Abstract
Standard cell proliferation assays use bulk media drug concentration to ascertain the potency of chemotherapeutic drugs; however, the relevant quantity is clearly the amount of drug actually taken up by the cell. To address this discrepancy, we have developed a flow cytometric clonogenic assay to correlate the amount of drug in a single cell with the cell’s ability to proliferate using a cell tracing dye and doxorubicin, a naturally fluorescent chemotherapeutic drug. By varying doxorubicin concentration in the media, length of treatment time, and treatment with verapamil, an efflux pump inhibitor, we introduced 105 – 1010 doxorubicin molecules per cell; then used a dye-dilution assay to simultaneously assess the number of cell divisions. We find that a cell’s ability to proliferate is a surprisingly conserved function of the number of intracellular doxorubicin molecules, resulting in single-cell IC50 values of 4 – 12 million intracellular doxorubicin molecules. The developed assay is a straightforward method for understanding a drug’s single-cell potency and can be used for any fluorescent or fluorescently-labeled drug, including nanoparticles or antibody-drug conjugates.
Keywords: Cancer chemotherapy, Pharmacodynamics, Drug effects, Efflux pumps, Cell lines
Introduction
Routinely used in vitro proliferation assays provide a high throughput method for evaluating the potency of chemotherapeutic drugs.1,2 Typical proliferation assays use the bulk media concentration to determine the drug potency (i.e. IC50 or IC90). However, the drug concentration on a media volumetric basis would need to be freely in equilibrium with the drug’s intracellular target for this to truly represent the drug’s intrinsic potency. This is not true in almost any case since drugs encounter membranous diffusion barriers and may be substrates for active uptake or efflux transporters.3 The amount of drug internalized into the cell is a more physiologically relevant basis for comparison than the bulk media concentration4,5 especially when considering drug delivery systems that involve endosomal transport and processing steps, such as antibody-drug conjugates or liposome drug delivery systems.6 It is now within the purview and capability of the drug designer to attempt to alter a drug’s interaction with these transport and processing machineries, in order to attain more efficient delivery on target. However, a key piece of information in such cases is the number of drug molecules on target necessary for the desired effect (e.g. how many doxorubicin molecules does it take to kill a cell?). This information is not directly available from potencies determined on a media-volume basis. The assay described herein uses the amount of drug in an individual cell as the basis for cellular response rather than the drug concentration in the cell growth media.
Standard chemotherapy potency assays include non-clonogenic assays that are based on changes in cell membrane permeability (LDH or Trypan Blue), mitochondrial function (MTT or WST-1 Assay), or markers for early (Annexin V) or late apoptosis (TUNEL or cytochrome C).7 In contrast, clonogenic assays measure a cell’s ability to proliferate after treatment. Traditionally, proliferation is measured by counting clones that have grown out after cells have been plated at low density.8 Clonogenic assays capture all types of cell death and include cell growth after reversible damage, whereas non-clonogenic assays measure acute cellular toxicity, often specific to one type of cell death. Since clonogenic assays capture the integrated effect of many different types of cellular response to drug treatment, we focused on this assay type.
Here, we develop a flow cytometric dye-dilution clonogenic assay to determine the relationship between the amount of drug in a single cell and the cell’s ability to proliferate. Flow cytometry enables high-throughput screening of thousands of individual cells, resulting in analysis on a single-cell level rather than a bulk population level. The assay uses a cell tracing dye and a fluorescent drug. A cell tracing dye is used to track cell proliferation via dye dilution. All cells are initially stained with dye and the dye is diluted in half with each cell division. A fluorescent drug is used in order to measure the amount of drug taken up by each cell.
In this work, we used doxorubicin, a standard chemotherapeutic drug9, which is also naturally fluorescent10, as a model drug to demonstrate application of the assay. Doxorobucin is known to bind DNA and inhibit topoisomerase II9 and is widely used as a front-line therapy for a number of different types of cancer.11
Methods
Cell Lines and Materials
Eight different cell lines were used in this work: BT-474, HCT-15, HT-29, IGROV-112, MDA-MB-231, NCI-N87, SK-BR-3, and T-47D. Cell Lines were purchased from American Type Culture Collection (ATCC). All cell lines except HT-29 and SK-BR-3 were grown in RPMI (Corning) supplemented with 10 % fetal bovine serum (FBS) and 5 % penicillin-streptomycin. HT-29 and SK-BR-3 cells were grown in Dulbecco's Modified Eagle Medium (DMEM, Corning) and McCoy’s 5A Medium Modified (Lonza), respectively, supplemented in the same way. Doxorubicin hydrochloride and verapamil were purchased from Sigma.
Assay Set-up
Cells were stained using CellTrace™ Violet Cell Proliferation Kit (Life Technologies) following the “Standard Method for Labeling Cells in Suspension” as described in the product manual. Then, 105 cells were plated per well in 6-well tissue culture plates (BD). The cells were treated with doxorubicin hydrochloride at concentrations ranging from 10 nM to 5 µM in standard growth media. Control cells that were either stained with CellTrace Violet only or unstained were plated at the same time. After 24 h, the cells were washed with PBS and the media was replaced with fresh growth media. After an additional 3 days, the cells were trypsinized and prepared for flow cytometry. Flow cytometry was performed using a BD FACSCanto II. The doxorubicin signal was measured using excitation with a 488 nm laser and detection with a 585 nm ± 42 nm filter. The CellTrace Violet signal was measured using excitation with a 405 nm laser and detection with a 450 nm ± 50 nm filter. We collected data for 10,000 cells (gated based on forward and side scatter) per condition, unless there were an insufficient number of cells remaining.
Treatment Length Study
For the dosing time study, MDA-MB-231 cells were plated as described above in the Assay Set-up section. Initially, the media either had a medium (0.3 µM) or high (5 µM) dose of doxorubicin. The cells were washed at various time points (12, 24, 48, 72, and 96 h) and the media was replaced with fresh growth media. All cells were read on the flow cytometer at the same time after a total of 4 days after plating.
Verapamil Treatment
Using HCT-15 cells, the study with verapamil treatment was set up as described above in the Assay Set-up section with 20 µM verapamil in the growth media. The replacement media after 24 h also contained 20 µM verapamil.
Data Analysis
The raw flow cytometry data were processed in the following manner in order to draw together the results from numerous single-cell measurements from different treatment conditions. FlowJo software and MATLAB (Mathworks) were used for data processing. First, the doxorubicin signal was calibrated as described in the “Calibration of Doxorubicin Signal” section. Next, we normalized the CellTrace signal with respect to the median CellTrace Signal for untreated cells as described in equation 1.
| (1) |
Note that the fluorescence signal from untreated cells is in the numerator of the expression in equation 1. Cells that did not proliferate at all have a high CellTrace signal because the CellTrace has not been diluted by growth. Thus, the proliferation factor is low for cells which had fewer cell divisions and is equal to one if cells were unaffected by treatment. The theoretical minimum for the proliferation factor with complete inhibition of growth is 2−n, where n is the number of doublings for untreated cells.
