Abstract
The apical membrane antigen 1 of Plasmodium falciparum is one of the leading candidate antigens being developed as a vaccine to prevent malaria. This merozoite transmembrane protein has an ectodomain that can be divided into three subdomains (D I, D II, and D III). We have previously expressed a major portion of this ectodomain and have shown that it can induce antibodies that prevent merozoite invasion into red blood cells in an in vitro growth and invasion assay. To analyze the antibody responses directed against the individual subdomains, we constructed six different genes that express each of the domains separately (D I, D II, or D III) or in combination with another domain (D I+II, D II+III, or D I+III). These proteins were purified and used to immunize rabbits to raise construct-specific antibodies. We demonstrated that D I+II induced a significant amount of the growth-inhibitory antibodies active in the growth and invasion assay.
Several malaria surface proteins expressed during different stages of the parasite's life cycle have been identified and are being developed as vaccine target antigens. It has been demonstrated that antibodies against membrane proteins of malaria merozoites can, in some cases, block parasite invasion of the erythrocyte (10, 11, 15, 23, 27, 30, 32). One such recombinant vaccine being developed is based on the asexual blood stage integral membrane protein apical membrane antigen 1 (AMA-1) because antibodies to the protein effectively inhibited parasite invasion of erythrocytes in vitro (26, 30, 31, 33). The role of AMA-1 in invasion is further supported by the fact that passive transfer of strain-specific anti-AMA-1 antibodies to Plasmodium chabaudi-infected mice protected against lethal parasitemia (1).
AMA-1 of P. falciparum is a highly conserved 83-kDa transmembrane protein containing cytoplasmic, transmembrane, and ectodomain regions (28). It is synthesized in merozoites, localized in the micronemes until merozoite release, and then rapidly translocated onto the merozoite surface (4, 25, 35). The amino acid sequence of the ectodomain contains 16 cysteine residues that are cross-linked by eight disulfide bonds. The disulfide bond structure suggests that the ectodomain is composed of three distinct subdomains, domain I, domain II, and domain III (D I, D II, and D III) (13). There is evidence that during translocation onto the merozoite surface, AMA-1 is proteolytically cleaved into smaller fragments (8, 16, 17, 25).
Studies of animal malarias have heightened interest in the development of AMA-1 as a vaccine for human malaria. Immunization with purified recombinant AMA-1 is protective against the simian malaria parasites P. knowlesi (6) and P. fragile (2) and the rodent malaria parasites P. chabaudi (1) and P. yoelii (26). However, the protection was parasite strain specific, suggesting that the protective immune responses were directed toward the polymorphic regions of AMA-1 (5). Protective antibody-mediated immune responses induced against AMA-1 have repeatedly been shown to be directed against conformational epitopes that are dependent on disulfide bond stabilized conformations (1, 2, 5-7, 14, 21, 26). While the amino acid divergence observed among different isolates of P. falciparum AMA-1 has been small (∼5%), the changes are significant enough, in most cases, to dramatically affect the cross-strain recognition by heterologous protein-induced antibodies (19). The emergence of these differences, at least in D I, has recently been shown to be correlated with symptomatic malaria cases (3).
We recently completed the production and purification of an antimalaria vaccine based on the AMA-1 ectodomain from P. falciparum (3D7) (7). Immunization of rabbits with purified protein induced the production of antibodies that significantly (>80%) inhibited parasites in an in vitro growth and invasion assay (GIA). The same level of inhibition in the GIA was observed with whole antibodies and Fab fragments of the antibodies (8). In addition, monoclonal antibodies (MAb) have been produced against the ectodomain that significantly block invasion of red blood cells by merozoites in the GIA (20). To better understand how antibodies to each of the subdomains of the ectodomain of AMA-1 contribute to the growth-inhibitory effect seen in the GIA, we have expressed subdomain constructs, in single and doublet combinations, in Escherichia coli, purified each of the proteins, and raised rabbit antibodies against them. These antibodies allowed us to closely examine the effect of domain-specific antibodies on the overall inhibition of invasion measured in the GIA.
MATERIALS AND METHODS
Cloning and expression of the recombinant AMA-1 subdomains.
Appropriate PCR primers with 5′ NcoI and 3′ NotI restriction sites were designed (Bioserve Biotechnologies, Laurel, Md.) for the gene fragments encoding subdomains I, II, III, I+II, II+III, and I+III (domains as defined by Hodder et al. [13] from the 3D7 isolate of P. falciparum). Gene sequences corresponding to each of these fragments were amplified from a synthetic E. coli codon-optimized AMA-1 ectodomain gene of the 3D7 isolate (encoding amino acids Gly83 to Glu531) (7) by using Taq DNA polymerase and appropriate primers (see Fig. 1). To make D I+III, gene segments dI and dIII were ligated in a separate ligation reaction before TA cloning. The PCR-amplified products were cloned into TA Cloning Vector pCRr2.1 (Invitrogen, Carlsbad, Calif.), and positive clones were selected by DNA restriction endonuclease analyses and further confirmed by nucleotide sequence analyses. The expression plasmid (7) was restriction digested with NcoI and NotI, and gel-purified gene inserts from the TA cloning vectors were ligated in before transformation into E. coli strain BL21(DE3). In all cases, insertion into the expression vector resulted in His 6 tags on both the amino- and carboxy-terminal ends of the proteins. Bacterial colonies containing expected fragments were picked and analyzed by restriction digestion following plasmid DNA preparation (Qiagen Inc., Valencia, Calif.). Selected clones were further confirmed by nucleotide sequencing of the plasmid DNA samples. Glycerol (8%) stocks were made for each bacterial clone from an overnight culture and stored at −80°C.
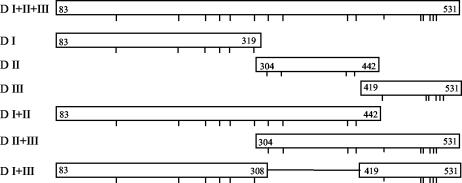
FIG. 1.
