Abstract
Oral streptococci play a large role in dental biofilm formation, and several types interact as early colonizers with the enamel salivary pellicle to form the primary biofilm, as well as to incorporate other bacteria on tooth surfaces. Interactions of surface molecules of individual streptococci with the salivary pellicle on the tooth surface have an influence on the etiological properties of an oral biofilm. To elucidate the molecular interactions of streptococci with salivary components, binding between surface protein (SspB and PAg) peptides of Streptococcus gordonii and Streptococcus sobrinus were investigated by utilizing BIAcore biosensor technology. The analogous peptide [change of T at position 400 to K in SspB(390-402), resulting in the SspB(390-T400K-402) peptide] from S. gordonii showed the greatest response for binding to salivary components and inhibited the binding of Streptococcus sanguis by more than 50% in a competitive inhibition assay in a comparison with other SspB and PAg peptides. This peptide also bound to the high-molecular-weight protein complex of salivary components and the agglutinin (gp340/DMBT1) peptide (scavenger receptor cysteine-rich domain peptide 2 [SRCRP 2]). In addition, the SspB(390-T400K-402) peptide was visualized by two surface positive charges in connection with the positively charged residues, in which lysine was a key residue for binding. Therefore, the region containing lysine may have binding activity in S. gordonii and S. sanguis, and the SRCRP 2 region may function as a receptor for the binding. These findings may provide useful information regarding the molecular mechanism of early biofilm formation by streptococci on tooth surfaces.
Oral streptococci are present in large numbers in dental plaque and account for approximately 20% of the total number of salivary bacteria (25). Furthermore, several types of these bacteria interact with the enamel salivary pellicle to form a biofilm on tooth surfaces. Early biofilm formation occurs by attachment and colonization of a variety of streptococci and is dependent on both the species involved and the surface composition (2, 12, 26, 33). The subsequent accumulation and growth of attached bacteria result in microcolonies that increase in size and eventually form a towering pillar- or mushroom-shaped biofilm (4). There is a strong relationship between the attachment, detachment, and aggregation of these organisms in plaque on tooth surfaces and the presence of dental decay in humans (14, 24).
Members of the mitis group of streptococci, which includes Streptococcus gordonii, Streptococcus mitis, Streptococcus oralis, Streptococcus parasanguis, and Streptococcus sanguis (16), are prominent components of the human oral microbiota and play a significant role as pioneer colonizers in the development of dental plaque (9, 34). Mutans streptococci (Streptococcus mutans and Streptococcus sobrinus) are also known to be primarily involved when the bacterial flora forms on the tooth surface. S. sanguis has been shown to have a strong association with saliva components; however, it was easily dissociated, compared with other streptococci, by using a BIAcore system, and although this organism showed a higher binding resonance unit (RU) value than S. mutans for association, it could be easily dissociated from saliva components (17).
S. mutans, S. sobrinus, and S. gordonii produce surface protein antigens (PAc, AgI/II, B, P1, SpaP, and MSL-1), PAg (SpaA), and SspB (SspA), which have molecular masses of approximately 190 kDa (6, 10, 18, 32, 36, 38), 170 kDa (22, 44), and 180 kDa (5, 15), respectively, and interact with salivary components, including lysozyme (37, 40), amylase (37), 18,000- and 38,000-Da proline-rich proteins (32), and agglutinin (5). 365TYEAALKQYEADL377 of PAc(365-377) in the alanine-rich repeating region (residues 219 to 464, A region) of the PAc molecule is an important region for the adherence of S. mutans to the tooth surface (39, 43). Furthermore, the T- and B-cell epitopes overlap (39, 41), and the epitope XYXXXLXXYXXXX, an essential sequence in the antigenic epitopes of the PAc protein, is recognized specifically by the inhibiting antibody (42). In the N-terminal region (residues 1 to 429) sequenced from S. gordonii M5 (previously designated S. sanguis), the cell surface adhesins SspB and SspA showed extensive homology [63 and 60% identity, respectively, with SpaP (PAc)] (6). The A region is composed of three long and two incomplete repeating sequences (26). Therefore, each repeating sequence contains sequences that are homologous to the amino acid sequence 365TYEAALKQYEADL377 of PAc(365-377), which is an important region for the initial attachment of S. mutans to the tooth surface (31, 36). The differences between the alanine-rich sequence in S. gordonii and the alanine-rich sequence in S. mutans may have an effect on the different affinities of early colonizers, such as S. gordonii and S. sanguis, and S. mutans for interactions with salivary receptors.
In the present study, to define the molecular mechanisms of the surface proteins of S. gordonii and S. sobrinus for binding to salivary components, peptides homologous to PAc(365-377) in the N-terminal region of SspB (84% identity with SspA) (7) were elucidated and used for analysis of binding to salivary components in BIAcore biosensor experiments. Interactions between the peptides and salivary agglutinin as a receptor were also analyzed by biosensor and crystallographic methods. An understanding of these molecules and their receptors is important for elucidating the mechanisms of biofilm formation in vivo.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The strains used in this study were S. sanguis ATCC 10556 and S. gordonii ATCC 10558. All bacteria were grown anaerobically by using a GasPak system (Becton-Dickinson, Sparks, Md.) in brain heart infusion medium (Difco Laboratory, Detroit, Mich.).
Human saliva collection.
Whole saliva was stimulated by chewing paraffin gum, and the saliva was collected from four subjects (27 to 34 years old, females and males) in ice-chilled sterile bottles over a 5-min period and then clarified by centrifugation at 10,000 × g for 10 min at 4°C. Fresh saliva samples from subject A (see Fig. 5) were used in every experiment, while the other samples were stored until they were used in some of the experiments.
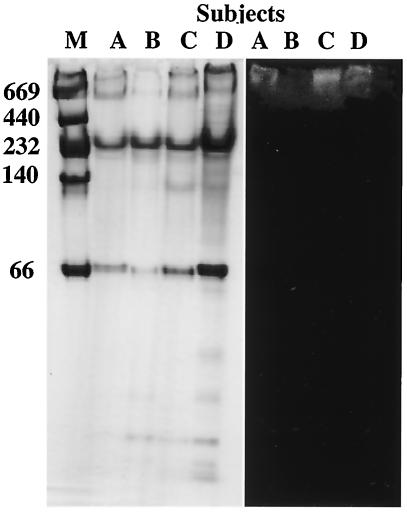
FIG. 5.