With both fluorescence signals converted, the cells were binned based on amount of intracellular doxorubicin. 100 bins were used with even logarithmic spacing from 104 to 1010 intracellular doxorubicin molecules. Any bin with fewer than 100 cells was omitted. For each bin of cells, median proliferation factor was plotted versus the median number of intracellular doxorubicin molecules resulting in a cellular response curve to doxorubicin treatment. The included plots show cellular response curves for either individual treatment conditions or for data from all treatment conditions concatenated into one response curve. When processing the fluorescence signal from control cells, half of the untreated cells appear as if they have doxorubicin signal despite never being treated with doxorubicin since the median doxorubicin signal for untreated cells was used to subtract out background fluorescence signal. In addition, the lower half of the control cells appear to have a negative number of doxorubicin cells based on the calibration of the doxorubicin signal and thus do not appear in the analysis plots since the plots are log-based.
Calculation of Standard and Single-Cell IC50
The standard IC50 is the media doxorubicin concentration required for 50 % of maximum reduction in proliferation factor. The single-cell IC50 is the number of intracellular doxorubicin molecules required for a 50% of maximum reduction in proliferation factor. The IC50 values were calculated from a nonlinear regression with the “log(inhibitor) vs. response (three parameter)” equation in GraphPad Prism software. Median proliferation factor for each treatment condition was used for standard IC50 and the median values from bins for the concatenated data for each cell line was used for single-cell IC50. Confidence intervals were also calculated in the GraphPad Prism software.
Calibration of Doxorubicin Signal
In order to convert the fluorescence signal from doxorubicin to numbers of intracellular doxorubicin molecules, we used calibration beads (Quantum™ Simply Cellular® anti-Human IgG, Bangs Laboratories, Inc.) and a Trastuzumab-doxorubicin conjugate (Tras-Dox). In future work, we will characterize the pharmacodynamics of Tras-Dox; however, for the purposes of the present discussion, it serves only as a useful calibration standard. BT-474, NCI-N87, and SK-BR-3 cells were used since they have a high expression of HER2, the antigen target for Trastuzumab. These cells were fixed using BD Cytofix™ Fixation Buffer following the recommended protocol. For each cell line, 1.25×105 cells were stained with 60 nM Alexa Fluor-647 labeled Trastuzumab (Tras-647) in staining buffer (PBS with 0.2 % bovine serum albumin and 0.09 % sodium azide). Another 1.25×105 cells were stained with 50 nM Tras-Dox in staining buffer. Cells were stained overnight at 37 °C. After staining, cells were washed twice with cold stain buffer before being read on the flow cytometer.
Calibration beads were stained with 100 nM Tras-647 following the manufacturer’s recommended protocol. The calibration beads and Tras-647 labeled cells were read on a BD Accuri C6 Flow Cytometer. Using the linear fit from calibration, the HER2 receptor expression level on each of the cell lines was determined from the fixed cells stained with Tras-647. Cells stained with Tras-Dox were read on a BD FACS Canto II. From the HER2 expression level, Tras-Dox drug-to-antibody ratio, and doxorubicin fluorescence intensity of cells saturated with Tras-Dox, the number of doxorubicin molecules per fluorescence signal unit was determined. This was used as a scaling factor to convert fluorescence signal from cells to number of intracellular doxorubicin molecules. The fluorescence signal above background (median fluorescence signal of untreated cells) was considered signal from doxorubicin.
Synthesis of Doxorubicin-SMCC
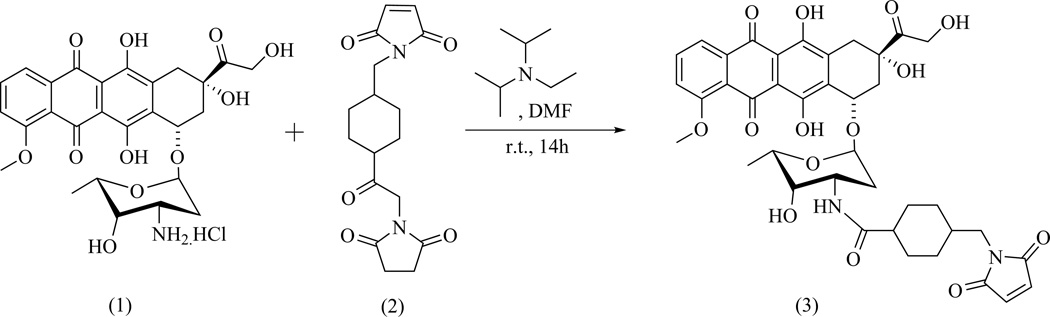
Doxorubicin-SMCC was prepared using a modification of a previously published procedure13 as illustrated in Scheme 1. Doxorubicin HCl salt (1, 0.050 g, 0.086 mmol, 1.0 equiv.), succinimidyl 4-[N-maleimidomethyl] cyclohexane-1-carboxylate (SMCC, 2, 0.035 g, 0.104 mmol, 1.2 equiv.), and diisoprpylethylamine (Hünig’s base, 0.065 mmol, 1.5 equiv.) were added to anhydrous DMF (3.0 mL) in a 100 mL round-bottomed flask. The reaction was allowed to run in the dark at room temperature under inert conditions overnight.
Scheme 1.
Synthesis of Doxorubicin–SMCC
After evaporation of the solvent under vacuum at 35 – 40 °C, the residue was dissolved in 15 mL dichloromethane and washed with brine three times (3×5 mL). The organic phases were pooled together and dried over anhydrous magnesium sulfate. After filtration and washing of the solid filter cake with additional dichloromethane (2×5 mL), the filtrate and washings were combined and concentrated in vacuo to viscous oil. Silica gel column chromatography of the residue (eluted with 0 – 5 % methanol/dichloromethane) yielded a red solid product at 89 % yield: MALDI-TOF MS m/z 785.559 [with Na+, the K+ adduct also observed at 801.528]; Rf 0.31 (95/5 CH2Cl2/MeOH); UV λmax 204, 251, 479, 490 nm. Major 1H NMR signals were found to be identical with previously reported13 compound (3): 1H NMR (400 MHz, DMSO-d6) δ = 0.91 (m), 1.19 (m), 1.62 (m, 4H), 1.84 (m, 1H), 2.20 (m), 3.21 (d, 2H), 3.81 (m, 1H), 4.21 (m, 4H), 4.62 (s, 2H), 4.80 (d, 1H), 4.94, 5.24 (s, 1H), 5.80 (s, 1H), 7.40 (d, 1H), 7.67 (d, 1H), 7.96 (m, 2H), 12.43 (s, 1H), 13.21 (s) ppm.
