Abstract
Long noncoding RNAs (lncRNAs) represent a newly discovered class of regulatory molecules that impact a variety of biological processes in cells and organ systems. In humans, it is estimated that there may be more than twice as many lncRNA genes than protein-coding genes. However, only a handful of lncRNAs have been analyzed in detail. In this review, we describe expression and functions of lncRNAs that have been demonstrated to impact innate and adaptive immunity. These emerging paradigms illustrate remarkably diverse mechanisms that lncRNAs utilize to impact the transcriptional programs of immune cells required to fight against pathogens and maintain normal health and homeostasis.
Keywords: Long non-coding RNA, Whole genome RNA-sequencing, Immune system, Epigenetics, Transcriptional regulation
1. Introduction
The first draft of the sequence of the human genome was released in 2001 (Lander et al., 2001; Venter et al., 2001). Shortly thereafter in 2003, the Encyclopedia of DNA Elements (ENCODE) consortium was founded, whose goal was, as the name implies, to discover and functionally define all DNA elements in the human genome (Consortium et al., 2007; Euskirchen et al., 2007; Gerstein et al., 2007; Korbel et al., 2007; Rozowsky et al., 2007; Consortium, 2012; Harrow et al., 2012; Guo et al., 2014). One of the major surprises that emerged from these studies was that the vast majority of the human genome is transcribed in some cell type at some stage during development (Amaral et al., 2008; Dinger et al., 2009; Trapnell et al., 2010; Cabili et al., 2011; Chu et al., 2011; Derrien et al., 2012; Djebali et al., 2012). In contrast, only about 2% of the genome contains genes that are transcribed into mRNAs and translated into proteins.
From these studies, a new class of RNA molecule has emerged termed long non-coding RNA or lncRNA (Cabili et al., 2011; Dinger et al., 2008a; Guttman et al., 2009; Mercer et al., 2009; Orom et al., 2010; Rinn and Chang, 2012; Hangauer et al., 2013a; Morris and Mattick, 2014). LncRNAs are operationally defined as greater than 200 bp in length to distinguish them from short RNAs such as microRNAs. Studies reveal that the majority of lncRNAs discovered thus far have a 5′ cap structure, are polyadenylated, and are spliced just like mRNAs. Epigenetic studies that examine histone modifications at genomic loci have also aided the discovery of lncRNAs. For example, the H3K4-trimethylation and H3K36-trimethylation marks are common tags at the transcription initiation sites and the gene bodies of actively transcribed genes that encode mRNAs, respectively. In fact, one major aid in the discovery of lncRNAs was chromatin immunoprecipitation-whole genome sequencing, ChIP-seq (Euskirchen et al., 2007), studies that demonstrated that many sites in the genome possessed these marks but were not associated with any known protein-coding gene. The discovery of lncRNAs came from the recognition via whole genome RNA-sequencing or other methods that RNA molecules were actually transcribed from these genomic loci containing these characteristic epigenetic marks. The major difference between mRNAs and lncRNAs is that lncRNAs are littered with translational stop codons and thus have little if any protein-coding potential. Both computational and experimental approaches are available to test protein-coding potential of a novel RNA transcript. Computational approaches include PhyloCSF or phylogenetic analysis of multi-species genome alignments and CPAT or coding-potential assessment tool (Wang et al., 2013a; Lin et al., 2011a). Experimental approaches include ribosome footprinting, analysis of the association of lncRNAs of interest with polysomes and in vitro transcription and translation procedures (Guttman et al., 2013; Ingolia et al., 2011). However, a concern with these computational and experimental approaches is that they only produce a negative finding so improved approaches to positively identify lncRNAs are being actively sought (Dinger et al., 2008b). According to current estimates, there are approximately equivalent numbers of genes encoding lncRNAs as genes encoding mRNAs in the human genome that have been discovered thus far (Fig. 1A). Since the study of lncRNAs is a relatively new field, it seems that many more lncRNA genes will be discovered and the number of genes encoding lncRNAs is likely to far exceed the number of genes in the human genome that encode mRNAs.
Fig. 1.
Comparison of non-coding and coding genes across different genomes. (A) Venndiagram illustrates numbers and percentages of known genes in human and mouse genomes (adapted from GENCODE, 2015 update). (B) Increased complexity of organisms is associated with marked expansion of genome size and non-coding RNA. X-axis is genome size in bp and Y-axis is size of the genome encoding mRNA or ncRNA (adapted from: Liu, G, Mattick, JS, Taft, RJ. ‘A meta-analysis of the genomic and transcriptomic composition of complex life’. Cell Cycle 12:2061–2072, 2013).
Evolutionary studies demonstrate that humans, as well as other vertebrates have approximately the same number of protein-coding genes (∼20,000) as nematodes or round worms (Kapusta and Feschotte, 2014). However the sizes of the genomes in these two organisms are vastly different and a consequence of this is that the fraction of the genome devoted to synthesis of non-coding RNA has increased dramatically during evolution (Fig. 1B). Since most of the human genome is transcribed into various non-coding RNA species, one interpretation of these data is that that lower species such as zebrafish, nematodes, fruit flies, and yeast contain a much lower number of lncRNAs and lower ratios of lncRNAs to protein coding genes than higher species such as primates, mice, cows, etc. (Liu et al, 2013; Clark et al, 2013). However, a caution is that annotations of the genome in these lower species are not as extensive as they are in human genomes. As we will discuss, lncRNAs play important roles in many developmental processes and exhibit cell type specific expression patterns and functions. As such, it may be that as an organism becomes developmentally more complex, it acquires more lncRNA genes to help guide this developmental complexity (Mattick et al, 2009; Guttman et al., 2011; Ulitsky and Bartel, 2013).
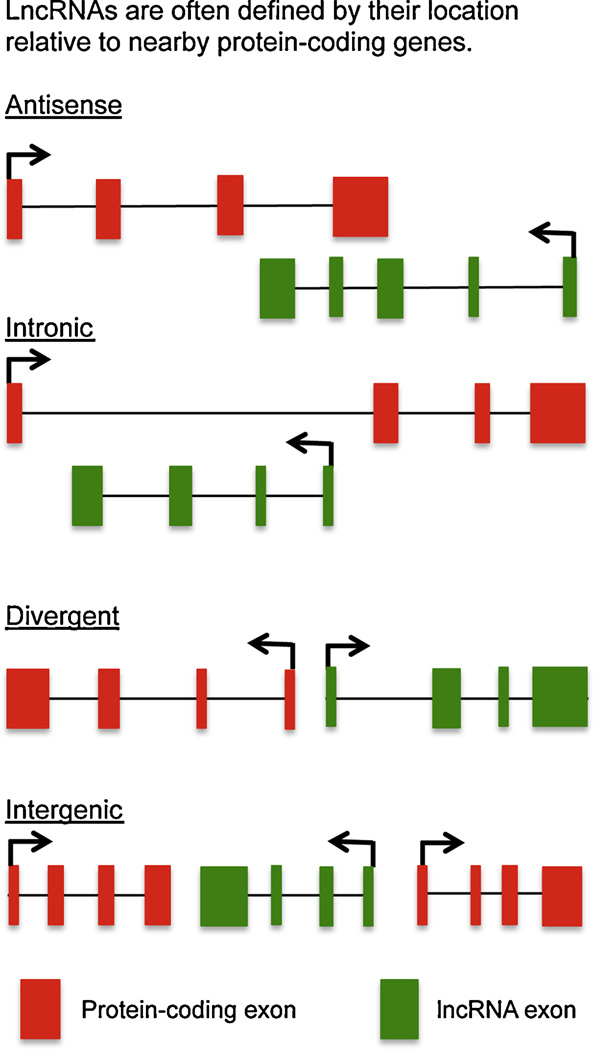
LncRNAs are often named according to their gene location in genomes relative to protein coding genes (Fig. 2). LncRNA genes that are located between two protein-coding genes with no overlap with protein-coding genes are often referred to as long or large intergenic or intervening ncRNAs abbreviated lincRNA LncRNA genes may also be totally located within one intron of a protein-coding gene with no overlap with protein-coding exon. These lncRNA genes are referred to as ‘intronic’. LncRNA genes are also found that overlap with exons and introns of protein coding genes but also utilize novel exons not located within the protein-coding gene body and are oftentimes transcribed from the opposite DNA strand. These are referred to as antisense lncRNAs. Divergent lncRNAs initiate in a divergent fashion or from the opposite strand of a protein-coding gene and transcriptional initiations sites of these two genes are oftentimes within a few hundred bp of each other (Sigova et al., 2013). LncRNAs are often named according to their proximity to the nearest protein-coding gene. In part, this convention has been adopted because some lncRNAs often regulate transcription of the neighboring protein-coding gene. A couple of examples would be the PANDA (p21 associated ncRNA DNA damage activated) lncRNA and FENDRR (FOXF1 adjacent non-coding developmental regulatory RNA) (Hung et al, 2011; Grote et al, 2013). As we will see later, this is not a hard and fast rule as many lncRNAs do not affect transcription of their nearest protein-coding gene neighbor but this naming convention is still quite common.
Fig. 2.
Anatomy of long noncoding RNA (lncRNA) loci. Antisense lncRNAs initiate inside or 3′ of a protein-coding gene, are transcribed in the opposite direction of proteincoding genes, and overlap at least one coding exon (THRIL, Fas-AS1, IL-1b–AS, TH2-LCR lncRNA). Intronic lncRNAs initiate inside of an intron of a protein-coding gene in either direction and terminate without overlapping exons (NRON). Divergent lncRNAs initiate in a divergent fashion from the promoter of a protein-coding gene; the precise distance cutoff constituting bidirectionality is not defined but is generally within a few hundred base pairs (GATA3-AS1). Intergenic lncRNAs (also termed long intergenic noncoding RNAs or lincRNAs) are lncRNAs with separate transcriptional units from protein-coding genes (IFNG-AS1 (NeST, Tmevpg1), lincRNA-Cox2, Lethe, lnc-DC, Linc-Ccr2-5′AS, PACER, linc-MAF-4, lincRNA-p21 (adapted from reference Rinn and Chang (2012)).
