Abstract
The development of the cortex is an elaborate process which integrates a plethora of finely tuned molecular processes ranging from carefully regulated gradients of transcription factors, dynamic changes in the chromatin landscape or formation of protein complexes to elicit and regulate transcription. Combined with cellular processes such as cell type specification, proliferation, differentiation and migration, all of these developmental processes result in the establishment of an adult mammalian cortex with its typical lamination and regional patterning. By examining in-depth the role of one transcription factor, Pax6, on the regulation of cortical development, its integration in the regulation of chromatin state and its regulation by cis-regulatory elements, we aim to demonstrate the importance of integrating each level of regulation in our understanding of cortical development.
Introduction
The mammalian cortex is a region with an exceedingly complicated cytoarchitecture. Distinct regions of the adult cortex execute discrete cortical functions (i.e. visual processing) that are central to cognition. Moreover, it is organized into six layers, which differ in their cellular constituents and connectivity. The cortex possesses two main categories of neurons: excitatory projection neurons, that extend their axons over long distances to cortical and subcortical targets; and inhibitory interneurons, which generally have short axons and regulate local circuits. Excitatory neurons are generated by cortical progenitors, whereas inhibitory neurons are generated by subcortical progenitors in the ganglionic eminences. Dysregulation of cortical histogenesis is central to developmental disorders such as epilepsy, mental deficiency, autism and schizophrenia. Thus, understanding how the cortex develops in non-disease conditions is the first critical step to gaining insights into these disease-states.
Neural progenitors in the ventricular zone (VZ) of the pallium (dorsal telencephalon) generate the projection neurons of the cerebral cortex and hippocampus. Regional patterning of the cerebral cortex occurs during the earliest stages of its development. The dorsal midline (roof plate) and paramedial parts of the telencephalon, produces gradients of WNTs and BMPs regulating multiple processes such as cortical hem formation, choroid plexus induction and dorsal cortical patterning (Monuki et al., 2001; Cheng et al., 2006). In turn, the cortical hem, a region that secretes WNT and BMP subtypes and which is interposed between the hippocampal neuroepithelium and the choroid plexus anlage (reviewed by Subramanian and Tole, 2009), further regulates cortical patterning (Caronia-Brown et al., 2014). The cortical hem also generates Cajal-Retzius cells, an early-generated transient neuron that resides in layer I (Meyer et al., 2002; Bielle et al., 2005). Cajal-Retzius cells secrete the extracellular matrix glycoprotein, Reelin, which regulates radial migration of immature excitatory neurons (Del Río et al., 1997; Förster et al., 2002). FGF signalling (Fgf8, 15 and 17) from the rostral patterning center specifies rostral cortical identity in part through repressing expression of transcription factors that promote caudal fate (e.g. CoupTFI) (Fukuchi-Shimogori and Grove, 2001; Garel et al., 2003; Cholfin and Rubenstein, 2007; 2008). These forebrain-patterning centres are the source of cellular and molecular cues controlling initial cortical patterning.
As development progresses and the pallium is gradually subdivided, and differentiates into discrete regions (reviewed by Mallamaci and Stoykova, 2006). This process is regulated by a host of transcription factors such as CoupTF1, Emx2, Lef1, Lhx2, Pax6 and Sp8, which are expressed in graded patterns along the rostral/caudal (R/C) or ventral/dorsal (V/D) axes of the developing VZ (Bishop et al., 2000; Galceran et al., 2000; Yun et al., 2001; Mallamaci and Stoykova, 2006; Armentano et al., 2007; Sahara et al., 2007; Faedo et al., 2008; Mangale et al., 2008; Chou et al., 2009; Borello et al., 2014). These TFs are among the key patterning molecules of the developing cortex and act to set-up the regionalisation of the cortex. In fact, loss of function of Pax6 and Sp8 lead to the “caudalisation” of the cortex both through the loss of anterior regions of the cortex as well as through an increase in the area occupied by the visual cortex (Bishop et al., 2000; O'Leary et al., 2007; Zembrzycki et al., 2007). By contrast, the loss of Coup-TF1 or Emx2 lead to a reduction in caudal cortical areas in favour of the expansion of frontal regions such as the motor cortex (Bishop et al., 2000; Mallamaci et al., 2000; Bishop et al., 2003; Armentano et al., 2007). Moreover, loss of both Pax6 and Emx2 leads to the drastic reduction of cortical structures in favour of the choroidal roof and subpallium (Muzio et al., 2002a). Regional identity is then translated from the VZ to a secondary progenitor domain called the subventricular zone (SVZ) and then to the cortical plate (CP). Initially, the CP also exhibits gradients of TFs (Rubenstein et al., 1999; i.e. Bhlhb5, Id2, Tbr1 and Tbr2) that are gradually converted to patterns with regional boundaries that are correlated with anatomical and functional regions; there is evidence that these TFs also regulate regional fate (Alfano et al., 2013; Joshi et al., 2008; Bedogni et al., 2010; Elsen et al., 2013; Greig et al., 2013). Later, thalamic afferents contribute to refining cortical areal properties (Chou et al., 2013; Pouchelon et al., 2014).
A central unanswered question is how are the gradients of TFs in the VZ translated into positional information in the SVZ and CP, to generate cortical regions and cortical layers? How are neuronal subtype-specific profiles of gene expression established both at the right time and at the right location? How are cellular subtypes able to establish these complex gene expression profiles in response to extracellular signals and cell intrinsic programs? Transcription regulation undoubtedly plays a key role in regulating these processes. Thus, it is important to define the transcription factors (TF) and the network of chromosomal components that they bind to and/or regulate (enhancers, promoters, and downstream transcription units) that participate in the transcriptional circuits that control corticogenesis. In order to elucidate this circuitry, it is essential to identify both the TFs and gene regulatory elements that control gene expression at various stages of cellular maturation. Furthermore, the identification of enhancer elements is essential for elucidating potential loci where human mutations may impact cortical development and lead to neuropsychiatric disorders. Herein, we will use Pax6 as an example to address how TFs exert their effect such as the regulation of cortical development. We will next review current knowledge of how chromatin dynamics and other epigenetic mechanisms play a role in regulating the expression and the downstream effects of TFs such as Pax6. Finally, we will discuss how cis-regulatory elements control TF expression (i.e. Pax6) and how they are in turn regulated by said TF.
1. Pax6 and evolution
Transcription factors are DNA-binding proteins that play a variety of roles in cortical development including establishing patterning, regulating neurogenesis and conferring cell subtype identity. They act in a cell-type specific manner to either promote or repress the transcription of their target genes.
Members of the paired-box (Pax) gene family are part of an evolutionarily conserved family of transcription factors that possess a DNA-binding domain known as the Paired domain (reviewed by Gruss and Walther, 1992). A subgroup of the Pax genes also encode a homeodomain (e.g. Pax6). The Pax family is comprised of 9 members (Pax1–9) which are expressed in several organs where they play roles in multiple processes including cell fate determination and cell proliferation (Chi and Epstein, 2002; Eccles et al., 2002). Pax6, in particular, seems to have evolved in higher metazoans from an ancestral gene termed PaxB (Kozmik et al., 2003). This ancestral gene contained a Pax2-like paired domain and octapeptide as well as Pax6-like homeodomain; its duplication may have given rise to Pax2 and Pax6 (Kozmik et al., 2003).
Pax6 was first studied for its role in ocular formation when it was shown that human aniridia could be attributed to a member of the paired-box family (Ton et al., 1991). This same gene was subsequently shown to be one and the same as the gene causing a loss of eye phenotype in the various spontaneous “small-eye” mouse mutants Sey (Hill et al., 1991). Pax6 homologues have been found in all Bilaterians that have been studied thus far including insects (i.e. eyeless and twin of eyeless in Drosophila), fishes (i.e. Pax(zf-a) in zebrafish) and mammals as well as nematodes, squids and annelids (reviewed by Gehring, 2004). Pax6 is a highly evolutionarily conserved protein, with over 90% conservation between the amino acid sequence of the DNA-binding domains of Pax6 and eyeless (Quiring et al., 1994). The association of Pax6 and its orthologues with eye development has been extensively studied and reviewed (Gehring and Ikeo, 1999; Kozmik et al., 2003; Gehring, 2004). In the embryonic eye, Pax6 is essential in the earliest stages of eye induction in vertebrates and invertebrates - and ectopic expression of Pax6 elicits lens formation in aberrant tissues (Altmann et al., 1997; Chow et al., 1999). Moreover, induction of an ectopic eye in Drosophila can be established using Pax6 homologues from other species thus underlining the molecular conservation of this transcription factor (Kozmik et al., 2003). Although Pax6 is often credited as a contributor to the evolution of the eye, it is important to keep in mind that a homologue of Pax6 has been discovered in the sea urchins, which do not have eyes or a brain (Czerny and Busslinger, 1995; Agca et al., 2011).
Moreover, Pax6 regulates the development of a variety of tissue types apart from the eye including the cortex, diencephalon, spinal cord, and pancreas. Pax6 has been shown to be required for the development of other structures in the anterior head. In Drosophila, which have four Pax6-like genes, mutation of one of them (Toy), fails to develop a head (Kronhamn et al., 2002). Likewise, in the nematode C. elegans, mutants for the Pax6 orthologue, vab-3, exhibit gross head dysmorphorgenesis (Chisholm and Horvitz, 1995). Since heterozygous mutations in PAX6 are at the heart of a number of abnormal phenotypes both in humans and in mice, the molecular functions of Pax6 and its different mutants have been the focus of many studies in the last 30 years (See Figure 1). Reviews on the roles of Pax6 were recently published (Osumi et al., 2008; Georgala et al., 2011a; Shaham et al., 2012) and thus, in this article, we will focus on a few recent studies to highlight how Pax6 acts to regulate cortical development.
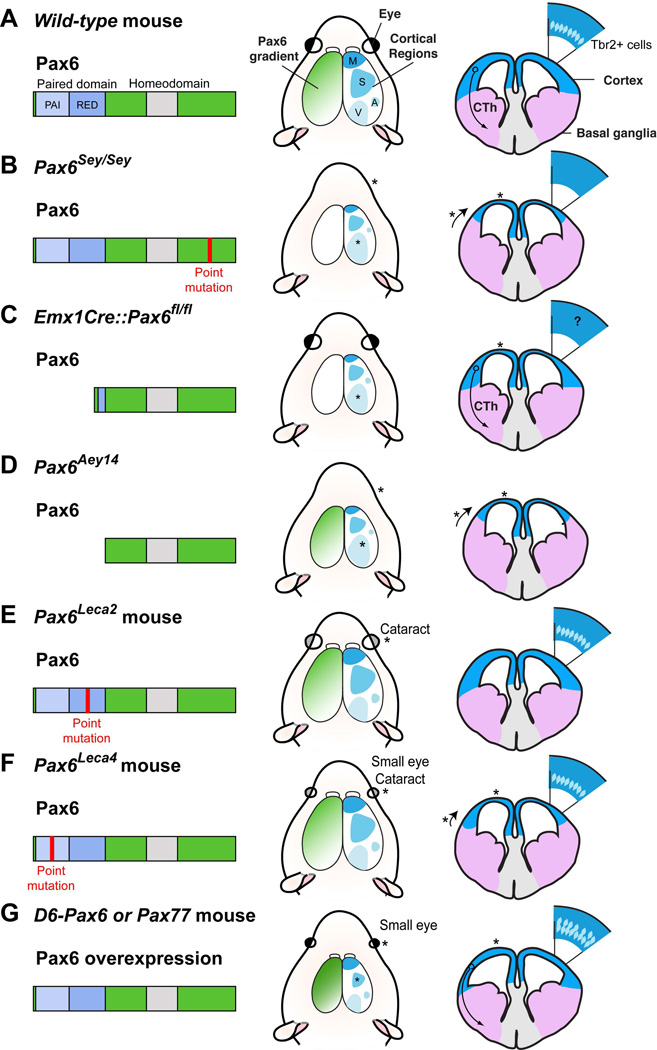
Figure 1.
A) In wild-type mice, the Pax6 protein possesses a homeodomain and a paired domain which can be subdivided into PAI and RED subdomains. The transcription factor is present in the cortex in a graded expression pattern which is necessary for appropriate cortical development. This includes the patterning of cortical regions such as motor (M), somatosensory (S), auditory (A) and visual (V) cortex. Moreover, the ventro-dorsal pallial (blue in drawing on right) and subpallial (pink) boundary is appropriately positioned. Finally, a corticothalamic projection (CTh) and Tbr2+ basal progenitors can be observed. Note also the correct formation of the eye in the presence of Pax6. B–E) Pax6 mutants have mutations in different subdomains (depicted on the Pax6 structure schematic) which result in a large array of cortical and ocular phenotypes (denoted by asterisks). B) The Pax6Sey/Sey mouse has a smaller and thinner cortex, patterning defects of both cortical regions and the pallial-subpallial boundary and a loss of the CTh projection as well as of Tbr2+ cells in the SVZ (Hill et al., 1991; Stoykova et al., 1996). C) The Pax6Aey14 mouse has the same phenotypes as the Pax6Sey/Sey mouse although it is unclear whether the CTh and Tbr2+ cells are also affected (Haubst et al., 2004). D) The Pax6Leca2 mutant has no obvious cortical phenotype (though the CTh projection was not described) but has some ocular defects in the form of cataracts (Walcher et al., 2013). E) The Pax6Leca4 mutant has a thinner cortex and some ventral cortical patterning defects as well as a small eye phenotype (Walcher et al., 2013). F) Both D6-Pax6 and Pax77 mice overexpress Pax6 and have a thinner cortex, an increase in Tbr2+ cells/expression and a small eye phenotype (Manuel et al., 2007; Sansom et al., 2009; Georgala et al., 2011b). In addition, the Pax77 mouse has a smaller somatosensory area and retains the CTh projection (Manuel et al., 2007).
