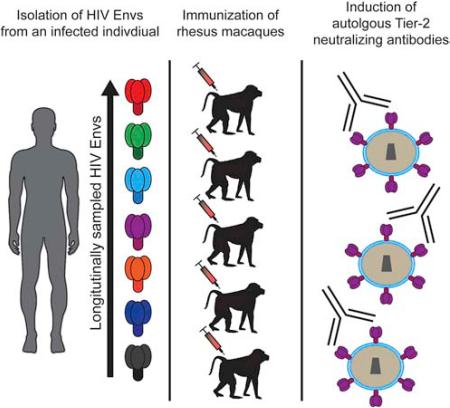
Summary
Antibodies that neutralize autologous transmitted/founder (TF) HIV occur in most HIV-infected individuals and can evolve to neutralization breadth. Autologous neutralizing antibodies (nAbs) against neutralization-resistant (Tier-2) viruses are rarely induced by vaccination. While broadly neutralizing antibody (bnAb)-HIV-Envelope structures have been defined, the structures of autologous nAbs have not. We show here that immunization with TF mutant Envs gp140 oligomers induced high-titer, V5-dependent, plasma neutralization for a Tier-2 autologous TF evolved mutant virus. Structural analysis of autologous nAb DH427 revealed binding to V5, demonstrating the source of narrow nAb specificity, and explaining the failure to acquire breadth. Thus, oligomeric TF Envs can elicit autologous nAbs to Tier-2 HIVs, but induction of bnAbs will require targeting of precursors of B cell lineages that can mature to heterologous neutralization.
Graphical Abstract
INTRODUCTION
The HIV-1 envelope protein (Env) is the primary target of neutralizing antibodies (nAbs) (Wyatt and Sodroski, 1998; Zhou et al., 2007). One major obstacle to developing an effective HIV-1 vaccine is finding an immunogen that can elicit broadly neutralizing antibodies (bnAbs) with the capacity to overcome variability of the virus and to retain neutralizing activity for most circulating HIV-1 strains (Burton et al., 2012; Mascola and Haynes, 2013).
Between 3 and 12 months after HIV-1 transmission, most infected individuals develop autologous, strain-specific, nAbs to the transmitted/founder (TF) virus and TF variants (Ariyoshi et al., 1992; Richman et al., 2003; Wei et al., 2003). The autologous nAb response drives viral escape and stimulates additional specificities of nAbs that neutralize escape viruses (Richman et al., 2003; Wei et al., 2003). This antibody-virus co-evolution persists throughout infection, and in ~20% of individuals, it leads, after years of infection, to development of high levels of bnAbs (Doria-Rose et al., 2010; Gray et al., 2011; Liao et al., 2013a; Tomaras et al., 2011; Walker et al., 2011).
Two recent studies mapped the ontogeny of bnAbs and TF viruses from the time of transmission to bnAb development and showed that bnAbs arise from autologous nAb B cell clonal lineages but that only a small number of the autologous nAb lineages ultimately evolve to neutralization breadth (Doria-Rose et al., 2014; Liao et al., 2013a). Identification of immunogens that can induce nAbs against autologous, neutralization-resistant (Tier-2) viruses is a major challenge for HIV vaccine design, and examples of vaccine-matched, Tier-2 nAb responses elicited by vaccination in primates are few (Sanders et al., 2015; Willey et al., 2003). Moreover, no vaccine-induced Tier-2 nAbs have yet been isolated and characterized, nor have structures of their Env complexes been determined.
CAP206 is an HIV-infected African individual who later developed gp41-targeted bnAbs (Gray et al., 2009a; Morris et al., 2011). As a critical first step in HIV vaccine design, we sought to map the autologous nAb response and to elicit Tier-2 nAbs that mimicked this early autologous nAb response by immunization with Env proteins isolated over the course of infection. We report here that immunization of rhesus macaques with HIV-1 TF variants from CAP206 induced strain-specific nAbs to vaccine-matched Tier-2 autologous viruses in 3 of 6 animals. After only two immunizations, one macaque had a high-titer nAb response that targeted the CD4 binding site (bs) and mimicked the autologous nAb response observed in CAP206. We isolated a vaccine-induced nAb clonal lineage (DH427) that potently neutralized the Tier-2 CAP206 6-month virus and recapitulated the observed plasma neutralization response. A crystal structure of DH427 in complex with HIV Env showed that DH427 bound close to the CD4bs, but also interacted with variable regions in the HIV Env (Loop E and V5-loop), explaining the restricted neutralization breadth and the failure of DH427 to evolve to heterologous neutralization.
RESULTS
Immunization of rhesus macaques elicits Tier-2 autologous neutralization
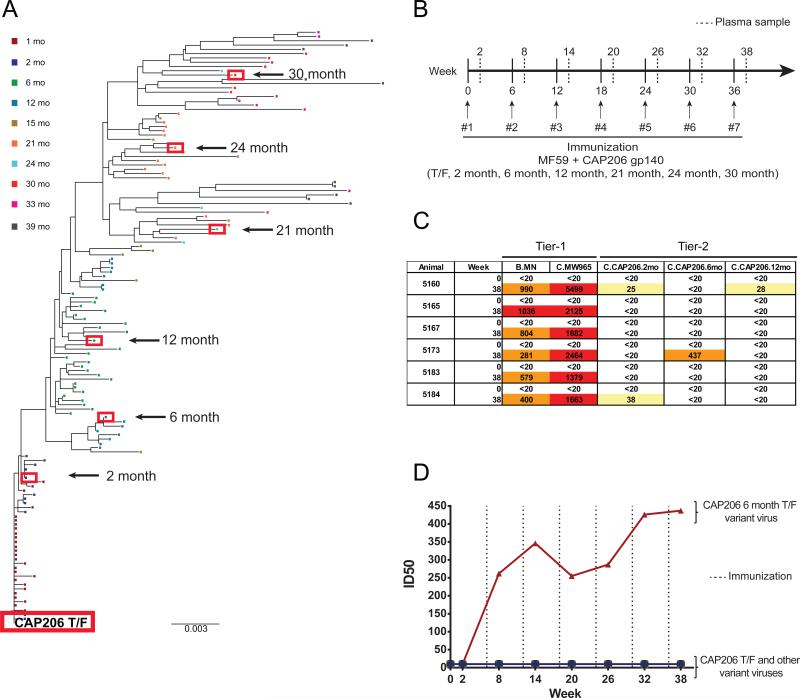
We tracked the evolution of the CAP206 env gene from the TF virus until 39 months after transmission (Fig. 1A). We selected the CAP206 TF and 6 additional representative mutant env genes from 2, 6, 12, 21, 24, and 30 month timepoints, and produced them as recombinant gp140 oligomers that were predominately trimers (Fig. S1A). The antigenic and functional epitopes expressed on each of the recombinant CAP206 Envs were determined by SPR assays (Fig. S1B). All Envs bound to CD4 and mAb A32 which binds well to uncleaved trimers, and magnitude of CD4 binding increased in Envs isolated from later CAP206 timepoints. (Fig. S1B). The 7 Envs showed binding to a panel of neutralizing antibodies and bound to 17b, an antibody that binds to the CD4-induced conformation of Env, in the absence of CD4 (Fig. S2). Some Envs lacked binding of bnAbs that target the CD4bs, and others only reacted weakly, indicating disruption of canonical CD4bs bnAb epitopes in the CAP206 Envs.
Figure 1. Immunization with T/F Envs Elicits Tier-2 Neutralization.