Preparation of Trastuzumab-Doxorubicin Conjugate (Tras-Dox)
A Trastuzumab antibody mutant with an engineered cysteine site for conjugation, Trastuzumab A114C, was prepared according to literature procedure.14 Trastuzumab A114C antibody (7 mg/mL final concentration) was reacted with 6 equivalents of doxorubicin-SMCC in PBS with 15 % dimethylacetamide. The reaction tube was incubated in a Thermomixer (Eppendorf) at 25 °C and 700 rpm for 2 h. Initial clean-up and concentration of the reaction mixture were completed using a PD-10 desalting column (GE Healthcare) and an Amicon centrifugal filter unit (EMD Millipore), respectively. An AKTA size exclusion chromatography system (GE Healthcare) was employed for purification. The final conjugate had a drug-to-antibody ratio (DAR) of 2, based on LC-MS characterization results.
Preparation of Trastuzumab-Alexa Fluor 647 Conjugate (Tras-647)
A Trastuzumab antibody mutant with an engineered cysteine site for conjugation, Trastuzumab A114C, was prepared according to literature procedure.14 Trastuzumab A114C antibody (1 mg/mL final concentration) was reacted with 5 equivalents of NHS-Alexa Fluor 647 (Life Technologies) in PBS with 10 % 1 M borate buffer (pH 9). The reaction tube was affixed to a rotator at room temperature for 2.5 h. Initial clean-up and concentration of the reaction mixture were completed using a PD-10 desalting column (GE Healthcare) and an Amicon centrifugal filter unit (EMD Millipore), respectively. An AKTA size exclusion chromatography system (GE Healthcare) was employed for purification. The final conjugate had a fluorophore-to-antibody ratio of 1.4, based on LC-MS characterization results.
Results
Data Processing for Flow Cytometric Clonogenic Assay
For meaningful comparisons, the raw flow cytometry data were processed as illustrated in Fig. 1. The doxorubicin signal was converted to number of intracellular doxorubicin molecules using the calibration method as described in the methods section. The doxorubicin signal from calibration beads (105 antibody binding sites) saturated with Tras-Dox was too dim to use for calibration (data not shown). Instead, we measured the signal for saturated, fixed cells (106 antibody binding sites) stained with Tras-Dox and correlated this signal with the known number of doxorubicin molecules per antibody and number of HER2 receptors per cell, which was measured using calibration beads and Alexa Fluor 647 labeled Trastuzumab (Tras-647). This calibration yielded a scaling factor for the doxorubicin signal observed and number of intracellular doxorubicin molecules. With our flow cytometry settings, 100,000 ± 19,000 (standard error of the mean) doxorubicin molecules result in one flow cytometric fluorescence unit. See Supplemental Table 1 for the flow cytometry data used to reach this conversion factor.
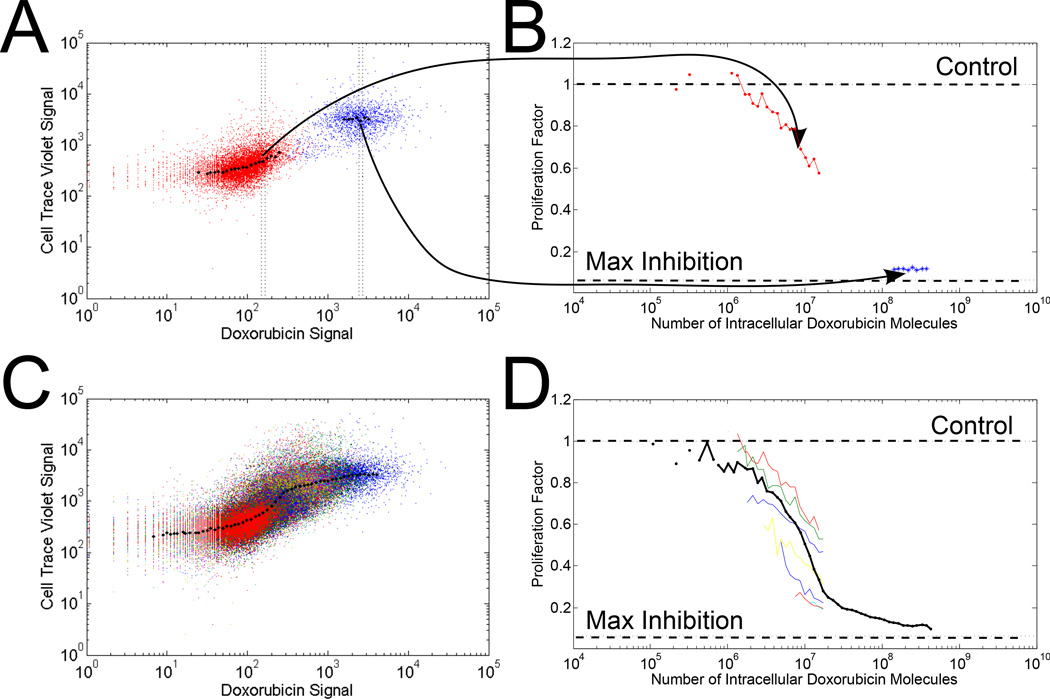
Figure 1.
Data processing steps (A) The raw flow cytometry results for the control population (stained with CellTrace, no doxorubicin treatment) are shown in red and the highest doxorubicin treatment population (5 µM doxorubicin in media) are shown in blue. (B) Conversion of fluorescence signals to number of intracellular doxorubicin molecules and the proliferation factor are shown for cell populations shown in A. The horizontal lines illustrate growth equivalent to untreated cells and maximum inhibition with a cell doubling rate of 24 h for untreated cells. (C) and (D) demonstrate the same transformation as in (A) and (B) with the full range (10 nM – 5 µM) of media doxorubicin treatment concentrations. In (D), each color represents an individual doxorubicin media concentration. The solid black line is the resulting response curve from the concatenation of all cell treatment data.
Fig. 1A and 1B illustrate the data processing for untreated control cells and cells treated with the highest doxorubicin treatment (5 µM doxorubicin in media). As expected, the cells treated with doxorubicin did not proliferate at the rate of control cells, and consequently did not dilute the tracer dye to as great an extent. The control cells are included as a reference for background signal and variability.