(Left panel) Native PAGE of saliva samples collected from subjects A, B, C, and D stained with Coomassie blue. (Right panel) Western blot analysis of the samples performed with biotin-labeled SspB(390-T400K-402) peptide. Adherence to the peptide was detected by chemiluminescence. The positions of molecular weight markers (lane M) are indicated on the left.
Peptide synthesis.
The sequence of PAc(365-377) (TYEAALKQYEADL) was derived from the sequence of the PAc gene from S. mutans MT8148, which corresponds to a portion of the A region, as described by Okahashi et al. (32) (Table 1). The SspB peptides in the A region of the surface protein (SspB) derived from S. gordonii (5), which are homologous to the PAc(365-377) and SspA peptides, and the PAg peptides derived from S. sobrinus (44), used as a control, are also listed in Table 1. The results shown in Fig. 1 and Fig. 7 are based on the supposition that lysine is an important amino acid residue for the binding of SspB(390-402) peptides to salivary components. Therefore, a peptide analogous to the SspB peptide [change of T at position 400 to K in SspB(390-402), resulting in the SspB(390-T400K-402) peptide] was also synthesized. Scavenger receptor cysteine-rich domain peptide 2 (SRCRP 2) (QGRVEVLYRGSWGTVC) on agglutinin/gp340/DMBT1 (1) was used as a receptor for these bacterial peptides, which were synthesized by a stepwise solid-phase procedure at Asahi Techno Glass Co. Inc. (Tokyo, Japan). The synthesized peptide samples were subsequently purified by reverse-phase high-performance liquid chromatography on a TSK-GEL column (1 by 30 cm; TOSO, Tokyo, Japan) with a 10 to 45% acetonitrile gradient in 0.1% trifluoroacetic acid, which was developed over 50 min at a flow rate of 5 ml/min. The purity in each tube was determined to be greater than 95% by high-performance liquid chromatography analysis. To confirm the amino acid sequences of these synthetic peptides, several samples were randomly selected and then analyzed by using a System 7300 amino acid analyzer (Beckman, Fullerton, Calif.) and a model 477A protein sequencer (Applied Biosystems, Foster City, Calif.).
TABLE 1.
Levels of homology of amino acid sequences of SspB and PAg to PAc(365-377)
| Bacterial strain (surface protein) | Amino acid sequence | Position | % Homology
|
|
|---|---|---|---|---|
| PAc(365-377) | SspB(390-T400K-402) | |||
| S. mutans MT8148 (PAc) | TYEAALKQYEADL | A2, 365-377 | 100.0 | 38.5 |
| S. gordonii M5 (SspB) | TYEAAMKQYEADL | A1, 283-295 | 92.3 (92.3)a | 30.8 |
| TYDAAVKKYEADL | A2, 365-377 | 76.9 (76.9) | 30.8 | |
| DYEAKLAKYEADL | A3, 447-459 | 69.2 (100.0) | 69.2 | |
| EYEAKLAQYQKDL | 201-213 | 61.5 (100.0) | 69.2 | |
| DYQNKLSAYQTEL | A1, 226-238 | 30.8 (92.3) | 76.9 | |
| DYQAKLAAYQTEL | A2, 308-320 | 38.5 (92.3) | 92.3 | |
| DYQAKLAAYQTEL | A3, 390-402 | 38.5 (92.3) | 92.3 | |
| DYQAKLAAYQKEL | 390-T400K-402 | 38.5 (84.6) | 100.0 | |
| S. sobrinus MT3791 (PAg) | DYEAKLAQYEKDL | A1, 286-298 | 69.2 | 69.2 |
| NYEAKLAQYQKDL | A2, 368-380 | 61.5 | 61.5 | |
| DYELKLSKYQEEL | A3, 450-462 | 38.5 | 61.5 | |
| DYEAKLAQYQKDL | 204-216 | 61.5 | 76.9 | |
| AYAAAKEAYDKEL | A1, 229-241 | 38.5 | 61.5 | |
| DYQAKKAAYEQEL | A2, 311-323 | 38.5 | 76.9 | |
| DYQEKLAAYEKEL | A3, 393-405 | 38.5 | 84.6 | |
The values in parentheses are the percentages of homologous sequences in the corresponding SpaA peptides.
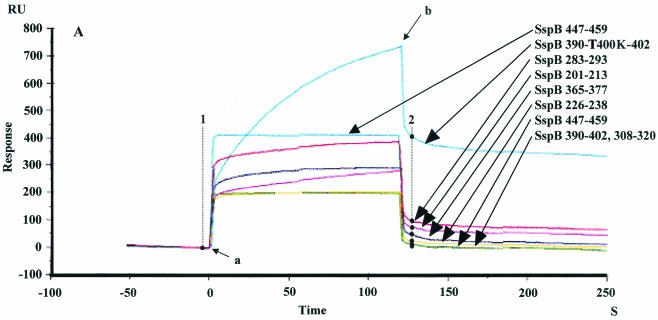
FIG. 1.
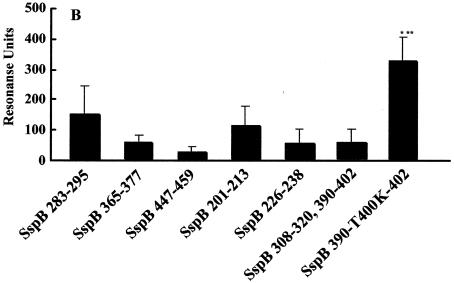
(A) Sensorgram illustrating the binding of SspB peptides to immobilized salivary components. SspB(390-T400K-402) (blue), SspB(283-295) (red), SspB(201-213) (pink), SspB(365-377) (purple), SspB(226-238) (yellow), SspB(447-459) (light blue), and SspB(308-320) and SspB(390-402) (green) peptides were applied to the sensor chip. The sensorgram for immobilization of the salivary components showed an increase of 2,267.8 RU. The arrows indicate the start of each injection. Injection of 20 μl of peptide (arrow a) was followed by injection of PBS (arrow b). The increases of 398, 94, 72, 43, 25, 12, and 11 RU between points 1 and 2 correspond to the binding of the SspB(390-T400K-402), SspB(283-295), SspB(201-213), SspB(355-377), SspB(226-213), SspB(365-377), SspB(226-238), SspB(447-459), and SspB(308-320) peptides to salivary components. (B) Binding of various SspB peptides to immobilized salivary components. Twenty microliters of a peptide solution (1.0 mg/ml) was applied to the sensor chip. The results are expressed as means ± standard deviations for three independent assays. The asterisks indicate significant differences, as follows: one asterisk, P < 0.05 compared with the SspB(283-295) peptide; two asterisks, P < 0.01 compared with other peptides.