Since lncRNAs can alter the transcription of protein-coding genes either via direct or indirect mechanisms, one approach to identify these target genes is to manipulate levels of the lncRNA of interest, either positively or negatively, and analyze global changes in gene expression via some method such as whole-genome RNA sequencing (RNA-seq) or microarrays to identify potential protein-coding gene targets. The number of lncRNAs studied in detail is relatively small compared to the total number of lncRNAs known to exist. Of the lncRNAs that have been described, many of which are nuclear-enriched lncRNAs, some general themes apply and the mechanism that each lncRNA employs typically involves DNA, protein, or RNA elements (Satpathy and Chang, 2015). They are (1) lncRNA expression, like mRNA expression, is regulated by intracellular signaling pathways and the combinatorial actions of signaling responsive and basal transcription factors, (2) lncRNAs associate with chromatin-modifying enzymes and are able to recruit them to target gene loci; these target gene loci may be either adjacent protein-coding genes (regulation in cis), or distant and possibly multiple protein-coding genes (regulation in trans), (3) lncRNAs also associate with transcription factors and titrate them away from target gene loci to negatively impact transcription or stabilize transcription factors to allow increased accumulation and activity, (4) lncRNAs form ribonucleoprotein complexes that positively or negatively affect transcription of target genes; the heterogeneous ribonucleoproteins or hnRNPs are one such example (Table 1) (Mercer et al, 2009; Mattick et al., 2009; Khalil et al., 2009; Nagano and Fraser, 2011; Guttman and Rinn, 2012; Hu et al., 2012) and (5) lncRNA-cytoplasmic RNA interactions target the stability of messenger RNAs and alter translation (Satpathy et al., 2015; Carrieri et al., 2012 ). Less is known about cytoplasmic incRNA-RNA interactions. Thus, some common themes of mechanisms of action of lncRNAs have emerged but undoubtedly, additional mechanisms of action of this diverse population of RNA species will be discovered in the not too distant future.
Table 1.
Some common mechanisms of regulation and action of lncRNAs.
| LncRNA genes are regulated by combinatorial activities of transcription factors and promoters and distal enhancers just like protein-coding genes, IFNG-AS1 (NeST, Tmevpg1) |
| LncRNAs act as ‘guides’ or ‘scaffolds’ and form ribonucleoprotein complexes with a) histone modifying enzymes to write the ‘histone code’ at target loci in cis (IFNG-AS1, linc-MAF-4) or trans (THRIL), b) form complexes with hnRNPs to affect transcription of target protein-coding genes (lincRNA-p21, lincRNA-Cox2), orc) transcription factors to help guide them to target genomic loci (THRIL). |
| LncRNAs bind to mRNAs to alter rates of translation (lincRNA-p21) |
| LncRNAs bind to proteins to affect their activity, stability and intracellular levels (NRON, lincRNA-p21) |
| LncRNAs act as ‘decoys’ to sequester transcription factors away from promoters/enhancers to block transcription of target protein-coding genes (Lethe) Adapted from reference 22. |
2. T and B lymphocytes and lncRNAs
Emerging evidence suggests that the majority of lncRNAs are expressed in a cell-type specific fashion suggesting that each cell lineage may express its own unique constellation of lncRNAs. Also, lncRNAs are thought to play key roles in the developmental programs that give rise to different cells and organs of the body (Paralkar et al., 2014; Sunwoo et al., 2009). It is also thought that those organs that exhibit the greatest complexity, such as the brain, also contain the most diverse lncRNA transcriptome (Mercer et al., 2007; Mercer et al., 2008a,b; Sauvageau et al., 2013). This feature may contribute to the many diverse functions of the central and peripheral nervous systems. By extension, these lncRNAs may be critically important for maintaining cell identity. Innate and adaptive immune systems arise from hematopoietic stem cells and give rise to an array of different cell types during development (Hollander et al., 2006; Kurobe et al., 2006; Steinman and Hemmi, 2006; Takahama, 2006; Boehm, 2008; Medzhitov and Horng, 2009; Pepper and Jenkins, 2011; Murray and Smale, 2012; Merad et al., 2013; Murphy, 2013; Youngblood et al., 2013; Diefenbach et al., 2014; Farber et al., 2013; McKenzie et al., 2014; Shih et al., 2014). Certain lineages, such as B cells, T cells, and monocytes are very distinct in their roles in the immune system while others, such as NK cells, NKT cells, and T cells exhibit both shared and unshared properties and functions. Additional examples are the conventional T cells that express traditional a/b T cell receptors, T cells that express g/d T cell receptors, and the recently identified innate lymphoid cells that express many T cell markers but do not express T cell receptors. T lymphocytes also differentiate in the thymus through distinct developmental stages from the CD4, CD8 double negative (DN) stage, DN1, DN2, DN3, DN4, to the CD4, CD8 double positive (DP) stage, DP1, DP1, to the CD4 or CD8 single positive stage prior to emigration to the periphery. In the periphery, conventional CD4+ T cells can undergo further differentiation to become stable effector cells, TH1, TH2, or TH17, defined by their ability to express genes that encode the cytokines, IFN-γ, IL-4, IL-13, and IL-5, or IL-17, respectively. Similarly, there are independent lineages of innate lymphoid cells that selectively express IFN-γ, IL-4, IL-13, and IL-5, or IL-17, respectively. T follicular helper cells also develop in the periphery from naïve CD4+ T cells in response to differentiating stimuli. The T regulatory compartment can also be divided by developmental lineages, termed nTreg or natural T regulatory cells that develop within the thymus and iTreg or induced T reg cells that develop in the periphery. These are distinct cells but share certain features and functions. Several studies have employed whole genome RNA-seq to identify lncRNAs in human and mouse lymphocytes (Pang et al., 2009; Hu et al., 2013; Ranzani et al., 2015; Spurlock et al., 2015). In general, these studies have identified thousands of unique lncRNAs expressed by CD4+ T cells at different stages of development and differentiation. In keeping with the ideas about the complexity of the brain and the greater diversity of expression of lncRNAs, it is possible that the complexity of the immune system may also be represented by similarly complex patterns of lncRNA expression. This idea has been examined in a rather comprehensive study (Table 2) (Hu et al., 2013). Of the lncRNAs expressed at the DN stages of thymocyte development, the DP and SP stages of thymocyte development, and the different lineages of T effector cells, iTreg, TH1, TH2, and TH17, the majority of lncRNAs are expressed in a lineage specific manner and a minority expressed by T cells at all stages of development and differentiation. For example, during the DN stage of thymocyte development, >50% of expressed lncRNAs are found at only one DN stage while ∼10% of expressed lncRNAs are found at all DN stages. In marked contrast, this is not the case for mRNAs. Thus, only 8% of mRNAs are expressed at only one DN stage while >75% of expressed mRNAs are found at all DN stages. Overall, T cells express most mRNAs in a non-lineage specific manner at all stages of development and differentiation and only small numbers of mRNAs exhibit cell-type specific expression.
Table 2.
LncRNAs display cell lineage specific expression patterns.
| THYMUS | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| DN1 | DN2 | DN3 | DN4 | DP1 | DP2 | CD4 | CD8 | tTreg | |
| Lineage specific lncRNAs | 135 | 29 | 69 | 32 | 104 | 17 | 3 | 11 | 143 |
| Common lncRNAs | 61 | 79 | |||||||
| Lineage specific mRNAs | 728 | 107 | 180 | 196 | 210 | 94 | 30 | 142 | 412 |
| Common mRNAs | 11,005 | 11,008 | |||||||
| PERIPHERY | |||||||||
| naïve CD4 | iTreg | TH1 | TH2 | TH17 | |||||
| Lineage specific lncRNAs | 79 | 37 | 101 | 63 | 27 | ||||
| Common lncRNAs | 100 | ||||||||
| Lineage specific mRNAs | 148 | 139 | 343 | 166 | 128 | ||||
| Common mRNAs | 10,874 |
Numbers indicate total unique lineage specific or cell type specific lncRNAs or common lncRNAs and total lineage specific mRNAs or common mRNAs found in the indicated classes of double negative (DN) and double positive (DP) and CD4+, CD8+ and iTreg in the thymus and the indicated classes of naïve and effector peripheral T cells (adapted from reference Farber et al. (2013)).
Besides the effector T cell compartment, which is thought to be short-lived, naïve T cells also develop into T cell memory compartments, both central memory, Tcm, and effector memory, Tem, by acquiring and sustaining new transcriptional programs. How these altered transcriptional programs are maintained for long periods of time and how this information is transmitted to daughter cells is not well understood but it seems that lncRNAs are likely candidates to play critical roles in these processes, which are key to the ability of the host to sustain long-lived immunity to a foreign pathogen after initial infection. Below are specific examples of lncRNAs that contribute to both T helper cell differentiation and function and B lymphocyte function that have been characterized in some detail.
2.1. IFNG-AS1
IFNG-AS1 was originally discovered using genetic studies as a gene adjacent to IFNG in both mouse and human genomes that encodes a lncRNA implicated in the control of Theiler’s virus persistence perhaps by transcriptional regulation of the gene encoding IFN-γ (Vigneau et al., 2003). This lncRNA is encoded on the DNA strand opposite to the IFNG coding strand. Originally named TMEVPG1 (Theiler’s murine encephalomyelitis virus persistence candidate gene 1), this gene has also been named NeST (NEt-toie Salmonella pas Theiler’s (cleanup Salmonella not Theiler’s)) (Gomez et al., 2013) and more recently IFNG-AS1 since it is transcribed on the opposite DNA strand to IFNG. CD4+ T cells, CD8+ T cells and NK cells express this lncRNA, making it one of the first lncRNAs discovered that is expressed by cells of the immune system. For simplicity, we will use the IFNG-AS1 nomenclature. Naïve CD4+ T cells do not express IFNG-AS1 lncRNA but in response to TH1 differentiation signals, this lncRNA is induced via Stat4 and T-bet dependent pathways (Collier et al., 2012). IFNG-AS1 and IFNG utilize distinct enhancer elements that both respond to the TH1 lineage specific transcription factors, Stat4 and T-bet, as well as the non-lineage specific transcription factors, NF-κB and Ets1 (Collier et al, 2014; Chang and Aune, 2007; Collins et al., 2010; Collins et al., 2012). This ability to respond to NF-κB probably contributes to markedly increased expression of IFNG-AS1 in effector TH1 cells in response to only T cell receptor stimulation in the absence of the initial TH1 differentiation signals. During early differentiation of TH1 cells, IFNG-AS1 lncRNA also cooperates with T-bet to stimulate IFNG transcription. It is not known if IFNG transcription at later stages of TH1 differentiation or by effector memory T cells also depends upon functional cooperativity between IFNG-AS1 lncRNA and T-bet or is dependent only upon IFNG-AS1 lncRNA. This cooperativity appears to be achieved by the ability of IFNG-AS1 lncRNA to associate with histone H3K4 methyltransferase enzyme complexes and recruit these to the IFNG locus to establish H3K4 tri-methylation at the IFNG promoter and intronic regions, thus creating a permissive transcriptional environment (Gomez et al, 2013). Importantly, ability to produce IFNG-AS1 lncRNA confers resistance to lethal infection by a bacterial pathogen, thus establishing a critical role for an lncRNA in one of the major functions of the adaptive immune system (Fig. 3).
Fig. 3.