(A) Neighbor-joining phylogenic tree of isolated env sequences from South African HIV-1 infected individual CAP206 from the time of transmission to 39 months post infection. Red boxes indicate envs selected for production of recombinant immunogens. (B) Immunization regimen; 6 rhesus macaques immunized 7 times every 6 weeks with a swarm of 7 CAP206 Envs. Plasma samples collected 2 weeks post immunization. (C) Plasma neutralization of heterologous Tier-1 and autologous Tier-2 viruses before immunization (week 0) and 2 weeks after the last immunization (week 38) measured as the plasma dilution at which relative luminescence units (RLUs) were reduced 50% compared to control wells in the TZM-bl neutralization assay (Yellow, ID50 > 20;Orange, ID50 > 200; Red, ID50>1000). (D) Plasma neutralization over the course of immunization for rhesus monkey number 5173 of the CAP206 autologous Tier-2 viruses (T/F, 2-month, 6-month, 12-month, 21-month, 24-month, and 30-month) in the TZM-bl neutralization assay. Dashed line indicates immunization timepoints. See also Figures S1, S2 and S3.
We immunized six rhesus macaques with a cocktail of all 7 CAP206 recombinant gp140 oligomers, and collected plasma and PBMCs before the first immunization and 2 weeks after each subsequent immunization (Fig. 1B). We tested plasma from Env-immunized animals for the presence of anti-HIV neutralizing activity using the TZM-bl pseudovirus assay. Plasmas collected before the first immunization (week 0) and 2 weeks after the last immunization (week 38) were tested for neutralization of heterologous, neutralization-sensitive (Tier-1), isolates MN (clade B) and MW965 (clade C), and of autologous, Tier-2, vaccine-matched isolates of the seven CAP206 Envs (Fig. 1C & S3A). We observed potent neutralization titers against the Tier-1 MN and MW965 viruses in all animals. Low levels of neutralizing activity against Tier-2 autologous viruses were present in two animals (Fig. 1C & S3B). One animal, rhesus macaque 5173, had potent plasma neutralization activity against the Tier-2 autologous 6-month CAP206 TF variant virus (Fig. 1C & S3B).
Neutralization of the 6-month virus emerged in 5173 plasma 2 weeks after the second immunization, and was boosted by repetitive immunization and persisted throughout the remainder of the immunization regimen (Fig. 1D). Plasma neutralization responses waned 20 months after the last immunization (data not shown). Thus, immunization with CAP206 Envs effectively elicited potent Tier-1 heterologous virus neutralization in all 6 animals and low levels of Tier-2 nAbs in 2 of them; in one animal, it rapidly induced an early and robust neutralization response to an autologous Tier-2 virus.
Immunization-induced neutralization mimics early autologous CAP206 plasma neutralization response
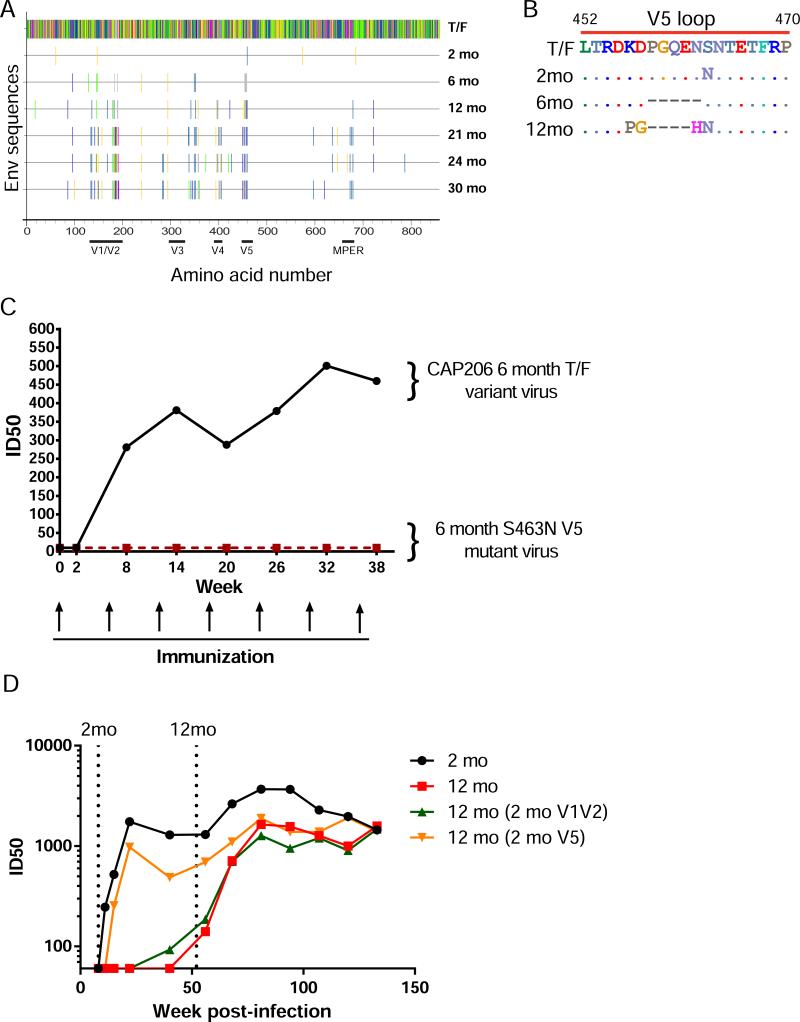
Autologous Tier-2 neutralization in macaque 5173 was specific for the CAP206 6-month virus. Env sequences that were isolated from later timepoints post-infection from CAP206 contained numerous mutations that were candidate sites of immune pressure. Two of the very first observed Env sequence changes retained in the 6-month Env were in the V1/V2 and V5 regions (Fig. 2A).
Figure 2. Glycosylation of the V5 Loop Blocks Autologous Tier-2 Neutralization.
(A) Highlighter plot showing mutations in the Env that occur in the longitudinal immunogens from the T/F sequence. (B) Amino acid alignment of the CAP206 V5 regions. (C) Macaque 5173 plasma neutralization of wild-type and mutant CAP206 6-month viruses over the course of immunization measured as the plasma dilution at which relative luminescence units (RLUs) were reduced 50% compared to control wells in the TZM-bl neutralization assay. (D) CAP206 plasma neutralization of autologous 2-month and 12-month viruses compared with 12-month reverted mutant viruses over the course of infection.
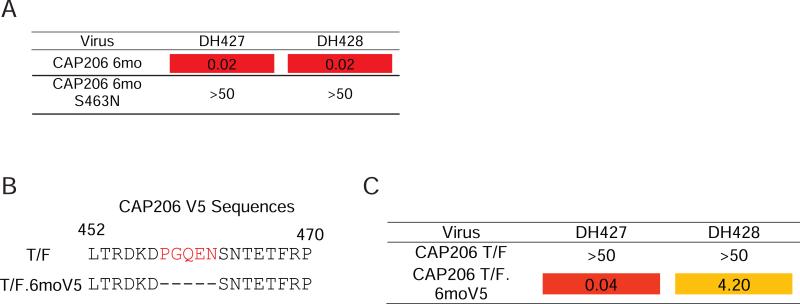
Closer inspection of the V5 sequences revealed extensive diversity in this region among longitudinal CAP206 Envs. Specifically, the 6-month Env had a 5 amino acid deletion and lacked a predicted glycosylation site at position 463 (Fig. 2B). To determine if 5173 neutralization of the 6-month CAP206 virus targeted V5, we engineered a mutation in the 6-month virus that introduced a glycosylation site at position 463 (S463N). We found persistent plasma neutralization of the wild-type 6-month virus for samples across the full immunization period, but no neutralization of the glycosylation site variant (Fig. 2C).
To examine the role of mutations in the V5 loop in early autologous escape of the infecting virus in CAP206, we tested CAP206 plasma isolated post-infection for the ability to neutralize the autologous 2-month and 12-month viral isolate. Early plasma samples exclusively neutralized the 2-month virus, and only later plasma samples acquired neutralization activity for the 12-month virus (Fig. 2D). When we reverted the 12-month V5 region back to the 2-month sequence, the chimeric virus became sensitive to neutralization by early CAP206 plasma timepoints. Reverting mutations in the V1V2 in the 12-month back to the 2-month did not have any effect on early neutralization. Thus, the V5 loop was a target for autologous nAbs in CAP206 after infection and a V5-dependent autologous nAb response was elicited in a rhesus macaque by immunization with a CAP206 TF variant.