FIG. 7.
Schematic representation of SspB and PAg peptides. Models of the secondary structure and surface charges (positive [blue] and negative [red]) in the peptides were constructed by using the MOE software with Chemical Computing Graphics. Some of the amino acid residues on the surfaces of the peptides are indicated.
Modeling of the secondary structure with surface charge and hydrophobicity.
The A regions of PAc and PAg are thought to assume an α-helical structure (5, 32, 44). Recently, new experimental evidence has shown that A regions from the binding proteins (SspA and SpaP) of S. gordonii and S. mutans that bind to salivary components are highly stable and α-helical (8). However, the A region of SspB is less stable than the corresponding A regions of SspA and SpaP. Furthermore, the secondary structures of the regions of the three proteins are very similar, as each of the regions is comprised of approximately 60% α-helix, 7% β-sheet, and 33% random structures. These peptides, which are homologous to the PAc(365-377) peptide in the three proteins, include the amino acid sequence XYXXXLXXYXXXX or XYXXXXXXYXXXX, so that the monoclonal antibodies react with an α-helix or coiled-coil determinant of PAc (41, 42). Therefore, modeling of the secondary structures with surface charges and determination of hydrophobicity in SspB and PAg peptides were performed on the basis of an α-helical structure by using the MOE software (Ryoka Systems Inc., Tokyo, Japan).
Immobilizing the ligand.
The binding of peptides to salivary components was determined by using a BIAcore biosensor system (BIAcore AB), which has been shown to provide real-time interactive analysis of bacteria and salivary components (17, 43). The results for various peptides interacting with salivary components in the BIAcore system were compared in detail in separate stages of association and dissociation, which is different from the comparison with conventional methods that analyze coupled stages of association and dissociation. In the present study, we used a standard CM5 sensor chip and a pioneer sensor chip, F1 (BIAcore AB), which has a shorter dextran matrix than other standard chips and is useful for working with large analyses, such as analyses of cells and virus particles. The carboxymethylated dextran-coated gold surface of the sensor chip was activated by injecting 70 μl of a solution containing 400 mM N-ethyl-N′-(3-diethylaminoprophyl)carbodiimide and 100 mM N-hydroxysuccinimide at a flow rate of 10 μl/min. Following activation, 70 μl of 1:20-diluted whole saliva or 1 mg of SRCRP 2 peptide per ml in 10 mM sodium acetate buffer (pH 5.0) was applied to and immobilized on the surface. Residual N-hydroxysuccinimide esters were then inactivated with 70 μl of 1 M ethanolamine hydrochloride. The flow rate of phosphate-buffered saline (PBS) (pH 7.4) was maintained at 10 μl/min throughout the immobilization procedure.
Interaction of the peptides with salivary components.
The peptide solutions (0.6 and 1.0 mg/ml) were exposed to the immobilized CM5 sensor chip (flow rate, 10 μl/min), and the dissociation phase was followed by injection of PBS at a rate of 10 μl/min. All binding experiments were conducted at 25°C (17, 43). At the end of each binding cycle, the surface of the sensor chip was regenerated by exposure to 50 mM glycine-NaOH (pH 9.5) for 60 s. The detection system of the BIAcore utilizes surface plasmon resonance, a quantum-mechanical phenomenon that represents changes in optical properties at the surface of the sensor chip and eliminates the need to label the interactants. The resonance angle depends on the refractive index in the vicinity of the surface, which changes as the concentration of molecules on the surface is modified, and the signal is expressed in RU. The surface plasmon resonance signal obtained in each binding cycle was recorded as a sensorgram that was a real-time pattern with a sampling interval of 0.2 to 0.5 s plotted in RU versus time. The amount of binding was expressed as the increase in RU between the start and end of each binding cycle. A response of 1,000 RU corresponded to a shift of 0.1° in the response angle, which in turn represented a change in the surface protein concentration of about 1 ng/mm2.
S. sanguis ATCC 10556 and S. gordonii ATCC 10558 were diluted with PBS after centrifugation at 5,000 × g for 10 min at 4°C, and ultrasonic processing (60 W, 10 min) was carried out to disrupt the bacterial chains. The bacterial concentration was adjusted by using fresh PBS and a spectrophotometer (UV-2200A; Shimazu, Kyoto, Japan) to an optical density at 550 nm of 1.0. After application of the peptide to the immobilized salivary components on the F1 chip, a suspension of S. sanguis or S. gordonii was injected (flow rate, 20 μl/min) for a competitive inhibition BIAcore assay. The percent inhibition of the binding of S. sanguis or S. gordonii to salivary components by the SspB peptides was calculated as follows: 100 × (RU for S. sanguis or S. gordonii plus saliva − RU for S. sanguis or S. gordonii plus peptide-treated saliva)/RU for S. sanguis or S. gordonii plus saliva. In the assay for binding of the SspB and PAg peptides to SRCRP 2, SspB(283-295), SspB(447-459), and Ssp(390-T400K-402), PAg(450-462), and PAg(311-323) solutions were diluted to a concentration of 1 mg/ml of PBS, adjusted by the above-mentioned methods, and exposed to an SRCRP 2-immobilized CM5 sensor chip surface at a flow rate of 10 μl/min.
Native PAGE and Western blot analyses.