2. Pax6: A master regulator of cortical development
Pax6 is necessary for proper cortical development where it regulates patterning (Stoykova et al., 1996; Toresson et al., 2000; Yun et al., 2001), neuronal migration (Schmahl et al., 1993; Chapouton et al., 1999; Nomura and Osumi, 2004), proliferation (Gotz et al., 1998; Heins et al., 2002), neurogenesis (Heins et al., 2002; Hack et al., 2004; Haubst et al., 2004) and, by preventing an early-depletion of the cortical progenitor pool, the specification of upper-layer neurons (Tarabykin et al., 2001; Tuoc et al., 2009). Moreover, as for eye specification, Pax6 appears to be sufficient to specify neuronal lineage. Indeed, expression of Pax6 in non-neuronal cell types, such as astroglia, can reprogram these cells to form neurons (Heins et al., 2002; Berninger et al., 2007). In addition, Pax6 in conjunction with Foxg1, another regulator of cortical development, was shown to be able to induce a direct and rapid conversion of “primed” mouse embryonic fibroblasts into neural precursor cell-like (NPC-like) cells (Raciti et al., 2013).
In terms of patterning, Pax6 is expressed in the progenitor cells, or radial glia cells, of the VZ in the developing pallium in rostral to caudal and ventral to dorsal gradients where its presence is necessary for conferring a rostral-ventral identity to the developing cortex. Pax6 mutants (Pax6Sey/Sey) have a point mutation in the Pax6 gene that generates a null allele. As a result, they die perinatally with a plethora of abnormalities including in the telencephalon (Hill et al., 1991) (Stoykova et al., 1996) (See Figure 1). Their phenotype is similar to that of Pax6 null animals and therefore, both Pax6−/− and Pax6Sey/Sey can be used interchangeably to study the effect of Pax6 loss (St-Onge et al., 1997; Jones et al., 2002). In the absence of a functional PAX6 protein, the establishment of the pallial-subpallial boundary is compromised (Carney et al., 2009). Therefore, in Pax6 knockouts, genes characteristic of subpallial domains (such as Gsx2, Mash1, Dlx) are expressed ectopically in the dorsal forebrain and neurons that normally become glutamatergic adopt a GABAergic cell-fate (Stoykova et al., 2000; Toresson et al., 2000; Yun et al., 2001; Kroll and O'Leary, 2005). Moreover, rostrocaudal patterning of the pallium is disorganised in the Sey mouse (Bishop et al., 2000) (Muzio et al., 2002b). In fact, in the Pax6 mutants, the caudal areas shift forward whereas the rostral areas are very reduced, in contrast to Emx2 and CoupTFI mutants (Muzio et al., 2002a; b; Armentano et al., 2007; Faedo et al., 2008; Borello et al., 2014). Moreover, when Pax6 is specifically removed in the cortex in Emx1::Cre; Pax6Fl/Fl mice, cortical arealization is perturbed with a general rostral shift of the visual/somatosensory and somatosensory/motor boundaries (Piñon et al., 2008). Moreover, the cortex of the conditional knockouts exhibited abnormalities in lamination and the somtatosensory map in the barrel fields of the mutants was both reduced in size and incomplete (Piñon et al., 2008; Zembrzycki et al. 2013). However, despite these gross regionalisation defects, both the thalamocortical and corticofugal connections appeared to form correctly (Piñon et al., 2008). Interestingly, the selective reintroduction of Pax6 in the cortex of the Emx1::Cre; Pax6Fl/Fl mice was able to partially rescue the size and positioning of part of the cortical body map demonstrating that Pax6 is crucial for the specification of S1-area identity in cortical progenitors (Zembrzycki et al. 2013).
Lastly, the Pax77 mutant, which carries several copies of the human PAX6 locus (including its regulatory upstream and downstream regions) in the endogenous mouse region, does not display any overt aeralization phenotype, except for a reduction in the size of the somatosensory cortical area, though the mutant reveals that overexpression of Pax6 does lead to a dysregulation of cell cycle late in corticogenesis with an increase in cell cycle length and cell cycle exit (See Figure 1) (Manuel et al., 2007; Georgala et al., 2011b). The latter studies, amongst others, demonstrate that in order to exert its effects appropriately, the dosage of Pax6 must be tightly regulated (Schedl et al., 1996; Sansom et al., 2009). It therefore demonstrates why heterozygous mutations of Pax6 cause disease (Prosser and van Heyningen, 1998).
The Pax6 protein contains a paired domain and a homeodomain that recognise different consensus binding sites (Ton et al., 1991). It was previously shown that the homeodomain is crucial in regulating lens formation and retinal specification but has no clear effect on forebrain development (Haubst et al., 2004; Ninkovic et al., 2010). Conversely, Pax6Aey18 mice that have a deletion of the paired domain severely affect several aspects of forebrain development, including cortical patterning, thereby providing evidence that the paired domain functions to regulate the development of the telencephalon (See Figure 1)(Haubst et al., 2004). The paired domain can be subdivided into a C-terminal RED subdomain and an N-terminal PAI subdomain (Epstein et al., 1994; Yamaguchi et al., 1997). The N-terminal PAI subdomain is functionally blocked in an alternative transcript of Pax6 known as Pax6(5a) in which exon5a is expressed in lieu of the traditional exon5 (Haubst et al., 2004) During early neurogenesis in the developing telencephalon (E12.5), “conventional” Pax6 trancsripts have been shown to be around six times more abundant than Pax6(5a) transcripts (Pinson et al., 2005). These ratios drop as the cortex matures and by E14.5, there is a 1:2 ratio of Pax6 to Pax6(5a) transcripts suggesting that these two isoforms play different roles in cortical processes. In fact, both subdomains of the paired box (RED and PAI) have previously been shown to bind to different consensus sequences (P6CON and 5aCON respectively) while the consensus paired domain can bind both (Epstein et al., 1994; Yamaguchi et al., 1997). Interestingly overexpression of Pax6(5a), using retroviral infection of cortical progenitors cells in an In vitro model, inhibited cell proliferation but didn’t affect cell fate (Haubst et al., 2004). A recent study took advantage of the following homozygous mutants to continue dissecting out the roles of the Pax6 paired domain: Pax6Sey mutants, Pax6Leca4 with a substitution in the PAI domain, and Pax6Leca2 with a substitution in the RED subdomain (See Figure 1) (Walcher et al., 2013). This study showed that mutation in the PAI domain reduced cell proliferation whereas mutations in the RED domain had the opposite effect. The opposite effects of these neighbouring subdomains most likely contribute to the fine-tuning of proliferation by Pax6 in cortical progenitors and possibly explains why some studies have shown that Pax6 promotes or inhibits cell proliferation in the developing cortex (Holm et al., 2007; Osumi et al., 2008; Sansom et al., 2009). Moreover the Pax6Leca4 mouse showed similar impairment in neurogenesis as in the Pax6Sey mutants. Interestingly, neither Pax6Leca2 nor Pax6Leca4 mutant had the severe cortical patterning defects that are observed in the Pax6 complete loss of function alleles. This suggests that the function of the remaining Pax6 subdomain, either RED or PAI, is sufficient to promote the correct patterning of the cerebral cortex despite proliferation and neurogenesis defects. In addition, deletion of Pax6(5a) had no overt effect on neurogenesis, proliferation or patterning of the cortex, possibly because Pax6 transcripts are overexpressed in Pax6(5a)−/− mice (Haubst et al., 2004). Taken together, the studies of the Pax6 subdomains show that its modular structure enables Pax6 to perform distinct (and at times opposing) functions using discrete domains and subdomains for regulating patterning, neurogenesis and proliferation (See Figure 1). Recently, the Cvelk lab pursued the dissection of the roles of the different Pax6 subdomains by studying the binding abilities of several missense PAX6 and PAX6(5a) mutants to bind to a panel of Pax6 consensus sequences recognised by different Pax6 subdomains (Xie et al., 2014). This study showed that, depending on the combination of mutations and binding sequences, Pax6 mutants acted as gain-, neutral- or loss-of function mutations thereby highlighting the complexity of this TF in regulating gene transcription.
A surprising study uncovered that, in the forebrain, in order to regulate cortical development, the coding region of Pax6 could be replaced by that of Pax2 or Pax6(5a) and still rescue cortical phenotypes associated with Pax6 knockout (though eye phenotypes were not rescued in either scenario) (Carbe et al., 2013). When Pax6(5a) was expressed in lieu of Pax6, dorsoventral patterning and progenitor proliferation defects were normalised though defects at the ventral boundary persisted. Moreover, inserting Pax2, which lacks a homeodomain, in the Pax6 locus resulted in a rescue of forebrain patterning and neurogenesis. This study further strengthened the case for the necessity of a functional “paired-domain” for proper cortical development (Carbe et al., 2013).
Taken together, the studies described above show that a given TF, such as Pax6, exerts different effects depending on its DNA binding domain and moreover, that the regulation of its expression is crucial to ensuring that its effects are targeted. That being said, endogenously, Pax6 is present both in the developing eye and in the developing forebrain. Why doesn’t Pax6 activate genes specific for eye development in the forebrain? Clearly, the regulation of its expression is not sufficient to explain the differential transcriptional networks activated in different tissue types. In addition to tissue-specific expression of other TFs, this difference may in part be explained by differential access of Pax6 and other TFs to gene promoters and cis-regulatory elements in each tissue. As it happens, a study by Xie and colleagues revealed that Pax6 preferentially binds to the promoter regions of its target genes in a tissue-specific manner (Xie et al., 2013). In fact, when comparing Pax6-bound regions, as identified by ChIP-Chip in the neonatal lens and prenatal forebrain and pancreas, an overlap of only 20% of bound-regions was found between these tissue types, suggesting that most Pax6 downstream targets are tissue-specific (Xie et al., 2013).
3. How is the chromatin landscape remodelled to allow or inhibit access by a given TF?
In eukaryotes, genomic DNA is tightly wrapped around a histone protein core to form a macromolecule known as chromatin, which serves to condense DNA and regulate its transcription. This chromatin can dynamically switch conformations from a structurally open state associated with transcriptional activation and a structurally condensed state linked to transcriptional repression. This was shown elegantly in a recent study that combined fluorescence in situ hybridisation with super-resolution structured illumination microscopy (Patel et al., 2013). In this study, the authors found that, as development of the neural tube progresses, chromatin around the Pax6 locus exhibited signs of decompaction coinciding with transcriptional onset. Further, in nearby somites, which do not express Pax6, this chromatin decompaction did not occur.
The regulation of the dynamic switch of chromatin states is encoded by epigenetic mechanisms (reviewed by Nord et al., 2015). These include DNA cytosine methylation as well as post-translational chemical modifications of histones, that alters their tertiary structure, by such as processes as acetylation, methylation, phosphorylation, sumoylation, and ubiquitylation (reviewed by Kouzarides, 2007 and Nord et al., 2015). In addition, small non-coding RNAs can also modify transmissible changes in gene expression without affecting the actual DNA sequence. Cortical development is rife with examples wherein epigenetic processes play a decisive role in regulating neurogenesis, proliferation, cell specification and other important functions (reviewed by Hsieh and Gage, 2004; Wu and Sun, 2006; Mehler, 2008; Kim and Rosenfeld, 2010; Mahgoub and Monteggia, 2013; MuhChyi et al., 2013).
3.1. Chromatin remodelling
Several studies have shown that certain chromatin regulators strongly impact neuronal specification during cortical development, such as histone-lysine N-methyltransferases (e.g. Ezh2) (Pereira et al., 2010) or histone deacetylases (Juliandi et al., 2012). Fewer studies have so far looked at the effect of transcription factors on chromatin modification. Yet, Pax6 has been shown to bind several chromatin modifiers. Indeed, in the pancreas, Pax6 interacts with the transcriptional co-activator p300 (a histone acetyltransferase) to promote transcription of proglucagon by transactivating its G1 promoter element (Hussain and Habener, 1999). This interaction is stabilised by the binding of another TF known as Cdx2. The adapter protein p300, and its closely related associate CBP, are histone acetyltransferases that open the chromatin structure around promoters and enhancers (reviewed by Sheikh, 2014). Moreover, CBP/p300 are able to recruit the basal transcriptional machinery including RNA polymerase II in order to directly promote transcription of a gene (reviewed by Sheikh, 2014). By binding p300, Pax6 could thus both influence the chromatin structure of its target genes and enable their transcription.
Further, Pax6 has interactions with several subunits of the vertebrate SWI/SNF (switch/sucrose nonfermentable) complex, or BAF (Brg1/Brm-associated factor) complex, which regulates chromatin remodelling (reviewed by Wen et al., 2009; MuhChyi et al., 2013; Ronan et al., 2013). The BAF complex contains a core ATPase subunit (Brg1 or Brm), invariant core subunits (BAF47, BAF155 or BAF170) and other subunits that are lineage-specific. In the developing cortex, co-immunoprecipiation experiments determined that Pax6 can bind to BAF170, BAF155 as well as Brm; it can also recruit BAF complexes containing these subunits to its target promoters (Tuoc et al., 2013). BAF170, was shown to specifically repress several genes typically activated by Pax6, such as Tbr2, Cux1 and Tle1, by recruiting the REST (RE1-silencing transcription factor)-corepressor complex (Tuoc et al., 2013). Pax6 was also shown to interact directly with BAF155 resulting in the elimination of REST-corepressor complex on Pax6 target gene promoters. More importantly, the presence of BAF155 was linked to the induction of euchromatin state at Pax6 target loci as measured by a decrease in DNA methylation and in the repressive histone mark H3K27Me3 and by an increase in the active histone mark H3K9Ac (Tuoc et al., 2013). During cortical development, BAF170 and BAF155 subunits are both present in early progenitors but, by E15.5, BAF170 is nearly absent from these progenitors while the expression BAF155 subunit is maintained. It therefore appears that the balance of Pax6-BAF170 to Pax6-BAF155 complexes can regulate the activation and repression of Pax6 targets in development. Pax6 regulates the specification of upper-layer neurons by regulating progenitor cell-cycle in order to maintain the cortical progenitor pool (Tarabykin et al., 2001; Tuoc et al., 2009). In this context, the substitution of repressive Pax6-BAF170 complexes by activating Pax6-BAF155 complexes during development can perhaps serve to regulate the timing of certain Pax6 target genes and in particular those genes necessary for upper-layer specification. In the cortex, Pax6 was not shown to recruit Brg1 complexes to its targets. In adult neural stem cells, Pax6 directly interacts with the Brg1-containing BAF complex and, in the absence of Brg1, Pax6 loses its neurogenic activity (Ninkovic et al., 2013). Similarly, in the developing lens, Pax6 directly recruits Brg1-complexes to the crystallin gene locus (Yang et al., 2006).