Immunization elicited CD4bs autologous neutralizing antibodies
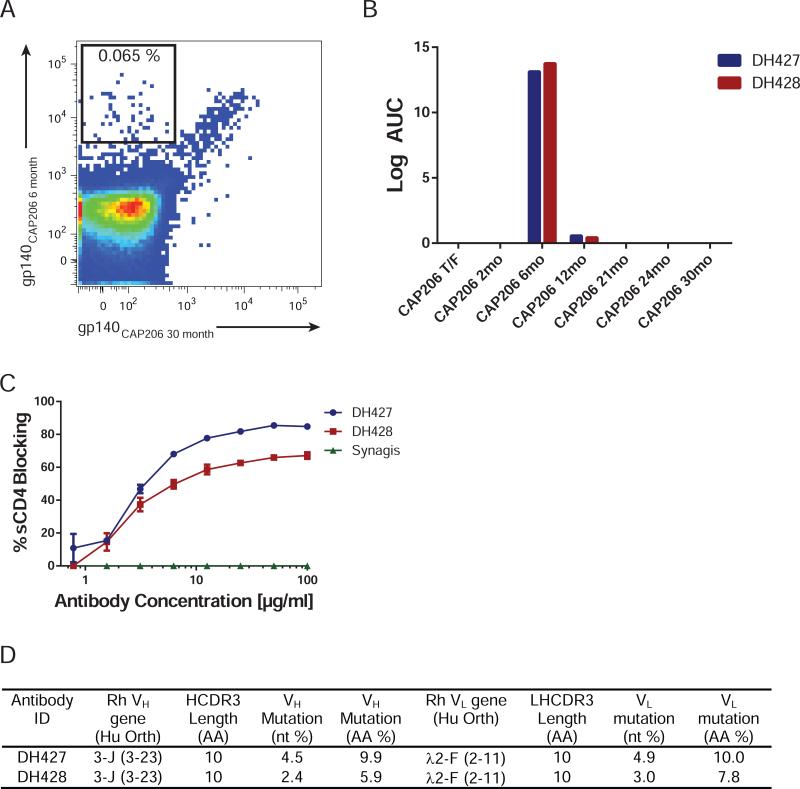
We isolated single Env-specific memory B-cells from macaque 5173 PBMC using tetramers of the neutralization-sensitive CAP206 6-month Env as well as the neutralization-resistant CAP206 30-month Env. Memory cells (CD20+, CD27+) that bound the 6-month but not the 30-month Env were sorted into individual wells of a 96-well plate (Fig. 3A). The frequency of 6-month Env-specific memory B cells was 0.07% (Fig. 3A). Single-cell PCR amplification and transient expression of immunoglobulin (Ig) genes of the sorted memory B cells identified two mAbs (DH427 and DH428) that specifically bound the CAP206 6-month Env in ELISA (Fig. 3B). As expected from their sensitivity to changes at position 463, DH427 and DH428 competed for binding of soluble CD4 to CAP206 6-month Env (Fig. 3C). Sequences of the heavy and light chain Ig genes showed that DH427 and DH428 are two members of the same B cell clonal lineage, which used the rhesus orthologs of the human VH 3-23 and Vλ 2-11 genes (Fig. 3D & S4). Members of the DH427 lineage have short HCDR3s (10aa); the variable heavy regions were mutated 4.5% in DH427 and 2.4% in DH428 (Fig. 3D).
Figure 3. Isolation of Antibodies that Specifically Target the CAP206 6-month Env.
(A) PBMCs from macaque 5173 at week 38 after the 7th immunization were used for sorting CD20+ CD27+ memory B cells that specifically reacted with the 6-month Env and not the 30-month Env (B) Binding of isolated mAbs DH427 and DH428 to the 7 CAP206 Env immunogen proteins by ELISA (C) Cross-competition of sCD4-Ig binding to the CAP206 6-month Env by DH427, DH428, and RSV antibody synagis determined by ELISA. (D) Genomic characteristics of DH427 and DH428. See also Figure S4.
DH427 CD4bs antibody lineage neutralized the Tier-2 CAP206 6-month virus
DH427 and DH428 neutralized the 6-month virus, but did not neutralize any of the other autologous or heterologous viruses tested (Fig. 4A & S5). Moreover, DH427 and DH428 did not neutralize the CAP206 6-month virus with an N463 mutation, recapitulating the pattern of neutralization observed in the 5173 plasma (Fig. 4A).
Figure 4. DH427 and DH428 Neutralize the Tier-2 CAP206 6-month Virus.
(A) Neutralization of the CAP206 6-month virus and engineered glycosylation site mutant measured as the antibody concentration at which relative luminescence units (RLUs) were reduced 50% compared to control wells (IC50) in the TZM-bl neutralization assay (μg/ml). (B) CAP206 T/F V5 sequence and mutated V5 sequence to match the 6-month Env. Red amino acids were deleted. (C) DH427 and DH428 neutralization of the T/F and T/F mutant that had a V5 that matched the 6-month Env in the TZM-bl neutralization assay (μg/ml; Orange, IC50 < 5.0; Red, IC50 < 1.0). See also Figure S5.
The CAP206 6-month Env lacked a glycosylation site in V5; introduction of a glycosylation site in V5 of the CAP206 6-month Env eliminated neutralization by both 5173 plasma and DH427 lineage antibodies. The CAP206 TF Env also lacked a glycosylation site in V5, but the loop was 5 amino-acid residues longer (Fig. 4B). This virus resisted neutralization by DH427 and DH428; deletion of the 5 residues in V5 of the CAP206 TF virus to match the 6-month virus made it sensitive to both antibodies (Fig. 4C). Thus, both the presence of glycosylation sites and the length of the V5 loop restricted CAP206 virus neutralization by DH427 and DH428.