Schematic diagram of regions of P. falciparum 3D7 AMA-1 ectodomain expressed for use in this study. Numbers refer to the amino acid residue of the P. falciparum AMA-1 sequence (Gen Bank accession number U65407.1); downward tick marks represent the relative positions of conserved cysteine residues. For D I+III, the construct was a chimera that linked D I residue# 308 to D III residue# 419. Forward and reverse primers, respectively, used for PCR amplification for each of the constructs were as follows: D I, GGAACCGGCGCCGCAG and CTGCAGGTTTTTACGCGGGCACAC; D II, GCGTAAAAACCTGCAGAAC and CGGAAAGTTGTTTTCCACTTC; D III, CTGATTAACAACAGCAGCTAT and GCGGCCGCTTCATCTTTAGA, D I+II, GGAACCGGCGCCGCAG and CGGAAAGTTGTTTTCCACTTC; D II+III, GCGTAAAAACCTGCAGAAC and TGTGGCGGCCGCTTCATCTTTAGA, D I+III, GGAACCGGCGCCGCAG- (-CTGATTAACAACAGCAGCTAT) and CTGCAGGTTTTTACGCGGGCACAC- (-GCGGCCGCTTCATCTTTAGA), where the sequences in parentheses indicate the end of DI and the beginning of DIII, respectively.
For expression in shake flasks, cells were grown to an optical density at 600 nm of 0.5 and then induced with a final concentration of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were harvested after 2 h of induction by centrifugation at 6,200 × g and 4°C for 30 min. Expression in a 10-liter bioreactor (New Brunswick Scientific, Edison, NJ) used Terrific Broth media (1.2% tryptone, 2.4% yeast extract, 72 mM K2HPO4, 28 mM KH2PO4 [pH 7.2]) containing 0.8% glycerol and 100 μg of ampicillin per ml. The 10 liters of media in the bioreactor was inoculated with 100 ml of an overnight culture, and growth was allowed to proceed at 37°C. pH 7.2 was maintained by controlled addition of either HCl or NaOH. At an optical density at 600 nm of 7 to 8, the temperature was reduced to 25°C and then IPTG was added to a final concentration of 0.5 mM. At 2 h after induction, cells were harvested by centrifugation at 5,000 × g and 4°C for 30 min. Under these conditions, all six constructs had similar growth characteristics. The cell pastes were stored at −80°C. Aliquots were taken from these for developing the purification conditions for each of the subdomain proteins.
Purification of the subdomain fragments.
The E. coli cell pastes were solubilized in a buffer containing 15 mM Na2HPO4, 5.1 mM KH2PO4, 450 mM NaCl, and 2.5% sodium N-lauroylsarcosine (Sarkosyl) (pH 7.4) at a concentration of 10 ml of buffer/g of cell paste. The cells were disrupted by a single pass through a high-pressure microfluidizer (model 1109; Microfluidic Corp., Newton, Mass.). Following clarification by centrifugation (12,000 × g, for 45 min at 4°C), supernatants containing the recombinant proteins were incubated by a batch method with nickel nitrilotriacetic acid (Ni-NTA)-agarose chelating resin (0.4 ml of resin/g of cell paste) (Qiagen Inc.) at room temperature (RT) (∼22°C) for 1 h in the presence of 35 mM imidazole. The resin was then loaded into a fritted column, and the unbound proteins were allowed to flow through. The resin was washed with a minimum of 40 column volumes (cv) of 20 mM NaHPO4-450 mM NaCl-10 mM imidazole-0.125% Sarkosyl (pH 7.4) followed by 10 cv of 20 mM NaHPO4-15 mM imidazole (pH 8.0). Bound proteins were eluted with 500 mM imidazole in 20 mM phosphate buffer containing 0.125% Sarkosyl (pH 8.0). Proteins eluted from the Ni-NTA-agarose were rapidly diluted to 30 to 40 μg/ml in a redox-coupled GSH-GSSG solution and allowed to fold overnight as described previously (7). After the refolding period, the protein solution was pH adjusted (to 5.8 for D I and D II, 6.4 for D III, and 6.0 for all double domains) and passed through preequilibrated SP Sepharose (Amersham Pharmacia Biotech) (∼0.3 ml of resin/g of cell paste). The resin-bound protein was washed with a minimum of 50 cv of 20 mM NaHPO4-1mM EDTA containing 100 mM NaCl (for double-domain constructs) or 250 mM NaCl (for single-domain constructs) at pH 6.0. The bound protein was eluted with 20 mM NaHPO4-1 mM EDTA (pH 7.4) plus 150 mM NaCl (pH 6.0) (for single-domain constructs) 300 mM NaCl (pH 6.0) (for D I+II), or 400 mM NaCl (pH 6.0) (for D I+III and D II+III).
Purity.
Purity was analyzed on precast 4 to 12% Bis-Tris gels (NuPage; Invitrogen, Carlsbad, Calif.) run as recommended by the manufacturer and stained with Coomassie blue. Additional analysis was carried out by reversed-phase high-performance liquid chromatography (HPLC) using a Waters-510 HPLC pump connected to a Waters-712 WISP auto sampler and controlled by Millennium (Release 3.2) chromatographic software. A Waters 2487 dual λ absorbance detector was used to monitor the elution profile at 215 and 280 nm. Reversed-phase chromatographic analysis was done with a Symmetry 300 C4 column (pore size, 5 μm; 4.6 by 150 mm) (Waters Corp., Milford, Mass.) at a flow rate of 3 ml min−1 and 10 μg of protein per load. Solvent A was 0.1% trifluroacetid acid in H2O; solvent B was 0.1% trifluoroacetic acid in acetonitrile. The solvent gradient consisted of 90% solvent A for 5 min, 90% to 10% solvent A over 30 min, 10 to 90% solvent A over 5 min, and reequilibration at 90% A for 10 min. Bovine serum albumin (BSA) and RNase A were used as standards for column resolution.
Molecular mass, reduction and alkylation, and free thiol analyses.
Purified domain protein samples were analyzed for mass by matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectrometry (Voyager Biospectrometry PR System; Applied Biosystems) with a sinapinic acid matrix. Lysozyme and cytochrome c were used as mass standards. Protein reduction was achieved by incubation with a 100-fold molar excess of dithiothreitol over cysteines in the presence of 4 M urea, and alkylation was carried out with a 1,000-fold molar excess of iodoacetamide over cysteines for 1 h at room temperature in the dark. Free sulfhydryl groups were estimated in the presence and absence of 4 M guanidine-HCl by using Ellman's reagent (5,5′-dithio-bis-3-nitrobenzoic acid). l-Cystine was used to plot the standard curve.
SDS-PAGE and immunoblot analyses.