The ability of Pax6 to interact directly with chromatin modifiers (that are themselves regulated in a spatio-temporal manner) is crucial in determining which downstream targets are accessible to Pax6. At E15, microarray data indicate that the chromatin remodeller SatB2 and the DNA methyltransferase Dnmt3a are differentiatlly expressed in the cortex of Pax6Sey/Sey versus wild-type mice (Holm et al., 2007). Further work needs to be carried out to determine whether Pax6 directly controls the transcription of these genes or whether the observed differential regulation occurs through an indirect mechanism.
In addition, chromatin modifiers regulate Pax6 expression during brain and eye development. For instance, the histone demethylase, Jarid1b, controls the levels of H3K4me3, a histone modification that is associated with active transcription (reviewed by Kouzarides, 2007). In the absence of Jarid1b in embryogenesis, H3K4me3 marks accumulate on developmental genes that should not be active (Albert et al., 2013). For instance, there is a global increase of H3K4me3 on Pax6 in Jaridb1 knockout brains and a concomitant increase in Pax6 expression. Moreover, Jaridb1 null animals have eye defects reminiscent of those present in Pax6Sey mice. In addiiton, during human pluripotent cell differentiation, one of the subunits of the BAF chromatin remodeller complex, SNF5, plays the role of activating OCT4-repressed target genes such as Pax6, thus regulating the balance between pluripotency and differentiation (You et al., 2013).
Interestingly, in the cortex, new evidence has uncovered that an early response transcription factor binds in an activity-dependent manner to cis-regulatory elements to regulate transcription of late-response genes (Malik et al., 2014). Indeed, in postnatal cortical neurons, membrane depolarization induces a rapid change in the number of H3K27Ac peaks (Malik et al., 2014). Moreover, increased H3K27Ac around gene loci such as the Fos locus correlated with an increase in transcription from these loci including an increase in Fos mRNA thus raising the possibility that, for activity-dependent genes, an increase in H3K27Ac is necessary to activate their enhancers and promote their transcription. This study reveals a potential mechanism whereby gene transcription is regulated rapidly, at the chromatin level, by extrinsic cues and in the context of development, it serves as a proof of concept that as differentiation progresses, chromatin state and transcription can undergo rapid alterations.
3.2. DNA methylation
DNA methylation is a type of epigenetic modification that can regulate gene expression (typically repression). A recent study compared epigenetic marks of TF occupied DNA sequences (OSs) and their orthologue sequences in a variety of cell lines (Cheng et al., 2014). When TF occupancy between the OS and its orthologues was comparable, methylation profiles were also similar; however, when TF occupancy was not conserved in OSs and their orthologues, methylation levels were significantly increased in the unbound orthologous sequences (Cheng et al., 2014). This confirms the general theory that DNA methylation is usually indicative of a transcriptionally repressed state whereas hypomethylation denotes a transcriptionally active state.
A recent focus on DNA methylation profiles in the cortex has been driven by the realization that in patients with disorders such as schizophrenia and depression, methylation profiles are altered in the cerebral cortex (reviewed by Akbarian, 2010; Nabeshima and Kim, 2013). The Pax6 promoter is frequently methylated in a variety of cancers including bladder, gastric and invasive breast cancer; and this is often associated with the silencing of PAX6 and poor prognosis for the patient (Salem et al., 2000; Moelans et al., 2011; Yang et al., 2013). To our knowledge, the methylation profile of Pax6 has not yet been assessed in the developing cortex. Nonetheless, a recent study looked at Pax6 expression and methylation in a model in which mouse embryonic stem cell are differentiated into radial glial cells (Gao et al., 2011). They found that Pax6 expression was regulated by CTCF, a zinc finger protein, which acts as a barrier between the Pax6 promoter and an adjacent cis-regulatory element by binding to either one of five functional and specific motifs in a repressor element located within its promoter region (Wu et al., 2006). CTCF was previously shown to negatively regulate Pax6 levels in ocular tissues (Li et al., 2006). The regulation of Pax6 by CTCF was dependent on DNA methylation; in fact as differentiation into radial glia progressed, the repressor element within the Pax6 promoter was methylated which inversely correlated with CTCF occupancy (Gao et al., 2011). In this case, since methylation occurred on a repressor region of the Pax6 locus, methylation acted to increase transcription of Pax6. Further work will need to focus on examining the methylation profile of the Pax6 locus in radial glial cells during cortical development and specifically dissect out its effect on the specific cis-regulatory elements present within the Pax6 regulatory region.
Pax6’s ability to transcriptionally regulate its downstream targets is enhanced by its capacity to interact with several chromatin modifiers such as p300 and members of the BAF complex (see above, section 2.1). Being able to regulate the methylation/demethylation of its targets would further add to the arsenal of tools that this TF possesses to exert its effects on pallial development. Remarkably, upon electroporation of BAF170 into E13.5 cortices, CpG methylation of two Pax6 downstream targets, Tle1 and Cux1, was increased (Tuoc et al., 2013). This effect was largely rescued upon electroporation of BAF170/BAF155, indicating that BAF155 can act to negatively regulate BAF170-promoted DNA methylation. Since Pax6 and BAF155 can interact, perhaps Pax6 can also indirectly regulate the methylation state of its downstream targets.
3.3. Regulation by non-coding RNA
Pax6 exerts its effects by binding to enhancers but also by regulating non-coding RNA elements. In fact, in the developing lens, Pax6 was shown to co-regulate Tprm3 and the intronic microRNA gene mir-204, which is a negative regulator of gene expression during eye development (Shaham et al., 2013). Micro-RNAs are involved in the post-transcriptional repression of transcripts that possess an miRNA-specific sequence in their untranslated regions (Grishok et al., 2001; Lewis et al., 2005). By regulating mir-204, Pax6 could in effect rapidly repress a large number of transcripts possessing the appropriate miRNA-specific sequence. In addition, in a neuroblastoma cell line, N2A, a long non-coding RNA (lncRNA) named Paupar was shown to negatively regulate Pax6 expression (Vance et al., 2014). Paupar is located adjacent to the Pax6 locus and is ultraconserved across species. Interestingly, not only does Paupar regulate Pax6 expression, it also appears to generally associate with many other loci in the genome. These include loci whose transcription Paupar regulates; and at some of which there appears to be Paupar-Pax6 co-occupancy (Vance et al., 2014). This study provides evidence that this lncRNA might be crucial for neural development and it will be noteworthy to uncover its in vivo relevance to Pax6 function and cortical development. Finally, Pax6 was shown to regulate the expression of mir-135b during differentiation of human ESCs into neuroectodermal cells and mir-135b was subsequently shown to promote neural differentiation of humans ESCs (Bhinge et al., 2014).
Pax6 may in turn be regulated by non-coding RNA. Indeed, a bioinformatics analysis of mir-92b binding sites predicted that this mRNA could bind to the 3’UTR of Pax6 in the embryonic cortex (Nowakowski et al., 2013). Postnatally, Pax6 protein continues to be expressed in a high dorsal-low ventral gradients in progenitors situated along the walls of the lateral ventricles which generate olfactory bulb interneurons (de Chevigny et al., 2012). This Pax6 gradient is necessary for the correct generation of olfactory dopaminergic interneurons and it was shown that it is regulated at the post-translational level by mir-7a. Mir-7a, which is expressed in a low dorsal-high ventral gradient, binds to the 3’UTR of Pax6 post-transcriptionally and inhibits the synthesis of the PAX6 protein in vivo (de Chevigny et al., 2012).
Thus the epigenetic mechanisms that regulate transcription through altering chromatin state or DNA methylation, play major roles in regulating development. Furthermore, Pax6 can modulate the chromatin state of its target genes. In addition Pax6, in concert with DNA methylation and non-coding RNA can regulate Pax6 and its downstream targets. As research progresses, it will become increasingly apparent that the regulation networks that control epigenetic mechanisms in development are exceedingly complex and probably involve multiple checkpoints and partially redundant pathways. This tight control of chromatin state and of transcription is necessary to ensure that TFs can bind to their downstream targets (the enhancers and promoters of the genes that they control) in a temporal and spatially restricted manner so that appropriate developmental programs are enabled and off-target effects are limited.
4. Identification of enhancers that are active during cortical development
Enhancers are regulatory sequences (non-coding DNA) that can be located long distances from promoters and that selectively regulate gene transcription. The discovery of enhancers that are active during cortical development has gained momentum as whole genome sequences became available (enabling comparative genomic analyses that identifies conserved non-coding domains), and following the success of next-generation sequencing technologies (e.g. ChIP-Seq for active chromatin marks, such as AcH3K27). Enhancer sequences are bound by multiple transcription factors and thus serve as modules to regulate transcription of their associated gene in a finely tuned spatiotemporal manner. In addition, mutations in enhancers are implicated in human diseases (reviewed by Nord et al., 2015).
4.1. Pax6 binds to promoters and enhancers of downstream targets
Gene expression changes in the Pax6 mutants, assayed by in situ or microarray experiments, have shown that Pax6 directly regulates or indirectly affects the transcription of a large number of downstream targets including TFs, cell adhesion molecules, signal transduction proteins, and axon guidance molecules (Holm et al., 2007; Visel et al., 2007). A sizeable proportion of TFs that are dysregulated in the mutants’ cortex at E12 included TFs involved in neurogenesis, patterning and differentiation, such as Ngn2, AP2-γ, Tbr2, Tbr1, SatB2 and NeuroD6 (Holm et al., 2007). Hierarchical clustering of the annotated expression patterns of in situs in the developing E14.5 mouse cortex, revealed a potential Pax6 regulatory network comprised of 80 genes (Visel et al., 2007). Within this set, 14 genes (collectively possessing 27 consensus Pax6 binding sites) were shown by electromobility shift assays to bind to Pax6 including six TFs: Arx, NeuroD1, NeuroD4, Neurod6, Ngn1 and Ngn2.
A ChIP-Chip study by Sansom et al. (2009) revealed that Pax6 was bound to 1560 promoters in the mouse cortex at E12.5 (Sansom et al., 2009). A further ChIP-on-ChIP study established that Pax6 occupies approximately 2484 promoters in the mouse cortex at E15.5 (Xie et al., 2013). At E12.5, gene ontology analysis of the Pax6-bound genes showed enrichment for genes involved in cell cycle regulation, neurogenesis, proliferation and neuronal differentiation (Sansom et al., 2009). Interestingly, Pax6 was found to bind to the promoters of many TFs expressed in radial glia progenitors including the patterning genes CoupTF2, Emx2 and Pax6 itself (Sansom et al., 2009). Additionally, Pax6 bound the promoters of TFs expressed in basal progenitors such as Tbr2 and Neurod1 as well Hmga2, a member of a protein complex that regulates transcription at enhancers. The authors then compared this dataset with microarray datasets obtained on E12.5 cortices in gain-of-function D6-Pax6 mice (which constitutively overexpress Pax6 in the VZ of the developing cortex) and loss-of-function Pax6Sey/Sey mice. For the large part (78% of cases), the binding of Pax6 to a promoter region did not correlate with changes in the genes expression in gain- and loss-of-function mutants and vice-versa. This indicates that the transcription of some genes are regulated indirectly or that they might be regulated by binding of Pax6 to more distal enhancers (Sansom et al., 2009).
As described in Visel et al. (2013), we and our collaborators discovered and characterized 145 enhancers that are active in the E11.5 telencephalon. We fate mapped the activity of 14 of the cortical enhancers, using CreER stable transgenic lines that we generated. The results provide evidence for a progenitor zone protomap of the cortex (including the frontal cortex) that is of higher resolution than of the expression patterns of known transcription factors (Pattabiraman et al., 2014). Using ChIP-Seq, we identified COUPTF1 and PAX6 binding sites on many of these enhancers (Pattabiraman et al., 2014). ChIP-Seq for Pax6 yielded a few thousand peaks at E12.5 and E13.5 and amongst those, approximately 9% were within 5kb of the nearest gene’s transcriptional start site. The distribution of Pax6 binding sites on the genome therefore suggests that Pax6 predominantly binds to distal regulatory elements rather than to promoter regions. Further analysis was carried out on some of the previously characterized enhancers (Visel et al., 2013) and it was shown that four enhancers were activated by Pax6 in luciferase assays: hs636, hs643, hs840 and hs1172; these are enhancer elements located near Rrsrc1, Dmrta1, EphA5 and CoupTFI respectively (Pattabiraman et al., 2014). Moreover, when crossed with Pax6Sey/Sey, the activity of hs840 and hs636 was greatly reduced in the developing palllium providing further evidence that these enhancers are downstream targets of Pax6 in vivo (Pattabiraman et al., 2014).
Pax6 was also found to bind and transactivate a number of other enhancers. For instance, the LATE enhancer upstream of Neurogenin1 requires Pax6 in order to maintain its telencephalic expression in mice (Blader et al., 2004). Moreover, the Neurogenin2 enhancer, E1, also requires Pax6 to retain its endogenous expression in the lateral cortex and in the spinal cord (Scardigli et al., 2001). In addition, this enhancer was shown to possess a low-affinity Pax6 binding site which, when mutated, also led to the loss of E1 expression in the developing cortex (but not in the spinal cord) (Scardigli, 2003). Remarkably, inserting a high-affinity binding site for Pax6 in E1, or increasing the levels of Pax6, resulted in ectopic E1 expression with an expanded domain of expression in the developing cortex (Scardigli, 2003). Both Neurogenin1 and Neurogenin2 had previously been shown to depend on Pax6 for expression in cortical progenitors (Stoykova et al., 2000; Toresson et al., 2000; Yun et al., 2001). Finally, Pax6 was also shown to bind to a FoxP2 enhancer, ECR1, using ChIP in 28-hpf zebrafish embryos (roughly equivalent to mouse E11.5) (Coutinho et al., 2011). The ECR1 enhancer drove reporter expression in the fish forebrain in the same pattern as FoxP2 and in the mouse at E11.5. This expression was downregulated by mutating the Pax6-binding site in ECR1, by morpholino injection of Pax6a and Pax6b, zebrafish paralogs of Pax6, as well as in Pax6 null mice (Coutinho et al., 2011).