Prior to electrophoretic analysis, the saliva samples were diluted with an equal volume of native polyacrylamide gel electrophoresis (PAGE) sample buffer (0.06 M Tris-HCl [pH 6.8], 20% glycerol, 0.0012% bromphenol blue). The samples were subjected to 10% PAGE, and electrophoretic separation of the proteins was carried out in a native PAGE gel for 30 min at 100 mA. High-molecular-mass markers (thyroglobulin [669 kDa], ferritin [440 kDa], catalase [232 kDa], lactate dehydrogenase [140 kDa], and albumin [66 kDa]; Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom) were used as molecular mass markers. When electrophoresis was complete, proteins in the gels were stained with 0.1% Coomassie blue. For the Western blot analysis, proteins in the gels were transferred to nitrocellulose membranes (ATTO Corp., Tokyo, Japan). The membranes were washed in buffer containing 25 mM Tris-HCl (pH 7.4), 0.5 M NaCl, and 0.1% (vol/vol) Tween 20 (TTBS), and specific binding was blocked by incubating the blots for 1 h in 0.3% skim milk in TTBS. The transfers were then incubated for 1 h in TTBS containing 100 μg of biotin-labeled SspB(390-T400K-402) peptide per ml and washed in TTBS three times for 5 min. Each membrane was then incubated for 1 h in 1/500 streptoavidin-horseradish peroxidase conjugate and again washed in TTBS three times for 5 min. The specific binding of the SspB 390-402, 400(K) peptide was finally detected with ECL Plus, a Western blot detection system (Amersham Pharmacia Biotech), by using a chemiluminescent reaction and Lumishoot (ATTO) to produce the image.
Statistical analysis.
Statistical analysis was performed by analysis of variance. A P value of 0.05 or less was considered to indicate statistical significance.
RESULTS
Binding of SspB and PAg peptides to salivary components.
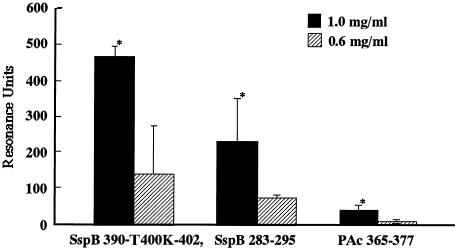
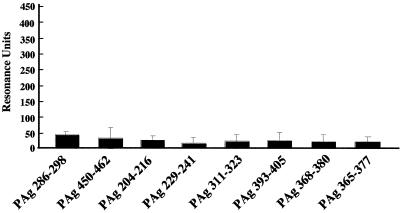
The sensorgram results for immobilization of salivary components showed increases of 2,000 to ∼4,300 RU, which involved covalent binding of approximately 2.0 to 4.3 ng to the activated carboxymethylated dextran coating on the biosensor surface. Each peptide (1 mg/ml) was applied to the immobilized salivary components. The increases in the RU indicated the levels of response of the peptides to the salivary components, as shown in the sensorgrams in Fig. 1A. Thus, approximately 325.1 ± 72.3 RU was required for the binding of the SspB(390-T400K-402) peptide to the immobilized salivary components on the sensor chip surface (Fig. 1B). The SspB(283-295) and SspB(201-213) peptides showed moderate reactivities (147.3 ± 91.7 and 107.2 ± 64.3 RU, respectively), while the responses of other peptides were less than 100 RU. Furthermore, dose-dependent binding of the SspB(390-T400K-402) peptide was found, which was greater than that of the SspB(283-295) and PAc(365-377) peptides (Fig. 2). All of the PAg peptides showed poor binding to the salivary components (Fig. 3).
FIG. 2.
Dose-dependent binding of SspB peptides to immobilized salivary components. SspB(390-T400K-402), SspB(283-295), and PAc(365-377) at a concentration of 0.6 or 1.0 mg/ml were applied to the sensor chip. The sensorgram for immobilization of the salivary components showed an increase of 2,582.1 RU. Twenty microliters of peptide solution (0.6 or 1.0 mg/ml) was applied to the sensor chip. The results are expressed as means ± standard deviations for three independent assays. An asterisk indicates that there was a significant difference (P < 0.01) between the data for 1.0 mg of peptide per ml and the data for 0.6 mg of peptide per ml.
FIG. 3.
Binding of PAg peptides to immobilized salivary components. PAg(286-298), PAg(450-462), PAg(204-216), PAg(368-380), PAg (311-323), PAg(393-405), and PAg(229-241) were applied to the sensor chip. The sensorgram for immobilization of the salivary components showed an increase of 2,077.1 RU. Twenty microliters of each peptide solution (1.0 mg/ml) was applied to the sensor chip. The results are expressed as means ± standard deviations for three independent assays.
Inhibition of S. sanguis binding to salivary components.
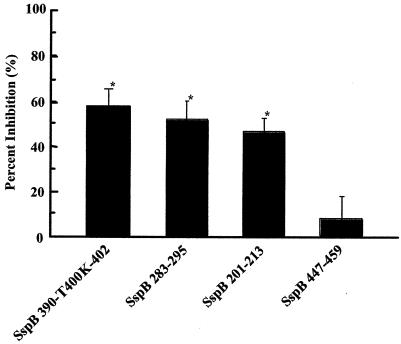
Binding of S. sanguis ATCC 10556 was inhibited by more than 50% by SspB(390-T400K-402) peptide treatment (Fig. 4), and the inhibition of S. gordonii ATCC 10558 binding was also significant (data not shown). The level of inhibition by the SspB(390-T400K-402) peptide was similar to the levels of inhibition by the SspB(283-295) and SspB(201-213) peptides, each of which showed moderate responses to the salivary components in Fig. 1 (Fig. 4). In contrast, the level of inhibition by the SspB(447-459) peptide was low.
FIG. 4.
Binding of S. sanguis ATCC 10556 in a competitive inhibition assay with SspB peptides. After immobilization of the salivary components, solutions containing 1.0 mg of SspB(390-T400K-402) per ml, 1.0 mg of SspB(283-295) per ml, 1.0 mg of SspB(201-213) per ml, and 1.0 mg of SspB(447-459) per ml were applied to the sensor chip. In addition, S. sanguis suspensions in PBS (optical density at 550 nm, 1.0) were applied to the sensor chip with and without treatment with SspB peptides. The sensorgram for immobilization of the salivary components showed an increase of 4,212.7 RU. The results are expressed as means ± standard deviations for three independent assays. An asterisk indicates that there was a significant difference (P < 0.01) for a comparison with SspB(447-459).