Cartoon model of IFNG-AS1 regulation and function (human genome is shown). TH1 differentiation signals, e.g. Stat4, T-bet, etc., cooperatively activate genomic proximal and distal enhancers selective for either IFNG or IFNGAS1. T-bet and IFNG-AS1 (blue arrow) cooperate to stimulate TH1-dependent IFNG transcription; IFNG-AS1 RNA recruits H3K4 histone methyltransferases to IFNG locus helping to maintain locus in a transcriptionally favorable state. Presence of IFNG-AS1 RNA is essential for robust expression of IFNG and resistance to Salmonella infection, in vivo, in mice.
2.2. TH2-LCR lncRNA
We have also performed whole genome RNA-sequencing (RNA-seq) to identify additional lncRNAs expressed in a TH1, TH2, or TH17 lineage specific manner in human T cells (Spurlock et al, 2015). We divided the analysis to search for both lncRNAs previously annotated in the human genome and novel lncRNAs that were not previously identified in the human genome. We identified both previously annotated and novel lncRNAs expressed in a TH1, TH2, and TH17 lineage specific fashion by human T cells. Most lineage specific protein coding genes in the genome were adjacent to genes that encode lineage specific lncRNAs and these mRNAs were co-expressed with these lncRNAs. One interpretation is that these lineage specific lncRNAs shape the lineage specific mRNA transcriptional profile that is essential for effector TH1, TH2, and TH17 cells to carry out their unique immune functions.
One lncRNA expressed in a TH2 lineage specific manner is co-expressed with the IL4, IL5, and IL13 genes. This lncRNA actually represents four novel lncRNA isoforms with both shared and unique RNA sequences. Using the nomenclature described above, this lncRNA cluster can be considered an antisense lncRNA because it is transcribed antisense to the RAD50 gene and partially shares RAD50 exons and introns but also employs exons outside of the RAD50 locus between RAD50 and IL13 and between IL13 and IL4. Interestingly, this same genomic region in mice functions as a locus control region or LCR (Lee et al., 2005). In general, LCRs are operationally defined by their ability to control expression of linked genes, usually genes that are members of a gene family, such as the globin genes, genes that encode the growth hormone family, or in this case, genes that encode the key TH2 cytokines, IL-4, IL-13, and IL-5. For these reasons, we named this lncRNA cluster, TH2-LCR lncRNA. Depletion of the TH2-lncRNA cluster by siRNA targeting a shared sequence among the different isoforms abrogates expression of the TH2 cytokines, IL4, IL-13, and IL-5. The TH2-LCR lncRNA associates with the WDR5 component of the histone H3K4 methyltransferase enzyme complex. Depletion of the TH2-LCR lncRNA cluster abrogates recruitment of WDR5 to IL4 and IL13 genomic promoter and enhancer elements and blocks H3K4 tri-methylation at these sites. Thus, as with certain other activating lncRNAs, the TH2-LCR lncRNA appears to act by recruiting the H3K4 methyltransferase to establish a positive transcriptional environment by formation of histone H3K4 di- and tri-methylation marks at loci around the genes that encode TH2 cytokines, IL-4, IL-13, and IL-5 (Fig. 4).
Fig. 4.
Cartoon model of the TH2-LCR lncRNA. TH2 differentiation signals induce expression of TH2-LCR lncRNA isoforms. TH2-LCR lncRNA associates with the H3K4 methyltransferase enzyme complex and facilitates recruitment to IL4, IL13, and IL5 (perhaps) gene loci to establish H3K4 trimethylation marks at the promoters and distal enhancers of these genes creating a stimulatory transcriptional environment. The TH2-LCR lncRNA is required for expression of TH2 cytokines, IL-4, IL 13, and IL-5.
Roles for TH2-LCR lncRNA in animal models have yet to be described. It is clearly established that TH2 differentiation and expression of IL-4, IL-13, and IL-5 exacerbate animal models of asthma and allergy and promote immune responses to certain pathogens, such as helminths, and suppress certain autoimmune models, such as type 1 diabetes and multiple sclerosis. One might predict that depletion of TH2-LCR lncRNA may inhibit allergic responses and asthma and that increased expression of TH2-LCR lncRNA may enhance TH2 immunity and inhibit certain autoimmune diseases in animal models and that similar results may be achieved in humans. However, this remains to be demonstrated experimentally. The fact that TH2-LCR lncRNA regulates expression of all three TH2 cytokines, IL-4, IL-13, and IL-5, may make it an attractive therapeutic target for asthma or severe allergic disease as opposed to targeting individual cytokines.
Another point is that historically, LCRs have been functionally defined using various genetic manipulations, such as transgenic mice, to delete genomic elements and test for loss of function (Kim and Dean, 2012). On a mechanistic basis, it is not totally clear how LCRs function. Although somewhat speculative, our results may suggest that LCR genomic regions actually contain genes that encode lncRNAs and the lncRNAs actually regulate transcription of their target protein-coding genes to achieve developmental or lineage specific patterns of mRNA expression of these linked genes.
2.3. Linc-Ccr2–5′AS
The lncRNA, linc-Ccr2–5′AS, is a mouse lncRNA also originally discovered by employing RNA-seq methods (Hu et al., 2013). The gene that encodes this lncRNA is located on mouse chromosome 9 in the Ccr1, Ccr3, Ccr2, and Ccr5 chemokine receptor cluster between Ccr3 and Ccr2. Thus, linc-Ccr2–5′AS fulfills the criteria of an intergenic lncRNA. As are the Ccr1, Ccr3, Ccr2, and Ccr5 genes, linc-Ccr2–5′AS is selectively expressed in the TH2 lineage and this is dependent upon the presence of the TH2 ‘master regulator’, GATA3. Depletion of linc-Ccr2–5′AS using shRNA methodology lowers expression of Ccr1, Ccr3, Ccr2, and Ccr5 genes in cells of the TH2 lineage. In a cell transfer model, depletion of linc-Ccr2–5′AS inhibits migration of TH2 effector cells into the lung of recipient mice. In contrast to lncRNAs such as IFNG-AS1 and TH2-LCR lncRNA, linc-Ccr2–5′AS does not appear to modulate histone H3K4 di- and tri-methylation at Ccr1, Ccr3, Ccr2, and Ccr5 gene loci. Thus the precise mechanism by which linc-Ccr2–5′AS regulates Ccr1, Ccr3, Ccr2 and Ccr5 expression is not known at present and will most certainly be the subject of future investigations. However, it is noteworthy that this lncRNA plays a functional role in a murine model, in vivo.
2.4. Linc-MAF-4
Linc-MAF-4 is another lncRNA that is a TH lineage specific lncRNA discovered by RNA-seq methods that plays a key role in TH lineage speciation but employs a mechanistic strategy some-what distinct from some of the other lncRNAs (Ranzani et al., 2015). The transcription factor MAF is expressed in a TH2 lineage specific manner and promotes expression of genes encoding TH2 cytokines. The gene that encodes linc-MAF-4 is located in the genome on chromosome 16 greater than 150 kb from the gene that encodes MAF. Linc-MAF-4 is expressed in a TH1 lineage specific manner and promotes TH1 differentiation and IFNG expression apparently by inhibiting expression of MAF in effector TH1 cells, which, in turn is thought to inhibit opposing TH2 differentiation thus promoting TH1 differentiation. To do so, a long distance physical interaction is established between linc-MAF-4 and MAF genomic regions, linc-MAF-4 lncRNA associates with the chromatin modifiers LSD1 and EZH2 and presumably facilitates formation of H3K27 tri-methylation marks at MAF genomic loci, thus producing a repressive chromatin environment to inhibit transcription of MAF.
In a sense, this scenario is somewhat similar to mechanisms other lncRNAs utilize to affect gene expression. What seems unique is that linc-MAF-4 is expressed in a TH1 lineage specific manner and suppresses expression of MAF, thus making expression of MAF TH2 lineage specific. The end result is that linc-MAF-4 enhances TH1 responses and IFNG transcription by dampening the TH2 response thus, shaping overall TH1/TH2 responses. It seems likely that this type of strategy may be employed by many lncRNAs that play key roles in determining cell fate choice during development.
2.5. GATA3-AS1
GATA3-AS1 fulfills the definition of a divergent lncRNA (Sigova et al., 2013). The transcriptional start site of the GATA3-AS1 gene is positioned a few hundred bp from the transcriptional start site of GATA3 and GATA3-AS1 is transcribed from the opposite DNA strand. GATA3-AS1 is a TH2 lineage specific lncRNA (Zhang et al., 2013). It is also expressed at increased levels by CD4+ T cells from allergic subjects in response to allergens. Although the function of GATA3-AS1 is unknown, these results raise the possibility that this lncRNA may play either a positive or negative role mediating allergic responses in humans.
2.6. NRON
The transcription factor NFAT was initially described in activated T cells as a critical transcriptional activator of IL2 expression. Further studies have shown that there are multiple NFAT isoforms and these are a critical transcriptional regulators of many genes in many different cell types or organ systems. In resting cells, NFAT is sequestered in the cytoplasm by phosphorylation. Dephosphorylation of NFAT by calcineurin results in translocation of NFAT to the nucleus leading to transcriptional activation of NFAT target genes, including IL2. The complex that retains NFAT in the cytoplasm also contains a lncRNA termed NRON or noncoding RNA repressor of NFAT (Willingham et al., 2005; Sharma et al., 2011). Included in this ribonucleoprotein complex are also NFAT kinases that keep NFAT phosphorylated. Additional proteins are also present in this ribonucleoprotein scaffold that contribute to keep NFAT retained in the cytoplasm and inactive. Depletion of NRON appears to disrupt this complex resulting in dephosphorylation of NFAT, and translocation to the nucleus allowing NFAT to act as a transcriptional activator of its target genes. In contrast to some of the other lncRNAs described above, NRON is located in the cytoplasm rather than the nucleus and controls gene expression by inhibiting the activity of a transcription factor rather than by targeting transcription of individual genes via epigenetic mechanisms. Further, the gene that encodes NRON is not located in the genome nearby the genes that encode NFAT or other components of the NFAT-NRON ribonucleoprotein scaffold.
2.7. Fas-AS1
The TNF receptor superfamily member, Fas (TNFRSF6, APO-1, CD95), which is expressed by T cells, B cells and a variety of tumor cells as well as other normal human tissues, has a major role in the extrinsic pathway of apoptosis. Fas is normally membrane bound. Alternate splicing leads to elimination of a single exon and produces a soluble form of Fas. This soluble form of Fas termed sFas binds Fas ligand, FasL, and inhibits binding of FasL to membrane-bound Fas, thus inhibiting apoptosis mediated by FasL. Levels of the alternate spliced forms of Fas and sFas are controlled by an antisense lncRNA named Fas-AS1 (Sehgal et al., 2014). Fas-AS1 acts by binding to the RNA binding protein, RBM5, and inhibiting exon skipping during Fas mRNA transcription, thus altering Fas:sFas ratios resulting in increased FasL-mediated apoptosis. Importantly, resistance to FasL-mediated apoptosis promotes growth and/or survival of certain primary B cell lymphomas and these lymphomas exhibit elevated levels of sFas and depressed levels of Fas-AS1. Thus, this lncRNA may be linked to lymphoma survival and resistance to apoptosis. Fas and FasL also play critical roles in regulating apoptosis via the extrinsic pathway at various stages of lymphocyte development and immune responses. The role that Fas-AS1 may play in these events is not known.