In other tissue types, Pax6 was found to bind enhancers of several genes. In the, optic vesicle and lens placodal surface area, Pax6 bound to the N3 enhancer of Sox2 (Inoue et al., 2007), in the mid-gestational neural tube Pax6 bound to the D and E elements of the Hoxd4 cis-regulatory region (Nolte et al., 2006) and Pax6 bound to several sites of the Six6 enhancer in the prenatal diencephalon and several ocular structures (Tétreault et al., 2009). In the latter study, Pax6 and Lhx2 were both shown to synergistically activate Six6. Other studies have also shown that Pax6 requires a binding partner in order to exert its effects. For instance, Pax6 and Pdx1 form a complex to bind and activate the somatostatin enhancer in the pancreas (Andersen et al., 1999). Moreover, Pax6 also forms a molecular complex with Sox2 to bind to the δ-crystallin lens enhancer, DC5 (Kamachi et al., 2001). In addition, Pax6 binds to an ectoderm-specific enhancer (LE9) of the Pax6 locus in order to regulate its own expression (Aota et al., 2003). Indeed, Aota and colleagues (2003) showed that Pax6 could bind to the enhancer alone to elicit its activity, but that Pax6 could form complexes with Sox2 and Sox3 to enhance the regulation of this enhancer(Aota et al., 2003)
4.2. Pax6 cortical enhancers
The strict spatio-temporal control of PAX6 expression is regulated by multiple enhancer elements located around the Pax6 locus. The first cis-regulatory elements controlling PAX6 expression were described in human patients with aniridia bearing chromosomal breakpoints and rearrangements downstream of Pax6 (Fantes et al., 1995; Lauderdale et al., 2000; Kleinjan et al., 2001). Several cis-regulatory elements that regulate Pax6 expression in the eye have since been described (Williams et al., 1998; Xu et al., 1999; Griffin et al., 2002; Kleinjan et al., 2004; 2008; McBride et al., 2011; Bhatia et al., 2013; 2014). An innovative approach recently identified two new cis-regulatory elements around the Pax6 locus by looking at the conservation of ancient conserved non-coding elements (aCNES) which are defined as evolutionarily conserved sequences found in multiple contemporary species that are evolutionarily very divergent (Bhatia et al., 2014). In this study, the scientists compared the Pax6 locus of an elephant shark and that of humans and discovered new ocular enhancers for Pax6.
The first Pax6 cortical enhancer was discovered in its promoter region, between the two alternative promoter regions P0 and P1 (Kammandel et al., 1999). This 5kb fragment is active in the developing mouse cortex, hindbrain and spinal cord. By extending the upstream and downstream sequence of this enhancer, the authors showed that the enhancer could more accurately recapitulate the embryonic brain expression of Pax6 (Kammandel et al., 1999). This cis-regulatory element is highly conserved between Fugu and mouse. More recently, McBride et al. (2011) described other Pax6 enhancers, that drives expression in the mouse cortex (along with several other tissues) (McBride et al., 2011). One dubbed ultra-conserved E60 element (E60UCS) is located 25kb downstream of the Pax6 3’UTR in an intron of a neighbouring gene, Elp4 (McBride et al., 2011). An adjacent element named E60A is also evolutionarily conserved. Both these elements drive expression of a reporter in the entire telencephalon, in addition to other tissues (McBride et al., 2011). Building upon a study in which a YAC containing 420kb of the human Pax6 locus was used to rescue deficits in the Pax6Sey mutant (Schedl et al., 1996), Mi and colleagues investigated the cis-regulatory elements of this construct (Mi et al., 2013). Reporter assays indicated that the large DNA fragment drives expression of a TauGFP reporter in the telencephalon in a pattern largely resembling that of Pax6. Moreover, the researchers narrowed down a core regulatory element within 20kb downstream of the downstream regulatory element of Pax6 which drives reporter expression in the cortex and thalamus.
4.3. Enhancers for other cortical patterning genes
Multiple strategies can be employed to detect enhancer elements in the developing cortex. In our collaboration with the Lawrence Berkeley National Lab, we used sequence conservation and ChIP-Seq for the acetyltransferase p300 on E11.5 mouse forebrain tissue to identify potential forebrain enhancers (Visel et al., 2013). Combining both methods, we identified 4656 noncoding regulatory sequences that were candidate forebrain enhancers. We then tested 329 of these elements, using the human sequence, in transient transgenic mouse assays and found that 105 of these elements were bona fide forebrain enhancers in vivo at E11.5. Additional forebrain enhancers can be found on the VISTA Enhancer Browser website: http://enhancer.lbl.gov.
Enhancers that are active in cortical development, that are associated with genes implicated in cortical development (Emx2, Fezf2, Neurogenin1/2, Sox2, Pax6,Tbr2, and SatB2) are in Table 1. Upstream of Emx2 there is an enhancer that drives expression in the dorsal telencephalon; this DNA contains binding sites for effectors of the Wnt and Bmp signalling pathways, Tcf and Smad respectively (Theil et al., 2002). Deletion of this enhancer resulted in a downregulation, but not abolition, of Emx2 expression in the telencephalon suggesting that additional enhancers drive telencephalic Emx2 expression as well as to confer on it the particular graded expression along the rostrocaudal axis (Suda, 2010). Similarly, two enhancers of Fezf2 have been studied so far: the E4 (or 434) enhancer and the 1316 enhancer (Shim et al., 2012; Eckler et al., 2014). The E4 downstream enhancer is a cortex-specific enhancer of Fezf2 which drives expression in cortical progenitors; its absence is characterized by a reduction in Fezf2 expression and an absence of the corticospinal tract which is reminiscent of a cortex-specific deletion of Fezf2 (Shim et al., 2012). This enhancer is positively regulated by SOX4 and SOX11 and negatively regulated by SOX5 (Shim et al., 2012) and can also bind to TBR1, FOXG1 and NF1B (Eckler et al., 2014). The Fezf2 upstream enhancer, enhancer 1316, shows activity in deep-layer neurons and high throughput chromosome conformation capture (Hi-C) data confirms its ability to bind the Fezf2 promoter (Eckler et al., 2014).
Table 1.
List of enhancers known to drive cortical expression of TFs in development
| Gene | Coordinates of enhancer |
Size of enhancer |
Forebrain expression pattern | Pertinent information | Reference |
|---|---|---|---|---|---|
|
Multiple genes |
Multiple enhancers | A large-scale analysis of in vivo enhancer elements in the forebrain detected over 145 enhancers that were active in the forebrain, many of which were active in the developing telencephalon. The patterns of expression of these forebrain enhancers (and others) can be found on the VISTA enhancer browser website: http://enhancer.lbl.gov. |
A follow-up study on 14 telencephalic enhancers revealed that enhancers in development drive expression in protodomains that fate map to distinct cortical regions in adulthood. These enhancers are regulated by TF which are expressed in graded expression in the developing pallium and can perhpas serve to translate these gradients of TFs into regional maps. |
{Visel:2013it, Pattabiraman:2014ic} | |
|
Multiple genes |
Multiple enhancers | Enhancers active in the developing cortex at E14.5 were detected using ChIP-Seq for P300. Eight enhancers expressed in a laminar pattern in the neocortex were further characterized. |
This study also discusses several properties of active enhancers for predictive purposes including enrichment of TF binding sites and conservation of enhancer sequences across species. |
{Wenger:2013jh} | |
| Cux2 | hs611 on Vista Browser |
854 | Mouse enhancer expressed in dorsomedial telencephalon and patchy expression in other tissues. |
Mutation of two TCF/Lef-binding sites did not abolish reporter expression. |
{HasenpuschTheil:2012gt}{Visel:2009jp} |
| Dmrt3 | Hs112 on Vista Browser |
1873 | Both mouse and human enhancer drive expression in dorsal telencephalon and nasal placode. |
Point mutations in the ultraconserved Tcf-binding site greatly reduced expression in telencephalon |
{HasenpuschTheil:2012gt}{Visel:2009jp} |
| Emx2 | DT1/θ = immediately downstream of Emx2 |
DT1/θ = 450 bp |
Expression in dorsal telencephalon with higher levels caudal/medial by E11.5 = analogous to Emx2. Aberrant expression in ventral diencephalon. |
Contains a binding site for Tcf + Smad that, when mutated, abolish telencephalic enhancer activity. Ectopic Wnt + BMP signalling leads to ectopic activation of enhancer. Also regulated by Otx2. When deleted, Emx2 expression is reduced that this enhancer regulates endogenous Emx2 expression but other cortical Emx2 enhancers must also exist. |
{Theil:2002ts}{Suda:2010dn} |
| ER81 | −2 kb to −1 kb of ER81 | Less than 1.8 kb |
Two constructs containing 2kb and 1.5 kb upstream of zebrafish ER81 drove reporter expression in the lateral mouse pallium, in a subset of ER81-positive cells. |
There is a Pax6 consensus binding site in this putative ER81 enhancer. |
{Langevin:2007ka} |
| Fezf2 | E4 = +7.3 kb of FezF2 TSS |
861 bp | The absence of E4 from a Fezf2-BAC transgene leads to a loss of reporter activity from the entire cerebral cortex at P0. Enhancer expression in VZ of cortex |
Deletion of this enhancer leads to a loss of corticospinal axons. Binds to p300 in vivo. Binds to Sox4 and Sox11; this binding is necessary for Fezf2 expression. Compete with repressive action of Sox5 |
{Shim:2012cl}{Visel:2009jp}{Eckler:2014jd} |
| 1316 = around 50kb away from Fezf2 (intronic to Cadps) |
2290 bp | Expression in deep-layer neurons at P0 + P12. | Some cells that expressed the enhancer at P0 also expressed markers such as SatB2 and TIP2. |
{Eckler:2014jd} | |
| Gli3 | hs111 on Vista Browser |
438 bp | Both mouse and human enhancer drive expression in telencephalon and dorsal thalamus. Mouse enhancer present in VZ cells but not differentiating neurons like Gli3. |
No enhancer activity is observed when the Tcf/LEF- binding site is mutated. |
{HasenpuschTheil:2012gt}{Visel:2009jp} |
| Gsh1 | Enh16 = intronic to Gsh1 |
282 | Active in mouse cortex at E14.5 | {Bery:2013gx} | |
| Hop | −7kb of Hop exon1 | 200 bp | Expression confined to Hop-positive cells = including medial pallium and cortical hem (as well as hindbrain and spinal cord). |
Enhancer expression could be induced ectopicallly in the lateral pallium when in proximity to roof plate explants. The enhancer contains a putative Oct6 binding site. |
{Muhlfriedel:2005jj} |
| Nfia | hs1450 on Vista Browser= downstream of Nfia |
823 bp | Expression in dorsomedial telencephalon = cortical hem + roof plate |
Mutation of Tcf/Lef-binding site abolishes activity in the cortical hem. |
{HasenpuschTheil:2012gt}{Visel:2009jp} |
| Ngn1 | 1) −8.4 kb of Ngn1 | 8.4 kb | Telencephalic expression in zebrafish and mice. | {Blader:2004cz} | |
| 2) LSE enhancer = −6.7 to −6.5 kb |
212 bp | Telencephalic expression in zebrafish and dorsal telencephalic expression in mice. |
|||
| 3) LATE enhancer = −1.8 to −1.4 kb |
407 bp | No telencephalic expression in zebrafish (but diencephalic + hindbrain) and lateral telencephalic expression in mouse. |
Requires Pax6 for telencephalic expression in mouse and for diencephalic expression in zebrafish. |
||
| Ngn2 | E1 enhancer = −9 kb of Ngn2 |
1758 bp | Drive expression to dorsal telencephalon at E10.5 (as well as spinal cord and other regions of endogenous Ngn2 expression). |
Pax6 required for E1 expression in dorsal elencephalon | {Scardigli:2001vh} |
| E3 enhancer = 3’ UTR of Ngn2 |
2616 bp | ||||
| Pax6 | E60UCS = +25 kb of Pax6 |
1400 bp | Drives expression to the entire cerebral cortex from E10.5 to E16.5. |
This is an ultra-conserved region between mouse and human. No autoregulation of enhancer expression in Pax6 Sey mutant. |
{Kleinjan:2008gy, McBride:2011kq} |
| E60A = downstream of E60UCS |
1732 bp | Drives expression to the entire cerebral cortex from E10.5 to E16.5. |
No autoregulation of enhancer expression in Pax6 Sey mutant. |
||
| Pou2f1 | Enh18 = chr1:167864977-167866022 |
1045 bp | Active in mouse cortex at E14.5 | Mutating a Sox consensus binding site or a Pou one or both in the enhancer led to a reduction in enhancer activity. Mostly active in VZ/SVZ. |
{Bery:2013gx} |
| Sall3 | enh15 = chr18 : 81448646-81449314 |
667 bp | Active in mouse cortex at E14.5 | Mostly active in the SVZ/CP. | {Bery:2013gx} |
| SatB2 | As021 SINE = −390 kb of SatB2 |
601 bp | Displays enhancer activity in mouse developing cortex in early-born SatB2+ neurons and in deep- layer neurons (but not superficial-layer neurons) at embryonic and postnatal stages. |
This enhancer is active in SatB2+ corpus callosal neurons. |
{Tashiro:2011jk} |
| Sox2 | N2 enhancer = −5.7 to −3.3 kb of Sox2 |
800 bp | Drives expression of reporter specifically to the dorsal forebrain in mouse and chicken = a subset of Sox2 domain of expression. |
Heterozygous deletion of this enhancer leads to loss of reporter in the dorsal forebrain. This enhancer has potential site for POU family proteins. |
{Zappone:2000vv} {Uchikawa:2003vn} |
| R1 enhancer = −3.8 to −3.4 kb of Sox2 |
389 bp | Drives expression to telencephalic neuroepithelial cells (contained within the N2 enhancer). |
In NSCs, this sequence binds to Brm and Brac1. In vivo, Gli2 is required to transactivate the R1 enhancer. |
{Takanaga:2009fr, Kondo:2004kt} | |
|
Tbr2 (Eomes) |
Eo5 = −150 kb of Tbr2 | 3658 bp | Drives telencephalic expression (and ectopic midbrain expression). |
This is a highly conserved element in mouse, human, opossum and chick. Also known as hs1557 in the screen by Visel et al. (2009). |
{Pernaute:2010ed} |
| −215 kb of Tbr2 | unknown | A breakpoint in this regulatory region leads to loss of Tbr2 transcripts in the cortex |
Baala et al {Baala:2007dw} |
The complete analysis of a cortical development enhancer element require defining: 1) its temporal and spatial activity pattern; 2) the gene(s) that it regulates; 3) the molecules that regulate the enhancer element (e.g. TFs, histone modifications, DNA methylation, miRNAs). Moreover, as we gather more information about cortical enhancers, perhaps we will find markers that predict their location such as a combination of histone marks and transcription factor motifs. For instance, in a variety of cell types including mouse embryonic fibroblasts, a unique signature of three chromatin marks can serve to predict whether these cells are amenable to being reprogrammed into induced neurons by one of the master regulators, Ascl1 (Wapinski et al., 2013).