Fig. 1C and 1D illustrate results for a typical experiment when cells were treated with a range (10 nM to 5 µM) of media doxorubicin concentrations. The data from all treatment conditions were combined and processed together, resulting in one cellular response curve, as illustrated in Fig. 1D (black line). For reference, the response curves for individual doxorubicin treatment conditions are also shown in Fig. 1D (each color represents one media doxorubicin concentration). The horizontal dashed lines shown in Fig. 1B and 1D indicate where the proliferation factor is equal to 1, when cells are proliferating as if they were untreated, as well as the theoretical lower limit (maximum inhibition) for the proliferation factor with an assumed cell doubling time of 24 h.
Cell Proliferation Response with a Range of Doxorubicin Treatment
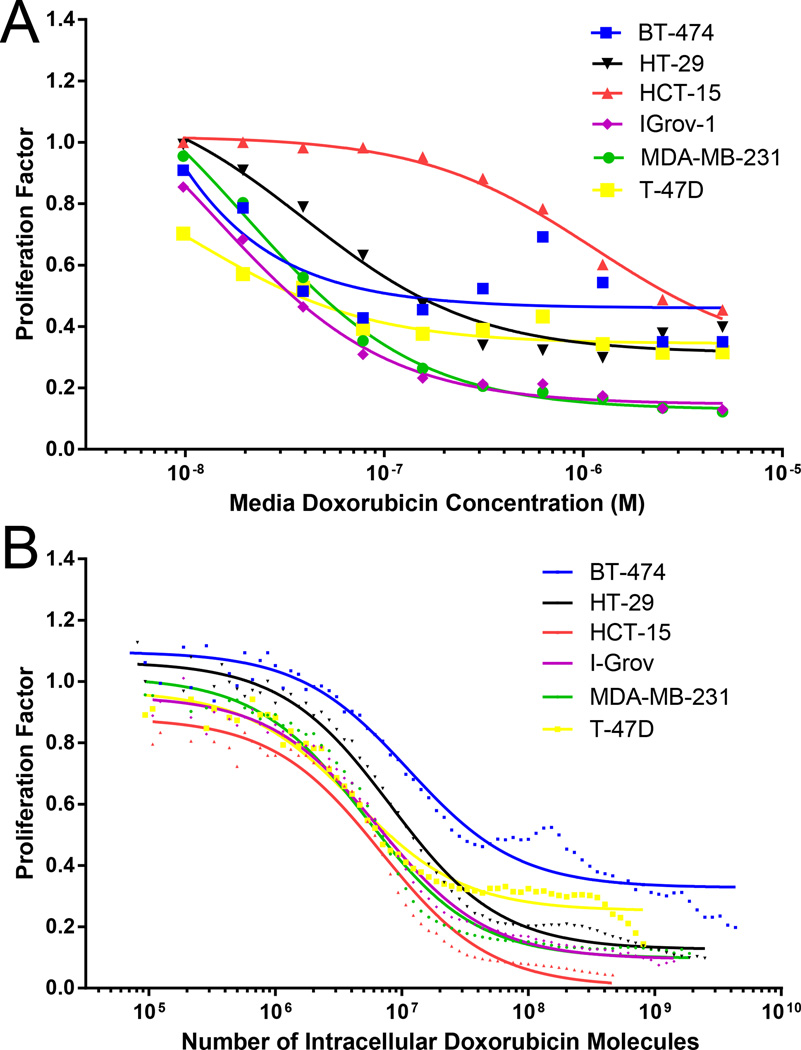
The growth response of six cell lines (BT-474, HCT-15, HT-29, IGROV-1, MDA-MB-231, and T-47D) to varying concentrations of doxorubicin was determined on the most commonly used culture volumetric basis for the drug (Fig. 2A). There appears to be substantial variation in potency of doxorubicin across these cell lines, ranging from 0.8 nM to 1.1 µM IC50 as listed in Table 1. This variation may be due to differences in transport efficiency of the drug to the nucleus amongst cell lines, or instead because there are differential responses to a given quantity of doxorubicin in the nucleus. From the information available in this plot, it is not possible to discriminate between these two fundamentally different possibilities.
Figure 2.
Cell proliferation response to doxorubicin treatment. Flow cytometric clonogenic assay results are shown with (A) media doxorubicin concentration as the basis and (B) intracellular doxorubicin concentration as the basis. Data were processed in same manner described in Fig. 1. Lines represent thecurves for IC50 value fits. In (A), the data points represent the median proliferation factor for a given media doxorubicin treatment, whereas in (B), the data points represent bins from the concatenation of all treatment conditions and replicates.
Table 1.
Doxorubicin IC50 Values (on a media concentration basis)
| Cell Line | IC50 (Media Doxorobucin Concentration, M) |
95% Confidence Interval on IC50 (M) |
|---|---|---|
| BT-474 | 1.8 × 10−8 | 1.3 × 10−10 – 2.7 × 10−6 |
| HCT-15 | 1.3 × 10−6 | 7.4 × 10−7 – 2.3 × 10−6 |
| HT-29 | 7.1 × 10−8 | 2.2 × 10−8 – 2.3 × 10−7 |
| IGrov-1 | 2.9 × 10−8 | 1.3 × 10−8 – 6.7 × 10−8 |
| MDA-MB-231 | 4.5 × 10−8 | 1.78 × 10−8 – 1.2 × 10−7 |
| T-47D | 1.1 × 10−8 | 1.8 × 10−9 – 7.2 × 10−8 |
We used our flow cytometric clonogenic assay to determine cell proliferation response to a particular number of intracellular doxorubicin molecules for the same six cell lines, as illustrated in Fig. 2B. Note that the response data points shown in Fig. 2B are the product of data concatenated from all treatment conditions from replicate experiments. Response curves for each individual treatment condition are shown in Supplemental Figure 1. More variability was seen in the BT-474 cell response to doxorubicin treatment than in the responses of other cell lines. The BT-474 cell line is known to have a variable response to chemotherapy,15 which may be due to protein fluctuations.16
Table 2 lists the single-cell IC50 for each cell line. Thus, cellular uptake of 4 – 12 million molecules of doxorubicin results in 50 % of maximum reduction in proliferation. It is noteworthy how similar the IC50 is on a per-cell basis, as opposed to a culture-volumetric basis, indicating that differential subcellular trafficking rather than differential drug responsiveness accounts for the apparent variation in sensitivity in Fig. 2A.
Table 2.