3. Innate immunity and lncRNAs
Similar whole genome lncRNA profiling using either RNA-seq or microarray based methods has also been performed with the goal of identifying lncRNAs that are uniquely expressed in cells of the innate immune system either in response to stimulation or induction of differentiation pathways. As above, many novel lncRNAs are induced or selectively expressed under these conditions. These studies also begin to illustrate the patterns of lncRNAs induced in response to a variety of stimuli, for example multiple toll-like receptor (TLR) agonists or by single stimuli, for example a single TLR agonist. The pro-inflammatory, pro-survival transcription factor, NF- κB, is also central to many pro-inflammatory responses and these studies begin to identify diverse mechanisms by which lncRNAs regulate NF-κB activity. A few lncRNAs have been studied in detail and these studies illustrate the critical role lncRNAs play in stimulation and differentiation pathways that cells of the innate immune system undergo. These studies also support that general concept that lncRNAs tend to be expressed under both cell-type and stimulus-specific conditions.
3.1. lincRNA-Cox2
Genome-wide transcriptome profiling (RNA-seq) has identified lncRNAs expressed by stimulated macrophages of the innate immune system. One such lncRNA, lncRNA-Cox2 is induced in murine bone marrow derived macrophages after stimulation of the Toll-like receptor 2 (TLR2) by lipopolysaccharide, by the synthetic bacterial lipoprotein, Pam3CSK4 (also a TLR2 agonist), and by the TLR7/8 agonist, R848 (Carpenter et al, 2013). Macrophages infected with Listeria monocytogenes or splenocytes from mice infected with Listeria also exhibit increased levels of lncRNA-Cox2 transcripts indicating that this lncRNA is induced after bacterial infection, both in tissue culture and in vivo. This lncRNA gene is an example of a non-overlapping or intergenic lncRNA gene and is so named because it is located in the genome adjacent to the Cox2 (Ptgs2) gene. Induction of Cox2 and lincRNA-Cox2 genes both depend upon the Myd88 pathway, which is central to many TLR signaling paths, and the NF-κB transcription factor. However, in contrast to many lncRNAs, such as some of those described above, lncRNA-Cox2 does not regulate transcription of Cox2. Rather, lncRNA-Cox2, either directly or indirectly, increases or decreases expression of an array of immune response genes, including genes that encode chemokines, chemokine receptors, TLR1, IL-6, and IL23a, as well as a number of genes induced by IFN-α, often referred to as IFN-response genes.
As described above, lncRNAs can localize to either cytoplasmic or nuclear compartments. In the nucleus, they may preferentially associate with the genome or may be found in other nuclear structures and this information can be useful to guide further mechanistic studies. Besides binding to enzyme complexes that write the epigenetic code, many lncRNAs bind to heterogeneous ribonucleoproteins or hnRNPs. Approximately twenty unique hnRNPs are known in humans. These proteins possess common amino acid sequence as well as amino acid sequences unique to each hnRNP and are involved in different functions of RNA biology. In the case of lincRNA-Cox2, this lncRNA is known to associate with hnRNP-A/B and hnRNP-A2/B1 and these hnRNPs are necessary for lncRNA-Cox2 to mediate its functions, which appear to result, in part, by the regulation of recruitment of RNA polymerase II to the promoters of target genes. However, it is worth noting that its association with these hnRNPs does not account for all the functions of lincRNA-Cox2. Thus, additional mechanisms of action are likely and remain to be defined.
3.2. PACER
There is also a second lncRNA gene adjacent in the genome to the Cox2 gene (Krawczyk and Emerson, 2014). This lncRNA has been named PACER for p50-associated COX-2 extragenic RNA and PACER, in contrast to lincRNA-Cox2, is a positive regulator of Cox2 transcription. The chromatin factor CCCTC-binding factor or CTCF, which is thought to function as an insulator to define open and closed chromatin boundaries, binds to a genomic region upstream of Cox2 to promote PACER lncRNA expression. PACER appears to promote transcription of Cox2 by binding to NF-κB p50/p50 homodimers, which are transcriptional repressor of the NF-κB p50/p65 transcriptional activator, and thus titrates these repressor complexes away from the Cox2 promoter that contains NF-κB binding sites. This favors binding of active NF-κB heterodimers, p65/p50, to the NF-κB binding sites in the Cox2 promoter and stimulates Cox2 transcription. It is worth noting that the p65/p50 NF-κB heterodimer is a strong transcriptional activator of many genes including many genes whose protein products play key roles in the innate immune system. However, prevailing views are that the actions of PACER are exclusively involved in the regulation of Cox2 in monocyte-macrophage cells and epithelial cells. Genomic CTCF binding sites are also involved in establishing three-dimensional chromosome conformations. It is possible that the CTCF binding sites around the PACER gene may be utilized to bring the PACER gene in close proximity to the Cox2 gene thus allowing PACER lncRNA to be produced in the immediate vicinity of the Cox2 promoter and enable PACER lncRNA to specifically titrate inhibitory NF-κB p50/p50 homodimer complexes away from the Cox2 promoter. This may also limit its ability to target NF-κB binding elements of other enhancers in the genome and increase transcription of other NF-κB target genes. Additional studies will be required to test this hypothesis.
3.3. lnc-DC
Another lncRNA expressed by cells of the monocyte/macrophage lineage is lnc-DC (Wang et al., 2014). This lncRNA is expressed exclusively by conventional human dendritic cells and is required for differentiation of human monocytes into conventional dendritic cells and differentiation of mouse bone marrow cells into conventional dendritic cells, in vivo. lnc-DC is not expressed by other lymphoid and myeloid cells including plasmacytoid dendritic cells but is expressed by conventional DC from skin and blood.
The mechanism by which lnc-DC promotes differentiation of monocytes to conventional dendritic cells is by binding to the STAT3 transcription factor and inhibiting dephosphorylation of STAT3 by the protein tyrosine phosphatase, SHP1, thus driving sustained translocation to the nucleus and sustained transcriptional activity of STAT3; see also refs (Steinman and Hemmi, 2006; Murray and Smale, 2012; Merad et al., 2013; Murphy, 2013; Guermonprez et al., 2013; Laouar et al., 2003). In so doing, lnc-DC shapes the transcriptional program required for differentiation of monocytes into conventional dendritic cells, to maintain the differentiated state of conventional dendritic cells, and to carry out the unique and critical functions of conventional dendritic cells including antigen processing and presentation and production of critical cytokines, such as IL-12, that shape the both the innate and adaptive immune response.
3.4. THRIL
THRIL or TNF-α and heterogenous nuclear ribonucleoprotein L (hnRNPL) related immunoregulatory LincRNA is a lncRNA whose levels are repressed in cells of the monocyte macrophage lineage by stimulation with the TLR1/2 agonist, Pam3CSK4 (Li et al., 2014). In the basal state, THRIL is also expressed in many but not all tissues. THRIL is required for induction of TNF gene expression in THP1 macrophages by Pam3CSK4. THRIL is also required for induction of expression of a number of immune-response genes including genes that encode different cytokines and chemokines. It is not entirely certain the extent to which this is due to a direct effect of THRIL on these other genes or an indirect effect due to its ability to regulate TNF gene expression. THRIL is another lncRNA that associates with hnRNPs, in this case, hnRNPL. The THRIL-hnRNPL complex binds to the TNF promoter/enhancer suggesting that the complex acts by stimulating transcriptional activity of the promoter/enhancer resulting in increased TNF gene expression. It is not known if this THRIL-hnRNPL complex contains other components such as transcription factors like NF-κB that are known to be required for TNF gene expression. Interestingly, high levels of TNF-α expression repress THRIL levels suggesting there is also a negative feedback loop to further regulate levels of TNF-α expressed by cells in response to inflammatory stimuli. THRIL levels have also been examined in the blood of children with Kawasaki disease. Kawasaki disease is an inflammatory disease of unknown cause that usually affects children under the age of five and is associated with elevated levels of TNF-α during the acute phase of the disease and reduction of TNF-α levels during convalescence. In contrast, THRIL levels in blood are depressed during the acute phase of disease and elevated during the convalescent phase of disease. This mirrors the negative feedback loop in cell-based models whereby TNF-α represses THRIL levels. These data raise the possibility that THRIL may play a role in inflammation due to infection. The possibility that THRIL levels may also play a role in the ‘sterile inflammation’ produced by autoimmune disease has not been examined.
3.5. Lnc-IL7R
Another lncRNA that is induced in response to LPS stimulation is named lnc-IL7R because the gene that encodes lnc-IL7R overlaps with the 3′ untranslated region of the IL7R gene (Cui et al., 2014). In contrast to many lncRNAs, lnc-IL7R is transcribed from the same 5′ to 3′ direction as is IL7R but similar to many lncRNAs, lnc-IL7R has a 5′ CAP and is polyadenylated at the 3′ end. Lnc-IL7R is also induced by the TLR2 agonist, Pam3CSK4, but not by the TLR3 agonist, poly I/C. Similar to other lncRNAs, lnc-IL7R is localized in the nucleus. Lnc-IL7R does not regulate expression of IL7R even though LPS and Pam3CSK4 also induce IL7R. Rather, it functions as a negative regulator of the LPS-induced inflammatory response in tissue culture models by repressing induction of a number of pro-inflammatory genes including those that encode E-selectin, VCAM-1, IL-6 and IL-8. Lnc-IL7R appears to act by regulating levels H3K27-trimethylation at the promoters of these genes as knockdown of lnc-IL7R results in decreased H3K27-trimethylation at these promoters and H3K27-trimethylation is associated with transcriptional silencing. Thus, lnc-IL7R appears to act by limiting the inflammatory response induced by TLR2 and TLR4 agonists. The extent to which this lncRNA regulates inflammation, in vivo, in response to pathogen infections or in autoimmune settings will be important to determine.
3.6. Lethe
Pseudogenes are thought to arise from duplication of protein-coding genes. Accumulation of subsequent mutations is thought to render them transcriptionally inactive. As such, the general view has been that pseudogenes are not transcribed into RNAs. However, recent studies from the ENCODE project have challenged this general view. Current estimates now suggest that >10,000 pseudogenes exist in the human genome and >10% are actually transcribed into lncRNAs. As with other estimates of lncRNAs, these numbers will probably increase as the annotation of the human genome improves with addition of different organ systems and cell types undergoing various stages of differentiation and responding to different stimuli.