Binding of the SspB(390-T400K-402) peptide to salivary high-molecular-weight components.
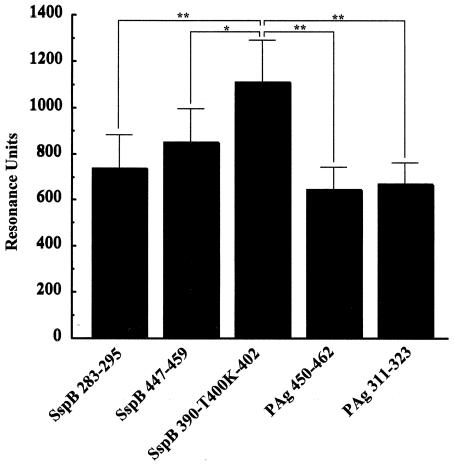
Whole saliva samples from all four subjects were electrophoretically separated, and the protein components were revealed by Coomassie brilliant blue staining (Fig. 5). Examples of the binding patterns observed when blots of the salivary glycoproteins were overlaid with biotin-labeled SspB(390-T400K-402) peptide are shown in Fig. 5. Binding to the high-molecular-weight components containing salivary components with low mobility on the saliva smears was detected in samples from all four subjects. Agglutinin is a candidate component for the binding of S. gordonii (5), and high binding activities of SRCRP 2 to various streptococci were recently reported. Therefore, we investigated the reactivity of SspB and PAg peptides to SRCRP 2 and found that the highest binding activity to SRCRP 2 was that of the SspB(390-T400K-402) peptide (1,105.5 ± 183.3 RU) (Fig. 6).
FIG. 6.
Binding of SspB and PAg peptides to immobilized SRCRP 2. SspB(283-295), SspB(447-459), SspB(390-T400K-402) PAg(450-462), and PAg(311-323) peptide solutions at a concentration of 1.0 mg/ml were applied to the sensor chip. Immobilization of SRCRP 2 showed an increase of 16,537.6 RU in the sensorgram. The results are expressed as means ± standard deviations for three independent assays. Asterisks indicate significant differences (one asterisk, P < 0.05; two asterisks, P < 0.01).
Secondary structure and surface charge.
To clarify the mechanisms of binding of the peptides which we investigated to salivary components, the secondary structures and surface charges of the peptides were analyzed visually by using computer software (Fig. 7). The SspB(390-T400K-402) peptide, which had the highest binding activity to salivary components (Fig. 1), and SRCRP 2 (Fig. 6) had two surface positive charges at residues 390 (D) and 400 (K) in connection with the positively charged residues, and this was analogous to the situation in the SspB(390-402) peptide. Aspartic acid is not positively charged; however, the main chain (amidohydrogen) in D was charged positively and partially on the surface of the α-helical peptide structure in the crystallographic analysis (Fig. 7). In contrast, the SspB(390-402) and SspB(308-320) peptides exhibited lower binding activity and were not charged positively on the surface of residues 400 and 318, respectively. SspB(283-295), which exhibited moderate binding activity, had two surface positive charges at residues 288 (M) and 289 (K) in connection with the positively charged residues. Methionine is nonpolar; however, the main chain (amidohydrogen) in M was charged positively and partially on the surface of the α-helical peptide structure (Fig. 7). Furthermore, the PAc(365-377) peptide, which had lower binding activity and was analogous to SspB(283-295) [M-288 in SspB(283-295) was replaced by L at position 370 in PAc(365-377)], was not charged positively on the surface of residue 370 in connection with the positively charged residues. The SspB(201-213) peptide, which had moderate binding activity, showed two surface positive charges at residues 201 (E) and 211 (K). Glutamic acid is not positively charged; however, the main chain (amidohydrogen) in E was also charged positively and partially on the surface of the α-helical peptide structure (Fig. 7). SspB(447-459) showed two surface positive charges, whereas residue 454 (K) was charged positively on a different side than residue 451 (K) and also showed a lower level of binding activity. The other peptides, including the PAg peptides, did not show two surface positive charges in connection with positively charged residues.
DISCUSSION
S. sanguis is an early colonizer of the salivary pellicle, and S. mutans colonizes later; however, the abilities of each organism to bind to salivary proteins and glycoproteins are important in biofilm development (3, 45), and the differences in the affinity to the salivary pellicle between these two bacteria may be relevant for the specificity of biofilm conditions. There is a lack of comparative research results for precisely analyzing the differences, although the abilities to bind to salivary components have been reported by various investigators (21, 28, 35, 46).
In the present study, we found that specific residues within sequences SspB(390-T400K-402), SspB(283-295), and SspB(201-213) of SspB, the surface protein of S. gordonii (previously designated S. sanguis), contribute to receptor binding, and we also demonstrated that a typical application of a synthetic peptide, which forms adhesion epitopes, may selectively contribute to binding with S. gordonii and S. sanguis. Furthermore, we investigated the inhibition of S. sanguis affinity to salivary components by SspB peptides in real time. The affinity of S. sanguis ATCC 10556 or S. gordonii ATCC 10558 to whole salivary components was inhibited by more than 50% following SspB(390-T400K-402) peptide treatment of the salivary components in BIAcore competitive inhibition assays. In addition, another analogue of PAc(365-377) and a variant peptide [SspB(283-295) and SspB(201-213), respectively] caused inhibition at the same level as the SspB(390-T400K-402) peptide.
We considered the possibility that the binding of S. sanguis to salivary components might compete with the salivary component binding of S. mutans. A variant [SspB(390-T400K-402), SspB(283-295), and SspB(201-213) peptide] of the PAc(365-377) peptide may have an influence on the binding of S. sanguis to salivary components, which is similar to the binding receptor of S. mutans. The differences in secondary structure between PAc(365-377) and the SspB and PAg peptides were visualized on the basis of α-helical structure, and the surface charges were plotted by chemical computing graphics (Fig. 7). The SspB(390-402) peptide did not produce high binding activity or two surface positive charges, in contrast to the SspB(390-T400K-402) peptide. The replacement of M288 with leucine [SspB(283-295) peptide] resulted in moderate binding activity and two surface positive charges in connection with the positively charged residues, which was different from observations made with the PAc(365-377) peptide. The SspB(201-213) peptide also showed moderate binding activity and two surface charges. These results indicate that the presence of two positive surface charges may have a role in the binding of S. gordonii and S. sanguis to salivary components.