Doxorubicin Single-cell IC50 Values
| Cell Line | Single-Cell IC50 (# of Intracellular Doxorubicin molecules) |
95% Confidence Interval on Single-Cell IC50 |
|---|---|---|
| BT-474 | 11.3 × 106 | 8.6 × 106 to 14.8 × 106 |
| HCT-15 | 6.9 × 106 | 5.6 × 106 to 8.4 × 106 |
| HT-29 | 8.2 × 106 | 7.3 × 106 to 9.1 × 106 |
| IGrov-1 | 6.5 × 106 | 5.9 × 106 to 7.3 × 106 |
| MDA-MB-231 | 5.2 × 106 | 4.5 × 106 to 6.0 × 106 |
| T-47D | 4.2 × 106 | 3.5 × 106 to 5.1 × 106 |
Proliferation Response as a Function of Length of Treatment
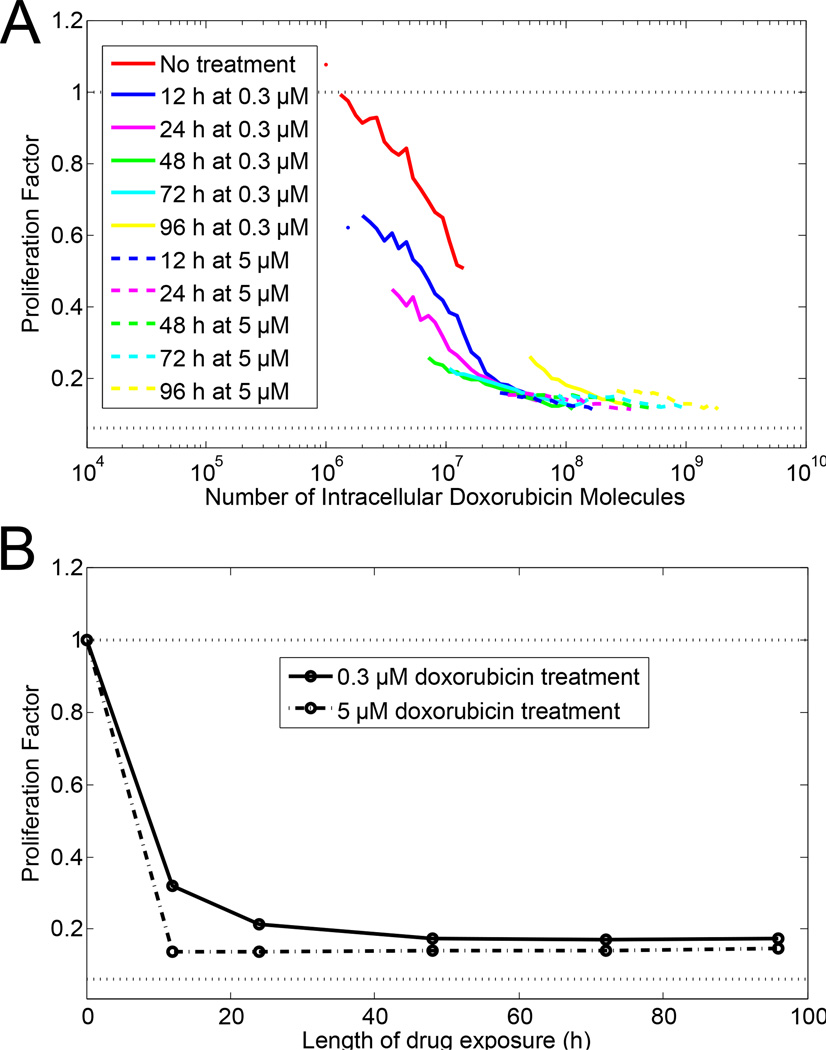
Next, we considered doxorubicin dose rate effects on cell proliferation. All experiments described up to this point involved a one-day treatment period followed by a three-day growth period. Fig. 3 illustrates how cells respond to varying treatment period length. At the high dose (5 µM doxorubicin), cells take up sufficient drug to stop proliferation within the first 12 h. Additional treatment time resulted in increased uptake of doxorubicin, but no change in proliferation. However, at the moderate dose (0.3 µM doxorubicin), cell proliferation decreased in response to increased doxorubicin treatment time in a manner dependent on the amount of doxorubicin in the cell. The cellular response to doxorubicin is one continuous function of the amount of doxorubicin in the cell, independent of the rate at which that level of doxorubicin was obtained. This effect is not observed when the length of treatment was used as the metric to evaluate cell proliferation instead of the amount of doxorubicin in the cells, as in Fig. 3B.
Figure 3.
Effect of length of drug exposure on cell proliferation. MDA-MB-231 cells were treated with a moderate drug concentration (0.3 µM, solid lines) or high drug concentration (5 µM, dashed lines). (A) Flow cytometric clonogenic assay with cell proliferation as a function of amount of doxorubicin in cells. Length of doxorubicin exposure is given by different colors as described in legend. (B) Proliferation as a function of the length of doxorubicin exposure with either 0.3 µM media doxorubicin concentration (solid line) or 5 µM media doxorubicin concentration (dashed line).
Doxorubicin Uptake and Cell Proliferation in Pgp Expressing Cells
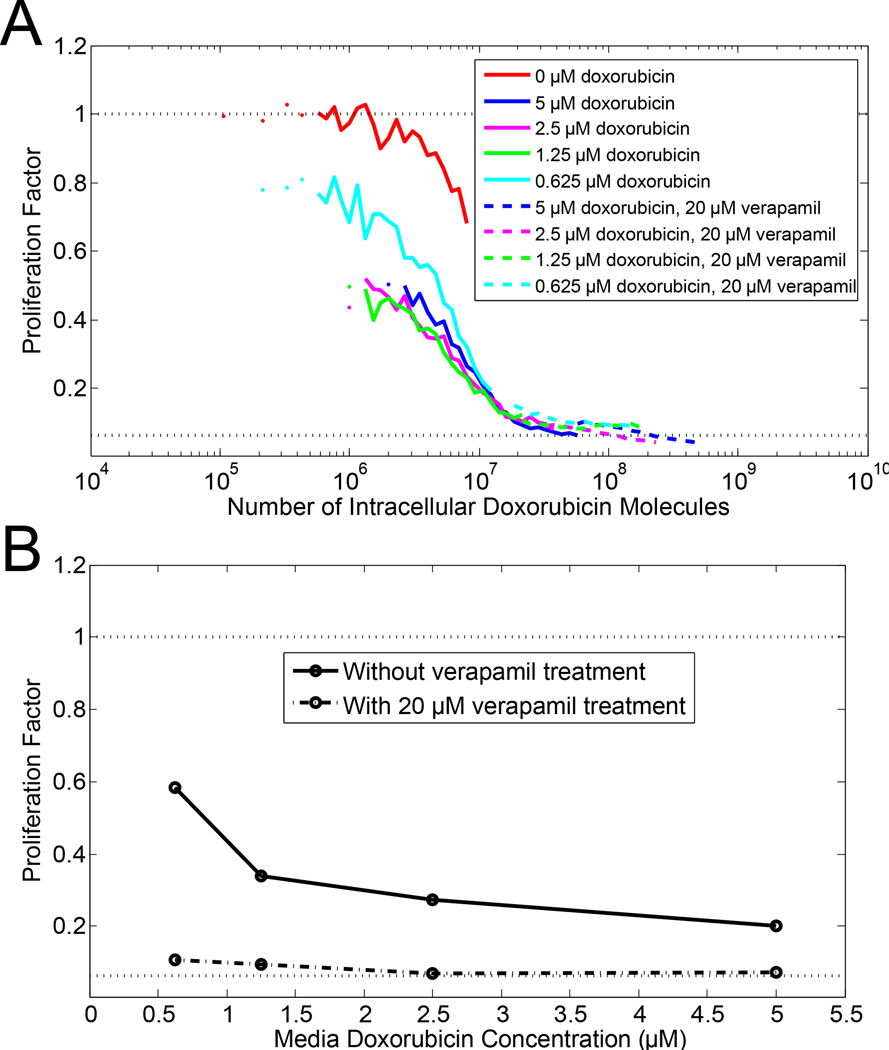
Doxorubicin treatment is known to induce multidrug resistance via expression of the drug efflux pump P-glycoprotein (Pgp).17–19 In Fig. 4, we observed how treatment with a Pgp inhibitor, verapamil, affected doxorubicin uptake and cell proliferation in Pgp-expressing cells. Of the cell lines that we tested, only HCT-15 cells expresses Pgp without any pre-treatment with doxorubicin.19 Again, the cell proliferation response to intracellular doxorubicin is one continuous response curve, rather than different response curves in the presence or absence of verapamil treatment. In contrast, the cell proliferation response curves with respect to the media doxorubicin concentration are two distinct curves in the presence or absence of verapamil treatment, as shown in Fig. 4B. Thus, the key factor for HCT-15 cell proliferation is the amount of doxorubicin in the cell. It is irrelevant in terms of cell proliferation whether the amount of doxorubicin in the cell resulted from a higher media doxorubicin concentration or from a lower media doxorubicin concentration with verapamil treatment.
