hicklin D, Lemischka IR. Hematopoietic activity of a stromal cell transmembrane protein containing epidermal growth factor-like repeat motifs. Proc Natl Acad Sci USA. 1997;94:4011–4016. doi: 10.1073/pnas.94.8.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chou S, Lodish HF. Fetal liver hepatic progenitors are supportive stromal cells for hematopoietic stem cells. Proc Natl Acad Sci USA. 2010;107:7799–7804. doi: 10.1073/pnas.1003586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka M, Okabe M, Suzuki K, Kamiya Y, Tsukahara Y, Saito S, Miyajima A. Mouse hepatoblasts at distinct developmental stages are characterized by expression of EpCAM and DLK1: Drastic change of EpCAM expression during liver development. Mech Dev. 2009;126:665–676. doi: 10.1016/j.mod.2009.06.939. [DOI] [PubMed] [Google Scholar]
- 16.Sheridan JM, Taoudi S, Medvinsky A, Blackburn CC. A novel method for the generation of reaggregated organotypic cultures that permits juxtaposition of defined cell populations. Genesis. 2009;47:346–351. doi: 10.1002/dvg.20505. [DOI] [PubMed] [Google Scholar]
- 17.Kim I, He S, Yilmaz OH, Kiel MJ, Morrison SJ. Enhanced purification of fetal liver hematopoietic stem cells using SLAM family receptors. Blood. 2006;108:737–744. doi: 10.1182/blood-2005-10-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu X, Bergles DE, Nishiyama A. NG2 cells generate both oligodendrocytes and gray matter astrocytes. Development. 2008;135:145–157. doi: 10.1242/dev.004895. [DOI] [PubMed] [Google Scholar]
- 19.Voehringer D, Liang HE, Locksley RM. Homeostasis and effector function of lymphopenia-induced “memory-like” T cells in constitutively T cell-depleted mice. J Immunol. 2008;180:4742–4753. doi: 10.4049/jimmunol.180.7.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowie MB, Kent DG, Copley MR, Eaves CJ. Steel factor responsiveness regulates the high self-renewal phenotype of fetal hematopoietic stem cells. Blood. 2007;109:5043–5048. doi: 10.1182/blood-2006-08-037770. [DOI] [PubMed] [Google Scholar]
- 21.Passegué E, Wagers AJ, Giuriato S, Anderson WC, Weissman IL. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J Exp Med. 2005;202:1599–1611. doi: 10.1084/jem.20050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR, Ma’ayan A. ChEA: Transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26:2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldberger AL, Amaral LA, Hausdorff JM, Ivanov PCh, Peng CK, Stanley HE. Fractal dynamics in physiology: Alterations with disease and aging. Proc Natl Acad Sci USA. 2002;99(Suppl 1):2466–2472. doi: 10.1073/pnas.012579499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christensen JL, Wright DE, Wagers AJ, Weissman IL. Circulation and chemotaxis of fetal hematopoietic stem cells. PLOS Biol. 2004;2:e75. doi: 10.1371/journal.pbio.0020075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolber FM, Leonard E, Michael S, Orschell-Traycoff CM, Yoder MC, Srour EF. Roles of spleen and liver in development of the murine hematopoietic system. Exp Hematol. 2002;30:1010–1019. doi: 10.1016/S0301-472X(02)00881-0. [DOI] [PubMed] [Google Scholar]
- 27.Hahn HK, Georg M, Peitgen H-O. Fractals in Biology and Medicine. chap 5. Birkhäuser; Basel: 2005. pp. 55–66. [Google Scholar]
- 28.Mignone JL, Kukekov V, Chiang AS, Steindler D, Enikolopov G. Neural stem and progenitor cells in nestin-GFP transgenic mice. J Comp Neurol. 2004;469:311–324. doi: 10.1002/cne.10964. [DOI] [PubMed] [Google Scholar]
- 29.Sheridan JM, Taoudi S, Medvinsky A, Blackburn CC. A novel method for the generation of reaggregated organotypic cultures that permits juxtaposition of defined cell populations. Genesis. 2009;47:346–351. doi: 10.1002/dvg.20505. [DOI] [PubMed] [Google Scholar]
- 30.Fiala JC. Reconstruct: A free editor for serial section microscopy. J Microsc. 2005;218:52–61. doi: 10.1111/j.1365-2818.2005.01466.x. [DOI] [PubMed] [Google Scholar]
- 31.Liu F, Lee JY, Wei H, Tanabe O, Engel JD, Morrison SJ, Guan JL. FIP200 is required for the cell-autonomous maintenance of fetal hematopoietic stem cells. Blood. 2010;116:4806–4814. doi: 10.1182/blood-2010-06-288589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller CL, Dykstra B, Eaves CJ. Current Protocols in Immunology. Unit 22B. Chapter 22. Wiley; 2008. p. 22. [DOI] [PubMed] [Google Scholar]
- 33.Hu Y, Smyth GK. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 34.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anders S, Pyl PT, Huber W. HTSeq–A Python framework to work with high-throughput sequencing data. Bioinformatics. 2014;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan CM, Chen EY, Dannenfelser R, Clark NR, Ma’ayan A. Network2Canvas: Network visualization on a canvas with enrichment analysis. Bioinformatics. 2013;29:1872–1878. doi: 10.1093/bioinformatics/btt319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen EY, Xu H, Gordonov S, Lim MP, Perkins MH, Ma’ayan A. Expression2Kinases: mRNA profiling linked to multiple upstream regulatory layers. Bioinformatics. 2012;28:105–111. doi: 10.1093/bioinformatics/btr625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu L, Timmers C, Maiti B, Saavedra HI, Sang L, Chong GT, Nuckolls F, Giangrande P, Wright FA, Field SJ, Greenberg ME, Orkin S, Nevins JR, Robinson ML, Leone G. The E2F1-3 transcription factors are essential for cellular proliferation. Nature. 2001;414:457–462. doi: 10.1038/35106593. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.