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using precast 4 to 12% Bis-Tris gels and Western blotted as described previously (7), except that the blots were washed with phosphate-buffered saline plus 0.5% Tween 20 (PBST) and developed with BM Blue POD or nitroblue tetrazolium/5-bromo-4-chloro-3-indolylphosphate (NBT/BCIP; Roche, Indianapolis, Ind.) substrate as recommended by the manufacturer.
Preparation of rabbit sera.
Groups of New Zealand White rabbits (n = 3) were immunized subcutaneously with 100 μg of each double-domain protein construct emulsified in Montanide ISA-720 (Seppic Inc., Paris, France) as described previously (7). Research was conducted in compliance with the Animal Welfare Act and other Federal statutes and regulations relating to animals and experiments involving animals. All procedures were reviewed and approved by the Walter Reed Army Institute of Research Institutional Animal Care and Use Committee and performed in a facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, International.
ELISA.
Antibody responses were evaluated by enzyme-linked immunosorbent assay (ELISA). Ninety-six-well microtiter plates (Dynax, Chantilly, Va.) were incubated overnight at 4°C with 50 μl of a 2-μg/ml solution of either AMA-1 ectodomain (7) or individual domain fragments. The plates were blocked at RT for 1 h with PBST containing 5% casein (Sigma, St. Louis, Mo.) and then washed with PBST. Consecutive two-fold dilutions of individual rabbit sera were incubated for 2 h at RT. The plates were washed and incubated with 1:5,000-diluted horseradish peroxidase/alkaline phosphatase-conjugated secondary antibody for 1 h. They were then washed and developed for 30 min with NBT-BCIP-peroxidase substrate (Kirkegaard & Perry Laboratories, Gaithersburg, Md.). The absorbance at 405 nm was recorded, and comparative ELISA titers were calculated using regression analysis on the titration curve. The ELISA was repeated three times for each individual serum sample, in triplicate wells; on separate days.
Competition ELISA.
Competition ELISA was done using sera from rabbits immunized with AMA-1 ectodomain (7). This was diluted 1:32,000 and preincubated, with constant mixing, overnight at 4°C with 1 μM AMA-1 ectodomain, D I, D II, D III, D I+II, D II+III, DI+III or BSA. The tubes were centrifuged at 18,500 × g for 15 min, and the supernatants were analyzed by ELISA with AMA-1 ectodomain as the capture antigen.
Indirect IFA.
Recognition of P. falciparum 3D7 schizonts by anti-AMA1 domain antibodies was analyzed by an indirect immunofluorescence assay (IFA) as described previously (7).
Parasite culture and GIA.
P. falciparum (3D7) cultures and GIA were performed as described previously (7, 12). Briefly, cultures were grown in 48-well plates and kept in suspension culture by rotation on a platform or under static conditions. Control serum or immunoglobulin G (IgG) (dialyzed into RPMI-NaOH) was added to a final hematocrit of 4%. To assess the antigen specificity of the antibody-mediated inhibitions, antigens (D I, D II, D III, D I+II, D II+III, D I+III, AMA-1 ectodomain, or BSA) were added to the IgG preparations (0.5 mg/ml) before the GIA was performed. The final concentration of each test protein in the GIA was 0.5 μM. Merozoites were released after approximately 34 h, and developing ring stages were harvested 14 h postinvasion, stained with Hoechst dye 33342, and analyzed by flow cytometry. The fluorescence signal was determined for a minimum of 40,000 erythrocytes gated on forward scatter. The fluorescent signal of ring-infected erythrocytes was about 20 times that of uninfected erythrocytes, and the signal of schizont-infected erythrocytes, if present, was about another 20-fold above that. Almost all (>99%) of the parasites harvested from the assays were ring forms or early trophozoites stages, as confirmed by spot checks of Giemsa-stained thin smears. The percent inhibition was calculated from the mean parasitemia of triplicate test and control wells as 100% − (test/control). Sera from rabbits immunized with the adjuvant and PBS were used as controls in the GIA. Prebleeds from individual rabbits were tested, and the results were used as controls in the GIA.
To assess the antigen specificity of the antibody-mediated inhibitions, proteins (D I, D II, D III, D I+II, D II+III, D I+III, AMA-1 ectodomain, or BSA) were added to the IgG preparations (0.5 mg/ml) before the GIA was performed. The final concentration of each test protein in the GIA was 0.5 μM.
Statistical analysis.
Microsoft Excel was used to calculate the P values for the two-tailed t tests and the correlation coefficients (r2). For the data presented in Fig. 6 and 7, estimates of means, confidence intervals, and hypothesis tests were generated using bootstrap. Each of the available data sets was resampled (bootstrapped) 10,000 times. The observed bootstrap probability estimates reported here are consistent with estimates obtained by assuming that bootstrap means and variances are asymptotically normal. The S language for statistical programming (R v1.8.0) was used to develop the bootstrap estimates. Specific language elements used included the “boot” library and the “Marsaglia-Multicarry” random-number generator.
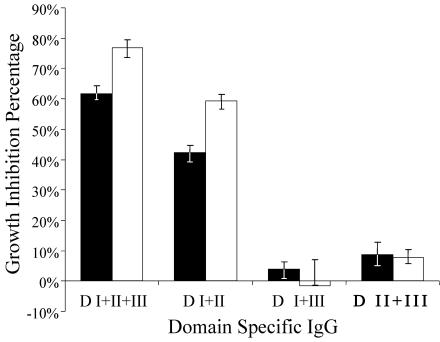
FIG. 6.
GIA of purified IgG from rabbit sera immunized with each of the multidomain constructs. Solid bars, IgG at 0.5 mg/ml; open bars, IgG at 1.0 mg/ml. Error bars represent the upper and lower confidence intervals of the bootstrap analysis.
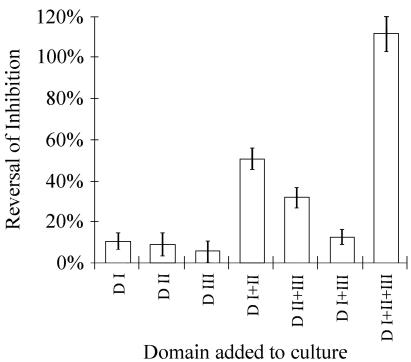
FIG. 7.