One of the main pitfalls so far in enhancer studies is the lack of evidence that the so-called enhancer regions bind to the promoter of the gene they are supposedly regulating. To obtain this information, it is useful to define chromatin conformation to assess whether the enhancer is in proximity to the promoter of the gene that it regulates. It is known that beyond sharing a conservation of primary DNA sequences, syntenic regions in mouse and human also share a similarity in their higher order chromatin structure (Chambers et al., 2013). A recent study set out to identify enhancer-promoter units (EPUs) and provide information about the chromatin structure (Shen et al., 2012). These authors were interested in expanding upon the finding that enhancers may not always target their closest promoter. Using a combination of ChIP-Seq data commonly used to identify cis-regulatory elements in several tissue types, the authors developed an algorithm enabling them to identify probable EPUs. They then verified their postulate by showing that, as predicted, five potential enhancer-promoter complexes were associated in the cortex but not in ES cells using chromosome conformation capture (3C) to detect long distance looping interactions (Shen et al., 2012).
4.4. Moving the protomap onwards: from the VZ to the cortical plate
It will be important to determine the gene regulatory networks important for conferring appropriate patterning to the cortex as it develops. We hypothesise that in order to pass along the information encoded in gradients of TFs present in the VZ, enhancers for genes important in cell specifications will be bound by some of the TFs located in the VZ such as Pax6, Emx2, sp8 etc (see Figure 2)… We postulate that enhancers serve as protein-binding modules that translate gradients of TFs in cortical progenitors into region-specific expression in cortical neurons.
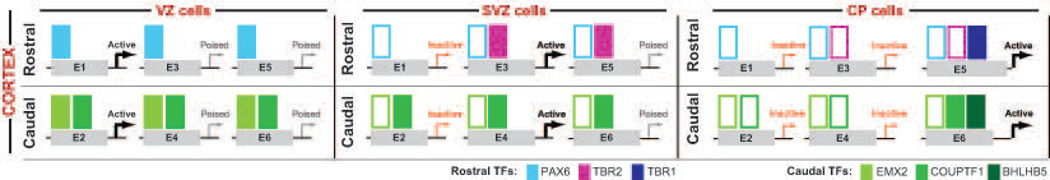
Figure 2.
A central unanswered question is how are the gradients of TFs in the VZ translated into positional information in the SVZ and CP, to generate cortical regions? We hypothesize that enhancers active in the VZ {E1, E2}, SVZ {E3, E4} and CP {E5, E6} are differentially bound by TFs that drive expression of region/layer-specific genes in post-mitotic cortical neurons. The enhancers serve as protein-binding modules that translate rostrocaudal gradients of TFs in cortical progenitors into region-specific expression in cortical neurons. In this simplified model, enhancers active in the SVZ (E3, E4) and CP (E5, E6) are primed in the VZ by TFs. A filled colored box symbolizes TF binding to the enhancer; an empty box symbolizes that the TF, which is no longer expressed, had previously bound that enhancer, and modified its epigenetic state. For instance, binding of a PAX6 in rostral VZ cells (where its concentration is highest) preferentially activates rostral VZ enhancers (E1) and not caudal VZ enhancers (E2) that are activated by EMX2 and COUPTF1. We postulate that PAX6, EMX2 and COUPTF1 will prime certain SVZ and CP enhancers, marking them epigenetically with modified histone marks of poised or active chromatin, thus passing on positional information to enhancers that are active in the SVZ and CP. Propagation of the VZ protomap into the SVZ and CP is carried out by TBR2, COUPTF1, TBR1 and BHLHB5.
Pax6 was shown to control regionalisation through a downstream cascade beginning with the induction of Eomes (Tbr2) in the intermediate progenitors and propagated by Tbr1 to the cortical plate (Bedogni et al., 2010; Elsen et al., 2013). In this model, aeralization information encoded by the gradient of Pax6 in the VZ would be passed along in an “intermediate map” in the SVZ encoded by Tbr2. In the absence of Tbr2, a rostrocaudal shift occurs in the patterning of the developing cortex with shifts in the gene expression of a variety of genes reminiscent of changes seen in mutants in which Pax6 is exclusively removed in the cortex (Piñon et al., 2008; Elsen et al., 2013). In both cases, the thalamocortical distribution remains unperturbed indicating that the perturbation in arealization is not accompanied by a change in cortical identity. Blurring of the cortical area boundaries however does occur in the absence of Tbr2 indicating that it is necessary for the refinement of the cortical areal map. Pax6 can directly transactivate Tbr2 (Sansom et al., 2009) and in Pax6Sey mice, although intermediate progenitors are still found in the SVZ, Tbr2 expression is severely reduced in the proliferative zone of the cortex but it is still retained in the telencephalon of both Leca mutants discussed above (Quinn et al., 2007; Walcher et al., 2013). In fact, in Pax6−/− mice, Tbr2+ cells are nearly exclusively found in the preplate rather than in the SVZ, and the steepness of the Tbr2 gradient observed in wild-type mice is flattened in the mutants (Quinn et al., 2007). The regulation of Tbr2 by Pax6 occurs in a cell autonomous manner since Pax6−/−↔ Pax6+/+ cortical chimeras showed that cells derived from Pax6−/− genotype (as labelled by β-globin) did not express Tbr2 in the developing pallium even if they were surrounded by Tbr2+ cells (Quinn et al., 2007). This further substantiates the fact that Tbr2 is capable of propagating VZ-derived patterning information to the cortical plate. Finally, it was shown that one of the chromatin modifiers, BAF170, controls the accessibility of the Tbr2 locus to Pax6 (Tuoc et al., 2013). Moreover, in line with our model, it was shown that changing the level of CoupTFI expression in post-mitotic neurons of the caudal developing cortex was sufficient to change the arealization of the rostral cortex (Alfano et al., 2014). This indicates that TF levels in progenitors as well as in the SVZ and CP are important for controlling cortical regionalisation.
Currently, much work remains to understand the transcriptional network required to propagate patterning information from the VZ to the cortical plate, and an even more arduous enterprise will be required in identifying the cis-regulatory elements at play in this complex developmental process.
Conclusion
The advent of next-generation sequencing techniques (reviewed by Maze et al., 2014; Shin et al., 2014) should technically enable researchers to tie together the transcriptional networks, the chromatin regulators and the enhancer elements that are key in cortical development. In this review, we have highlighted the role of Pax6 at each level in order to provide concrete examples of the complexity of the biological processes occurring during cortical development. Of course, Pax6 is only one of many key TF for cortical development. Integrating the information garnered for each TF into a coherent model will indubitably be a major goal and hurdle in our understanding of cortical developmental processes. Major concerted efforts to determine the epigenetic marks present in different cell types at different time points, such as the large-scale project currently being carried out by ENCODE, are undeniably going to enable the community to begin understanding TF networks in the context of their epigenetic environment. Moreover, other efforts designed to systematically track down enhancers active in cortical tissue in development will also provide greatly needed resources for the neuroscience community (Visel et al., 2009; Kim et al., 2010; Visel et al., 2013; Wenger et al., 2013).
Acknowledgements
This work was supported by grants to J.L.R.R. from Nina Ireland, the National Institute of Neurological Disorders and Stroke (NINDS R01 NS34661), and the National Institute of Mental Health (NIMH R37 MH049428). A.R.Y. was supported by a postdoctoral fellowship from the Fondation Fyssen (Paris, France).
References
- Agca C, Elhajj MC, Klein WH, Venuti JM. Neurosensory and neuromuscular organization in tube feet of the sea urchin Strongylocentrotus purpuratus. J Comp Neurol. 2011;519:3566–3579. doi: 10.1002/cne.22724. [DOI] [PubMed] [Google Scholar]
- Akbarian S. The molecular pathology of schizophrenia--focus on histone and DNA modifications. Brain Research Bulletin. 2010;83:103–107. doi: 10.1016/j.brainresbull.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert M, Schmitz SU, Kooistra SM, Malatesta M, Morales Torres C, Rekling JC, Johansen JV, Abarrategui I, Helin K. The histone demethylase Jarid1b ensures faithful mouse development by protecting developmental genes from aberrant H3K4me3. PLoS Genet. 2013;9:e1003461. doi: 10.1371/journal.pgen.1003461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano C, Magrinelli E, Harb K, Hevner RF, Studer M. Postmitotic control of sensory area specification during neocortical development. Nat Commun. 2014;5:5632. doi: 10.1038/ncomms6632. [DOI] [PubMed] [Google Scholar]
- Altmann CR, Chow RL, Lang RA, Hemmati-Brivanlou A. Lens induction by Pax-6 in Xenopus laevis. Developmental Biology. 1997;185:119–123. doi: 10.1006/dbio.1997.8573. [DOI] [PubMed] [Google Scholar]
- Aota S-I, Nakajima N, Sakamoto R, Watanabe S, Ibaraki N, Okazaki K. Pax6 autoregulation mediated by direct interaction of Pax6 protein with the head surface ectoderm-specific enhancer of the mouse Pax6 gene. Developmental Biology. 2003;257:1–13. doi: 10.1016/s0012-1606(03)00058-7. [DOI] [PubMed] [Google Scholar]
- Armentano M, Chou S-J, Tomassy GS, Leingärtner A, O'Leary DDM, Studer M. COUP-TFI regulates the balance of cortical patterning between frontal/motor and sensory areas. Nat Neurosci. 2007;10:1277–1286. doi: 10.1038/nn1958. [DOI] [PubMed] [Google Scholar]
- Bedogni F, Hodge RD, Elsen GE, Nelson BR, Daza RAM, Beyer RP, Bammler TK, Rubenstein JLR, Hevner RF. Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proceedings of the National Academy of Sciences. 2010;107:13129–13134. doi: 10.1073/pnas.1002285107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berninger B, Costa MR, Koch U, Schroeder T, Sutor B, Grothe B, Gotz M. Functional Properties of Neurons Derived from In Vitro Reprogrammed Postnatal Astroglia. Journal of Neuroscience. 2007;27:8654–8664. doi: 10.1523/JNEUROSCI.1615-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bery A, Martynoga B, Guillemot F, Joly J-S, Rétaux S. Characterization of Enhancers Active in the Mouse Embryonic Cerebral Cortex Suggests Sox/Pou cis-Regulatory Logics and Heterogeneity of Cortical Progenitors. Cerebral Cortex. 2013;24:2822–2834. doi: 10.1093/cercor/bht126. [DOI] [PubMed] [Google Scholar]
- Bhatia S, Bengani H, Fish M, Brown A, Divizia MT, de Marco R, Damante G, Grainger R, van Heyningen V, Kleinjan DA. Disruption of autoregulatory feedback by a mutation in a remote, ultraconserved PAX6 enhancer causes aniridia. Am J Hum Genet. 2013;93:1126–1134. doi: 10.1016/j.ajhg.2013.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia S, Monahan J, Ravi V, Gautier P, Murdoch E, Brenner S, van Heyningen V, Venkatesh B, Kleinjan DA. A survey of ancient conserved non-coding elements in the PAX6 locus reveals a landscape of interdigitated cis-regulatory archipelagos. Developmental Biology. 2014;387:214–228. doi: 10.1016/j.ydbio.2014.01.007. [DOI] [PubMed] [Google Scholar]
- Bhinge A, Poschmann J, Namboori SC, Tian X, Jia Hui Loh S, Traczyk A, Prabhakar S, Stanton LW. MiR-135b is a direct PAX6 target and specifies human neuroectoderm by inhibiting TGF-/BMP signaling. EMBO J. 2014 doi: 10.1002/embj.201387215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielle F, Griveau A, Narboux-Nême N, Vigneau S, Sigrist M, Arber S, Wassef M, Pierani A. Multiple origins of Cajal-Retzius cells at the borders of the developing pallium. Nat Neurosci. 2005;8:1002–1012. doi: 10.1038/nn1511. [DOI] [PubMed] [Google Scholar]
- Bishop KM, Garel S, Nakagawa Y, Rubenstein JLR, O'Leary DDM. Emx1 and Emx2 cooperate to regulate cortical size, lamination, neuronal differentiation, development of cortical efferents, and thalamocortical pathfinding. J Comp Neurol. 2003;457:345–360. doi: 10.1002/cne.10549. [DOI] [PubMed] [Google Scholar]
- Bishop KM, Goudreau G, O'Leary DD. Regulation of area identity in the mammalian neocortex by Emx2 and Pax6. Science. 2000;288:344–349. doi: 10.1126/science.288.5464.344. [DOI] [PubMed] [Google Scholar]
- Blader P, Lam CS, Rastegar S, Scardigli R, Nicod J-C, Simplicio N, Plessy C, Fischer N, Schuurmans C, Guillemot F, Strähle U. Conserved and acquired features of neurogenin1 regulation. Development. 2004;131:5627–5637. doi: 10.1242/dev.01455. [DOI] [PubMed] [Google Scholar]