In peptides that showed positive binding, lysine was the common active site and likely the core residue for binding to salivary components (Fig. 7). In previous studies, lysine inhibited whole saliva- and salivary agglutinin-mediated aggregation of both S. mutans and S. sanguis (13), as well as the adherence of S. mutans to adhesin-promoting protein-coated hydroxyapatite (20). Several studies have also reported interactions between the sialic acid residues of human salivary agglutinin and SspB, SspA, and PAc (5, 8, 31, 37). Furthermore, Kelly et al. noted that E1037, located outside the A region, might be required for an AgI/II interaction with the sialic acid residue of salivary agglutinin and that the residue corresponding to E1037 is lysine in the sialic acid binding of the AgI/II homologue of S. gordonii (19).
The previous reports, along with our findings, support the notion that lysine has a role in the different interactions of S. mutans and S. sanguis with salivary components. The adherence of S. mutans and S. sanguis is also determined by such physicochemical properties as the hydrophobicity of the bacterial cell surface, in addition to the surface charge (21). Computer models of the peptides were made to analyze hydrophobicity; however, no positive relationships to binding activities were seen. For example, there were no differences between SspB(390-T400K-402), SspB(390-402), or SspB(283-285) and PAc(365-377) regarding hydrophobicity as a secondary conformation (data not shown). The features of binding of S. sanguis and S. mutans or S. sobrinus to salivary components in the association phase showed a correlation (17); however, the features of binding in the dissociation phase did not correlate with the binding features of the PAc(365-377) peptide or the SspB and PAg peptides in the present study. We cannot exclude the possibility that hydrophobicity is required for the different dissociation rates seen for S. sanguis and S. mutans, because an interaction with the synthetic peptide may not be related to all of the kinetics of the multiple binding of the organisms to salivary components. However, based on our results, as well as those of other workers, we considered the possibility that the positive surface charges on lysine and other amino acid residues might control the differences in the features of binding of S. mutans, S. gordonii, and S. sanguis to the salivary receptors.
S. sobrinus showed a low level of binding to salivary components based on the RU value (17), and its ability to associate with salivary agglutinin is also considered to be low. Furthermore, PAg peptides, which showed greater than 60% homology to SspB peptides (Table 1), showed poor responses to salivary components, possibly supporting the hypothesis that a mechanism is present in the biofilm itself. However, PAg peptides from S. sobrinus did not exhibit a definite response to the salivary components in the real-time biosensor. Two homologous PAg sequences [PAg(286-298) (69.2%) and PAg(393-405) (84.6%)] were found in the long repeating sequences of PAg and differed in their amino acid sequences (286XXFXXXXQXEXDX298 and 393XXXEXXXXXEXXX405, respectively) from the SspB(390-T400K-402) peptide. In the models of the secondary structure, two positive surface charges in connection with the positively charged residues, such as those of the SspB peptide, were not found in the PAg(286-298) or PAg(393-405) peptide, as observed in the other PAg peptides. These variants may have an influence on the lower affinity of S. sobrinus to salivary components and characterize such species of oral bacteria.
Streptococcal adhesion is often the result of specific interactions between the carbohydrate portions of receptor glycoproteins and protein complexes, which are known as adhesins, on bacterial cell surfaces (5). In addition, salivary agglutinin is known for its S. mutans-agglutinating properties (37), while AgI/II, SspB, and SspA bind to 300- to 400-kDa salivary agglutinin (5, 8, 9, 37). In the present study, the binding receptor of the SspB peptide was demonstrated to be a salivary component with a high-molecular-weight complex. Salivary agglutinin, which is encoded by DMBT1, is identical to gp340, and is also a member of the scavenger receptor cysteine-rich (SRCR) superfamily, is known for its S. mutans-agglutinating properties (27, 29). Furthermore, it was recently reported that the SRCRP 2 peptide of the SRCR domain has a bacterial binding feature with a multivalent character (1). In the present experiments, the SspB(390-T400K-402) peptide was also able to induce the highest level of binding to the SRCRP 2 peptide. Recently, it was found that a recombinant peptide encompassing the A region of SspB did not interact with purified gp340 or competitively inhibit the interaction of gp340 with intact S. gordonii cells (8). In contrast, the corresponding A-region peptides from SspA and SpaP of S. mutans are effective inhibitors of gp340-mediated streptococcal aggregation (8). However, the SspB(283-295) and SspB(201-213) peptides that bound to salivary components are identical to the corresponding peptides of SspA (92.3 and 100% identity, respectively) (Table 1). Therefore, we considered that the present findings for the binding activities of SspB were likely to provide identical elements for binding of SspA to salivary components. Altogether, the binding activity of S. gordonii may be dependent on the interaction between two surface positive charges, as well as lysine and the SRCRP 2 peptide region.
The differences in binding of analogous and variant peptides to PAc(365-377) in the surface proteins of S. gordonii and S. sobrinus may explain one of the molecular mechanisms of early biofilm formation on teeth reported in previous epidemiological findings (11, 23, 30). Therefore, it is suggested that molecular interactions with salivary components and agglutinin are influenced by the etiological abilities of streptococci in the development of dental caries. Our findings also demonstrated that critical residues within the synthetic peptide adhesion epitope are involved with the development of competitive peptide inhibitors as novel antimicrobial agents.
Acknowledgments
This work was supported in part by a grant-in-aid for development of scientific research 1530571 from the Ministry of Education, Science, and Culture of Japan and by a grant from the Japan Health Science Foundation to H.S.