Reversal of the AMA-1 antibody growth inhibition activity by protein domains. AMA-1 domains were added to cultures of P. falciparum 3D7 grown in the presence of 0.5 mg of anti-D I+II+III rabbit IgG per ml. Reversal of inhibition by a specific domain was calculated as the difference of inhibition percentages between cultures containing anti-D I+II+III antibodies and cultures containing antibodies plus 0.5 μmol of that domain per liter. Assays were performed in triplicate, and data in the figure represent bootstrap estimates for 95% confidence intervals on mean reversal, assuming normal distribution. Error bars represent the upper and lower confidence intervals of the bootstrap analysis.
RESULTS
Expression and purification.
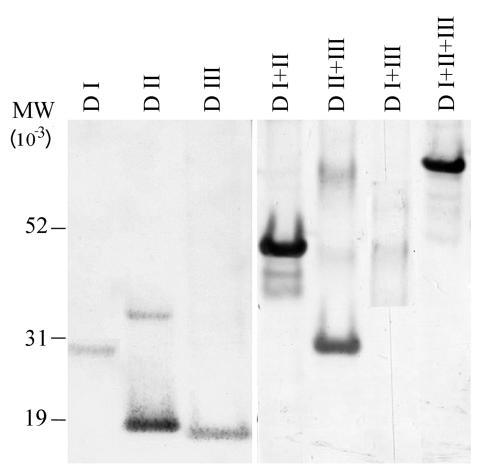
All six constructs made from the AMA-1 ectodomain were cloned and expressed in E. coli strain BL-21(DE3). The AMA-1 ectodomain construct, D I+II+III, was made as previously described (7). Figure 1 details the constructs used for individual domains. The biomass yield of a typical 10-liter fermentation was about 400 to 550 g of bacterial paste. All constructs had a His 6 tag, allowing Ni-NTA agarose affinity chromatography as a first step in purification. After refolding, an SP-Sepharose cation-exchange resin was used as a second step of purification, where protein monomers could be separated from multimers using appropriate pH (5.8 to 6.4) and salt concentrations in the wash and elution conditions. This two-step purification yielded >85% pure recombinant domain proteins, as determined from densitometric scans of SDS-PAGE (Fig. 2) and reversed-phase HPLC (data not shown) analyses. All the subdomain fragments of AMA-1 except D I+III resolved into single symmetric peak on a C4 reversed-phase HPLC column (data not shown). Proteins corresponding to DIII and D I+III had a tendency to precipitate at neutral pH when stored at concentrations above 120 μg/ml. The final yield of purified proteins was 15 to 20 mg/liter of starting culture medium for D I, D II, D I+II, and D II+III and 5 to 7 mg/liter for D III and D I+III.
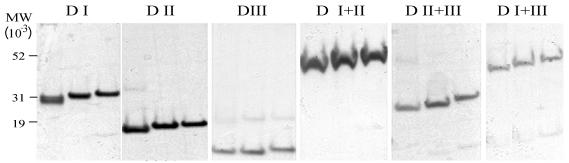
FIG. 2.
Coomassie blue-stained SDS-PAGE gel of purified protein from each domain construct, as listed above each panel. The proteins in each panel are (left to right) refolded, reduced, reduced and alkylated. MW, molecular weight.
Mass and disulfide bond confirmation.
The molecular masses of all the constructs were as predicted from the deduced amino acid sequence. The observed average masses of D I, D II, D III, D I+II, D II+III, D I+III, and D I+II+III were 28,895.6 (28,861.3), 18,999.3 (19,004.2), 16,099.4 (16,130.0), 44,113.5 (44,102.4), 29,331.8 (29,340), 41,879.9 (41,882), and 54,656 (54,633) respectively, where the values in parentheses are the predicted masses, and were all within the experimental-error limits of measurements of the MALDI-TOF mass spectrometer. The absence of free cysteines was confirmed by an Ellman test. The presence of substantial amounts of free cysteines in the recombinant products would have indicated incorrect folding. All proteins, except the two constructs D III and D I+III, were found to contain less than 0.1 μM free -SH per μmol of protein. The two D III-containing constructs could not be assayed using the Ellman test due to solubility problems at the concentration and pH required for the test. As a second test for disulfide bond-stabilized folding, the proteins were subjected to reduction and alkylation followed by SDS-PAGE analysis. The measurements from the Ellman test show that there were, as predicted from the deduced amino acid sequence of the genes, eight disulfide bonds in D I+II+III, six in D I+III, five each in D I+II and D II+III, three each in D I and D III, and two in D II. All constructs showed significant mobility shifts when subjected to reduction and alkylation (Fig. 2).
Recognition by antibodies.
We used several different experimental conditions to determine the extent of recognition of the domains by antisera made against the entire ectodomain (D I+II+III). All six recombinant constructs were recognized on Western blots at the appropriate molecular masses by antibodies in a polyclonal rabbit serum raised against D I+II+III (reference 7 and data not shown) and by a pool of P. falciparum-infected sera collected from western Kenya (Fig. 3). Constructs D I and D I+III were less reactive than the other domains. There was no recognition of these domains in analyses with normal human or rabbit sera at the same concentration (data not shown). MAb 4G2dcl, known to bind to a conformation-dependent epitope within the region encompassing D I+II (16), recognized recombinant D I+II, as shown by Western blotting and ELISA run under nonreduced conditions (data not shown). None of the other constructs were recognized by MAb 4G2dc1 either by ELISA or by Western blotting.
FIG. 3.
Western blot of each purified construct. A 100-ng amount of each purified nonreduced protein was subjected to SDS-PAGE, electrophoretically transferred to nitrocellulose membranes, and reacted with a pool of sera (diluted 1:1,000) from people who lived in a malaria-endemic area of Western Kenya. MW, molecular weight.
Immune analysis of P. falciparum AMA-1 subdomains.
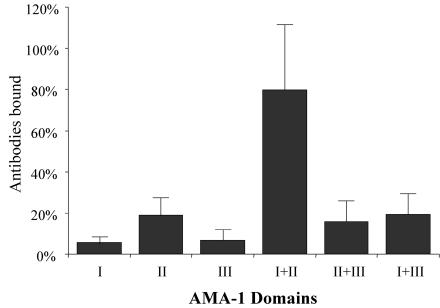
In an ELISA format, about 80% of the antibodies made against the ectodomain were directed against D I+II (Fig. 4). In contrast, D I and D II by themselves bound only 6 and 19% of the antibodies, respectively. The other double-domain constructs, D II+III and D I+III, each bound less than 20% of the total antibody.