Maturation of the DH427 clonal lineage
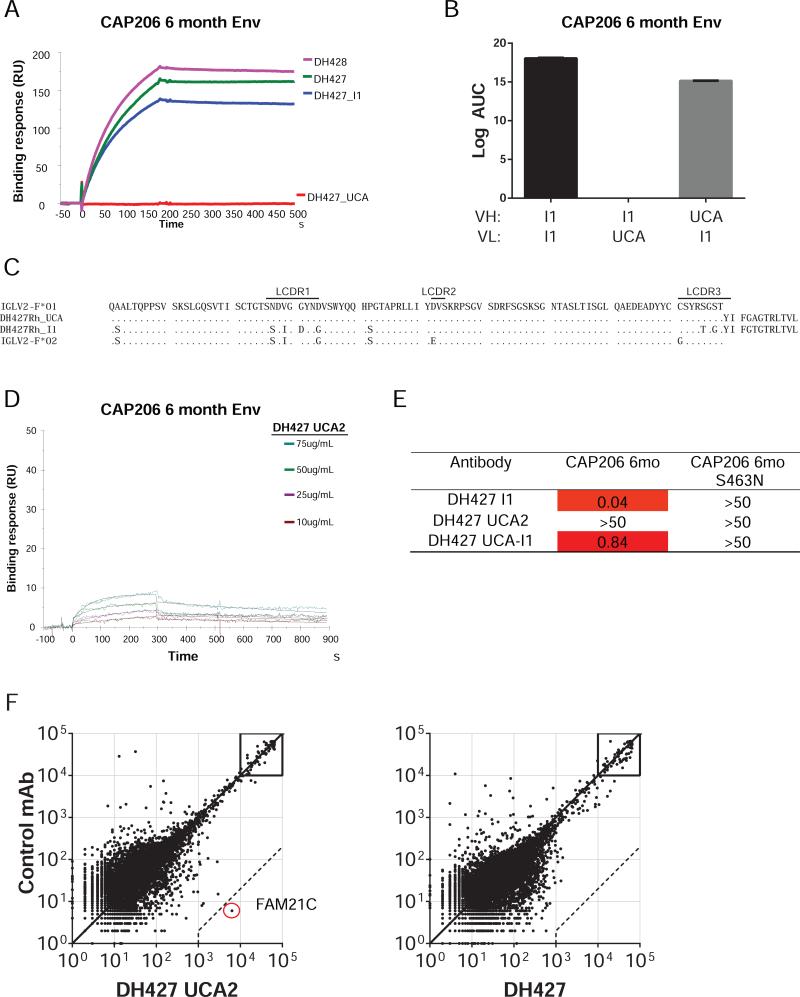
For an analysis of the evolution of the DH427 antibody lineage we inferred the intermediate (I1) and the unmutated common ancestor (UCA) antibodies of DH427 and DH428. The I1 and mature antibodies specifically and exclusively bound the CAP206 6-month Env, but there was no detectable binding of this particular UCA candidate antibody to any of the recombinant CAP206 Envs (Fig. 5A & S6). We showed that light-chain mutations between the UCA and I1 determined the difference in DH427/DH428 binding by pairing the UCA heavy chain with the I1 light-chain and vice versa. Only the former had detectable affinity (Fig. 5B). Failure of the inferred DH427 UCA to bind the immunogen led us to consider whether one or more of the amino acid residues considered to have come from somatic hypermutation (SHM) might instead be germline encoded in an as yet undiscovered rhesus macaque Vλ allelic variant, leading to an incorrect inference for the UCA.
Figure 5. Reconstruction of the DH427 Clonal Lineage.
(A) Binding of DH427, DH428 and the inferred intermediate (I1) and unmuated common ancestor (UCA) to the CAP206 6-month Env measure by SPR. Antibodies were immobilized on a CM5 chip and protein was flowed over at 100μg/ml. (B) Binding of the DH427 I1 and I1-UCA hybrids to the CAP206 6-month Env measured by ELISA.(C) Amino acid alignment of the germ-line IGVL2-F*01 with DH427 UCA and I1 antibodies compared with IGVL2-F*02 allele discovered by sequencing the IGVL2-F genomic DNA from animal 5173. (D) Binding of newly designed DH427 UCA (DH427 UCA2) to the CAP206 6-month Env by SPR. DH427 UCA2 was immobilized on a CM5 chip and protein was flowed over at the indicated concentrations. (E) Neutralization of DH427 I1, UCA2, and chimeric (UCA VH:I1VL) of the CAP206 6-month virus and glycosylation site mutant measured as the antibody concentration (μg/ml) at which relative luminescence units (RLUs) were reduced 50% compared to control wells (IC50) in the TZM-bl neutralization assay. (F) Reactivity of DH427UCA2 and DH427 on a panel of 9,400 human proteins tested using a ProtoArray microchip compared with a nonreactive rhesus mAb. Axis values are relative fluorescent signal intensity. Each dot represents an average of duplicate array proteins. Dashed lines indicate 500-fold signal/background ratio as cutoff for reactivity. See also Figures S6 and S7; Table S1.
We examined the germline IGLV2-F sequence in macaque 5173 by targeted genomic amplification and sequencing of extracted genomic DNA (Fig. S7). Sequencing of individual colonies detected an IGLV2-F allele (IGLV2-F*02) that shared 3 of the 4 residues that we had considered mutated in I1 (Fig. 5C). We then sequenced the IGLV2-F allele in the 5 remaining animals in the immunization study and found IGLV2-F*02 in the genomic DNA of all 5 (Fig. S6C). From these results, we designed a new UCA (UCA2) by incorporating the new germline light-chain allele. We found that inferred UCA2 bound the 6-month Env, indicating that UCA2 was the authentic initiator of the DH427/DH428 B cell lineage (Fig. 5D).
The UCA2 antibody did not neutralize the 6-month virus, but both DH471 I1 and the chimeric (UCA VH: I1VL) antibody did neutralize this virus (Fig. 5E). These results indicated that autologous neutralization activity required a specific sequence of light-chain affinity maturation.
DH427 UCA reacts with host protein FAM21C
Two macaques (5160 and 5184) had weak autologous nAb responses, one (5173) had a robust autologous nAb response, and three (5165, 5167, 5183) had no autologous nAb response, yet all had received the same vaccine immunogen. This variability in the response suggested that host factors limited high levels of responses in the other 5 animals. To examine the role of cross-reactivity with host antigens for the development of Tier-2 nAbs, we tested each immunized macaque's plasma pre- and post-immunization for reactivity to common auto-antigens recognized by individuals with autoimmune disease (Fig. S7D). We did not detect significant antibody polyreactivity in plasma for any of the auto-antigens tested.
Next, we tested the DH427 lineage for reactivity with the same auto-antigens and did not detect reactivity (Fig. S7E). We then tested DH427 and the DH427 UCA antibodies for reactivity to >9,400 human proteins on a microarray (Fig. 5F). We found that neither DH427 nor the UCA were polyreactive, but the UCA did react strongly with a single host protein Family of Sequence Similarity 21, Member C (FAM21C; Fig. 5F & Table S1). FAM21C is a member of the WASH complex that is involved in intracellular trafficking (Gomez and Billadeau, 2009). These data raise the hypothesis that reactivity with auto-antigens may have blocked the initial maturation and/or expansion of DH427-like lineages in the other 5 macaques while having the correct germline VL allele.
Structure of DH427 in complex with HIV-1 gp120
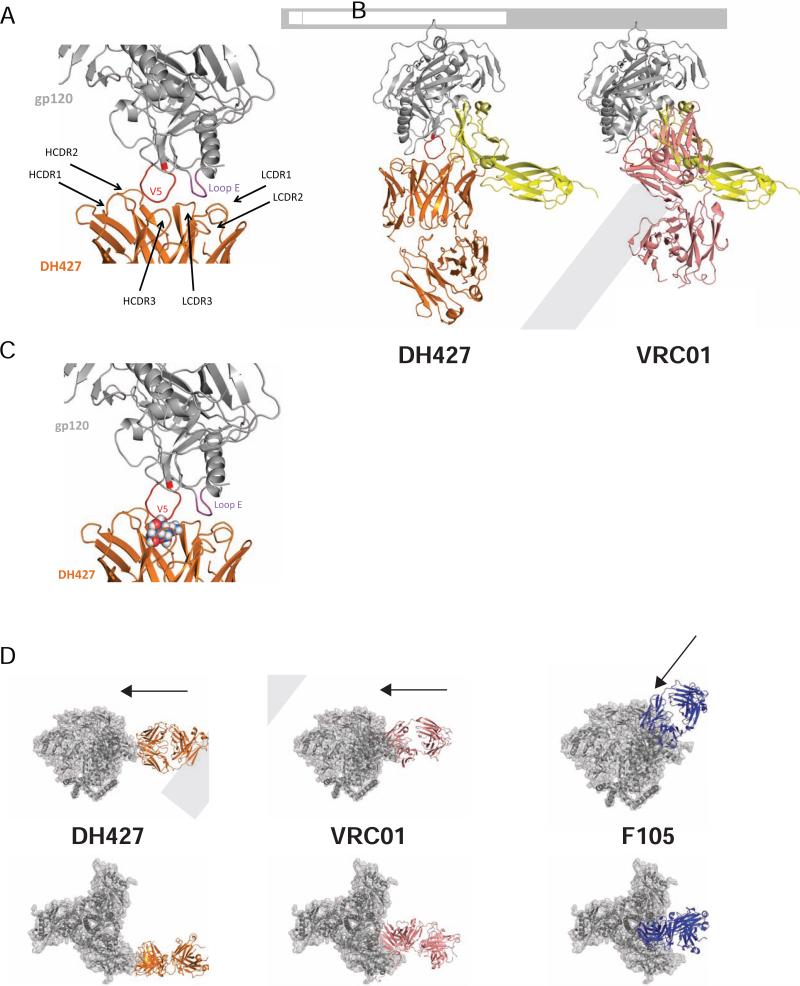
The DH427/DH428 vaccine-induced lineage blocked CD4 binding and neutralized an autologous Tier-2 virus, but its genetic characteristics differed from those of most CD4bs nAbs. We crystallized and determined at 6.6 Å resolution the structure of the DH427 Fab in complex with a recombinant variant of the ZM1766.66 gp120 that had the CAP206 6-month V5-loop sequence. We also determined the structures of the DH427 and DH428 Fabs at 2.69 Å and 2.37 Å resolution, respectively. (Table S2).