Editor: V. J. DiRita
REFERENCES
- 1.Bikker, F. J., A. J. Ligtenberg, K. Nazmi, E. C. Veerman, W. van't Hof, J. G. Bolscher, A. Poustka, A. V. Nieuw Amerongen, and J. Mollenhauer. 2002. Identification of the bacteria-binding peptide domain on salivary agglutinin (gp-340/DMBT1), a member of the scavenger receptor cysteine-rich superfamily. J. Biol. Chem. 277:32109-32115. [DOI] [PubMed] [Google Scholar]
- 2.Bowden, G. H. 1990. Microbiology of root surface caries in humans. J. Dent. Res. 69:1205-1210. [DOI] [PubMed] [Google Scholar]
- 3.Carlsson, J., H. Grahnen, and G. Jonsson. 1975. Lactobacilli and streptococci in the mouth of children. Caries Res. 9:333-339. [DOI] [PubMed] [Google Scholar]
- 4.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 5.Demuth, D. R., E. E. Golub, and D. Malamud. 1990. Streptococcal-host interactions. Structural and functional analysis of a Streptococcus sanguis receptor for a human salivary glycoprotein. J. Biol. Chem. 265:7120-7126. [PubMed] [Google Scholar]
- 6.Demuth, D. R., M. S. Lammey, M. Huck, E. T. Lally, and D. Malamud. 1990. Comparison of Streptococcus mutans and Streptococcus sanguis receptors for human salivary agglutinin. Microb. Pathog. 9:199-211. [DOI] [PubMed] [Google Scholar]
- 7.Demuth, D. R., Y. Duan, W. Brooks, A. R. Holmes, R. McNab, and H. F. Jenkinson. 1996. Tandem genes encode cell-surface polypeptide SspA and SspB which mediate adhesion of the oral bacterium Streptococcus gordonii to human and bacterial receptors. Mol. Microbiol. 20:403-413. [DOI] [PubMed] [Google Scholar]
- 8.Demuth, D. R., and D. C. Irvine. 2002. Structural and functional variation within the alanine-rich repetitive domain of streptococcal antigen I/II. Infect. Immun. 70:6389-6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ericson, T., and J. Rundegren. 1983. Characterization of a salivary agglutinin reacting with a serotype strain of Streptococcus mutans. Eur. J. Biochem. 133:255-261. [DOI] [PubMed] [Google Scholar]
- 10.Forester, H., N. Hunter, and K. W. Knox. 1983. Characteristics of a high molecular weight extracellular protein of Streptococcus mutans. J. Gen. Microbiol. 129:2779-2788. [DOI] [PubMed] [Google Scholar]
- 11.Gibbons, R. J., and J. van Houte. 1980. Bacterial adherence and the formation of dental plaques, p. 62-104. In E. H. Beachey (ed.), Bacterial adherence, receptors and recognition, series B, vol. 6. Chapman and Hall, London, United Kingdom. [Google Scholar]
- 12.Gibbons, R. J. 1996. Role of adhesion in microbial colonization of host tissues: a contribution of oral microbiology. J. Dent. Res. 75:866-870. [DOI] [PubMed] [Google Scholar]
- 13.Hajishengallis, G., T. Koga, and M. W. Russell. 1994. Affinity and specificity of the interactions between Streptococcus mutans antigen I/II and salivary components. J. Dent. Res. 73:1493-1502. [DOI] [PubMed] [Google Scholar]
- 14.Hamada, S., and H. D. Slade. 1980. Biology, immunology and cariogenicity of Streptococcus mutans. Microbiol. Rev. 44:331-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes, A. R., C. Gilbert, J. M. Wells, and J. F. Jenkinson. 1998. Binding properties of Streptococcus gordonii SspA and SspB (antigen I/II family) polypeptides expressed on the cell surface of Lactococcus lactis MG1363. Infect. Immun. 66:4633-4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawamura, Y., X. G. Hou, F. Sultana, H. Miura, and T. Ezaki. 1995. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int. J. Syst. Bacteriol. 45:406-408. [DOI] [PubMed] [Google Scholar]
- 17.Kawashima, M., N. Hanada, T. Hamada, J. Tagami, and H. Senpuku. 2003. Real-time interaction of oral streptococci with human salivary components. Oral Microbiol. Immunol. 18:220-225. [DOI] [PubMed] [Google Scholar]
- 18.Kelly, C., P. Evans, L. Bergmeier, S. F. Lee, A. Progulske-Fox, A. C. Harris, A. Aitken, A. S. Bleiweis, and T. Lehner. 1989. Sequence analysis of the cloned streptococcal cell surface antigen I/II. FEBS Lett. 258:127-132. [DOI] [PubMed] [Google Scholar]
- 19.Kelly, C. G., J. S. Younson, B. Y. Hikmat, S. M. Todryk, M. Czisch, P. I. Haris, I. R. Flindal, C. Newby, A. I. Mallet, J. K.-C. Ma, and T. Lehner. 1999. A synthetic peptide adhesion epitope as a novel antimicrobial agent. Nat. Biotechnol. 17:42-47. [DOI] [PubMed] [Google Scholar]
- 20.Kishimoto, E., D. I. Hay, and R. J. Gibbons. 1991. Inhibition of adhesion-promoting activity of a human salivary protein which promotes adhesion of Streptococcus mutans JBP to hydroxyapatite. FEMS Microbiol. Lett. 69:19-22. [DOI] [PubMed] [Google Scholar]
- 21.Koga, T., N. Okahashi, I. Takahashi, T. Kanamoto, H. Asakawa, and M. Iwaki. 1990. Hydrophobicity, adherence, and aggregation of cell surface protein antigen mutants of Streptococcus mutans serotype c. Infect. Immun. 58:289-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LaPolla, R. J., J. A. Haron, C. G. Kelly, W. R. Taylor, C. Bohart, M. Hendricks, J. P. Pyati, R. T. Graff, J. K. Ma, and T. Lehner. 1991. Sequence and structural analysis of surface protein antigen I/II (SpaA) of Streptococcus sobrinus. Infect. Immun. 59:2677-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liljemark, W. F., and R. J. Gibbons. 1972. Proportional distribution and relative adherence of Streptococcus miteor (mitis) or various surfaces in the human oral cavity. Infect. Immun. 6:853-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loesche, W. J. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50:353-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marsh, P. D. 1999. Microbiologic aspects of dental plaque and dental caries. Dent. Clin. N. Am. 43:599-614. [PubMed] [Google Scholar]