There are five rivers of Hades: Styx, Phlegethon, Acheron, Cocytus and Lethe. The river Lethe causes complete forgetfulness if an individual drinks its water and this lncRNA is so named because of its negative feedback function (Rapicavoli et al., 2013). Lethe was discovered in a search for lncRNAs induced by TNF-α stimulation in mouse embryonic fibroblasts (MEF). Since signaling pathways activated by TNF-α are largely conserved in distinct cell types and TNF-α is an important regulator of both adaptive and immune responses, it seems worthwhile to include discussion of this lncRNA here.
Lethe is a pseudogene of the ribosomal protein S15a (Rps15a) gene, is adjacent to the Gmeb1 gene in the mouse genome, and is induced by stimulation of MEFs with TNF-α. In contrast to Lethe, transcript levels of Rps15a and Gmeb1 are unchanged under these same stimulation conditions. Lethe is also induced by the glucocorticoid receptor agonist, dexamethasone. This is consistent with the fact that glucocorticoid receptors and NF-κB share common DNA binding sites and regulate similar genes and activation of NF-κB is a major signaling response to TNF-α stimulation of cells. NF-κB activity also increases with aging. In contrast, Lethe expression, which is largely limited to spleen, actually decreases with aging. Taken together, these results suggested the possibility that Lethe may regulate NF-κB function. Using a variety of approaches these investigators were able to show that Lethe inhibits NF-κB by binding to the RelA or p65 component of the active NF-κB p60/p65 heterodimer and thus specifically inhibits binding of RelA to DNA leading to inhibition of NF-κB transactivation. Thus, a second lncRNA has been discovered that regulates NF-κB activity. As described above, PACER is a positive regulator of NF-κB activity by titrating inhibitory NF-κB p50/p50 homodimers away from NF-κB DNA response elements while Lethe is a negative regulator of NF-κB activity by titrating activating RelA or p65 containing activating heterodimers away from NF-κB DNA response elements. At present, it is not known to what extent Lethe is expressed by other cell types, is induced by other stimuli, or plays a role in regulating of TNF-α responses to pathogen infection or the ‘sterile inflammation’ seen in autoimmune diseases.
4. LincRNA-p21
LincRNA-p21 is included in this review because its level of expression is decreased in the inflammatory disease, rheumatoid arthritis and it possesses anti-inflammatory properties (Spurlock 3rd et al., 2014). LincRNA-p21 was originally discovered by examination of the cellular response to DNA damage in mouse embryonic fibroblasts (Huarte et al., 2010). The tumor suppressor protein, p53, is a major regulator of the DNA damage response resulting in either p53-mediated cell cycle arrest or apoptosis depending upon the degree of DNA damage. p53 induces transcription of the cell cycle arrest protein, p21, and induces transcription of lincRNA-p21. Genes encoding lincRNA-p21 and p21 are adjacent in both mouse and human genomes. lincRNA-p21 inhibits expression of a number of genes, either directly or indirectly, that encode proteins with pro-apoptotic functions. LincRNA-p21 associates with one of the hnRNP proteins, hnRNP-K, to carry out these functions. Thus, lincRNA-p21 contributes to DNA damage responses by lowering the threshold for apoptosis.
Studies in human HeLa cells reveal an additional mechanism of action of lncRNA-p21 (Yoon et al., 2012). lincRNA-p21 regulates levels of certain proteins, Jun B, beta catenin, at the translational level. Specifically, lincRNA-p21 binds to these mRNAs by forming sequence-specific RNA-RNA duplexes and prevents their association with the protein synthesis machinery thus preventing translation of these mRNAs into proteins.
A third function of linc RNA-p21 is in the cellular response to hypoxia, known as the Warburg effect so named after its discoverer, Otto Warburg. The Warburg effect was originally discovered when studies were undertaken to determine how cells in solid tumors survived hypoxia (Warburg, 1956). To do so, cells shift from oxidative phosphorylation to glycolysis to generate energy. This shift is achieved by induction of the transcription factor; hypoxia inducing factor 1-alpha or HIF1α and HIF1α induces a new transcriptional program to allow cells to employ glycolysis to generate energy in this anaerobic environment (Iyer et al., 1998). The tumor suppressor von Hippel Lindau protein or VHL controls levels of HIF1 α protein (Maxwell et al., 1999). VHL binds to HIF1 α resulting in its ubiquitination and subsequent degradation by the proteasome (Jaakkola et al., 2001). LincRNA-p21 is induced by hypoxia and inhibits the HIF1α — VHL interaction and prevents ubiquitination of HIF1 α and degradation by the proteasome allowing levels of HIF1a to accumulate (Yang et al., 2014). Therefore, in response to hypoxia, lincRNA-p21 functions as a tumor promoter by enabling tumor cells to shift to anaerobic glycolysis as their energy source and to survive and proliferate in the hypoxic environment of a solid tumor.
As cited above, levels of lincRNA-p21 are reduced in PBMC from subjects with rheumatoid arthritis (RA) compared to healthy controls. Basal levels of activity of the transcription factor NF-κB are also elevated in RA CD4+ T lymphocytes. Further analysis shows that lincRNA-p21 is a negative regulator of basal NF-κB activity in T cells and depressed levels of lincRNA-p21 in RA contribute to elevated basal NF-κB activity in RA. LincRNA-p21 associates with RelA (p65) mRNA encoding one heterodimer of active NF-κB resulting in a reduction in RelA protein levels. Association between RelA mRNA and lincRNA-p21 is presumed to inhibit RelA mRNA translation resulting in a reduction of RelA protein and lower overall NF-κB activity.
Mechanisms that underlie reduced lincRNA-p21 levels in RA are not known. Reduced levels of lincRNA-p21 in PBMC are not observed in a number of other inflammatory autoimmune diseases and thus may be a unique feature of RA. Interestingly, low-dose methotrexate, one of the most common and effective therapies for RA restores depressed levels of lincRNA-p21 in subjects with RA. In tissue culture, treatment of cells with low concentrations of methotrexate markedly induces lincRNA-p21. Although methotrexate also induces p53 in these models, induction of lincRNA-p21 is not p53-dependent but rather is DNA-PKcs-dependent, which is a sentinel of DNA damage. The exact mechanism by which methotrexate activates DNA-PKcs is not known nor is it known how activation of DNA-PKcs results in increased levels of lincRNA-p21. Taken together, these results indicate that lincRNA-p21 has anti-inflammatory properties and that reduced levels of lincRNA-p21 may contribute to chronic inflammation seen in RA. Thus, anti-inflammatory effects of methotrexate may be derived, in part, by its ability to restore lincRNA-p21 levels to normal.
In summary, these studies might seem somewhat paradoxical. LincRNA-p21 levels are associated with a major inflammatory autoimmune disease, rheumatoid arthritis, and respond to a major treatment, methotrexate. In this setting, lincRNA-p21 may be considered anti-inflammatory. LincRNA-p21 is also induced by cellular responses to DNA damage and to hypoxia. It seems somewhat curious that in the setting of the DNA damage response, lincRNA-p21 seems to have properties of a tumor suppressor because it promotes apoptosis while in the setting of hypoxia, lincRNA-p21 seems to have properties of a tumor promoter because it enhances survival and growth of tumor cells under conditions of low oxygen tension that are found in a solid tumor thus providing tumor cells with a growth advantage. LincRNA-p21 also has multiple modes of action. First, it directly targets gene expression via a hnRNP-K dependent mechanism. Second, lincRNA-p21 forms sequence specific RNA-RNA hybrids with certain mRNAs to inhibit translation. Third, lincRNA-p21 directly binds to proteins to affect their functions. Thus, the current state our understanding of lincRNA-p21 biology is that it is induced by different stimuli in different cell types and has a multitude of functions some of which are even opposing functions. At this point, we do not know if these effects might be mediated by different unique lincRNA-p21 isoforms or by a single lincRNA-p21 species or if there maybe a common mechanism that may explain these diverse and seemingly context-dependent activities. It is also not known if the ability to exert these multiple functions is a unique property of lincRNA-p21 or if other lncRNAs also possess diverse activities and modes of action.
5. GWAS and lncRNAs
By analyzing single nucleotide polymorphisms (SNP), genome-wide association studies (GWAS) have significantly advanced our understanding of the contribution of genetic variation to complex human traits including susceptibility to a diverse array of complex diseases. However, mechanistic understanding of how these SNPs may contribute to genetic risk is complicated by two important factors (Farh et al., 2015). First is the fact that >90% of GWAS identified SNPs are present in regions of the genome that do not code for proteins making it not obvious how these SNPs may contribute to phenotypic traits. Second, many identified SNPs are present in regions of the genome that exhibit high levels of linkage disequilibrium thus making it difficult to distinguish among the SNPs that actually contribute to disease susceptibility, causative SNPs, and those that are simply in high linkage disequilibrium with the causal SNP. Nevertheless, these studies suggest that the non-coding region of the genome may contribute to genetic susceptibility for the development of complex diseases and current and future studies will be devoted to developing an improved mechanistic understanding of these relationships.
One approach to develop a better understanding of GWAS data has been to examine the association between SNPs and expression of mRNAs in cells or tissues of interest. These genome-wide studies have clearly shown that given disease-associated and trait-associated SNPs are also associated with mRNA expression levels, termed expression Quantitative Trait Loci or eQTL. It is relevant to note that some of these eQTL associations are between SNPs and eQTL on the same chromosome, termed cis-eQTL, but other associations are between SNPs and eQTL on different chromosomes, termed trans-eQTL. Recognizing that lncRNAs are among most abundant class of RNA molecules transcribed from the human genome, investigators have begun to examine the relationships among disease associated SNPs and expression levels of distinct lncRNAs in different human primary tissues, focusing primarily upon SNPs associated with autoimmune diseases and expression profiles of lncRNAs in lymphoid and myeloid cells as malfunction of these cells are thought to be major contributors to development of autoimmune disease (Kumar et al., 2013; Hrdlickova et al., 2014).
These studies clearly show the association of disease associated SNPs with expression of nearby lncRNAs in the genome termed lin-cRNA cis-eQTLs. Additionally, these SNPs are not associated with differences in expression of neighboring protein-coding genes in the genome. In keeping with the idea that many if not most lncRNAs exhibit very restricted cell or tissue expression patterns, these correlations between disease-associated SNPs and lincRNA cis-eQTLS are also very tissue-dependent. Although studies such as these are still in their infancy, it is likely that significant knowledge will be gained. Since lncRNAs tend to exhibit cell lineage specific expression patterns, it may be possible to infer which lymphoid and myeloid cells contribute to pathogenesis of a given autoimmune disease by determining the lncRNAs that are differentially expressed in an autoimmune disease of interest and which cell expresses the given lncRNA and associations between differentially expressed lncRNAs and disease associated SNPs. Additionally, be examining co-expression of lncRNAs and protein-coding genes using such methods as pathways analyses, it may be possible to infer defective biological processes that may be contributory for a given autoimmune disease. Although somewhat speculative, these types of studies are actually ongoing and have produced promising results.