The DH427 Fab associates with HIV gp120 along an outer rim of the CD4-binding surface. CDRH2 and CDRH3 contact the V5 loop, and CDRL1 and CDRL3 contact loop E (Fig. 6A). The limited resolution did not allow detailed description of side-chain interactions. We superposed onto the gp120 core domain from our complex the structure of the gp120 core domain bound with soluble CD4. The bulk of the Fab was large enough to block CD4 binding, even though their overlapping footprints differed substantially (Fig. 6B). In contrast, the CD4bs bnAb VRC01 contacts conserved residues in the V5 loop and has more contacts in the CD4bs than DH427, allowing for more effective CD4 blocking and broad antigen recognition (Fig. 6B). Mutability of V5, at the core of their contact, and a very frequently found glycosylation site at position 355 on loop E, clearly accounted for the restricted breadth of DH427 and DH428. For example, the glycan at position 463 would project directly into the face of the antibody (Fig. 6C). Thus, further affinity maturation would be unlikely to generate much greater neutralization breadth.
Figure 6. Structure of DH427 in Complex with HIV gp120.
(A) DH427 contact sites within the V5 loop and loop E (DH427, orange; gp120, gray; V5 loop, red; loop E, purple). (B) DH427 in complex with core gp120 with VRC01 and CD4 molecule superimposed (DH427, orange; gp120, gray; V5 loop, red; VRC01, pink; CD4 molecule, yellow). (C) Co-crystal structure of DH427 and HIV Env with N-acetylglucosamine (GlcNAc) displayed as spheres at the V5 glycan site that would interfere with DH427 heavy chain. (D) Modeling of DH427, VRC01, and F105 onto the high resolution cryo-EM trimer structure of BG505 SOSIP (BG505 SOSIP trimer, gray) side view (top) and top view (bottom). See also Table S2.
Superposition of the gp120 outer domain from its complex with DH427 onto the homologous domain in the BG505 SOSIP gp140 structure shows that the Fab would project laterally from the periphery of the trimer (Fig. 6D). DH427 is in almost the same orientation as the CD4bs bnAb VRC01, but displaced outward by about 25Å. This angle is also distinct from the prototypical Tier-1 neutralizing CD4bs mAb F105, which projects in a more axial direction, making it prone to steric hindrance on the “closed” trimers of Tier-2 viruses (Fig. 6D).
DISCUSSION
Recent studies show that bnAb lineages can evolve out of autologous nAb lineages present early in HIV-1 infection (Doria-Rose et al., 2014; Liao et al., 2013a; Wibmer et al., 2013). All HIV-1 infected individuals produce autologous Tier-2 nAbs months after transmission, but not all HIV-infected individuals go on to make bnAbs (Bar et al., 2012; Doria-Rose et al., 2010; Gray et al., 2009b; Kwong et al., 2013; Richman et al., 2003; Wei et al., 2003). Thus, eliciting strain-specific nAbs that target neutralization-resistant Tier-2 viruses can be a first step to develop bnAbs during vaccination. CAP206 is an HIV-infected individual that produced bnAbs targeting the MPER, but we found that the V5 loop was a target for autologous nAbs early after infection in CAP206. Thus, in CAP206, the autologous nAb response was unrelated to the bnAb response. Our finding that after only two immunizations with a swarm of CAP206 Envs, the CAP206 6-month variant of the TF Env induced in 1 of 6 rhesus macaques potent autologous Tier-2 nAbs that targeted a similar nAb epitope observed in CAP206 shows that vaccination with gp140 oligomers can, in principle, also produce such antibodies. However, that only three macaques had autologous nAbs, and only one with high titers, contrasts with HIV infection, in which virtually all HIV-infected individuals make autologous nAbs. These studies raise several important issues for HIV vaccine development.
First, our structural studies of DH427 CD4bs autologous nAb demonstrated that DH427 neutralization was restricted to the autologous virus because antibody recognition centers on the V5 loop. Moreover, the mode of V5 contact prevented full engagement of the CD4bs and explains why this lineage is unlikely to achieve bnAb breadth with further affinity maturation.
Second, it is a conundrum that all HIV-infected individuals make autologous nAbs and up to 50% make some level of bnAbs, while no humans vaccinated with Envs have to date made either autologous nAbs or bnAbs. All the macaques in the group studied had the IGLV2-F germline allelic variant, and thus all had the required VL repertoire to respond to Env immunization. One hypothesis is that there may be host control mechanisms for autologous nAbs in both macaques and humans and that the responding macaque (5173) was permissive for release of antibodies that can recognize a closed or Tier-2 envelope. There was no reactivity with autoantigens common in autoimmune disease by the plasma or DH427 lineage antibodies, but the DH427 UCA antibody did react with host protein FAM21C. This reactivity may have limited expansion of this lineage in the other animals.
Broad neutralization appears only after years of persistent infection, in part because breadth requires high levels of SHM, even for those antibodies with relatively invariant Env contacts (Kepler et al., 2014; Kwong and Mascola, 2012; Scheid et al., 2009; Scheid et al., 2011). Driving high SHM by vaccination is particularly difficult, because many highly mutated antibodies are disfavored by the host immune system (Haynes and Verkoczy, 2014). DH427 lineage antibodies required SHM for neutralization, but they were less than 5% mutated and were induced rapidly after only 2 immunizations. Thus, the DH427 lineage achieved potent Tier-2 autologous neutralization with minimal SHM.
Finally, HIV Env shifts from a “closed” to an “open” conformation when CD4 binds (Liu et al., 2008). In that transition, it exposes epitopes concealed in the closed state. Any particular Env is in equilibrium between closed and open (and probably one or more intermediate) states (Julien et al., 2013; Pugach et al., 2015). The key structural correlate of the distinction between Tier-1 and Tier-2 is whether this equilibrium is strongly on the side of the closed conformation (Tier-2, primarily bnAb site accessible) or more frequently toward the open conformation (strain-specific and non-neutralizing sites accessible). A recent study demonstrated that a stabilized recombinant native-like “closed” trimer Env (BG505 SOSIP.664) elicited nAbs to the sequenced-matched Tier-2 virus in rabbits and in 3 of 4 macaques (Sanders et al., 2015). In the Sanders et al. study in macaques, the titers of autologous nAbs ranged from 32 to 168, similar to the titers observed in an earlier study by Willey et al. (40-113) and in the present study (25-437).The immunogen used here was a trimeric Env, but not a variant (such as BG505 SOSIP.664) locked into a closed state by intentional modification. Nonetheless, it elicited potent autologous nAbs, and in at least the one lineage we have studied, those nAbs recognized a site accessible of the surface of a Tier-2 virus, and thus by inference, on the surface of a “closed” trimer. Although the DH427 nAb Env epitope appears to be too variable, both in the length of the loops and in their potential for glycosylation, to be a target for bnAbs, its proximity to the CD4bs and its dependence on similar conformational parameters suggest that related vaccine immunogens could also induce the germline precursors of CD4bs bnAbs. Indeed, while most Env immunogens fail to engage germline-reverted CD4bs-directed bnAbs, including germline-reverted VRC01 (McGuire et al., 2013; Zhou et al., 2010), eliminating glycosylation sites in loop D and V5, as in the CAP206 6-month variant that induced the DH427 lineage, allowed DH427 germline-Env binding and B cell lineage development. Incorporating these sites, along with truncation or deletion of V5, into future Env immunogens may have a useful role in priming B cell responses to the CD4bs. Moreover, modifying the CAP206 Env to have a more closed structure by SOSIP modification may also increase immunogenicity. Such structural considerations coupled with understanding host control of induction of neutralizing antibodies should lead to design of strategies for inducing autologous nAbs to Tier-2 HIV with greater heterologous neutralization breadth (Liao et al., 2013a).