FIG. 4.
Percentage of antibodies (y axis) made against the P. falciparum 3D7 AMA-1 ectodomain (D I+II+III) that bind to individual domain constructs. Most of the antibodies generated by D I+II+III were directed to epitopes present in the D I+II region (P < 0.00008). The 100% point is equivalent to the ELISA value of anti-D I+II+III antibody that bound to D I+II+III protein. Error bars represent the variation observed in triplicate wells in two experiments.
In a solution competition experiment, the AMA-1 ectodomain (D I+II+III) could deplete 93% of antibodies made against itself while D I+II could deplete 72% of the anti-D I+II+III antibodies. The other double-domain constructs, D II+III and D I+III, could deplete 45 and 51%, respectively. Of the single domains, D I had the highest depletion (∼35%), while D II and D III depleted 28 and 26%, respectively.
Recombinant subdomains induce high-titer antibody responses in rabbits.
Immunization of rabbits (n = 3) with each of the three double-subdomain fragments induced high-titer antibodies (Table 1). On indirect IFA, all of these antibodies reacted with schizont forms from P. falciparum 3D7 at a dilution of 1:5,000. The reactivity of rabbit anti-double-domain antibodies with P. falciparum AMA-1 was confirmed by immunoblot analyses. Rabbit antibodies obtained by immunization with all three double-domain constructs (D I+II, D II+III, and D I+III) predominantly recognized two bands (Fig. 5) corresponding to the previously identified 83- and 66-kDa full-length and processed forms of AMA1 in P. falciparum (4, 8, 16, 25).
TABLE 1.
Summary of ELISA and IFA results for rabbits immunized with AMA-1 double domains
| Domains | Rabbit code | ELISA titera | Parasite IFA intensityb |
|---|---|---|---|
| D I+II | V05 | 138,126 | ++ |
| V24 | 86,449 | + | |
| V25 | 121,103 | ++ | |
| D II+III | V20 | 48,640 | + |
| V30 | 208,819 | + | |
| V44 | 38,579 | + | |
| D I+III | V27 | 288,744 | + |
| V27 | 107,106 | + | |
| V28 | 34,578 | + |
Reciprocal dilution (mean of three experiments) of sera that gave an optical density at 414 nm of 1.0 with the immunizing antigen coated on the plate.
Relative IFA intensity at a dilution of 1:5,000.
FIG. 5.
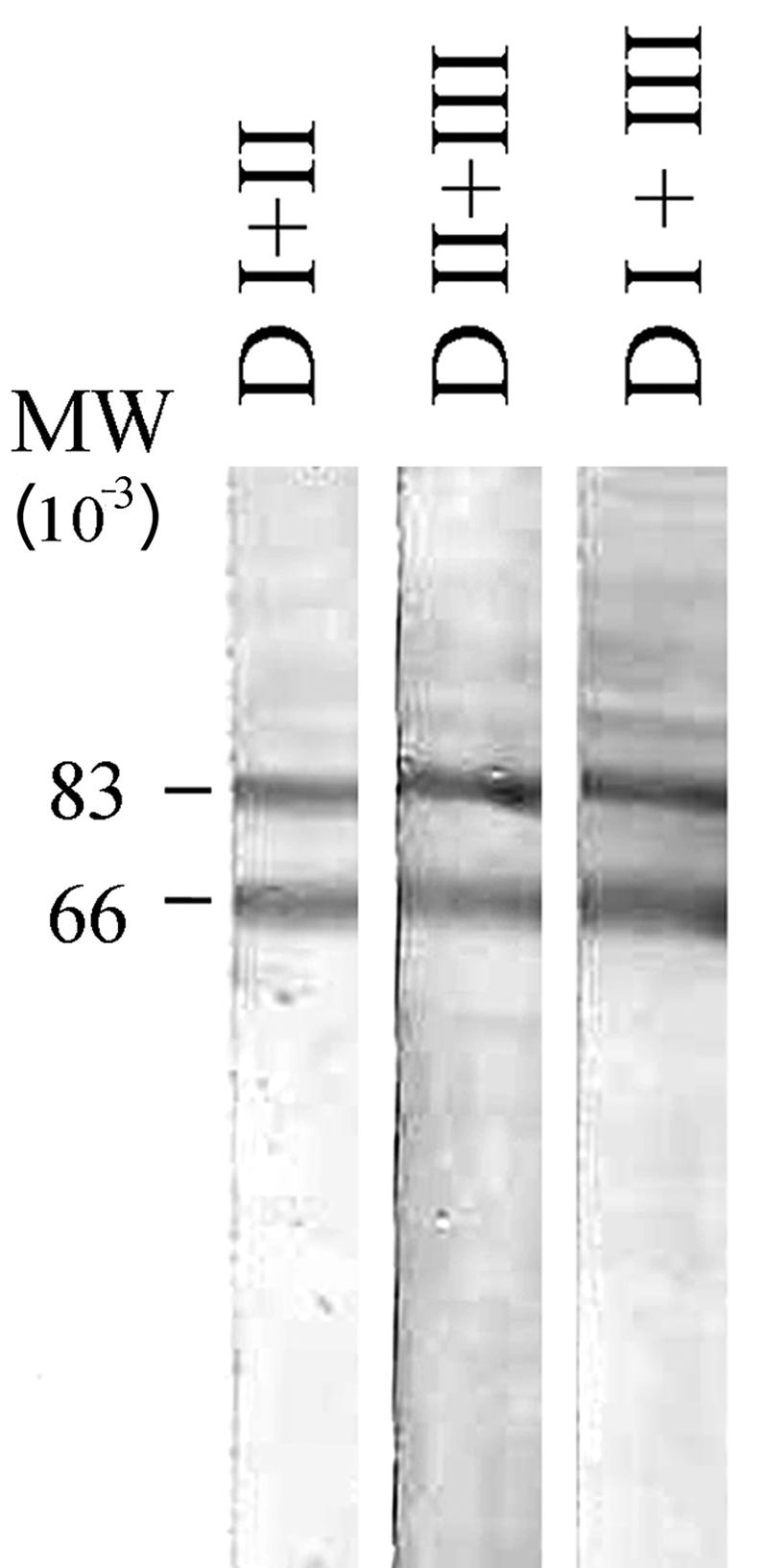
Western blot of total parasite extract probed with rabbit antisera made against domain constructs D I+II, D II+III or D I+III. P. falciparum 3D7 schizonts were extracted with 10% SDS, subjected to SDS-PAGE under nonreducing conditions, and transferred to a nitrocellulose membrane. The blot was treated with postimmune rabbit serum against each of the three double domains, as indicated. Prebleed serum from each rabbit was negative (data not shown).