Figure 4.
Effect of verapamil treatment on cell proliferation. HCT-15 cells, which express the drug efflux pump P-glycoprotein (Pgp), were treated with doxorubicin in the presence (dashed lines) and absence (solid lines) of 20 µM verapamil, a Pgp inhibitor. (A) Flow cytometric clonogenic assay with cell proliferation as a function of amount of intracellular doxorubicin in cells. Doxorubicin media concentration is given by different colors as described in legend. (B) Proliferation as a function of the media doxorubicin concentration with (dashed line) or without (solid line) 20 µM verapamil.
Discussion
In this work, a flow cytometric clonogenic assay was developed and the application of this assay was demonstrated with the model drug doxorubicin. Intracellular doxorubicin, rather than doxorubicin concentration in the cell growth media, was demonstrated as the key determinant of cell proliferation inhibition. Whether the amount of intracellular doxorubicin was achieved by varying doxorubicin media concentration, treating cells with verapamil, or increasing treatment length was irrelevant. These results are consistent with previous studies of doxorubicin cellular pharmacodynamics.20–24
The doxorubicin single-cell IC50 values were 4 – 12 million intracellular doxorubicin molecules. To put this number in context, this represents ~ 3% loading of the total number of binding sites on doxorubicin’s primary intracellular target, DNA, based on one doxorubicin molecule binding per ten DNA base pairs25 and approximately three billion base pairs per cell in humans.26 It is important to note that the quantification of intracellular doxorubicin molecules we report here is at the time of analysis, which is three days after ending doxorubicin treatment. During this incubation time, doxorubicin may efflux from the cell, be diluted by growth, or be degraded. Doxorubicin fluorescence is known to be quenched once bound to DNA, resulting in a 30–40 fold decrease in fluorescence signal.10,27,28 It has also been shown that some doxorubicin degradation products fluoresce in the same emission wavelength window as doxorubicin.27,29 Single-cell IC50 values provide a snapshot of the order of magnitude of intracellular doxorubicin molecules, but the interpretation of the exact quantitative values should consider these factors.
In this work, we assume that the intracellular doxorubicin is all available to bind to DNA; however, there may be a fraction of drug bound to proteins and lipids in the cell. The free drug (i.e. not bound to proteins or lipids) is the relevant quantity in relation to efficacy.30 Since doxorubicin is a relatively hydrophilic drug (log D7.4 = −1.98),31 this cellular protein bound drug accounts for less of the intracellular drug than for more hydrophobic drugs, where drug bound to cellular proteins could account for more than 90% of the intracellular drug.32 Plasma protein binding of drugs could be used as a correlate for cellular protein binding. Methods are established for plasma protein binding as well as published values for doxorubicin and a number of other drugs.33,34,32 Hydrophobicity can also affect a drug’s ability to diffuse into cells, which drives the difference between intracellular and extracellular drug concentrations.
Another key consideration when interpreting results from this assay is the fact that cells that have died by the time of analysis are not seen in the flow cytometry results. (The absence of dead cells is intrinsic to all clonogenic assays.) Thus, the reported intracellular doxorubicin values are for cells that are still intact. Likely, for many treatment conditions, there were cells that took up more doxorubicin and died before analysis was completed. Lower cell number and altered cell morphology as indicated by forward and side scatter patterns were observed for cells treated at the highest doxorubicin concentrations.
Previous studies that have quantified intracellular doxorubicin report 107 – 5×109 intracellular doxorubicin molecules after 2 – 12 h treatment periods.6,35–39 Many of these reported values are higher than the single-cell IC50 values shown here, because the respective studies use higher doxorubicin media concentrations for treatment and the intracellular doxorubicin was quantified immediately after treatment rather than after a three-day growth period as in this assay. Most of the previous intracellular doxorubicin quantification studies were not correlated with cell proliferation. However, Kerr et al. reported a LD90 (lethal dose for 90% of colonies) of 39 million intracellular doxorubicin molecules.22 This value corresponds well with single-cell IC50 values reported here.
Delivery of tens of millions of doxorubicin molecules to a cell, although easily accomplished via diffusion of the free drug, could be harder to achieve with an antibody-drug conjugate (ADC). ADCs are designed to carry a drug payload specifically to cancer cells and release the drug once internalized into the cell. Consider a cell with high antigen expression at 106 surface antigens per cell and an internalization half time of 12 h. With a drug-to-antibody ratio of 4, a cell would internalize ~ 4 million drug molecules per day. This estimated delivery is below many of the reported single-cell IC50 values for doxorubicin and doesn’t take into account the additional processing steps between ADC internalization and doxorubicin binding to DNA. The single-cell IC50 values measured in this work for doxorubicin support the generally accepted rule that ADCs need a potent drug to be successful.40–43 This assay provides a method for determining the required amount of drug that must be delivered intracellularly via an ADC.