6. Pipeline for lncRNA discovery by RNA-sequencing
Use of lncRNA microarray panels or next generation sequencing, such as RNA-seq, facilitates discovery of lncRNAs (Sanchez et al., 2014; Yu et al., 2012). A major limitation of pre-printed microarrays is the inability to identify novel lncRNA while RNA-seq facilitates discovery of both known and unknown lncRNAs (Fatica and Bozzoni, 2014). Paired-end RNA sequencing is preferred as these sequencing data provide information about novel splice junctions and rearrangement events such as insertions, deletions and inversions. Paired-end sequencing also allows for de novo transcript assembly and detection of novel short and/or long noncoding RNAs. An important consideration is strand orientation and preserving both strands for downstream analyses.
Generating RNA-seq data is not as difficult as the analysis required to derive meaningful results from large datasets. The computational and technical infrastructure required presents a barrier to research that many investigators without access to bioinformatics core services will encounter. The file deliverable from an RNA-sequencing project is often a series of raw, pooled barcoded data that have been de-multiplexed into individual FASTQ files. These files are analyzed for quality control using one of many programs, such as the FASTQC package that fuels rapid assessment of data and generates HTML based reports for quick visualization of problem areas. This is a standalone application, but it can be integrated into the workflow for most RNA-sequencing analysis pipelines. Other useful tools are FASTX-Toolkit, QC3 and NGS QC Toolkit (Patel and Jain, 2012; Guo et al, 2014a; Schmieder and Edwards, 2011). One important experimental consideration derived from these software packages will be the number of reads collected per sample, referred to as the total counts by lane, the base quality versus sequencing cycle, the nucleotide distribution versus cycle, and GC content by lane. These should be consistent across each experimental sample. Read depth is also of paramount importance and should be consistent across all samples wherever possible. A general guide is approximately 30 million reads per sample for mRNA analysis and 40 million reads for lncRNA analysis. Greater read depth is important for lncRNA analyses since these RNAs are expressed at rather low abundance compared to mRNAs. Once a quality control check has been performed, individual sequences are aligned to a reference genome, such as human genome 19 (hg19). This alignment, and further downstream analysis, can be performed using programs such as the Tuxedo Suite that includes Bowtie, TopHat, Cufflinks, and CummeRbund (Trapnell et al, 2012). The first step is to align the reads or map against a reference genome using Bowtie/Bowtie2 or TopHat/TopHat2. TopHat aligns the short reads to a reference genome but also discovers splice sites. The output is a SAM (sequence alignment map format) or BAM file, the binary counterpart to a SAM file. BAM files are preferred, as they take up less disk space. These files are then converted to sorted BAM files using a program called SAMtools that places the aligned reads in coordinate order by chromosome (Li et al., 2009). The sorted BAM file is then subjected to additional analysis using any one of many transcript-counting solutions available. These include, but are not limited to, HTSeq or Cufflinks. HTSeq will provide a simple read count of transcripts whereas, Cufflinks provides FPKM (fragments per kilobase of transcript per million mapped reads) values for indicated genes that are either supplied or detected through a ‘discovery’ option selectable in the command line (Trapnell et al., 2010; Anders et al., 2015). FPKM values are normalized and are one of the preferred normalization methods, especially for paired-end RNA sequencing. Supplied locations are typically in the GTF file format and can be downloaded from online resources such as Gencode (http://www.gencodegenes.org/). The next step involves normalization of these datasets and testing for derivation of differentially expressed genes. Experimental comparisons are made using software packages such as DESeq, baySeq, edgeR, NBPSeq, TSPM or Cuffdiff. These software packages are available through online repositories such as gitHub (https://github.com) or are provided as executable modules downloadable through R statistical computing and graphics software (e.g., edgeR) (Anders and Huber, 2010; Hardcastle and Kelly, 2010; Robinson et al., 2010; Di et al., 2011; Kvam et al., 2012; Anders et al., 2013). When testing for differentially expressed genes in the analysis pipeline, it is important to understand that no two packages deliver the same results (Anders et al., 2013). Therefore, many groups have adopted a multi-comparison approach that uses more than one package to test for differentially expressed genes, such as the inclusion of DESeq, edgeR, and baySeq, in a combined program called MultiRankSeq to combine these differentially expressed gene outputs and create a list to ranks agreement across all three software packages (Kvam et al., 2012; Guo et al., 2014b). Packages such as MultiRankSeq and CummeRbund, part of the Tuxedo Suite, include visualization software that can be modified for inclusion in publications. Gene Ontology analysis or pathways analysis using PANTHER can also facilitate creation of lists of overlapping genes and reveal pathways over- or under-represented across different conditions (Merad et al., 2013; Ashburner et al., 2000). Programs such as IGV allow users to view next generation sequencing reads, and often is the preferred method for visualizing read counts generated by RNA-sequencing (Thorvaldsdottir et al., 2013; Robinson et al., 2011). GViz, available on Bioconductor, is also useful for visualizing RNA-seq histogram plots for a given gene and provides a high quality image file (Dander et al., 2014).
7. Functional analysis of lncRNAs
Not all lncRNAs are polyadenylated so lncRNA discovery may be best achieved using libraries prepared from both poly(A) selection and stranded total RNA, such as the Illumina TruSeq Stranded Total RNA kit with ribo-zero. The analysis pipeline for de novo assembly can involve a number of different software platforms including Cufflinks in ‘discovery’ mode, the Trinity pipeline, or the recently published Bridger framework (Haas et al., 2013; Chang et al., 2015). These pipelines allows for the collection of novel gene lists. Analysis of coding potential of the RNA transcript can be determined computationally using such tools as GetORF (Alvarez-Dominguez et al, 2014; Rice et al., 2000), CPAT, the Coding Potential Assessment Tool, and the Coding Potential Calculator (CPC) (Wang et al., 2013b; Kong et al., 2007). These two methods use support vector machine modeling to analyze the sequence of novel transcripts and assign a coding probability score that is either a yes or no. Another popular application is PhyloCSF. PhyloCSF leverages multiple taxonomic alignments to examine codon substitution frequencies across different species in the tree of life, such as 29 different placental mammals. Given the lack of conservation of noncoding RNAs and lncRNAs, in particular, lack of conservation is a relatively efficient way to determine if a transcript or genomic region is noncoding or if it merely represents a novel exon of a previously described protein-coding gene (Lin et al, 2011b). Strand specific PCR can be performed to confirm expression measurements of lncRNAs, especially intragenic lncRNAs that overlap with known protein-coding gene exons and to preclude the possibility of detection of alternatively spliced exons resulting from mRNA transcription (Hangauer et al., 2013b; Zhao et al., 2008; Pandey et al., 2008).
Once the expression of the lncRNA is confirmed, a number of techniques can be employed to ascribe function to a new lncRNA. RNA interference is perhaps the most versatile tool. Using our own investigations of the TH2-LCR lncRNA as point of reference, our tissue culture experiments found that abrogation of individual, unique sequences for each TH2-LCR lncRNA transcript did not significantly alter IL-4, IL-5, or IL-13 expression patterns. While we saw modest affects for these proteins by flow cytometry, and >50% reduction of the individual siRNA transcript target, our greatest effects on expression of these TH2-specific cytokines were observed when the shared sequence for all four alternatively spliced TH2-LCR transcripts was depleted. Greatest primary cell and transformed cell RNA interference was observed using a combination of Silencer Select siRNA duplexes that contain a locked nucleic acid modification or Stealth siRNAs and Lipofectamine RNAiMax reagents from Life Technologies (Spurlock et al., 2015; Monnier et al., 2013). Protein immunoprecipitation can be used to identify protein binding partners of individual lncRNAs (Alvarez-Dominguez et al., 2014; Wei et al., 2011). Another approach, which is a variation of the ChIP assay involves chromatin isolation by RNA purification (ChIRP) and can be combined with next generation DNA sequencing to produce genome-wide association maps of lncRNA/protein and or RNA/DNA binding (Muers, 2011). Briefly, sample preparation includes glutaraldehyde fixation of cells of interest to cross-link proteins, DNA and RNA, sonication to shear DNA to an average fragment size of 500–1000 bp, hybridization of samples to labeled RNA tiling oligonucleotides with a biotin tag, immunoprecipitation, elution and reversal of crosslinks, proteinase K digestion to isolate DNA, and sequencing to determine genomic RNA-DNA interactions or mass spectrometry to identify protein-lncRNA binding partners. Fig. 5 highlights a rudimentary pipeline that investigators could use to further their exploration of these new molecules.
Fig. 5.
Example pipeline for identification and characterization of annotated and novel lncRNAs using whole genome sequencing.
8. Concluding remarks
One of the major surprises of the ENCODE project was the recognition that the majority of the human genome is transcribed. This created a debate whether this transcription was a result of spurious binding of RNA polymerases to the genome and pervasive transcription or whether this transcription or the RNA molecules produced had functional roles in biology. The discovery of lncRNAs as a class of RNA molecules supports the idea that at least portions of these RNA molecules have biological functions. However, it is important to note that numbers of known protein-coding genes are roughly equivalent to numbers of lncRNA genes in the human genome thus far discovered. Therefore, numbers of lncRNA genes plus protein-coding genes thus far discovered that are transcribed from the human genome still probably occupy a relatively small percentage of total genome space and do not totally account for the pervasive transcription noted by the ENCODE project. This strongly argues that there are many more lncRNA genes that remain to be discovered, that additional classes of RNA molecules remain to be discovered or that much of this transcription still may be spurious. It is also noteworthy that these lncRNAs as a class are critical regulators of expression and function of protein-coding genes and utilize a vast array of different mechanisms to establish this additional layer of regulation.
One general notion that has emerged is that more lncRNAs are expressed in a cell-type specific manner than are mRNAs. Of organ systems, the central nervous system probably is populated by the greatest cellular diversity and is also thought to express the greatest diversity of lncRNAs. The immune system may be second to the nervous system in cellular complexity. Lymphocytes comprise an extensive array of cellular identities that are revealed during development, lineage differentiation in the periphery, persistence of memory cells of which their may be multiple types, and the capacity to take up residence in different tissues. Similarly, cells of the myeloid lineage undergo multiple paths of development and differentiation and also reside in different tissues. The extent to which these processes are informed by lncRNAs is largely unknown. Examples included here (Table 3) predict that cells of the immune system express many unique lncRNAs that will add additional layers of gene regulation to innate and adaptive immunity. Also important will be definition of the functions of lncRNA in animal models of immunity and autoimmunity as well as the contributions of lncRNAs to immune responses to pathogens in humans as well as the role they may play in initiation and perpetuation of autoimmune diseases in humans (Heward and Lindsay, 2014).
Table 3.