Inhibition of P. falciparum invasion of erythrocytes.
Purified IgG from the pool of double-domain-immunized rabbit sera was used in the GIA. At 1.0 mg/ml, anti-D I+II+III inhibited 77% of parasite development and anti-D I+II resulted in 59% inhibition (Fig. 6). At 0.5 mg/ml, these antibodies resulted in 62 and 42% inhibition, respectively. Antibodies against the other domains or IgG purified antibodies from the preimmune sera did not significantly inhibit invasion in the GIA. To investigate the domain specificity of growth-inhibitory antibodies, the GIA was performed with purified anti-D I+II+III IgG with the addition of recombinant domain proteins to the culture medium (Fig. 7). The addition of DI+II+III protein caused >90% reversal of inhibition in a GIA, and the addition of D I+II and D II+III reversed the growth inhibition of the antisera by 62 and 30%, respectively. There was no significant reversal of growth inhibition in the GIA on addition of D I+III, D I, D II, or D III.
DISCUSSION
Several reports suggest that AMA-1 could be an important part of a multicomponent vaccine to combat malaria. AMA-1 from different Plasmodium species are targets of a host immune response to the parasite (18, 26). Knockout of the AMA-1 gene results in inability of the parasites to invade red blood cells, suggesting that it is a pivotal molecule in all Plasmodium species (34). Previously we had reported the production of a recombinant AMA-1 ectodomain protein containing all three domains (D I+II+III). Antibodies produced in rabbits to this product recognized the native parasite by indirect IFA and inhibited P. falciparum growth by >90% in a one-cycle GIA. To analyze the immune responses to the individual domains and to determine if particular regions of the molecule are better targets for vaccine development efforts, we produced recombinant polypeptides that represented different regions of the AMA-1 ectodomain. While it is impossible to determine the tertiary structure without having crystallographic information, we decided to follow the convention determined by Hodder et al. (13) and divided the ectodomain into three domains based on the loops formed by the cysteine cross-linkages. We have cloned the DNA delineating the domains, either singly or in combination with another domain, to generate six gene constructs (dI, dII, dIII, dI+II, dII+III, and dI+III), and expressed them in E. coli BL-21(DE3) cells.
Using two chromatographic columns, procedures were developed that yielded recombinant proteins of >85% purity. The final yield of recombinant protein per gram of bacterial biomass was two- to threefold higher for D I+II than for D I+II+III (7), most probably a reflection of the simpler disulfide bond formations in the smaller construct. It has been show that the disulfide bond-restrained conformation is critical for AMA-1 to induce growth-inhibitory antibodies after vaccination of rabbits (7). MAb 4G2dc1 has been used as a marker to identify a disulfide bond-dependent conformational epitope in AMA-1 (D I+II+III). This MAb bound to D I+II but not to the other single or double domains, indicating that the critical conformational epitope was maintained within D I+II. Our laboratory has produced seven MAbs against D I+II+III, all of which were directed against epitopes that required D II. Similar to 4G2dc1, these antibodies were epitope reduction sensitive (A. Barbosa, unpublished data). Although we do not have data about conformation specific MAbs that recognize D I, D III, or D I+III, recognition of these proteins by immune sera, mobility shifts on SDS-PAGE after reductive alkylation, sharp-and symmetric peaks on RP HPLC analysis, and the absence of free cysteine content in these preparations indicated that they were homogeneous and correctly folded.
Our data indicate that, using our adjuvant with the proteins in rabbit immunizations, D I+II is the most immunogenic region within the P. falciparum 3D7 AMA-1 D I+II+III, while D II is the most immunogenic domain among the three single domains. It is evident from direct and competition ELISA that single domains on their own are less immunogenic than the whole ectodomain. This was further confirmed by the reversal of growth inhibition in the GIA. The addition of the D I+II protein to purified IgG made against the entire ectodomain of AMA-1 resulted in significant reversal of the growth-inhibitory capacity of that IgG preparation. The effect was negligible when D I, D II, D III, D II+III, or D I+III was added. The results suggested that single domains, on their own, do not induce significant inhibitory antibodies and that the combination of D I+II plays large role in the total inhibitory activity of anti-D I+II+III antibodies.
To test the potential of domain-specific antibodies to inhibit invasion in the GIA, we immunized rabbits with the purified double-domain proteins D I+II, D II+III, and D I+III. A positive indirect IFA result with late-stage schizonts of P. falciparum 3D7 parasites and Western blot recognition of the native AMA-1 from parasite lysate with these antibodies suggested a close similarity of these recombinant preparations to the native parasite antigens and the recombinant vaccine construct containing D I+II+III (7). The ELISA titers were equally high for all three double domains; however, only antibodies from rabbits immunized with the D I+II construct could significantly inhibit parasite growth in the GIA. A low level of inhibition was measured with antiserum to D II+III, and, although D I+III induced high-titer antibodies (titer, 106), these antibodies were unable to inhibit parasite growth in vitro. These data demonstrated that a portion of the region defined within the recombinant D I+II induced a majority of the antibodies that are active in the GIA. Hodder et al. (14) suggested that most inhibitory antibodies were directed against D I because that region contained a majority of the amino acid differences seen among three P. falciparum strains (3D7, D10, and HB3) that were inhibited to different degrees in a GIA. Recent studies of D I, the most variable region in AMA-1 (9, 22, 29), suggested that naturally acquired protective immunity targets domain I (3). D I, in the context of D I+III, did not induce any inhibitory antibody, did not significantly inhibit anti-D I+II+III antisera in the competition ELISA, and did not reverse the growth-inhibitory qualities of anti-D I+II+III. While the D I amino acid sequence in the D I+III construct is 11 amino acid residues shorter then the D I domain alone (due to construction restraints encountered during PCR linking of the gene segments), the data still suggest that only in combination with the D II protein does D I express the structured epitopes that potentially can be blocked by antibody to prevent parasite invasion.