This flow cytometric clonogenic assay can be generalized to study any fluorescent drug or fluorescently-tagged drug. Fluorescent analogs of drugs can be very useful tools for tracking cellular uptake of a drug in assays such as this flow cytometric assay and others.44,45 However, fluorescent analogs may be difficult to synthesize, have reduced potency, and have different trafficking behavior when compared to the parent drug. Drug delivery systems that are taken up by cells, such as liposomes, nanoparticles, and ADCs, can also be used in this clonogenic assay with a fluorescent tag on the delivery system as proxy for how much drug was taken up, rather than using a fluorescent drug or fluorescent analog of a drug.
In conclusion, we have presented here a straightforward flow cytometric clonogenic assay using a cell tracing dye and a fluorescent drug. The first application of the assay revealed the single-cell potency of doxorubicin via single-cell IC50 values of 4 – 12 million intracellular doxorubicin molecules for the cancer cell lines evaluated. Single-cell potency is a key parameter for molecular design of targeting agents where subcellular trafficking and processing can be affected by altered design attributes.
Supplementary Material
Acknowledgements
We thank Lindsay King, Nahor Haddish-Berhane, and members of the Wittrup Lab for their technical suggestions and review. K.F.M. was supported by a Hertz Foundation Fellowship and a National Science Foundation Graduate Research Fellowship. C.K. was supported by the Pfizer Worldwide Research & Development Post-Doctoral Program. This work was also supported by a research grant from Pfizer and in part by the Koch Institute Support (core) Grant P30-CA14051 from the National Cancer Institute. We thank the Koch Institute Swanson Biotechnology Center for technical support, specifically the Flow Cytometry Core.
References
- 1.Fotakis G, Timbrell Ja. In vitro cytotoxicity assays: comparison of LDH, neutral red, MTT and protein assay in hepatoma cell lines following exposure to cadmium chloride. Toxicol. Lett. 2006;160:171–177. doi: 10.1016/j.toxlet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Weyermann J, Lochmann D, Zimmer A. A practical note on the use of cytotoxicity assays. Int. J. Pharm. 2005;288:369–376. doi: 10.1016/j.ijpharm.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 3.Gottesman MM. Mechanisms of cancer drug resistance. Annu. Rev. Med. 2002;53:615–627. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- 4.Pirie CM, Hackel BJ, Rosenblum MG, Wittrup KD. Convergent potency of internalized gelonin immunotoxins across varied cell lines, antigens, and targeting moieties. J. Biol. Chem. 2011;286:4165–4172. doi: 10.1074/jbc.M110.186973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sundman-Engberg B, Tidefelt U, Liliemark J, Paul C. Intracellular concentrations of anti cancer drugs in leukemic cells in vitro vs in vivo. Cancer Chemother. 1990:252–256. doi: 10.1007/BF00684881. [DOI] [PubMed] [Google Scholar]
- 6.Eliaz RE. Determination and Modeling of Kinetics of Cancer Cell Killing by Doxorubicin and Doxorubicin Encapsulated in Targeted Liposomes. Cancer Res. 2004;64:711–718. doi: 10.1158/0008-5472.can-03-0654. [DOI] [PubMed] [Google Scholar]
- 7.Sumantran VN. Cellular chemosensitivity assays: An overview. Methods Mol. Biol. 2011;731:219–236. doi: 10.1007/978-1-61779-080-5_19. [DOI] [PubMed] [Google Scholar]
- 8.Franken N, Rodermond H, Stap J. Clonogenic assay of cells in vitro. Nat. Protoc. 2006;1:2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 9.Minotti G, Menna P, Salvatorelli E, Cairo G, Gianni L. Anthracyclines : Molecular Advances and Pharmacologic Developments in Antitumor Activity and Cardiotoxicity. 2004;56:185–229. doi: 10.1124/pr.56.2.6. [DOI] [PubMed] [Google Scholar]
- 10.Karukstis K, Thompson E. Deciphering the fluorescence signature of daunomycin and doxorubicin. Biophys. 1998;73:249–263. doi: 10.1016/s0301-4622(98)00150-1. [DOI] [PubMed] [Google Scholar]
- 11.Saltiel E, McGuire W. Doxorubicin (Adriamycin) Cardiomyopathy—A Critical Review. West. J. Med. 1983;139:332–341. [PMC free article] [PubMed] [Google Scholar]
- 12.Bénard J, et al. Characterization of a Human Ovarian Adenocarcinoma Line , IGROV1 , in Tissue Culture and in Nude Mice Characterization of a Human Ovarian Adenocarcinoma Line , IGROV1 , in Tissue Culture and in Nude Mice1. Cancer R. 1985;45:4970–4979. [PubMed] [Google Scholar]
- 13.Chen Q, Gabathuler R. Efficient Synthesis of Doxorubicin Melanotransferrin p97 Conjugates Through SMCC Linker. Synth. Commun. 2004;34:2407–2414. [Google Scholar]
- 14.Junutula JR, et al. Site-specific conjugation of a cytotoxic drug to an antibody improves the therapeutic index. Nat. Biotechnol. 2008;26:925–932. doi: 10.1038/nbt.1480. [DOI] [PubMed] [Google Scholar]
- 15.Holliday DL, Speirs V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011;13:215. doi: 10.1186/bcr2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459:428–432. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottesman M, Pastan I. Biochemistry of multidrug resistance mediated by the multidrug transporter. Annu. Rev. Biochem. 1993;62:385–427. doi: 10.1146/annurev.bi.62.070193.002125. [DOI] [PubMed] [Google Scholar]
- 18.Shen F, et al. Quantitation of Doxorubicin Uptake , Efflux , and Modulation of Multidrug Resistance (MDR) in MDR Human Cancer Cells. Pharmacology. 2008;324:95–102. doi: 10.1124/jpet.107.127704. [DOI] [PubMed] [Google Scholar]
- 19.Lai G-M, Chen Y-N, Mickley La, Fojo AT, Bates SE. P-glycoprotein expression and schedule dependence of adriamycin cytotoxicity in human colon carcinoma cell lines. Int. J. Cancer. 1991;49:696–703. doi: 10.1002/ijc.2910490512. [DOI] [PubMed] [Google Scholar]
- 20.Durand R, Olive P. Flow cytometry studies of intracellular adriamycin in single cells in vitro. Cancer Res. 1981;41:3489–3494. [PubMed] [Google Scholar]