Overview of lncRNAs in the immune system.
| LncRNA | Expression | Target(s) | +/− | Mechanism | In vivo | Refs. |
|---|---|---|---|---|---|---|
| Adaptive Immunity | CD4 &CD8 | IFNG | + | epigenetic | yes | 70,71 |
| IFNG-AS1 (78) | CD4TH2 | IL4, IL13, IL5 | + | epigenetic | yes | 68 |
| TH2-LCR lncRNA | CD4TH2 | Ccr1, Ccr3, Ccr2, Ccr5 | + | ‡ | 66 | |
| Linc-Ccr2-5′AS | CD4TH1 | MAF | − | epigenetic | 67 | |
| Linc-MAF-4 | CD4TH2 | ‡ | ‡ | ‡ | 78 | |
| GATA3-AS1 | T cells | NFAT protein | − | sequesters NFAT in cytoplasm | 79,80 | |
| NRON | B cells | soluble FAS | − | splicing | 81 | |
| Fas-AS1 | DNA damage | multiple genes | − | binds hnRNP | 90 | |
| lincRNA-p21 | multiple (also in T cells) | RelA, JunB | − | binds target mRNA & inhibit translation | 91,92 | |
| hypoxia | HIF-1α | + | prevents VHL directed ubiquitination | 93 | ||
| Innate Immunity | ||||||
| lincRNA-Cox2 | monocyte | multiple | − | binds hnRNP-A2/B1 & A/B | 82 | |
| PACER | monocyte | COX2 | + | sequesters NF-κB p50/p50 | 83 | |
| Lnc-DC | dendritic cells | STAT3 | + | binds STAT3 protein | 84 | |
| THRIL | monocyte | TNF, IL6 | − | promoter binding | 87 | |
| Lnc-IL7R | monocytes | IL-6, VCAM-1,IL8, IL7R E-selectin | − | epigenetic | 88 | |
| Lethe | multiple | IL6, IL8, SOD2 | − | sequesters RelA | 89 |
Legend: +/− is positive (+) or negative (−) regulator; ‡ = unknown.
Finally, gains made in recent years in our understanding of gene regulation are really quite remarkable. Not that long ago, gene regulation was thought to be achieved by activating or silencing of one or two evolutionarily conserved enhancers and gene promoters by transcription factors. Currently, we recognize that functions of transcription factors, interpretation of ‘writing’ and ‘reading’ the epigenetic code, both histone marks and epigenetic modification of DNA, a dynamic chromosome conformation, expression of lncRNAs in both cis and trans, and functions of enhancer RNAs all contribute to transcriptional regulation of protein-coding genes, central processes to establishing cell identify and carrying out cellular functions. Given the dynamic nature of the immune system and the requirement to rapidly respond to pathogen infection, it is easy to see why these process need to be under very tight control as an inefficient immune response can lead to infection and even death from infection while an immune response that is too robust can lead to diseases such as sepsis and death. Failure to properly regulate the immune response also leads to an array of autoimmune diseases that also produce disability and even premature death. Hopefully, a better understanding of the functions of lncRNAs in the immune system will not only increase our basic knowledge but will also contribute to our ability to control the immune system in both positive and negative ways to contribute to better treatments for individuals with immune-related diseases.
Acknowledgements
We apologize for omission of work by colleagues due to oversight or limitations in space. This work was supported in part by the National Institutes of Health (R01 AI044924, R21 AR063846) and the National Science Foundation Graduate Research Fellowship Program (DGE0909667). Sponsors had no role in study design, data collection and interpretation in writing the report and submission of the article for publication.
Abbreviations
- lncRNA
long noncoding RNA
- eQTL
expression quantitative trait loc
- ENCODE
encyclopedia of DNA elements
- TH
T helper cell
- RNA-seq
whole genome RNA sequencing
- MEF
mouse embryonic fibroblasts
Contributor Information
Thomas M. Aune, Email: tom.aune@vanderbilt.edu.
Charles F. Spurlock, III, Email: chase.spurlock@vanderbilt.edu.
References
- Alvarez-Dominguez JR, Hu WQ, Yuan BB, Shi JH, Park SS, Gromatzky AA, et al. Global discovery of erythroid long noncoding RNAs reveals novel regulators of red cell maturation. Blood. 2014;123(4):570–581. doi: 10.1182/blood-2013-10-530683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;31:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 2013;8(9):1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm T. Thymus development and function. Curr. Opin. Immunol. 2008;20(2):178–184. doi: 10.1016/j.coi.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341(6147):789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491(7424):454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- Chang S, Aune TM. Dynamic changes in histone-methylation ‘marks’ across the locus encoding interferon-gamma during the differentiation of T helper type 2 cells. Nature Immunol. 2007;8(7):723–731. doi: 10.1038/ni1473. [DOI] [PubMed] [Google Scholar]
- Chang Z, Li GJ, Liu JT, Zhang Y, Ashby C, Liu DL, et al. Bridger: a new framework for de novo transcriptome assembly using RNA-seq data. Genome Biol. 2015;16 doi: 10.1186/s13059-015-0596-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell. 2011;44(4):667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MB, Choudhary A, Smith MA, Taft RJ, Mattick JS. The dark matter rises: the expanding world of regulatory RNAs. Essays Biochem. 2013;54:1–16. doi: 10.1042/bse0540001. [DOI] [PubMed] [Google Scholar]
- Collier SP, Collins PL, Williams CL, Boothby MR, Aune TM. Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J. Immunol. 2012;1:20. doi: 10.4049/jimmunol.1200774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier SP, Henderson MA, Tossberg JT, Aune TM. Regulation of the Th1 genomic locus from Ifng through Tmevpg1 by T-bet. J. Immunol. 2014;193(8):3959–3965. doi: 10.4049/jimmunol.1401099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins PL, Chang S, Henderson M, Soutto M, Davis GM, McLoed AG, et al. Distal regions of the human IFNG locus direct cell type-specific expression. J. Immunol. 2010;185(3):1492–1501. doi: 10.4049/jimmunol.1000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins PL, Henderson MA, Aune TM. Diverse functions of distal regulatory elements at the IFNG locus. J. Immunol. 2012;188(4):1726–1733. doi: 10.4049/jimmunol.1102879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium EP, Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Xie N, Tan Z, Banerjee S, Thannickal VJ, Abraham E, et al. The human long noncoding RNA lnc-IL7R regulates the inflammatory response. Eur. J. Immunol. 2014;44(7):2085–2095. doi: 10.1002/eji.201344126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dander A, Baldauf M, Sperk M, Pabinger S, Hiltpolt B, Trajanoski Z. Personalized Oncology Suite: integrating next-generation sequencing data and whole-slide bioimages. Bmc Bioinf. 2014;15 doi: 10.1186/1471-2105-15-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di YM, Schafer DW, Cumbie JS, Chang JH. The NBP. Negative binomial model for assessing differential gene expression from RNA-Seq. Stat. Appl. Genet. Mol. 2011;10:1. [Google Scholar]
- Diefenbach A, Colonna M, Koyasu S. Development, differentiation, and diversity of innate lymphoid cells. Immunity. 2014;41(3):354–365. doi: 10.1016/j.immuni.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008a;18(9):1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Pang KC, Mercer TR, Mattick JS. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput. Biol. 2008b;4(11):e1000176. doi: 10.1371/journal.pcbi.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Pang KC, Mercer TR, Crowe ML, Grimmond SM, Mattick JS. NRED. A database of long noncoding RNA expression. Nucleic Acids Res. 2009;37:D122–D126. doi: 10.1093/nar/gkn617. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;4:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euskirchen GM, Rozowsky JS, Wei CL, Lee WH, Zhang ZD, Hartman S, et al. Mapping of transcription factor binding regions in mammalian cells by ChIP: comparison of array- and sequencing-based technologies. Genome Res. 2007;17(6):898–909. doi: 10.1101/gr.5583007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber DL, Yudanin NA, Restifo NP. Human memory T cells: generation, compartmentalization and homeostasis. Nat. Rev. Immunol. 2013;14(1):24–35. doi: 10.1038/nri3567. 20;14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518(7539):337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nature Rev. Gen. 2014;15(1):7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- Gerstein MB, Bruce C, Rozowsky JS, Zheng D, Du J, Korbel JO, et al. What is a gene, post-ENCODE? History and updated definition. Genome Res. 2007;17(6):669–681. doi: 10.1101/gr.6339607. [DOI] [PubMed] [Google Scholar]
- Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell. 2013;152(4):743–754. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Cell Dev. 2013;24(2):206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guermonprez P, Helft J, Claser C, Deroubaix S, Karanje H, Gazumyan A, et al. Inflammatory Flt3l is essential to mobilize dendritic cells and for T cell responses during Plasmodium infection. Nature Med. 2013;19(6):730–738. doi: 10.1038/nm.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Ye F, Sheng Q, Clark T, Samuels DC. Three-stage quality control strategies for DNA re-sequencing data. Brief. Bioinform. 2014;15(6):879–889. doi: 10.1093/bib/bbt069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Zhao SL, Sheng QH, Ye F, Li J, Lehmann B, et al. Multi-perspective quality control of Illuminaexome sequencing data using QC3. Genomics. 2014a;103(5–6):323–328. doi: 10.1016/j.ygeno.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Zhao SL, Ye F, Sheng QH, Shyr Y. MultiRankSeq. Multiperspective approach for RNAseq differential expression analysis and quality control. Biomed. Res. Int. 2014b doi: 10.1155/2014/248090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154(1):240–251. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nature Protoc. 2013;8(8):1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013a;9(6):e1003569. doi: 10.1371/journal.pgen.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangauer MJ, Vaughn IW, McManus MT. Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs. PLoS Gen. 2013b;9(6) doi: 10.1371/journal.pgen.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardcastle TJ, Kelly KA. baySeq: empirical Bayesian methods for identifying differential expression in sequence count data. Bmc Bioinf. 2010;11:422. doi: 10.1186/1471-2105-11-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22(9):1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heward JA, Lindsay MA. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014;35(9):408–419. doi: 10.1016/j.it.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander G, Gill J, Zuklys S, Iwanami N, Liu C, Takahama Y. Cellular and molecular events during early thymus development. Immunol. Rev. 2006;209:28–46. doi: 10.1111/j.0105-2896.2006.00357.x. [DOI] [PubMed] [Google Scholar]