Polyclonal antibodies and MAbs against AMA-1 block the invasion of erythrocytes by merozoites. A majority of the epitopes that induce these inhibitory antibodies are conformational and are destroyed on reduction of the disulfide bonds. Due to the problems of disulfide bond formation with proteins expressed in bacteria, the yield of correctly folded full-length ectodomain in this system is low. This study was initiated, in part, to examine the possibility of using smaller AMA-1 constructs, which may be expressed at high levels in bacteria yet still function as effective malaria vaccines. To that end, this study has demonstrated that subdomains of AMA-1 can be expressed at high levels in E. coli. In addition, we showed that a polypeptide encompassing a region defined by D I+II can bind most of the antibodies induced on immunization with D I+II+II and that this construct induced potent growth-inhibitory antibodies. However, the overall GIA activity was lower than that observed with antibodies to the D I+II+III construct. Thus, D III may also contain important protective epitopes. Indeed, recently a virosomal formulation containing a synthetic peptide corresponding to D III of AMA-1 has shown growth-inhibitory activity against parasites (24).
Acknowledgments
We thank Alan Thomas for the MAb 4G2dc1; E. Angov for the expression plasmid; Robin Anders for advice on domains; Douglas Smoot for flow cytometry of parasites in the GIA; and Laura Yergus and Meredith Vassell for technical assistance.
This work was performed while P.V.L. and S.D. held National Research Council Research Associate awards at WRAIR and was supported in part by a grant from USAID.
The views expressed here are those of the authors and should not be construed to represent those of the U.S. Department of the Army or the U.S. Department of Defense.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Anders, R. F., P. E. Crewther, S. Edwards, M. Margetts, M. L. Matthew, B. Pollock, and D. Pye. 1998. Immunization with recombinant AMA-1 protects mice against infection with Plasmodium chabaudi. Vaccine 16:240-247. [DOI] [PubMed] [Google Scholar]
- 2.Collins, W. E., D. Pye, P. E. Crewther, K. L. Vandenberg, G. G. Galland, A. J. Sulzer, D. J. Kemp, S. J. Edwards, R. L. Coppel, J. S. Sullivan, C. L. Morris, and R. F. Anders. 1994. Protective immunity induced in squirrel monkeys with recombinant apical membrane antigen-1 of Plasmodium fragile. Am. Trop. Med. Hyg. 51:711-719 [DOI] [PubMed] [Google Scholar]
- 3.Cortes, A., M. Mellombo, I. Mueller, A. Benet, J. C. Reeder, and R. F. Anders. 2003. Geographical structure of diversity and differences between symptomatic and asymptomatic infections for Plasmodium falciparum vaccine candidate AMA1. Infect. Immun. 71:1416-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crewther, P. E., J. G. Culvenor, A. Silva, J. A. Cooper, and R. F. Anders. 1990. Plasmodium falciparum: two antigens of similar size are located in different compartments of the rhoptry. Exp. Parasitol. 70:193-206. [DOI] [PubMed] [Google Scholar]
- 5.Crewther, P. E., M. L. Matthew, R. H. Flegg, and R. F. Anders. 1996. Protective immune responses to apical membrane antigen 1 of Plasmodium chabaudi involve recognition of strain-specific epitopes. Infect. Immun. 64:3310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deans, J. A., A. M. Knight, W. C. Jean, A. P. Waters, S. Cohen, and G. H. Mitchell. 1988. Vaccination trials in rhesus monkeys with a minor, invariant, Plasmodium knowlesi 66 kD merozoite antigen. Parasite Immunol. 10:535-552. [DOI] [PubMed] [Google Scholar]
- 7.Dutta, S., P. V. Lalitha, L. A. Ware, A. Barbosa, J. K. Moch, M. A. Vassell, B. B. Fileta, S. Kitov, N. Kolodny, D. G. Heppner, J. D. Haynes, and D. E. Lanar. 2002. Purification, characterization, and immunogenicity of the refolded ectodomain of the Plasmodium falciparum apical membrane antigen 1 expressed in Escherichia coli. Infect. Immun. 70:3101-3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dutta, S., J. D. Haynes, J. K. Moch, A. Barbosa, and D. E. Lanar. 2003. Invasion-inhibitory antibodies inhibit proteolytic processing of apical membrane antigen-1 of Plasmodium falciparum merozoites. Proc. Natl. Acad. Sci. USA 100:12295-12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Escalante, A. A., H. M. Grebert, S. C. Chaiyaroj, M. Magris, S. Biswas, B. L. Nahlen, and A. A. Lal. 2001. Polymorphism in the gene encoding the apical membrane antigen-1 (AMA-1) of Plasmodium falciparum. X. Asembo Bay Cohort Project. Mol. Biochem. Parasitol. 113:279-287. [DOI] [PubMed] [Google Scholar]
- 10.Good, M. F., D. C. Kaslow, and L. H. Miller. 1998. Pathways and strategies for developing a malaria blood-stage vaccine. Annu. Rev. Immunol. 16:57-87. [DOI] [PubMed] [Google Scholar]
- 11.Harnyuttanakorn, P., J. S. McBride, S. Donachie, H. G. Heidrich, and R. G. Ridley. 1992. Inhibitory monoclonal antibodies recognise epitopes adjacent to a proteolytic cleavage site on the RAP-1 protein of Plasmodium falciparum. Mol. Biochem. Parasitol. 55:177-186. [DOI] [PubMed] [Google Scholar]
- 12.Haynes, J. D., J. K. Moch, and D. S. Smoot. 2002. Malaria methods and protocols, p. 535-554. In D. L. Doolan (ed.), Methods in molecular medicine. The Humana Press, Inc., Totowa, N.J.