EXPERIMENTAL PROCEDURES
Donor subject
CAP206 is an HIV-1 subtype C chronically infected individual used for this study. This participant was part of the CAPRISA 002 Acute infection cohort whose antibody neutralization profile has been studied since the point of seroconversion (Gray et al., 2007; Gray et al., 2009b). This study was approved by the IRB's of the Universities of KwaZulu Natal and Witwatersrand in South Africa and Duke University. Written informed consent was obtained from all study participants.
Design and production of recombinant gp140 proteins derived from CAP206
The cloning and sequencing of env sequences was performed as described previously from longitudinal samples from CAP206 (Lynn Morris, unpublished;(Gray et al., 2007; Keele et al., 2008) . The single TF and 6 mutant clones from later time points (CAP206 TF, 2month, 6month, 12month, 21month, 24month, and 30month) were selected for production as soluble gp140 trimeric immunogens. The recombinant Envs were expressed in 293T cells and purified using lectin and size exclusion chromatography as described previously (Liao et al., 2013b).
Immunization of rhesus macaques
Six rhesus macaques each received 7 intramuscular immunizations at 6 week intervals with 7 CAP206 Env immunogens (100μg total) in MF59 adjuvant (Novartis). Blood samples were collected 2 weeks after each immunization. All rhesus macaques were housed at Bioqual, Inc, Rockville, MD. All rhesus macaques were maintained in accordance with the Association for Assessment and Accreditation of Laboratory Animals with the approval of the Animal Care and Use Committees of the National Institutes of Health and Harvard Medical School. Research was conducted in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animals and experiments involving animals and adheres to principles stated in the Guide for the Care and Use of Laboratory Animals, NRC Publication, 2011 edition.
Antibody isolation
CAP206 6 month gp140 and CAP206 30 month gp140 proteins were fluorescently labeled with AF647 and BV421 (Invitrogen), respectively. Peripheral blood mononuclear cells (PBMCs) from rhesus monkey 5173 at week 38 were stained with fluorescently labeled antibodies for cell surface markers and both CAP206 Envs. Memory B cells that were stained positive for the CAP206 6 month Env and not the CAP206 30 month Env were sorted into single wells of 96-well PCR plates containing RT-PCR buffer as previously described (Wiehe et al., 2014). Antibody variable heavy and variable light genes were amplified using nested PCR and purified and sequenced as described previously (Wiehe et al., 2014). VDJ arrangements, clonal relatedness, and identification of the intermediate and unmutated common ancestor were inferred using previously described computational methods (Kepler, 2013; Munshaw and Kepler, 2010).
Sequencing of germline variable region
Genomic DNA was isolated from all 6 animals from PBMCs at 2 weeks after the first immunization (QIAmp DNA Blood mini kit; Qiagen). IGLV2-F sequences were amplified using 2 independent primer sets. To ensure amplification of non-rearranged variable sequences, both primer sets reverse primers aligned to sequences present in the non-coding genomic DNA downstream the V-recombination site. The forward primer for set 1 resided in the IGV2-F leader sequence and upstream of the leader in set 2. The PCR fragments were cloned into a pcDNA2.1 (TOPO-TA kit; Life technologies) and transformed into bacteria for sequencing of individual colonies.
Expression of recombinant antibodies
Transient small-scale expression of antibodies was achieved by assembling VH, VK or VL sequences with linear cassettes that contain the CMV promoter, respective Ig constant region, and poly A signal sequence using overlapping PCR and transfection into 293T cells as described previously (Liao et al., 2009). Supernatants were directly used to screen for binding of Env antigens in ELISA.
For production of purified recombinant mAbs, the VH and VL genes from DH427 lineage antibodies were cloned into expression vectors and expressed and purified as described previously (Liao et al., 2011). Site-directed mutagenesis of antibody genes was performed using the Quikchange II lightening multi-site-directed mutagenesis kit (Agilent).
ELISA binding of transiently transfected supernatants and purified recombinant mAbs was performed as described previously (Liao et al., 2011).
Surface plasmon resonance
Surface plasmon resonance assays were performed on a BIAcore 4000 instrument and data analysis was performed with BIAevaluation 4.1 software (BIAcore Inc, Uppsala, Sweden). Anti-gp120 MAbs or sCD4 in 10 mM Na-acetate buffer (pH 4.5) were directly immobilized to CM5 sensor chips using a standard amine coupling protocol for protein immobilization. Purified CAP206 Env glycoproteins were flowed over CM5 sensor chips at concentrations of 2 to 100 μg/ml. Binding of CAP206 envelope proteins was monitored in real time at 25°C with a continuous flow of phosphate-buffered saline (150 mM NaCl, 0.005% surfactant P20 [pH 7.4]) at 10 to 30 μl/min (Alam et al., 2011; Gao et al., 2014).
Neutralization assays
Neutralization activities of animal plasma and purified antibodies were determined by the TZM-bl cell based neutralization assay (Sarzotti-Kelsoe et al., 2014). Tier phenotyping of the pseudoviruses was assayed by sensitivity to a pool of HIV infected serum as described previously (Seaman et al., 2010).
Antibody reactivity with host proteins
The polyreactivity of the DH427 lineage was assayed in the AtheNA multi-lyte system (Zeus scientific). The rhesus mAbs were also tested for reactivity with human host cellular antigens using ProtoArray 5 microchip (Life technologies) compared to a rhesus isotype-matched control antibody as described previously(Yang et al., 2013)
Expression and purification of proteins for crystallization
The heavy- and light-chain variable and constant domains of the DH427 and DH428 Fabs were cloned into the pVRC-8400 expression vector using Not1 and Nhe1 restriction sites and the tissue plasminogen activator signal sequence. The C terminus of the heavy-chain constructs contained a noncleavable 6x histidine tag. Fabs were expressed and purified as described previously (Fera et al., 2014).
The codon-optimized synthetic construct of the ZM176.66 HIV-1 subtype C gp120 containing a.a. 41–492 (HXB2 numbering) ΔV123 (core) was produced by GenScript with an N-terminal 6x-histidine tag and inserted into the pVRC- 8400 expression vector as described for Fabs. The V5 loop of this gp120 core was mutated to that of the CAP206 6 month envelope by site-directed mutagenesis using manufacturer's protocols (Stratagene) and expressed in 293S GnTi- suspension adapted cells using linear PEI. After 5 days of expression, supernatants were clarified by centrifugation, and passed over galanthus nivalis lectin resin pre-equilibrated with 1x PBS. Bound protein was washed with 1x PBS and eluted with 1x PBS and 500 mM methyl α-D mannopyranoside. The glycoprotein was then deglycosylated overnight at 37 °C with Endo H in a buffer of 50 mM sodium acetate, pH 6.0, 5mM EDTA, 500mM NaCl, 1 ug/ul leupeptin, and 1ug/ul aprotonin. Deglycosylated gp120 core was then purified by gel filtration chromatography in Buffer B (2.5 mM Tris, pH 7.5, 350 mM NaCl, 0.02% sodium azide) using a superdex 200 analytical column (GE Healthcare). To make the complex for co-crystallization, DH427 and the ZM176.66 gp120 mutant were mixed in a 1.2:1 molar ratio and incubated for at least 1 hour before passing over a superdex 200 preparatory column (GE Healthcare) in buffer B. Fractions corresponding to the complex were combined and concentrated to ~10 mg/ml for co-crystallization trials.