- Borello U, Madhavan M, Vilinsky I, Faedo A, Pierani A, Rubenstein J, Campbell K. Sp8 and COUP-TF1 reciprocally regulate patterning and Fgf signaling in cortical progenitors. Cerebral Cortex. 2014;24:1409–1421. doi: 10.1093/cercor/bhs412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbe C, Garg A, Cai Z, Li H, Powers A, Zhang X. An allelic series at the paired box gene 6 (Pax6) locus reveals the functional specificity of Pax genes. Journal of Biological Chemistry. 2013;288:12130–12141. doi: 10.1074/jbc.M112.436865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney RSE, Cocas LA, Hirata T, Mansfield K, Corbin JG. Differential regulation of telencephalic pallial-subpallial boundary patterning by Pax6 and Gsh2. Cerebral Cortex. 2009;19:745–759. doi: 10.1093/cercor/bhn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caronia-Brown G, Yoshida M, Gulden F, Assimacopoulos S, Grove EA. The cortical hem regulates the size and patterning of neocortex. Development [Internet] 2014;141:2855–2865. doi: 10.1242/dev.106914. Available from: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=24948604&retmode=ref&cmd=prlinks. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers EV, Bickmore WA, Semple CA. Divergence of mammalian higher order chromatin structure is associated with developmental loci. PLoS Comput Biol. 2013;9:e1003017. doi: 10.1371/journal.pcbi.1003017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapouton P, Gärtner A, Gotz M. The role of Pax6 in restricting cell migration between developing cortex and basal ganglia. Development. 1999;126:5569–5579. doi: 10.1242/dev.126.24.5569. [DOI] [PubMed] [Google Scholar]
- Cheng X, Hsu C-M, Currle DS, Hu JS, Barkovich AJ, Monuki ES. Central roles of the roof plate in telencephalic development and holoprosencephaly. Journal of Neuroscience. 2006;26:7640–7649. doi: 10.1523/JNEUROSCI.0714-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Ma Z, Kim B-H, Wu W, Cayting P, Boyle AP, Sundaram V, Xing X, Dogan N, Li J, Euskirchen G, Lin S, Lin Y, Visel A, Kawli T, Yang X, Patacsil D, Keller CA, Giardine B, The Mouse ENCODE Consortium. Kundaje A, Wang T, Pennacchio LA, Weng Z, Hardison RC, Snyder MP. Principles of regulatory information conservation between mouse and human. Nature. 2014;515:371–375. doi: 10.1038/nature13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi N, Epstein JA. Getting your Pax straight: Pax proteins in development and disease. Trends Genet. 2002;18:41–47. doi: 10.1016/s0168-9525(01)02594-x. [DOI] [PubMed] [Google Scholar]
- Chisholm AD, Horvitz HR. Patterning of the Caenorhabditis elegans head region by the Pax-6 family member vab-3. Nature. 1995;377:52–55. doi: 10.1038/377052a0. [DOI] [PubMed] [Google Scholar]
- Cholfin JA, Rubenstein JLR. Patterning of frontal cortex subdivisions by Fgf17. Proc Natl Acad Sci USA. 2007;104:7652–7657. doi: 10.1073/pnas.0702225104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholfin JA, Rubenstein JLR. Frontal cortex subdivision patterning is coordinately regulated by Fgf8, Fgf17, and Emx2. J Comp Neurol. 2008;509:144–155. doi: 10.1002/cne.21709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S-J, Babot Z, Leingärtner A, Studer M, Nakagawa Y, O'Leary DDM. Geniculocortical input drives genetic distinctions between primary and higher-order visual areas. Science. 2013;340:1239–1242. doi: 10.1126/science.1232806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S-J, Perez-Garcia CG, Kroll TT, O'Leary DDM. Lhx2 specifies regional fate in Emx1 lineage of telencephalic progenitors generating cerebral cortex. Nat Neurosci. 2009;12:1381–1389. doi: 10.1038/nn.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow RL, Altmann CR, Lang RA, Hemmati-Brivanlou A. Pax6 induces ectopic eyes in a vertebrate. Development. 1999;126:4213–4222. doi: 10.1242/dev.126.19.4213. [DOI] [PubMed] [Google Scholar]
- Coutinho P, Pavlou S, Bhatia S, Chalmers KJ, Kleinjan DA, van Heyningen V. Discovery and assessment of conserved Pax6 target genes and enhancers. Genome Res. 2011;21:1349–1359. doi: 10.1101/gr.124115.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerny T, Busslinger M. DNA-binding and transactivation properties of Pax-6: three amino acids in the paired domain are responsible for the different sequence recognition of Pax-6 and BSAP (Pax-5) Molecular and Cellular Biology. 1995;15:2858–2871. doi: 10.1128/mcb.15.5.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Chevigny A, Coré N, Follert P, Gaudin M, Barbry P, Béclin C, Cremer H. miR-7a regulation of Pax6 controls spatial origin of forebrain dopaminergic neurons. Nature Publishing Group. 2012;15:1120–1126. doi: 10.1038/nn.3142. [DOI] [PubMed] [Google Scholar]
- Del Río JA, Heimrich B, Borrell V, Förster E, Drakew A, Alcántara S, Nakajima K, Miyata T, Ogawa M, Mikoshiba K, Derer P, Frotscher M, Soriano E. A role for Cajal-Retzius cells and reelin in the development of hippocampal connections. Nature. 1997;385:70–74. doi: 10.1038/385070a0. [DOI] [PubMed] [Google Scholar]
- Eccles MR, He S, Legge M, Kumar R, Fox J, Zhou C, French M, Tsai RWS. PAX genes in development and disease: the role of PAX2 in urogenital tract development. Int J Dev Biol. 2002;46:535–544. [PubMed] [Google Scholar]
- Eckler MJ, Larkin KA, McKenna WL, Katzman S, Guo C, Roque R, Visel A, Rubenstein JLR, Chen B. Multiple conserved regulatory domains promote Fezf2 expression in the developing cerebral cortex. Neural Dev. 2014;9:6. doi: 10.1186/1749-8104-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsen GE, Hodge RD, Bedogni F, Daza RAM, Nelson BR, Shiba N, Reiner SL, Hevner RF. The protomap is propagated to cortical plate neurons through an Eomes-dependent intermediate map. Proceedings of the National Academy of Sciences. 2013;110:4081–4086. doi: 10.1073/pnas.1209076110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein J, Cai J, Glaser T, Jepeal L, Maas R. Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J Biol Chem. 1994;269:8355–8361. [PubMed] [Google Scholar]
- Faedo A, Tomassy GS, Ruan Y, Teichmann H, Krauss S, Pleasure SJ, Tsai SY, Tsai M-J, Studer M, Rubenstein JLR. COUP-TFI coordinates cortical patterning, neurogenesis, and laminar fate and modulates MAPK/ERK, AKT, and beta-catenin signaling. Cerebral Cortex. 2008;18:2117–2131. doi: 10.1093/cercor/bhm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantes J, Redeker B, Breen M, Boyle S, Brown J, Fletcher J, Jones S, Bickmore W, Fukushima Y, Mannens M. Aniridia-associated cytogenetic rearrangements suggest that a position effect may cause the mutant phenotype. Hum Mol Genet. 1995;4:415–422. doi: 10.1093/hmg/4.3.415. [DOI] [PubMed] [Google Scholar]
- Förster E, Tielsch A, Saum B, Weiss KH, Johanssen C, Graus-Porta D, Müller U, Frotscher M. Reelin, Disabled 1, and beta 1 integrins are required for the formation of the radial glial scaffold in the hippocampus. Proc Natl Acad Sci USA. 2002;99:13178–13183. doi: 10.1073/pnas.202035899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi-Shimogori T, Grove EA. Neocortex patterning by the secreted signaling molecule FGF8. Science. 2001;294:1071–1074. doi: 10.1126/science.1064252. [DOI] [PubMed] [Google Scholar]
- Galceran J, Miyashita-Lin EM, Devaney E, Rubenstein JL, Grosschedl R. Hippocampus development and generation of dentate gyrus granule cells is regulated by LEF1. Development. 2000;127:469–482. doi: 10.1242/dev.127.3.469. [DOI] [PubMed] [Google Scholar]
- Gao J, Wang J, Wang Y, Dai W, Lu L. Regulation of Pax6 by CTCF during induction of mouse ES cell differentiation. PLoS ONE. 2011;6:e20954. doi: 10.1371/journal.pone.0020954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel S, Huffman KJ, Rubenstein JLR. Molecular regionalization of the neocortex is disrupted in Fgf8 hypomorphic mutants. Development. 2003;130:1903–1914. doi: 10.1242/dev.00416. [DOI] [PubMed] [Google Scholar]
- Georgala PA, Carr CB, Price DJ. The role of Pax6 in forebrain development. Dev Neurobiol. 2011a;71:690–709. doi: 10.1002/dneu.20895. [DOI] [PubMed] [Google Scholar]
- Georgala PA, Manuel M, Price DJ. The generation of superficial cortical layers is regulated by levels of the transcription factor Pax6. Cerebral Cortex. 2011b;21:81–94. doi: 10.1093/cercor/bhq061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring WJ, Ikeo K. Pax 6: mastering eye morphogenesis and eye evolution. Trends in genetics. 1999 doi: 10.1016/s0168-9525(99)01776-x. [DOI] [PubMed] [Google Scholar]
- Gehring WJ. Historical perspective on the development and evolution of eyes and photoreceptors. Int J Dev Biol. 2004;48:707–717. doi: 10.1387/ijdb.041900wg. [DOI] [PubMed] [Google Scholar]
- Gotz M, Stoykova A, Gruss P. Pax6 controls radial glia differentiation in the cerebral cortex. Neuron. 1998;21:1031–1044. doi: 10.1016/s0896-6273(00)80621-2. [DOI] [PubMed] [Google Scholar]
- Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci. 2013;14:755–769. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin C, Kleinjan DA, Doe B, van Heyningen V. New 3' elements control Pax6 expression in the developing pretectum, neural retina and olfactory region. Mechanisms of Development. 2002;112:89–100. doi: 10.1016/s0925-4773(01)00646-3. [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Gruss P, Walther C. Pax in development. Cell. 1992;69:719–722. doi: 10.1016/0092-8674(92)90281-g. [DOI] [PubMed] [Google Scholar]
- Hack MA, Sugimori M, Lundberg C, Nakafuku M, Gotz M. Regionalization and fate specification in neurospheres: the role of Olig2 and Pax6. Mol Cell Neurosci. 2004;25:664–678. doi: 10.1016/j.mcn.2003.12.012. [DOI] [PubMed] [Google Scholar]
- Hasenpusch-Theil K, Magnani D, Amaniti E-M, Han L, Armstrong D, Theil T. Transcriptional analysis of Gli3 mutants identifies Wnt target genes in the developing hippocampus. Cerebral Cortex [Internet] 2012;22:2878–2893. doi: 10.1093/cercor/bhr365. Available from: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=22235033&retmode=ref&cmd=prlinks. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubst N, Berger J, Radjendirane V, Graw J, Favor J, Saunders GF, Stoykova A, Götz M. Molecular dissection of Pax6 function: the specific roles of the paired domain and homeodomain in brain development. Development. 2004;131:6131–6140. doi: 10.1242/dev.01524. [DOI] [PubMed] [Google Scholar]
- Heins N, Malatesta P, Cecconi F, Nakafuku M, Tucker KL, Hack MA, Chapouton P, Barde Y-A, Götz M. Glial cells generate neurons: the role of the transcription factor Pax6. Nat Neurosci. 2002;5:308–315. doi: 10.1038/nn828. [DOI] [PubMed] [Google Scholar]
- Hill RE, Favor J, Hogan BL, Ton CC, Saunders GF, Hanson IM, Prosser J, Jordan T, Hastie ND, van Heyningen V. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature. 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- Holm PC, Mader MT, Haubst N, Wizenmann A, Sigvardsson M, Götz M. Loss- and gain-of-function analyses reveal targets of Pax6 in the developing mouse telencephalon. Mol Cell Neurosci. 2007;34:99–119. doi: 10.1016/j.mcn.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Hsieh J, Gage FH. Epigenetic control of neural stem cell fate. Curr Opin Genet Dev. 2004;14:461–469. doi: 10.1016/j.gde.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Hussain MA, Habener JF. Glucagon gene transcription activation mediated by synergistic interactions of pax-6 and cdx-2 with the p300 co-activator. J Biol Chem. 1999;274:28950–28957. doi: 10.1074/jbc.274.41.28950. [DOI] [PubMed] [Google Scholar]
- Inoue M, Kamachi Y, Matsunami H, Imada K, Uchikawa M, Kondoh H. PAX6 and SOX2-dependent regulation of the Sox2 enhancer N-3 involved in embryonic visual system development. Genes Cells. 2007;12:1049–1061. doi: 10.1111/j.1365-2443.2007.01114.x. [DOI] [PubMed] [Google Scholar]
- Jones L, Lopez-Bendito G, Gruss P, Stoykova A, Molnár Z. Pax6 is required for the normal development of the forebrain axonal connections. Development. 2002;129:5041–5052. doi: 10.1242/dev.129.21.5041. [DOI] [PubMed] [Google Scholar]
- Joshi PS, Molyneaux BJ, Feng L, Xie X, Macklis JD, Gan L. Bhlhb5 Regulates the Postmitotic Acquisition of Area Identities in Layers II-Vof the Developing Neocortex. Neuron. 2008;60:258–272. doi: 10.1016/j.neuron.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliandi B, Abematsu M, Sanosaka T, Tsujimura K, Smith A, Nakashima K. Induction of superficial cortical layer neurons from mouse embryonic stem cells by valproic acid. Neurosci Res. 2012;72:23–31. doi: 10.1016/j.neures.2011.09.012. [DOI] [PubMed] [Google Scholar]
- Kamachi Y, Uchikawa M, Tanouchi A, Sekido R, Kondoh H. Pax6 and SOX2 form a co-DNA-binding partner complex that regulates initiation of lens development. Genes & Development. 2001;15:1272–1286. doi: 10.1101/gad.887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammandel B, Chowdhury K, Stoykova A, Aparicio S, Brenner S, Gruss P. Distinct cis-essential modules direct the time-space pattern of the Pax6 gene activity. Developmental Biology. 1999;205:79–97. doi: 10.1006/dbio.1998.9128. [DOI] [PubMed] [Google Scholar]
- Kim H-J, Rosenfeld MG. Epigenetic control of stem cell fate to neurons and glia. Arch Pharm Res. 2010;33:1467–1473. doi: 10.1007/s12272-010-1001-z. [DOI] [PubMed] [Google Scholar]