- 26.McEldowney, S., and M. Fletcher. 1987. Adhesion of bacteria from mixed cell suspension to solid surfaces. Arch. Microbiol. 148:57-62. [DOI] [PubMed] [Google Scholar]
- 27.Mollenhauer, J., S. Wiemann, W. Scheurlen, B. Korn, Y. Hayashi, K. K. Wilgenbus, A. von Deimling, and A. Poustka. 1997. BMBT1, a new member of the SRCR superfamily, on chromosome 10q25.3-26.1 is deleted in malignant brain tumours. Nat. Genet. 17:32-39. [DOI] [PubMed] [Google Scholar]
- 28.Newman, F., J. A. Beeley, and T. W. MacFarlane. 1996. Adherence of oral microorganisms to human parotid salivary protein. Electrophoresis 17:266-270. [DOI] [PubMed] [Google Scholar]
- 29.Nolmskov, U., P. Lawson, B. Teisner, I. Tornoe, A. C. Willis, C. Morgan, C. Koch, and K. B. Reid. 1997. Isolation and characterization of a new member of the scavenger receptor superfamily, glycoprotein-340 (gp-340), as a lung surfactant protein-D binding molecule. J. Biol. Chem. 272:13743-13749. [DOI] [PubMed] [Google Scholar]
- 30.Nyvad, B., and M. Kllian. 1990. Comparison of the initial streptococcal microflora on dental enamel in caries-active and in caries-inactive individuals. Caries Res. 24:267-272. [DOI] [PubMed] [Google Scholar]
- 31.Oho, T., H. Yu, Y. Yamashita, and T. Koga. 1998. Binding of salivary glycoprotein-secretory immunoglobulin A complex to the surface protein antigen of Streptococcus mutans. Infect. Immun. 66:115-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okahashi, N., C. Sasakawa, M. Yoshikawa, S. Hamada, and Koga T. 1989. Cloning of a surface protein antigen gene from serotype c Streptococcus mutans. Mol. Microbiol. 3:221-228. [DOI] [PubMed] [Google Scholar]
- 33.Pratt-Terpstra, I. H., A. H. Weekamp, and H. J. Busscher. 1989. The effects of pellicle formation on streptococcal adhesion to human enamel and artificial substrata with various surface free energies. J. Dent. Res. 68:463-467. [DOI] [PubMed] [Google Scholar]
- 34.Rosan, B., and R. J. Lamont. 2000. Dental plaque formation. Microbes Infect. 2:1599-1607. [DOI] [PubMed] [Google Scholar]
- 35.Rudney, J. D., K. L. Hickey, and Z. Ji. 1999. Cumulative correlations of lysozyme, lactoferrin, peroxidase, S-IgA, amylase, and total protein concentrations with adherence of oral viridans streptococci to microplates coated with human saliva. J. Dent. Res. 78:759-768. [DOI] [PubMed] [Google Scholar]
- 36.Russell, M. W., and T. Lehner. 1978. Characterisation of antigens extracted from cells and culture fluids of Streptococcus mutans serotype c. Arch. Oral Biol. 23:7-15. [DOI] [PubMed] [Google Scholar]
- 37.Russell, M. W., and B. Mansson-Rahemtulla. 1989. Interaction between surface protein antigens of Streptococcus mutans and human salivary components. Oral Microbiol. Immunol. 4:106-111. [DOI] [PubMed] [Google Scholar]
- 38.Russell, R. R. B. 1979. Wall-associated protein antigens of Streptococcus mutans. J. Gen. Microbiol. 114:109-115. [DOI] [PubMed] [Google Scholar]
- 39.Senpuku, H., T. Miyauchi, N. Hanada, and T. Nisizawa. 1995. An antigenic peptide inducing cross-reacting antibodies inhibiting the interaction of Streptococcus mutans PAc with human salivary components. Infect. Immun. 63:4695-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Senpuku, H., H. Kato, M. Todoroki, N. Hanada, and T. Nisizawa. 1996. Interaction of lysozyme with surface protein antigen of Streptococcus mutans. FEMS Microbiol. Lett. 139:195-211. [DOI] [PubMed] [Google Scholar]
- 41.Senpuku, H., T. Iizima, Y. Yamaguchi, S. Nagata, Y. Ueno, M. Saito, N. Hanada, and T. Nisizawa. 1996. Immunogenicity of peptides coupled with multiple T-cell epitopes of a surface protein antigen of Streptococcus mutans. Immunology 88:275-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Senpuku, H., H. Kato, H. Takeuchi, A. Noda, and T. Nisizawa. 1997. Identification of core B cell epitope in the synthetic peptide inducing cross-inhibiting antibodies to a surface protein antigen of Streptococcus mutans. Immunol. Investig. 26:531-548. [DOI] [PubMed] [Google Scholar]
- 43.Senpuku, H., K. Matin, M. A. Salam, I. Kurauchi, S. Sakurai, M. Kawashima, T. Murata, and N. Hanada. 2001. Inhibitory effects of monoclonal antibodies against a surface protein antigen in real-time adherence in vitro and recolonization in vivo of Streptococcus mutans. Scand. J. Immunol. 54:109-116. [DOI] [PubMed] [Google Scholar]
- 44.Tokuda, M., N. Okahashi, I. Takahashi, M. Nakai, S. Nagaoka, M. Kawagoe, and T. Koga. 1991. Complete nucleotide sequence of the gene for a surface protein antigen of Streptococcus sobrinus. Infect. Immun. 59:3309-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Houte, J., R. J. Gibbons, and S. B. Banghart. 1970. Adherence as a determinant of the presence of Streptococcus salivarius and Streptococcus sanguis on the human tooth surface. Arch. Oral Biol. 15:1025-1034. [DOI] [PubMed] [Google Scholar]
- 46.Whittaker, C. J., C. M. Klier, and P. E Kolenbrander. 1996. Mechanisms of adhesion by oral bacteria. Annu. Rev. Microbiol. 50:513-552. [DOI] [PubMed] [Google Scholar]