Crystallization, structure determination, and refinement
All His-tagged Fabs were crystallized at ~15 mg/mL and the DH427/ZM176.66 mutant gp120 core complex was crystallized at ~10 mg/mL. Crystals were grown in 96-well format using hanging drop vapor diffusion and appeared after 24–48 h at 20 °C. DH427 crystals were obtained in a condition of 20% polyethylene glycol (PEG) 2K monomethyl ether (MME), 100 mM sodium citrate, pH 4.0, and 100mM NaCl; and DH428 crystals were grown over a reservoir of 40% PEG 400, and 100 mM sodium acetate, pH 5.0. Initial crystals of DH427/gp120 core complex were obtained in a condition of 1.5 M ammonium sulfate and 100 mM Tris, pH 8.0 and were optimized to 24-well format to obtain larger crystals. All crystals were harvested and cryoprotected by the addition of 20–25% glycerol to the reservoir solution and then flash-cooled in liquid nitrogen. Crystals of complex were cryoprotected with a series of different cryoprotectants including 15-30% glycerol, 15-30% ethylene glycol, and 1-2 M sodium malonate, however the best diffraction was obtained with 20-30% glycerol or ethylene glycol as the cryoprotectant.
Diffraction data were obtained at 100 K from beam lines 24-ID-C and 24-ID-E at the Advanced Photon Source using a single wavelength. Datasets from individual crystals (two crystals for DH427, and one crystal for DH428) were processed with HKL2000 (Otwinowski and Minor, 1997) and XDS (Kabsch, 2010) for DH427 and DH428, respectively. Molecular replacement calculations for the free Fabs were carried out with PHASER (McCoy, 2007), using I3.2 from the CH103 lineage [Protein Data Bank (PDB) ID 4QHL] as the starting model. The I3.2 model was separated into its variable and constant domains for the DH427 and DH428 Fab structure determinations. Crystals of the DH427 and DH428 Fabs had 4 and 1 molecules per asymmetric unit, respectively. For both Fabs, subsequent refinement steps were carried out with PHENIX (Adams et al., 2010), and all model modifications were carried out with Coot (Emsley and Cowtan, 2004). During refinement, maps were generated from combinations of positional, group B-factor, and TLS (translation/ libration/screw) refinement algorithms. Secondary-structure restraints were included at all stages for all Fabs; noncrystallographic symmetry restraints were applied to the DH427 Fab throughout refinement.
Data from multiple crystals of the DH427/gp120 core complex were processed using HKL2000 (Otwinowski and Minor, 1997) and molecular replacement calculations were carried out with PHASER (McCoy, 2007), using the refined DH427 coordinates and gp120 core from the VRC01/gp120 complex [Protein Data Bank (PDB) ID 4LST] as the starting models. DH427 was separated into its variable and constant domains for structure determinations. There were 2 molecules per asymmetric unit. The resulting electron density map for the complex was further improved by solvent flattening, histogram matching, and noncrystallographic symmetry averaging using the program Parrot (Winn et al., 2011). Phase combination was disabled in these calculations. After density modification, rigid-body refinement was performed using Refmac in Coot.
Structure validations were performed periodically during refinement using the MolProbity server (Davis et al., 2007). The final refinement statistics are summarized in Table S1.
Protein Structure Analysis and Graphical Representations
The core domain of the chimeric ZM176.66 gp120 core mutant designed in our study was superposed on other gp120 cores from the PDB by least squares fitting in Coot. All graphical representations with protein crystal structures were made using PyMol.
Supplementary Material
HIGHLIGHTS.
HIV-1 TF Env immunization induced potent Tier-2 neutralizing antibodies (nAbs).
Vaccine-elicited nAbs target CD4bs and mimic autologous nAbs in infected individual.
Autologous nAb structure reveals mechanism of strain-specific neutralization.
ACKNOWLEDGMENTS
This work was supported by the Center for HIV/AIDS Vaccine Immunology-Immunogen Discovery (CHAVIID; UMI-AI100645) grant from NIH/NIAID/DAIDS. D.F. is supported by a F32 fellowship (1F32AI11635501) from the NIH. S.C.H. is an investigator with the Howard Hughes Medical Institute. We thank Dawn J. Marshall and John Whitesides for expert technical assistance with flow cytometry, and Kelly Soderberg and Samantha Bowen for project management. We also thank beam line staff at Advanced Photon Source 24-ID-C and 24-ID-E for support during data collection.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
T.B. isolated and characterized antibodies, designed assays, analyzed and interpreted data, and wrote and edited the manuscript. D.F. conducted structural and sequence analyses, protein expression, data analyses and interpretation, and edited the manuscript. J.B. performed CAP206 plasma analysis. S.S. assisted with animal study and analysis. K.A. and S.M.A. performed protein purification and SPR assays. R.Z, A.F. contributed to rhesus PCR and antibody production. X.L assisted with CAP206 Env protein production. H.X.L. lead protein and antibody production. X.N. and G.C. performed Protoarrays. K.E.L, C.S. and R.P. performed antibody binding assays. L.S., R.M.S. and C.M.B. performed rhesus immunizations. L.M. provided CAP206 Envs and sequences. S.A.K assisted with clinical protocols. S.B. provided vaccine adjuvant. A.M. and D.C.M. performed neutralization assays. T.B.K. designed software and performed computational analyses of antibody sequences and inferred UCAs. M.A.M. produced fluorophor-labeled Env proteins for memory B cell staining, S.C.H. led structural studies and analysis and edited the manuscript, and B.F.H. designed the study, oversaw all experiments, analyzed all data, and wrote and edited the manuscript.