- Kim T-K, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinjan DA, Bancewicz RM, Gautier P, Dahm R, Schonthaler HB, Damante G, Seawright A, Hever AM, Yeyati PL, van Heyningen V, Coutinho P. Subfunctionalization of duplicated zebrafish pax6 genes by cis-regulatory divergence. PLoS Genet. 2008;4:e29. doi: 10.1371/journal.pgen.0040029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinjan DA, Seawright A, Childs AJ, van Heyningen V. Conserved elements in Pax6 intron 7 involved in (auto)regulation and alternative transcription. Developmental Biology. 2004;265:462–477. doi: 10.1016/j.ydbio.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Kleinjan DA, Seawright A, Schedl A, Quinlan RA, Danes S, van Heyningen V. Aniridia-associated translocations, DNase hypersensitivity, sequence comparison and transgenic analysis redefine the functional domain of PAX6. Hum Mol Genet. 2001;10:2049–2059. doi: 10.1093/hmg/10.19.2049. [DOI] [PubMed] [Google Scholar]
- Kondo T, Raff M. Chromatin remodeling and histone modification in the conversion of oligodendrocyte precursors to neural stem cells. Genes & Development. 2004;18:2963–2972. doi: 10.1101/gad.309404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kozmik Z, Daube M, Frei E, Norman B, Kos L, Dishaw LJ, Noll M, Piatigorsky J. Role of Pax genes in eye evolution: a cnidarian PaxB gene uniting Pax2 and Pax6 functions. Developmental Cell. 2003;5:773–785. doi: 10.1016/s1534-5807(03)00325-3. [DOI] [PubMed] [Google Scholar]
- Kroll TT, O'Leary DDM. Ventralized dorsal telencephalic progenitors in Pax6 mutant mice generate GABA interneurons of a lateral ganglionic eminence fate. Proc Natl Acad Sci USA. 2005;102:7374–7379. doi: 10.1073/pnas.0500819102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronhamn J, Frei E, Daube M, Jiao R, Shi Y, Noll M, Rasmuson-Lestander A. Headless flies produced by mutations in the paralogous Pax6 genes eyeless and twin of eyeless. Development. 2002;129:1015–1026. doi: 10.1242/dev.129.4.1015. [DOI] [PubMed] [Google Scholar]
- Langevin LM, Mattar P, Scardigli R, Roussigné M, Logan C, Blader P, Schuurmans C. Validating in utero electroporation for the rapid analysis of gene regulatory elements in the murine telencephalon. Dev Dyn. 2007;236:1273–1286. doi: 10.1002/dvdy.21126. [DOI] [PubMed] [Google Scholar]
- Lauderdale JD, Wilensky JS, Oliver ER, Walton DS, Glaser T. 3' deletions cause aniridia by preventing PAX6 gene expression. Proc Natl Acad Sci USA. 2000;97:13755–13759. doi: 10.1073/pnas.240398797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li T, Lu Z, Lu L. Pax6 regulation in retinal cells by CCCTC binding factor. Investigative Ophthalmology & Visual Science. 2006;47:5218–5226. doi: 10.1167/iovs.06-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahgoub M, Monteggia LM. Epigenetics and psychiatry. Neurotherapeutics. 2013;10:734–741. doi: 10.1007/s13311-013-0213-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik AN, Vierbuchen T, Hemberg M, Rubin AA, Ling E, Couch CH, Stroud H, Spiegel I, Farh KK-H, Harmin DA, Greenberg ME. Genome-wide identification and characterization of functional neuronal activity-dependent enhancers. Nature Publishing Group. 2014;17:1330–1339. doi: 10.1038/nn.3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallamaci A, Muzio L, Chan CH, Parnavelas J, Boncinelli E. Area identity shifts in the early cerebral cortex of Emx2−/− mutant mice. Nat Neurosci. 2000;3:679–686. doi: 10.1038/76630. [DOI] [PubMed] [Google Scholar]
- Mallamaci A, Stoykova A. Gene networks controlling early cerebral cortex arealization. European Journal of Neuroscience. 2006;23:847–856. doi: 10.1111/j.1460-9568.2006.04634.x. [DOI] [PubMed] [Google Scholar]
- Mangale VS, Hirokawa KE, Satyaki PRV, Gokulchandran N, Chikbire S, Subramanian L, Shetty AS, Martynoga B, Paul J, Mai MV, Li Y, Flanagan LA, Tole S, Monuki ES. Lhx2 selector activity specifies cortical identity and suppresses hippocampal organizer fate. Science. 2008;319:304–309. doi: 10.1126/science.1151695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuel M, Georgala PA, Carr CB, Chanas S, Kleinjan DA, Martynoga B, Mason JO, Molinek M, Pinson J, Pratt T, Quinn JC, Simpson TI, Tyas DA, van Heyningen V, West JD, Price DJ. Controlled overexpression of Pax6 in vivo negatively autoregulates the Pax6 locus, causing cell-autonomous defects of late cortical progenitor proliferation with little effect on cortical arealization. Development. 2007;134:545–555. doi: 10.1242/dev.02764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Shen L, Zhang B, Garcia BA, Shao N, Mitchell A, Sun H, Akbarian S, Allis CD, Nestler EJ. Analytical tools and current challenges in the modern era of neuroepigenomics. Nature Publishing Group. 2014;17:1476–1490. doi: 10.1038/nn.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride DJ, Buckle A, van Heyningen V, Kleinjan DA. DNaseI hypersensitivity and ultraconservation reveal novel, interdependent long-range enhancers at the complex Pax6 cis-regulatory region. PLoS ONE. 2011;6:e28616. doi: 10.1371/journal.pone.0028616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehler MF. Epigenetic principles and mechanisms underlying nervous system functions in health and disease. Progress in Neurobiology. 2008;86:305–341. doi: 10.1016/j.pneurobio.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer G, Perez-Garcia CG, Abraham H, Caput D. Expression of p73 and Reelin in the developing human cortex. Journal of Neuroscience. 2002;22:4973–4986. doi: 10.1523/JNEUROSCI.22-12-04973.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi D, Huang Y-T, Kleinjan DA, Mason JO, Price DJ. Identification of genomic regions regulating Pax6 expression in embryonic forebrain using YAC reporter transgenic mouse lines. PLoS ONE. 2013;8:e80208. doi: 10.1371/journal.pone.0080208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moelans CB, Verschuur-Maes AHJ, van Diest PJ. Frequent promoter hypermethylation of BRCA2, CDH13, MSH6, PAX5, PAX6 and WT1 in ductal carcinoma in situ and invasive breast cancer. J Pathol. 2011;225:222–231. doi: 10.1002/path.2930. [DOI] [PubMed] [Google Scholar]
- Monuki ES, Porter FD, Walsh CA. Patterning of the dorsal telencephalon and cerebral cortex by a roof plate-Lhx2 pathway. Neuron. 2001;32:591–604. doi: 10.1016/s0896-6273(01)00504-9. [DOI] [PubMed] [Google Scholar]
- MuhChyi C, Juliandi B, Matsuda T, Nakashima K. Epigenetic regulation of neural stem cell fate during corticogenesis. Int J Dev Neurosci. 2013;31:424–433. doi: 10.1016/j.ijdevneu.2013.02.006. [DOI] [PubMed] [Google Scholar]
- Muzio L, Di Benedetto B, DiBenedetto B, Stoykova A, Boncinelli E, Gruss P, Mallamaci A. Conversion of cerebral cortex into basal ganglia in Emx2(−/−) Pax6(Sey/Sey) double-mutant mice. Nat Neurosci. 2002a;5:737–745. doi: 10.1038/nn892. [DOI] [PubMed] [Google Scholar]
- Muzio L, Di Benedetto B, DiBenedetto B, Stoykova A, Boncinelli E, Gruss P, Mallamaci A. Emx2 and Pax6 control regionalization of the pre-neuronogenic cortical primordium. Cereb Cortex. 2002b;12:129–139. doi: 10.1093/cercor/12.2.129. [DOI] [PubMed] [Google Scholar]
- Mühlfriedel S, Kirsch F, Gruss P, Stoykova A, Chowdhury K. A roof plate-dependent enhancer controls the expression of Homeodomain only protein in the developing cerebral cortex. Developmental Biology. 2005;283:522–534. doi: 10.1016/j.ydbio.2005.04.033. [DOI] [PubMed] [Google Scholar]
- Nabeshima T, Kim H-C. Involvement of genetic and environmental factors in the onset of depression. Exp Neurobiol. 2013;22:235–243. doi: 10.5607/en.2013.22.4.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninkovic J, Pinto L, Petricca S, Lepier A, Sun J, Rieger MA, Schroeder T, Cvekl A, Favor J, Götz M. The transcription factor Pax6 regulates survival of dopaminergic olfactory bulb neurons via crystallin αA. Neuron. 2010;68:682–694. doi: 10.1016/j.neuron.2010.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninkovic J, Steiner-Mezzadri A, Jawerka M, Akinci U, Masserdotti G, Petricca S, Fischer J, Holst von A, Beckers J, Lie CD, Petrik D, Miller E, Tang J, Wu J, Lefebvre V, Demmers J, Eisch A, Metzger D, Crabtree G, Irmler M, Poot R, Götz M. The BAF complex interacts with Pax6 in adult neural progenitors to establish a neurogenic cross-regulatory transcriptional network. Cell Stem Cell. 2013;13:403–418. doi: 10.1016/j.stem.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte C, Rastegar M, Amores A, Bouchard M, Grote D, Maas R, Kovacs EN, Postlethwait J, Rambaldi I, Rowan S, Yan Y-L, Zhang F, Featherstone M. Stereospecificity and PAX6 function direct Hoxd4 neural enhancer activity along the antero-posterior axis. Developmental Biology. 2006;299:582–593. doi: 10.1016/j.ydbio.2006.08.061. [DOI] [PubMed] [Google Scholar]
- Nomura T, Osumi N. Misrouting of mitral cell progenitors in the Pax6/small eye rat telencephalon. Development. 2004;131:787–796. doi: 10.1242/dev.00984. [DOI] [PubMed] [Google Scholar]
- Nord AS, Pattabiraman K, Visel A, Rubenstein JLR. Genomic Perspectives of Transcriptional Regulation in Forebrain Development. Neuron. 2015;85:27–47. doi: 10.1016/j.neuron.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski TJ, Fotaki V, Pollock A, Sun T, Pratt T, Price DJ. MicroRNA-92b regulates the development of intermediate cortical progenitors in embryonic mouse brain. Proceedings of the National Academy of Sciences. 2013;110:7056–7061. doi: 10.1073/pnas.1219385110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary DDM, Chou S-J, Sahara S. Area patterning of the mammalian cortex. Neuron. 2007;56:252–269. doi: 10.1016/j.neuron.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Osumi N, Shinohara H, Numayama-Tsuruta K, Maekawa M. Concise review: Pax6 transcription factor contributes to both embryonic and adult neurogenesis as a multifunctional regulator. Stem Cells. 2008;26:1663–1672. doi: 10.1634/stemcells.2007-0884. [DOI] [PubMed] [Google Scholar]
- Patel NS, Rhinn M, Semprich CI, Halley PA, Dollé P, Bickmore WA, Storey KG. FGF signalling regulates chromatin organisation during neural differentiation via mechanisms that can be uncoupled from transcription. PLoS Genet. 2013;9:e1003614. doi: 10.1371/journal.pgen.1003614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattabiraman K, Golonzhka O, Lindtner S, Nord AS, Taher L, Hoch R, Silberberg SN, Zhang D, Bin Chen, Zeng H, Pennacchio LA, Puelles L, Visel A, Rubenstein JLR. Transcriptional Regulation of Enhancers Activein Protodomains of the Developing Cerebral Cortex. Neuron. 2014;82:989–1003. doi: 10.1016/j.neuron.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira JD, Sansom SN, Smith J, Dobenecker M-W, Tarakhovsky A, Livesey FJ. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proceedings of the National Academy of Sciences. 2010;107:15957–15962. doi: 10.1073/pnas.1002530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernaute B, Cañon S, Crespo M, Fernandez-Tresguerres B, Rayon T, Manzanares M. Comparison of extraembryonic expression of Eomes and Cdx2 in pregastrulation chick and mouse embryo unveils regulatory changes along evolution. Dev Dyn. 2010;239:620–629. doi: 10.1002/dvdy.22176. [DOI] [PubMed] [Google Scholar]
- Pinson J, Mason JO, Simpson TI, Price DJ. Regulation of the Pax6 : Pax6(5a) mRNA ratio in the developing mammalian brain. BMC Dev Biol. 2005;5:13. doi: 10.1186/1471-213X-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñon MC, Tuoc TC, Ashery-Padan R, Molnár Z, Stoykova A. Altered molecular regionalization and normal thalamocortical connections in cortex-specific Pax6 knock-out mice. Journal of Neuroscience. 2008;28:8724–8734. doi: 10.1523/JNEUROSCI.2565-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouchelon G, Gambino F, Bellone C, Telley L, Vitali I, Lüscher C, Holtmaat A, Jabaudon D. Modality-specific thalamocortical inputs instruct the identity of postsynaptic L4 neurons. Nature. 2014;511:471–474. doi: 10.1038/nature13390. [DOI] [PubMed] [Google Scholar]
- Prosser J, van Heyningen V. PAX6 mutations reviewed. Hum Mutat. 1998;11:93–108. doi: 10.1002/(SICI)1098-1004(1998)11:2<93::AID-HUMU1>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Quinn JC, Molinek M, Martynoga BS, Zaki PA, Faedo A, Bulfone A, Hevner RF, West JD, Price DJ. Pax6 controls cerebral cortical cell number by regulating exit from the cell cycle and specifies cortical cell identity by a cell autonomous mechanism. Developmental Biology. 2007;302:50–65. doi: 10.1016/j.ydbio.2006.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiring R, Walldorf U, Kloter U, Gehring WJ. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- Raciti M, Granzotto M, Duc MD, Fimiani C, Cellot G, Cherubini E, Mallamaci A. Reprogramming fibroblasts to neural-precursor-like cells by structured overexpression of pallial patterning genes. Mol Cell Neurosci. 2013;57:42–53. doi: 10.1016/j.mcn.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet. 2013;14:347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein JL, Anderson S, Shi L, Miyashita-Lin E, Bulfone A, Hevner R. Genetic control of cortical regionalization and connectivity. Cereb Cortex. 1999;9:524–532. doi: 10.1093/cercor/9.6.524. [DOI] [PubMed] [Google Scholar]