- Hrdlickova B, Kumar V, Kanduri K, Zhernakova DV, Tripathi S, Karjalainen J, et al. Expression profiles of long non-coding RNAs located in autoimmune disease-associated regions reveal immune cell-type specificity. Genome Med. 2014;6(10):88. doi: 10.1186/s13073-014-0088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Alvarez-Dominguez JR, Lodish HF. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep. 2012;13(11):971–983. doi: 10.1038/embor.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Tang Q, Sharma S, Yu F, Escobar TM, Muljo SA, et al. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat. Immunol. 2013;14(11):1190–1198. doi: 10.1038/ni.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 2011;43(7):621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147(4):789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12(2):149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292(5516):468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Kapusta A, Feschotte C. Volatile evolution of long noncoding RNA repertoires: mechanisms and biological implications. Trends Gen. TIG. 2014;30(10):439–452. doi: 10.1016/j.tig.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U. S. A. 2009;106(28):11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A, Dean A. Chromatin loop formation in the beta-globin locus and its role inglobingene transcription. Mol. Cells. 2012;34(1):1–5. doi: 10.1007/s10059-012-0048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318(5849):420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk M, Emerson BM. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-kappaB complexes. eLife3. 2014:e01776. doi: 10.7554/eLife.01776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Westra HJ, Karjalainen J, Zhernakova DV, Esko T, Hrdlickova B, et al. Human disease-associated genetic variation impacts large intergenic non-coding RNA expression. PLoS Genet. 2013;9(1):e1003201. doi: 10.1371/journal.pgen.1003201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurobe H, Liu C, Ueno T, Saito F, Ohigashi I, Seach N, et al. CCR7-dependent cortex-to-medulla migration of positively selected thymocytes is essential for establishing central tolerance. Immunity 24(2) 2006:165–177. doi: 10.1016/j.immuni.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Kvam VM, Lu P, Si YQ. A Comparison of Statistical Methods for Detecting Differentially Expressed Genes from Rna-Seq Data. Am. J. Bot. 2012;99(2):248–256. doi: 10.3732/ajb.1100340. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Laouar Y, Welte T, Fu XY, Flavell RA. STAT3 is required for Flt3L–dependent dendritic cell differentiation. Immunity. 2003;19(6):903–912. doi: 10.1016/s1074-7613(03)00332-7. [DOI] [PubMed] [Google Scholar]
- Lee GR, Spilianakis CG, Flavell RA. Hypersensitive site 7 of the TH2 locus control region is essential for expressing TH2 cytokine genes and for long-range intrachromosomal interactions. Nature Immunol. 2005;6(1):42–48. doi: 10.1038/ni1148. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Chao TC, Chang KY, Lin N, Patil VS, Shimizu C, et al. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc. Natl. Acad. Sci. U. S. A. 2014;111(3):1002–1007. doi: 10.1073/pnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Jungreis I, PhyloC SF, Kellis M. a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011a;27(13):275–82. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Jungreis I, Kellis M. PhyloC, SF a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011b;27(13):1275–1282. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Mattick JS, Taft RJ. A meta-analysis of the genomic and transcriptomic composition of complex life. Cell cycle. 2013;12(13):2061–2072. doi: 10.4161/cc.25134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF. RNA regulation of epigenetic processes. Bioessays News Rev. Mol. Cell. Dev. Biol. 2009;31(1):51–59. doi: 10.1002/bies.080099. [DOI] [PubMed] [Google Scholar]
- Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399(6733):271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- McKenzie AN, Spits H, Eberl G. Innate lymphoid cells in inflammation and immunity. Immunity. 2014;41(3):366–374. doi: 10.1016/j.immuni.2014.09.006. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat. Rev. Immunol. 2009;9(10):692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- Merad M, Sathe P, Helft J, Miller J, Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annual Rev. Immunol. 2013;31:563–604. doi: 10.1146/annurev-immunol-020711-074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mariani J, Kosik KS, Mehler MF, Mattick JS. Noncoding RNAs in Long-Term Memory Formation. Neurosci. Rev. J. Bringing Neurobiol. Neurol. Psychiatry. 2008a;14(5):434–445. doi: 10.1177/1073858408319187. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. U. S. A. 2008b;105(2):716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Monnier P, Martinet C, Pontis J, Stancheva I, Ait-Si-Ali S, Dandolo L. H19 lncRNA controls gene expression of the Imprinted Gene Network by recruiting MBD1. Proc. Natl. Acad. Sci. U. S. A. 2013;110(51):20693–20698. doi: 10.1073/pnas.1310201110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV, Mattick JS. The rise of regulatory RNA. Nat. Rev. Genet. 2014;15(6):423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muers M. RNA Genome-wide views of long non-coding RNAs. Nature Rev. Gen. 2011;12:11. doi: 10.1038/nrg3088. [DOI] [PubMed] [Google Scholar]
- Murphy KM. Transcriptional control of dendritic cell development. Adv. Immunol. 2013;120:239–267. doi: 10.1016/B978-0-12-417028-5.00009-0. [DOI] [PubMed] [Google Scholar]
- Murray PJ, Smale ST. Restraint of inflammatory signaling by interdependent strata of negative regulatory pathways. Nat. Immunol. 2012;13(10):916–924. doi: 10.1038/ni.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145(2):178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, et al. Kcnq1ot1 antisense noncoding mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell. 2008;32(2):232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Pang KC, Dinger ME, Mercer TR, Malquori L, Grimmond SM, Chen W, et al. Genome-wide identification of long noncoding RNAs in CD8+ T cells. J. Immunol. 2009;182(12):7738–7748. doi: 10.4049/jimmunol.0900603. [DOI] [PubMed] [Google Scholar]
- Paralkar VR, Mishra T, Luan J, Yao Y, Kossenkov AV, Anderson SM, et al. Lineage and species-specific long noncoding RNAs during erythro-megakaryocytic development. Blood. 2014;123(12):1927–1937. doi: 10.1182/blood-2013-12-544494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel RK, Jain M. NGS QC. Toolkit: a toolkit for quality control of next generation sequencing data. PloS one. 2012;7(2):e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper M, Jenkins MK. Origins of CD4(+) effector and central memory T cells. Nature Immunol. 2011;12(6):467–471. doi: 10.1038/ni.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranzani V, Rossetti G, Panzeri I, Arrigoni A, Bonnal RJ, Curti S, et al. The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nat. Immunol. 2015;16(3):318–325. doi: 10.1038/ni.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge RM, Chang HY. A mammalian pseudogene lncRNA at the interface of inflammation and antiinflammatory therapeutics. eLife. 2013;2:e00762. doi: 10.7554/eLife.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS. The European molecular biology open software suite. Trends Gen. 2000;16(6):276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Ann. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nature Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozowsky JS, Newburger D, Sayward F, Wu J, Jordan G, Korbel JO, et al. The DART classification of unannotated transcription within the ENCODE regions: associating transcription with known and novel loci. Genome Res. 2007;17(6):732–745. doi: 10.1101/gr.5696007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y, Segura V, Marin-Bejar O, Athie A, Marchese FP, Gonzalez J, et al. Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nat. Commun. 2014;5:5812. doi: 10.1038/ncomms6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpathy AT, Chang HY. Long Noncoding RNA in Hematopoiesis and Immunity. Immunity. 2015;42(5):792–804. doi: 10.1016/j.immuni.2015.05.004. [DOI] [PubMed] [Google Scholar]
- Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27(6):863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal L, Mathur R, Braun FK, Wise JF, Berkova Z, Neelapu S, et al. FAS-antisense 1 lncRNA and production of soluble versus membrane Fas in B-cell lymphoma. Leukemia. 2014;28(12):2376–2387. doi: 10.1038/leu.2014.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Findlay GM, Bandukwala HS, Oberdoerffer S, Baust B, Li Z, et al. Dephosphorylationofthe nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA-protein scaffold complex. Proc. Natl. Acad. Sci. U. S. A. 2011;108(28):11381–11386. doi: 10.1073/pnas.1019711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih HY, Sciume G, Poholek AC, Vahedi G, Hirahara K, Villarino AV, et al. Transcriptional and epigenetic networks of helper T and innate lymphoid cells. Immunol. Rev. 2014;261(1):23–49. doi: 10.1111/imr.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigova AA, Mullen AC, Molinie B, Gupta S, Orlando DA, Guenther MG, et al. Divergent transcription of long noncoding RNA/mRNAgene pairs in embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A. 2013;110(8):2876–2881. doi: 10.1073/pnas.1221904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurlock CF, 3rd, Tossberg JT, Matlock BK, Olsen NJ, Aune TM. Methotrexate inhibits NF-kappaB activity via long intergenic (noncoding) RNA-p21 induction. Arthritis Rheumatol. 2014;66(11):2947–2957. doi: 10.1002/art.38805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurlock CF, 3rd, Tossberg JT, Guo Y, Collier SP, Crooke PS, 3rd, Aune TM. Expression and functions of long noncoding RNAs during human T helper cell differentiation. Nat. Commun. 2015;6:6932. doi: 10.1038/ncomms7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman RM, Hemmi H. Dendritic cells: translating innate to adaptive immunity. Cur. Topics Microbiol. Immunol. 2006;311:17–58. doi: 10.1007/3-540-32636-7_2. [DOI] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19(3):347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahama Y. Journey through the thymus: stromal guides for T-cell development and selection. Nat. Rev. Immunol. 2006;6(2):127–135. doi: 10.1038/nri1781. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings Bioinf. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28(5):1. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154(1):26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Vigneau S, Rohrlich PS, Brahic M, Bureau JF. Tmevpg1, a candidate gene for the control of Theiler’s virus persistence, could be implicated in the regulation of gamma interferon. J. Virol. 2003;77(10):5632–5638. doi: 10.1128/JVI.77.10.5632-5638.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Park HJ, Dasari S, Wang S, Kocher JP, Li W. CPAT. Coding-potential assessment tool using an alignment-free logistic regression model. Nucleic acids research. 2013a;41(6):e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Park HJ, Dasari S, Wang SQ, Kocher JP, Li W. CPAT Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013b;41:6. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, et al. The STATbinding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344(6181):310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Wei G, Abraham BJ, Yagi R, Jothi R, Cui K, Sharma S, et al. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity. 2011;35(2):299–311. doi: 10.1016/j.immuni.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309(5740):1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- Yang F, Zhang H, Mei Y, Wu M. Reciprocal regulation of HIF-1alpha and lincRNA-p21 modulates the Warburg effect. Mol. Cell. 2014;53(1):88–100. doi: 10.1016/j.molcel.2013.11.004. [DOI] [PubMed] [Google Scholar]
- Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, et al. LincRNA-p21 suppresses target mRNA translation. Mol. cell. 2012;47(4):648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblood B, Hale JS, Ahmed R. T-cell memory differentiation: insights from transcriptional signatures and epigenetics. Immunology. 2013;139(3):277–284. doi: 10.1111/imm.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Yao W, Wang J, Ma X, Xiao W, Li H, et al. LncRNAs expression signatures of renal clear cell carcinoma revealed by microarray. PLoS one. 2012;7(8):e42377. doi: 10.1371/journal.pone.0042377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Nestor CE, Zhao S, Lentini A, Bohle B, Benson M, et al. Profiling of human CD4+ T-cell subsets identifies the TH2-specific noncoding RNAGATA3-AS1. J. Allergy Clin. Immunol. 2013;132(4):1005–1008. doi: 10.1016/j.jaci.2013.05.033. [DOI] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb Proteins Targeted by a Short Repeat RNA to the Mouse X Chromosome. Science. 2008;322(5902):750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]