ACCESION NUMBERS
The Genbank accession numbers for the rhesus antibodies reported in this study are KU216183-KU216190 Coordinates and structure factors for DH427 and DH428 antibody and DH427 in complex with gp120 have been deposited in the Protein Data Bank with accession code T.B.D., T.B.D., and T.B.D., respectively.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam SM, Liao HX, Dennison SM, Jaeger F, Parks R, Anasti K, Foulger A, Donathan M, Lucas J, Verkoczy L, et al. Differential reactivity of germ line allelic variants of a broadly neutralizing HIV-1 antibody to a gp41 fusion intermediate conformation. Journal of virology. 2011;85:11725–11731. doi: 10.1128/JVI.05680-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariyoshi K, Harwood E, Chiengsong-Popov R, Weber J. Is clearance of HIV-1 viraemia at seroconversion mediated by neutralising antibodies? Lancet. 1992;340:1257–1258. doi: 10.1016/0140-6736(92)92953-d. [DOI] [PubMed] [Google Scholar]
- Bar KJ, Tsao CY, Iyer SS, Decker JM, Yang Y, Bonsignori M, Chen X, Hwang KK, Montefiori DC, Liao HX, et al. Early low-titer neutralizing antibodies impede HIV-1 replication and select for virus escape. PLoS pathogens. 2012;8:e1002721. doi: 10.1371/journal.ppat.1002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337:183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic acids research. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose NA, Klein RM, Daniels MG, O'Dell S, Nason M, Lapedes A, Bhattacharya T, Migueles SA, Wyatt RT, Korber BT, et al. Breadth of human immunodeficiency virus-specific neutralizing activity in sera: clustering analysis and association with clinical variables. Journal of virology. 2010;84:1631–1636. doi: 10.1128/JVI.01482-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica Section D, Biological crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fera D, Schmidt AG, Haynes BF, Gao F, Liao HX, Kepler TB, Harrison SC. Affinity maturation in an HIV broadly neutralizing B-cell lineage through reorientation of variable domains. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:10275–10280. doi: 10.1073/pnas.1409954111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Bonsignori M, Liao HX, Kumar A, Xia SM, Lu X, Cai F, Hwang KK, Song H, Zhou T, et al. Cooperation of B cell lineages in induction of HIV-1-broadly neutralizing antibodies. Cell. 2014;158:481–491. doi: 10.1016/j.cell.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez TS, Billadeau DD. A FAM21-containing WASH complex regulates retromer-dependent sorting. Developmental cell. 2009;17:699–711. doi: 10.1016/j.devcel.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray ES, Madiga MC, Hermanus T, Moore PL, Wibmer CK, Tumba NL, Werner L, Mlisana K, Sibeko S, Williamson C, et al. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. Journal of virology. 2011;85:4828–4840. doi: 10.1128/JVI.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray ES, Madiga MC, Moore PL, Mlisana K, Abdool Karim SS, Binley JM, Shaw GM, Mascola JR, Morris L. Broad neutralization of human immunodeficiency virus type 1 mediated by plasma antibodies against the gp41 membrane proximal external region. Journal of virology. 2009a;83:11265–11274. doi: 10.1128/JVI.01359-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray ES, Moore PL, Choge IA, Decker JM, Bibollet-Ruche F, Li H, Leseka N, Treurnicht F, Mlisana K, Shaw GM, et al. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. Journal of virology. 2007;81:6187–6196. doi: 10.1128/JVI.00239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray ES, Taylor N, Wycuff D, Moore PL, Tomaras GD, Wibmer CK, Puren A, DeCamp A, Gilbert PB, Wood B, et al. Antibody specificities associated with neutralization breadth in plasma from human immunodeficiency virus type 1 subtype C-infected blood donors. Journal of virology. 2009b;83:8925–8937. doi: 10.1128/JVI.00758-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Verkoczy L. AIDS/HIV. Host controls of HIV neutralizing antibodies. Science. 2014;344:588–589. doi: 10.1126/science.1254990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta crystallographica Section D, Biological crystallography. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepler TB. Reconstructing a B-cell clonal lineage. I. Statistical inference of unobserved ancestors. F1000Research. 2013;2:103. doi: 10.12688/f1000research.2-103.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepler TB, Liao HX, Alam SM, Bhaskarabhatla R, Zhang R, Yandava C, Stewart S, Anasti K, Kelsoe G, Parks R, et al. Immunoglobulin gene insertions and deletions in the affinity maturation of HIV-1 broadly reactive neutralizing antibodies. Cell host & microbe. 2014;16:304–313. doi: 10.1016/j.chom.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Mascola JR. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Mascola JR, Nabel GJ. Broadly neutralizing antibodies and the search for an HIV-1 vaccine: the end of the beginning. Nature reviews Immunology. 2013;13:693–701. doi: 10.1038/nri3516. [DOI] [PubMed] [Google Scholar]
- Liao HX, Chen X, Munshaw S, Zhang R, Marshall DJ, Vandergrift N, Whitesides JF, Lu X, Yu JS, Hwang KK, et al. Initial antibodies binding to HIV-1 gp41 in acutely infected subjects are polyreactive and highly mutated. The Journal of experimental medicine. 2011;208:2237–2249. doi: 10.1084/jem.20110363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Levesque MC, Nagel A, Dixon A, Zhang R, Walter E, Parks R, Whitesides J, Marshall DJ, Hwang KK, et al. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. Journal of virological methods. 2009;158:171–179. doi: 10.1016/j.jviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013a;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Tsao CY, Alam SM, Muldoon M, Vandergrift N, Ma BJ, Lu X, Sutherland LL, Scearce RM, Bowman C, et al. Antigenicity and immunogenicity of transmitted/founder, consensus, and chronic envelope glycoproteins of human immunodeficiency virus type 1. Journal of virology. 2013b;87:4185–4201. doi: 10.1128/JVI.02297-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR, Haynes BF. HIV-1 neutralizing antibodies: understanding nature's pathways. Immunological reviews. 2013;254:225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta crystallographica Section D, Biological crystallography. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire AT, Hoot S, Dreyer AM, Lippy A, Stuart A, Cohen KW, Jardine J, Menis S, Scheid JF, West AP, et al. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. The Journal of experimental medicine. 2013;210:655–663. doi: 10.1084/jem.20122824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris L, Chen X, Alam M, Tomaras G, Zhang R, Marshall DJ, Chen B, Parks R, Foulger A, Jaeger F, et al. Isolation of a human anti-HIV gp41 membrane proximal region neutralizing antibody by antigen-specific single B cell sorting. PloS one. 2011;6:e23532. doi: 10.1371/journal.pone.0023532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshaw S, Kepler TB. SoDA2: a Hidden Markov Model approach for identification of immunoglobulin rearrangements. Bioinformatics. 2010;26:867–872. doi: 10.1093/bioinformatics/btq056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pugach P, Ozorowski G, Cupo A, Ringe R, Yasmeen A, de Val N, Derking R, Kim HJ, Korzun J, Golabek M, et al. A native-like SOSIP.664 trimer based on an HIV-1 subtype B env gene. Journal of virology. 2015;89:3380–3395. doi: 10.1128/JVI.03473-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, et al. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015 doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarzotti-Kelsoe M, Bailer RT, Turk E, Lin CL, Bilska M, Greene KM, Gao H, Todd CA, Ozaki DA, Seaman MS, et al. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. Journal of immunological methods. 2014;409:131–146. doi: 10.1016/j.jim.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. Journal of virology. 2010;84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaras GD, Binley JM, Gray ES, Crooks ET, Osawa K, Moore PL, Tumba N, Tong T, Shen X, Yates NL, et al. Polyclonal B cell responses to conserved neutralization epitopes in a subset of HIV-1-infected individuals. Journal of virology. 2011;85:11502–11519. doi: 10.1128/JVI.05363-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Wibmer CK, Bhiman JN, Gray ES, Tumba N, Abdool Karim SS, Williamson C, Morris L, Moore PL. Viral escape from HIV-1 neutralizing antibodies drives increased plasma neutralization breadth through sequential recognition of multiple epitopes and immunotypes. PLoS pathogens. 2013;9:e1003738. doi: 10.1371/journal.ppat.1003738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiehe K, Easterhoff D, Luo K, Nicely NI, Bradley T, Jaeger FH, Dennison SM, Zhang R, Lloyd KE, Stolarchuk C, et al. Antibody light-chain-restricted recognition of the site of immune pressure in the RV144 HIV-1 vaccine trial is phylogenetically conserved. Immunity. 2014;41:909–918. doi: 10.1016/j.immuni.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willey RL, Byrum R, Piatak M, Kim YB, Cho MW, Rossio JL, Jr., Bess J, Jr., Igarashi T, Endo Y, Arthur LO, et al. Control of viremia and prevention of simian-human immunodeficiency virus-induced disease in rhesus macaques immunized with recombinant vaccinia viruses plus inactivated simian immunodeficiency virus and human immunodeficiency virus type 1 particles. Journal of virology. 2003;77:1163–1174. doi: 10.1128/JVI.77.2.1163-1174.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta crystallographica Section D, Biological crystallography. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- Yang G, Holl TM, Liu Y, Li Y, Lu X, Nicely NI, Kepler TB, Alam SM, Liao HX, Cain DW, et al. Identification of autoantigens recognized by the 2F5 and 4E10 broadly neutralizing HIV-1 antibodies. The Journal of experimental medicine. 2013;210:241–256. doi: 10.1084/jem.20121977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.