- Sahara S, Kawakami Y, Izpisua Belmonte JC, O'Leary DDM. Sp8 exhibits reciprocal induction with Fgf8 but has an opposing effect on anterior-posterior cortical area patterning. Neural Dev. 2007;2:10. doi: 10.1186/1749-8104-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem CE, Markl ID, Bender CM, Gonzales FA, Jones PA, Liang G. PAX6 methylation and ectopic expression in human tumor cells. Int J Cancer. 2000;87:179–185. doi: 10.1002/1097-0215(20000715)87:2<179::aid-ijc4>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Sansom SN, Griffiths DS, Faedo A, Kleinjan D-J, Ruan Y, Smith J, van Heyningen V, Rubenstein JL, Livesey FJ. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet. 2009;5:e1000511. doi: 10.1371/journal.pgen.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scardigli R, Schuurmans C, Gradwohl G, Guillemot F. Crossregulation between Neurogenin2 and pathways specifying neuronal identity in the spinal cord. Neuron. 2001;31:203–217. doi: 10.1016/s0896-6273(01)00358-0. [DOI] [PubMed] [Google Scholar]
- Scardigli R. Direct and concentration-dependent regulation of the proneural gene Neurogenin2 by Pax6. Development. 2003;130:3269–3281. doi: 10.1242/dev.00539. [DOI] [PubMed] [Google Scholar]
- Schedl A, Ross A, Lee M, Engelkamp D, Rashbass P, van Heyningen V, Hastie ND. Influence of PAX6 gene dosage on development: overexpression causes severe eye abnormalities. Cell. 1996;86:71–82. doi: 10.1016/s0092-8674(00)80078-1. [DOI] [PubMed] [Google Scholar]
- Schmahl W, Knoedlseder M, Favor J, Davidson D. Defects of neuronal migration and the pathogenesis of cortical malformations are associated with Small eye (Sey) in the mouse, a point mutation at the Pax-6-locus. Acta Neuropathol. 1993;86:126–135. doi: 10.1007/BF00334879. [DOI] [PubMed] [Google Scholar]
- Shaham O, Gueta K, Mor E, Oren-Giladi P, Grinberg D, Xie Q, Cvekl A, Shomron N, Davis N, Keydar-Prizant M, Raviv S, Pasmanik-Chor M, Bell RE, Levy C, Avellino R, Banfi S, Conte I, Ashery-Padan R. Pax6 regulates gene expression in the vertebrate lens through miR-204. PLoS Genet. 2013;9:e1003357. doi: 10.1371/journal.pgen.1003357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham O, Menuchin Y, Farhy C, Ashery-Padan R. Pax6: a multi-level regulator of ocular development. Progress in Retinal and Eye Research. 2012;31:351–376. doi: 10.1016/j.preteyeres.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Sheikh BN. Crafting the brain - role of histone acetyltransferases in neural development and disease. Cell Tissue Res. 2014;356:553–573. doi: 10.1007/s00441-014-1835-7. [DOI] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, Ren B. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S, Kwan KY, Li M, Lefebvre V, Šestan N. Cis-regulatory control of corticospinal system development and evolution. Nature. 2012;486:74–79. doi: 10.1038/nature11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Ming G-L, Song H. Decoding neural transcriptomes and epigenomes via high-throughput sequencing. Nature Publishing Group. 2014;17:1463–1475. doi: 10.1038/nn.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Onge L, Sosa-Pineda B, Chowdhury K, Mansouri A, Gruss P. Pax6 is required for differentiation of glucagon-producing alpha-cells in mouse pancreas. Nature. 1997;387:406–409. doi: 10.1038/387406a0. [DOI] [PubMed] [Google Scholar]
- Stoykova A, Fritsch R, Walther C, Gruss P. Forebrain patterning defects in Small eye mutant mice. Development. 1996;122:3453–3465. doi: 10.1242/dev.122.11.3453. [DOI] [PubMed] [Google Scholar]
- Stoykova A, Treichel D, Hallonet M, Gruss P. Pax6 modulates the dorsoventral patterning of the mammalian telencephalon. Journal of Neuroscience. 2000;20:8042–8050. doi: 10.1523/JNEUROSCI.20-21-08042.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian L, Tole S. Mechanisms Underlying the Specification, Positional Regulation, and Function of the Cortical Hem. Cereb Cortex. 2009;19 doi: 10.1093/cercor/bhp031. [DOI] [PubMed] [Google Scholar]
- Suda Y, Kokura K, Kimura J, Kajikawa E, Inoue F, Aizawa S. The same enhancer regulates the earliest Emx2 expression in caudal forebrain primordium, subsequent expression in dorsal telencephalon and later expression in the cortical ventricular zone. Development. 2010;137:2939–2949. doi: 10.1242/dev.048843. [DOI] [PubMed] [Google Scholar]
- Takanaga H, Tsuchida-Straeten N, Nishide K, Watanabe A, Aburatani H, Kondo T. Gli2 is a novel regulator of sox2 expression in telencephalic neuroepithelial cells. Stem Cells. 2009;27:165–174. doi: 10.1634/stemcells.2008-0580. [DOI] [PubMed] [Google Scholar]
- Tarabykin V, Stoykova A, Usman N, Gruss P. Cortical upper layer neurons derive from the subventricular zone as indicated by Svet1 gene expression. Development. 2001;128:1983–1993. doi: 10.1242/dev.128.11.1983. [DOI] [PubMed] [Google Scholar]
- Tashiro K, Teissier A, Kobayashi N, Nakanishi A, Sasaki T, Yan K, Tarabykin V, Vigier L, Sumiyama K, Hirakawa M, Nishihara H, Pierani A, Okada N. A mammalian conserved element derived from SINE displays enhancer properties recapitulating Satb2 expression in early-born callosal projection neurons. PLoS ONE. 2011;6:e28497. doi: 10.1371/journal.pone.0028497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tétreault N, Champagne M-P, Bernier G. The LIM homeobox transcription factor Lhx2 is required to specify the retina field and synergistically cooperates with Pax6 for Six6 trans-activation. Developmental Biology. 2009;327:541–550. doi: 10.1016/j.ydbio.2008.12.022. [DOI] [PubMed] [Google Scholar]
- Theil T, Aydin S, Koch S, Grotewold L, Rüther U. Wnt and Bmp signalling cooperatively regulate graded Emx2 expression in the dorsal telencephalon. Development. 2002;129:3045–3054. doi: 10.1242/dev.129.13.3045. [DOI] [PubMed] [Google Scholar]
- Ton CC, Hirvonen H, Miwa H, Weil MM, Monaghan P, Jordan T, van Heyningen V, Hastie ND, Meijers-Heijboer H, Drechsler M. Positional cloning and characterization of a paired box- and homeobox-containing gene from the aniridia region. Cell. 1991;67:1059–1074. doi: 10.1016/0092-8674(91)90284-6. [DOI] [PubMed] [Google Scholar]
- Toresson H, Potter SS, Campbell K. Genetic control of dorsal-ventral identity in the telencephalon: opposing roles for Pax6 and Gsh2. Development. 2000;127:4361–4371. doi: 10.1242/dev.127.20.4361. [DOI] [PubMed] [Google Scholar]
- Tuoc TC, Boretius S, Sansom SN, Pitulescu M-E, Frahm J, Livesey FJ, Stoykova A. Chromatin Regulation by BAF170 Controls Cerebral Cortical Size and Thickness. Developmental Cell. 2013;25:256–269. doi: 10.1016/j.devcel.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Tuoc TC, Radyushkin K, Tonchev AB, Piñon MC, Ashery-Padan R, Molnár Z, Davidoff MS, Stoykova A. Selective cortical layering abnormalities and behavioral deficits in cortex-specific Pax6 knock-out mice. Journal of Neuroscience. 2009;29:8335–8349. doi: 10.1523/JNEUROSCI.5669-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchikawa M, Ishida Y, Takemoto T, Kamachi Y, Kondoh H. Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Developmental Cell. 2003;4:509–519. doi: 10.1016/s1534-5807(03)00088-1. [DOI] [PubMed] [Google Scholar]
- Vance KW, Sansom SN, Lee S, Chalei V, Kong L, Cooper SE, Oliver PL, Ponting CP. The long non-coding RNA Paupar regulates the expression of both local and distal genes. EMBO J. 2014;33:296–311. doi: 10.1002/embj.201386225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Carson J, Oldekamp J, Warnecke M, Jakubcakova V, Zhou X, Shaw CA, Alvarez-Bolado G, Eichele G. Regulatory pathway analysis by high-throughput in situ hybridization. PLoS Genet. 2007;3:1867–1883. doi: 10.1371/journal.pgen.0030178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Taher L, Girgis H, May D, Golonzhka O, Hoch RV, McKinsey GL, Pattabiraman K, Silberberg SN, Blow MJ, Hansen DV, Nord AS, Akiyama JA, Holt A, Hosseini R, Phouanenavong S, Plajzer-Frick I, Shoukry M, Afzal V, Kaplan T, Kriegstein AR, Rubin EM, Ovcharenko I, Pennacchio LA, Rubenstein JLR. A high-resolution enhancer atlas of the developing telencephalon. Cell. 2013;152:895–908. doi: 10.1016/j.cell.2012.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walcher T, Xie Q, Sun J, Irmler M, Beckers J, Öztürk T, Niessing D, Stoykova A, Cvekl A, Ninkovic J, Götz M. Functional dissection of the paired domain of Pax6 reveals molecular mechanisms of coordinating neurogenesis and proliferation. Development. 2013;140:1123–1136. doi: 10.1242/dev.082875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski OL, Vierbuchen T, Qu K, Lee QY, Chanda S, Fuentes DR, Giresi PG, Ng YH, Marro S, Neff NF, Drechsel D, Martynoga B, Castro DS, Webb AE, Südhof TC, Brunet A, Guillemot F, Chang HY, Wernig M. Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell. 2013;155:621–635. doi: 10.1016/j.cell.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen S, Li H, Liu J. Epigenetic background of neuronal fate determination. Progress in Neurobiology. 2009;87:98–117. doi: 10.1016/j.pneurobio.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Wenger AM, Clarke SL, Notwell JH, Chung T, Tuteja G, Guturu H, Schaar BT, Bejerano G. The enhancer landscape during early neocortical development reveals patterns of dense regulation and co-option. PLoS Genet. 2013;9:e1003728. doi: 10.1371/journal.pgen.1003728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SC, Altmann CR, Chow RL, Hemmati-Brivanlou A, Lang RA. A highly conserved lens transcriptional control element from the Pax-6 gene. Mechanisms of Development. 1998;73:225–229. doi: 10.1016/s0925-4773(98)00057-4. [DOI] [PubMed] [Google Scholar]
- Wu D, Li T, Lu Z, Dai W, Xu M, Lu L. Effect of CTCF-binding motif on regulation of PAX6 transcription. Investigative Ophthalmology & Visual Science. 2006;47:2422–2429. doi: 10.1167/iovs.05-0536. [DOI] [PubMed] [Google Scholar]
- Wu H, Sun YE. Epigenetic regulation of stem cell differentiation. Pediatr Res. 2006;59:21R-5R. doi: 10.1203/01.pdr.0000203565.76028.2a. [DOI] [PubMed] [Google Scholar]
- Xie Q, Ung D, Khafizov K, Fiser A, Cvekl A. Gene regulation by PAX6: structural-functional correlations of missense mutants and transcriptional control of Trpm3/miR-204. Mol Vis. 2014;20:270–282. [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Yang Y, Huang J, Ninkovic J, Walcher T, Wolf L, Vitenzon A, Zheng D, Götz M, Beebe DC, Zavadil J, Cvekl A. Pax6 interactions with chromatin and identification of its novel direct target genes in lens and forebrain. PLoS ONE. 2013;8:e54507. doi: 10.1371/journal.pone.0054507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu PX, Zhang X, Heaney S, Yoon A, Michelson AM, Maas RL. Regulation of Pax6 expression is conserved between mice and flies. Development. 1999;126:383–395. doi: 10.1242/dev.126.2.383. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Sawada J, Yamada M, Handa H, Azuma N. Autoregulation of Pax6 transcriptional activation by two distinct DNA-binding subdomains of the paired domain. Genes Cells. 1997;2:255–261. doi: 10.1046/j.1365-2443.1997.1170315.x. [DOI] [PubMed] [Google Scholar]
- Yang Q, Shao Y, Shi J, Qu Y, Wu K, Dang S, Shi B, Hou P. Concomitant PIK3CA amplification and RASSF1A or PAX6 hypermethylation predict worse survival in gastric cancer. Clin Biochem. 2013 [PubMed] [Google Scholar]
- Yang Y, Stopka T, Golestaneh N, Wang Y, Wu K, Li A, Chauhan BK, Gao CY, Cveklová K, Duncan MK, Pestell RG, Chepelinsky AB, Skoultchi AI, Cvekl A. Regulation of alphaA-crystallin via Pax6, c-Maf, CREB and a broad domain of lens-specific chromatin. EMBO J. 2006;25:2107–2118. doi: 10.1038/sj.emboj.7601114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You JS, De Carvalho DD, Dai C, Liu M, Pandiyan K, Zhou XJ, Liang G, Jones PA. SNF5 is an essential executor of epigenetic regulation during differentiation. PLoS Genet. 2013;9:e1003459. doi: 10.1371/journal.pgen.1003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun K, Potter S, Rubenstein JL. Gsh2 and Pax6 play complementary roles in dorsoventral patterning of the mammalian telencephalon. Development. 2001;128:193–205. doi: 10.1242/dev.128.2.193. [DOI] [PubMed] [Google Scholar]
- Zappone MV, Galli R, Catena R, Meani N, De Biasi S, Mattei E, Tiveron C, Vescovi AL, Lovell-Badge R, Ottolenghi S, Nicolis SK. Sox2 regulatory sequences direct expression of a (beta)-geo transgene to telencephalic neural stem cells and precursors of the mouse embryo, revealing regionalization of gene expression in CNS stem cells. Development. 2000;127:2367–2382. doi: 10.1242/dev.127.11.2367. [DOI] [PubMed] [Google Scholar]
- Zembrzycki A, Griesel G, Stoykova A, Mansouri A. Genetic interplay between the transcription factors Sp8 and Emx2 in the patterning of the forebrain. Neural Dev. 2007;2:8. doi: 10.1186/1749-8104-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zembrzycki A, Chou S-J, Ashery-Padan R, Stoykova A, O'Leary DDM. Sensory cortex limits cortical maps and drives top-down plasticity in thalamocortical circuits. Nature Publishing Group. 2013;16:1060–1067. doi: 10.1038/nn.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]