- 21.El-Kareh AW, Secomb TW. Two-Mechanism Peak Concentration Model for Cellular Pharmacodynamics of Doxorubicin. Neoplasia. 2005;7:705–713. doi: 10.1593/neo.05118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerr DJ, Kerr aM, Freshney RI, Kaye SB. Comparative intracellular uptake of adriamycin and 4’-deoxydoxorubicin by non-small cell lung tumor cells in culture and its relationship to cell survival. Biochem. Pharmacol. 1986;35:2817–2823. doi: 10.1016/0006-2952(86)90195-4. [DOI] [PubMed] [Google Scholar]
- 23.Praet M, Stryckmans P, Ruysschaert J. Cellular uptake, cytotoxicity, and transport kinetics of anthracyclines in human sensitive and multidrug-resistant K562 cells. Biochem. Pharmacol. 1996;51:1341–1348. doi: 10.1016/0006-2952(96)00042-1. [DOI] [PubMed] [Google Scholar]
- 24.Yu T, Xiong Z, Chen S, Tu G. The use of models in ‘target’ theory to evaluate the survival curves of human ovarian carcinoma cell line exposure to adriamycin combined with ultrasound. Ultrason. Sonochem. 2005;12:345–348. doi: 10.1016/j.ultsonch.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 25.Tarasiuk J, Frézard F, Garnier-Suillerot a, Gattegno L. Anthracycline incorporation in human lymphocytes. Kinetics of uptake and nuclear concentration. Biochim. Biophys. Acta. 1989;1013:109–117. doi: 10.1016/0167-4889(89)90038-4. [DOI] [PubMed] [Google Scholar]
- 26.A Brief Guide to Genomics. April 14, 2014 at< http://www.genome.gov/18016863>. [Google Scholar]
- 27.Fiallo M, Laigle A. Accumulation of degradation products of doxorubicin and pirarubicin formed in cell culture medium within sensitive and resistant cells. Biochem. …. 1993;45:659–665. doi: 10.1016/0006-2952(93)90140-r. [DOI] [PubMed] [Google Scholar]
- 28.Hajian R, Shams N, Mohagheghian M. Study on the interaction between doxorubicin and Deoxyribonucleic acid with the use of methylene blue as a probe. J. Braz. Chem. Soc. 2009;20:1399–1405. [Google Scholar]
- 29.Hovorka O, et al. Spectral analysis of doxorubicin accumulation and the indirect quantification of its DNA intercalation. Eur. J. Pharm. Biopharm. 2010;76:514–524. doi: 10.1016/j.ejpb.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 30.Smith Da, Di L, Kerns EH. The effect of plasma protein binding on in vivo efficacy: misconceptions in drug discovery. Nat. Rev. Drug Discov. 2010;9:929–939. doi: 10.1038/nrd3287. [DOI] [PubMed] [Google Scholar]
- 31.Law V, et al. DrugBank 4.0: Shedding new light on drug metabolism. Nucleic Acids Res. 2014;42:1091–1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang F, Xue J, Shao J, Jia L. Compilation of 222 drugs’ plasma protein binding data and guidance for study designs. Drug Discov. Today. 2012;17:475–485. doi: 10.1016/j.drudis.2011.12.018. [DOI] [PubMed] [Google Scholar]
- 33.Mateus a, Matsson P, Artursson P. Rapid measurement of intracellular unbound drug concentrations. Mol. Pharm. 2013 doi: 10.1021/mp4000822. at< http://pubs.acs.org/doi/abs/10.1021/mp4000822>. [DOI] [PubMed] [Google Scholar]
- 34.Greene RF, Collins JM, Jenkins JF, Speyer JL, Myers CE. Plasma Pharmacokinetics of Adriamycin and Adriamycinol : Implications for the Design of in Vitro Experiments and Treatment Protocols Plasma Pharmacokinetics of Adriamycin and Adriamycinol : Implications for the Design of in Vitro Experiments and Treatment. 1983:3417–3421. [PubMed] [Google Scholar]
- 35.Anderson AB, Gergen J, Arriaga Ea. Detection of doxorubicin and metabolites in cell extracts and in single cells by capillary electrophoresis with laser-induced fluorescence detection. J. Chromatogr. B. Analyt. Technol. Biomed. Life Sci. 2002;769:97–106. doi: 10.1016/s1570-0232(01)00633-x. [DOI] [PubMed] [Google Scholar]
- 36.Deng B, et al. Analysis of doxorubicin uptake in single human leukemia K562 cells using capillary electrophoresis coupled with laser-induced fluorescence detection. Anal. Bioanal. Chem. 2011;401:2143–2152. doi: 10.1007/s00216-011-5315-6. [DOI] [PubMed] [Google Scholar]
- 37.Eichholtz-Wirth H. Dependence of the cytostatic effect of adriamycin on drug concenration and exposure time in vitro. Br. J. Cancer. 1980:886–891. doi: 10.1038/bjc.1980.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakai-Kato K, Saito E, Ishikura K, Kawanishi T. Analysis of intracellular doxorubicin and its metabolites by ultra-high-performance liquid chromatography. J. Chromatogr. B. Analyt. Technol. Biomed. Life Sci. 2010;878:1466–1470. doi: 10.1016/j.jchromb.2010.03.040. [DOI] [PubMed] [Google Scholar]
- 39.Xiong G, Chen Y, Arriaga Ea. Measuring the doxorubicin content of single nuclei by micellar electrokinetic capillary chromatography with laser-induced fluorescence detection. Anal. Chem. 2005;77:3488–3493. doi: 10.1021/ac0500378. [DOI] [PubMed] [Google Scholar]
- 40.Adair JR, Howard PW, Hartley Ja, Williams DG, Chester Ka. Antibody-drug conjugates - a perfect synergy. Expert Opin. Biol. Ther. 2012;12:1191–1206. doi: 10.1517/14712598.2012.693473. [DOI] [PubMed] [Google Scholar]
- 41.Alley SC, Okeley NM, Senter PD. Antibody-drug conjugates: targeted drug delivery for cancer. Curr. Opin. Chem. Biol. 2010;14:529–537. doi: 10.1016/j.cbpa.2010.06.170. [DOI] [PubMed] [Google Scholar]
- 42.Carter PJ, Senter PD. Antibody-drug conjugates for cancer therapy. Cancer J. 2008;14:154–169. doi: 10.1097/PPO.0b013e318172d704. [DOI] [PubMed] [Google Scholar]
- 43.Teicher Ba. Antibody-drug conjugate targets. Curr. Cancer Drug Targets. 2009;9:982–1004. doi: 10.2174/156800909790192365. [DOI] [PubMed] [Google Scholar]
- 44.Thurber GM, et al. Single-cell and subcellular pharmacokinetic imaging allows insight into drug action in vivo. Nat. Commun. 2013;4:1504. doi: 10.1038/ncomms2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang J, Chen H, Vlahov IR, Cheng J-X, Low PS. Evaluation of disulfide reduction during receptor-mediated endocytosis by using FRET imaging. Proc. Natl. Acad. Sci. U. S. A. 2006;103:13872–13877. doi: 10.1073/pnas.0601455103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.