- 13.Hodder, A. N., P. E. Crewther, M. L. Matthew, G. E. Reid, R. L. Moritz, R. J. Simpson, and R. F. Anders. 1996. The disulfide bond structure of Plasmodium apical membrane antigen-1. J. Biol. Chem. 271:29446-29452. [DOI] [PubMed] [Google Scholar]
- 14.Hodder, A. N., P. E. Crewther, and R. F. Anders. 2001. Specificity of the protective antibody response to apical membrane antigen 1. Infect. Immun. 69:3286-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holder, A. A. 1999. Malaria vaccines. Proc. Natl. Acad. Sci. USA 96:1167-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howell, S. A., C. Withers-Martinez, C. H. Kocken, A. W. Thomas, and M. J. Blackman. 2001. Proteolytic processing and primary structure of Plasmodium falciparum apical membrane antigen-1. J. Biol. Chem. 276:31311-31320. [DOI] [PubMed] [Google Scholar]
- 17.Howell, S. A., I. Well, S. L. Fleck, C. Kettleborough, C. R. Collins, and M. J. Blackman. 2003. A single malaria merozoite serine protease mediates shedding of multiple surface proteins by juxtamembrane cleavage. J. Biol. Chem. 278:23890-23898. [DOI] [PubMed] [Google Scholar]
- 18.Kappe, S. H., A. R. Noe, T. S. Fraser, P. L. Blair, and J. H. Adams. 1998. A family of chimeric erythrocyte binding proteins of malaria parasites. Proc. Natl. Acad. Sci. USA 95:1230-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kennedy, M. C., J. Wang, Y. Zhang, A. P. Miles, F. Chitsaz, A. Saul, C. A. Long, L. H. Miller, and A. W. Stowers. 2002. In vitro studies with recombinant Plasmodium falciparum apical membrane antigen 1 (AMA1): production and activity of an AMA1 vaccine and generation of a multiallelic response. Infect. Immun. 70:6948-6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kocken, C. H., A. M. van der Wel, M. A. Dubbeld, D. L. Narum, F. M. van de Rijke, G. J. van Gemert, X. van der Linde, L. H. Bannister, C. Janse, A. P. Waters, and A. W. Thomas. 1998. Precise timing of expression of a Plasmodium falciparum-derived transgene in Plasmodium berghei is a critical determinant of subsequent subcellular localization. J. Biol. Chem. 273:15119-15124. [DOI] [PubMed] [Google Scholar]
- 21.Kocken, C. H., C. Withers-Martinez, M. A. Dubbeld, A. Van Der Wel, F. Hackett, M. J. Blackman, and A. W. Thomas. 2002. High-level expression of the malaria blood-stage vaccine candidate Plasmodium falciparum apical membrane antigen 1 and induction of antibodies that inhibit erythrocyte invasion. Infect. Immun. 70:4471-4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marshall, V. M., L. Zhang, R. F. Anders, and R. L. Coppel. 1996. Diversity of the vaccine candidate AMA-1 of Plasmodium falciparum. Mol. Biochem. Parasitol. 77:109-113. [DOI] [PubMed] [Google Scholar]
- 23.Miller, L. H., and S. L. Hoffman. 1998. Research toward vaccines against malaria. Nat. Med. 4:520-524. [DOI] [PubMed] [Google Scholar]
- 24.Mueller, M. S., A. Renard, F. Boat, D. Vogel, M. Naegeli, R. Zurbriggen, J. A. Robinson, and G. Pluschke. 2003. Induction of parasite growth-inhibitory antibodies by a virosomal formulation of a peptidomimetic of loop I from domain III of Plasmodium falciparum apical membrane antigen 1. Infect. Immun. 71:4749-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narum, D. L., and A. W. Thomas. 1994. Differential localization of full length and processed form of PF83/AMA-1, an apical membrane antigen of Plasmodium falciparum merozoites. Mol. Biochem. Parasitol. 67:59-68. [DOI] [PubMed] [Google Scholar]
- 26.Narum, D. L., S. A. Ogun, A. W. Thomas, and A. A. Holder. 2000. Immunization with parasite-derived apical membrane antigen 1 or passive immunization with a specific monoclonal antibody protects BALB/c mice against lethal Plasmodium yoelii yoelii YM blood-stage infection. Infect. Immun. 68:2899-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pang, X. L., T. Mitamura, and T. Horii. 1999. Antibodies reactive with the N-terminal domain of Plasmodium falciparum serine repeat antigen inhibit cell proliferation by agglutinating merozoites and schizonts. Infect. Immun. 67:1821-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson, M. G., V. M. Marshall, J. A. Smythe, P. E. Crewther, A. Lew, A. Silva, R. F. Anders, and D. J. Kemp. 1989. Integral membrane protein located in the apical complex of Plasmodium falciparum. Mol. Cell. Biol. 9:3151-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polley, S. D., and D. J. Conway. 2001. Strong diversifying selection on domains of the Plasmodium falciparum apical membrane antigen 1 gene. Genetics 158:1505-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siddique, A. B., J. Iqbal, N. Ahlborg, B. Wahlin Flyg, P. Perlmann, and K. Berzins. 1998. Antibodies to nonrepeat sequences of antigen Pf155/RESA of Plasmodium falciparum inhibit parasite growth in vitro. Parasitol. Res. 84:485-491. [DOI] [PubMed] [Google Scholar]
- 31.Stowers, A. W., M. C. Kennedy, B. P. Keegan, A. Saul, C. A. Long, and L. H. Miller. 2002. Vaccination of monkeys with recombinant Plasmodium falciparum apical membrane antigen 1 confers protection against blood-stage malaria. Infect. Immun. 70:6961-6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Theisen, M., S. Soe, C. Oeuvray, A. W. Thomas, J. Vuust, S. Danielsen, S. Jepsen, and P. Druilhe. 1998. The glutamate-rich protein (GLURP) of Plasmodium falciparum is a target for antibody-dependent monocyte-mediated inhibition of parasite growth in vitro. Infect. Immun. 66:11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas, A. W., J. A. Deans, G. H. Mitchell, T. Alderson, and S. Cohen. 1984. The Fab fragments of monoclonal IgG to a merozoite surface antigen inhibit Plasmodium knowlesi invasion of erythrocytes. Mol. Biochem. Parasitol. 13:187-199. [DOI] [PubMed] [Google Scholar]
- 34.Triglia, T. J., S. Healer, R. Caruana, A. N. Hodder, R. F. Anders, B. S. Crabb, and A. F. Cowman. 2000. Apical membrane antigen 1 plays a central role in erythrocyte invasion by Plasmodium species. Mol. Microbiol. 38:706-718. [DOI] [PubMed] [Google Scholar]
- 35.Waters, A. P., A. W. Thomas, J. A. Deans, G. H. Mitchell, D. E. Hudson, L. H. Miller, T. F. McCutchan, and S. Cohen. 1990. A merozoite receptor protein from P. knowlesi is highly conserved and distributed throughout Plasmodium. J. Biol. Chem. 265:17974-17979. [PubMed] [Google Scholar]