Abstract
The gene lrrA, encoding a leucine-rich repeat protein, LrrA, that contains eight consensus tandem repeats of 23 amino acid residues, has been identified in Treponema denticola ATCC 35405. A leucine-rich repeat is a generally useful protein-binding motif, and proteins containing this repeat are typically involved in protein-protein interactions. Southern blot analysis demonstrated that T. denticola ATCC 35405 expresses the lrrA gene, but the gene was not identified in T. denticola ATCC 33520. In order to analyze the functions of LrrA in T. denticola, an lrrA-inactivated mutant of strain ATCC 35405 and an lrrA gene expression transformant of strain ATCC 33520 were constructed. Characterization of the mutant and transformant demonstrated that LrrA is associated with the extracytoplasmic fraction of T. denticola and expresses multifunctional properties. It was demonstrated that the attachment of strain ATCC 35405 to HEp-2 cell cultures and coaggregation with Tannerella forsythensis were attenuated by the lrrA mutation. In addition, an in vitro binding assay demonstrated specific binding of LrrA to a portion of the Tannerella forsythensis leucine-rich repeat protein, BspA, which is mediated by the N-terminal region of LrrA. It was also observed that the lrrA mutation caused a reduction of swarming in T. denticola ATCC 35405 and consequently attenuated tissue penetration. These results suggest that the leucine-rich repeat protein LrrA plays a role in the attachment and penetration of human epithelial cells and coaggregation with Tannerella forsythensis. These properties may play important roles in the virulence of T. denticola.
The oral spirochete Treponema denticola is strongly associated with the pathogenesis of periodontal diseases (3, 5, 19, 32, 41). This organism is frequently found in large numbers at diseased periodontal sites (45), and it expresses a variety of potential pathogenic properties, such as invasion of tissues, production of tissue-destructive enzymes, formation of cytotoxic products, and immunosuppression of host cell functions (3, 5, 19). The ability to attach to extracellular matrix molecules and/or bacterial partners may play an important role in the initiation of oral bacterial infections. The attachment of T. denticola to epithelial cells and extracellular matrix components was reported previously (1, 6, 14, 23-25, 38, 39, 48). In T. denticola, several surface proteins that mediate binding to proteins have been identified as putative virulence factors, i.e., fibronectin and collagen binding proteins (46, 47), Msp (8-10, 36), chymotrypsin-like protease complex (10), solute binding protein of an ATP-binding cassette-type peptide transporter, OppA (11), and hemin binding protein HbpA (49).
Leucine-rich repeats (LRRs) are protein interaction motifs of 20 to 29 residues consisting of a high proportion of leucine residues (20-22, 26, 27). These motifs are present in a large number of proteins with diverse functions and cellular locations in a variety of organisms and are mainly found in cell adhesion factors, hormone receptors, and enzyme inhibitors. Most of these proteins appear to be involved in protein-protein interactions. Recently, it was proposed that seven subfamilies of LRR proteins consist of different lengths and consensus sequences within the repeats (22, 27). In bacteria, at least three subfamilies of LRR extracellular proteins have been identified as bacterial, SDS22-like, and TpLRR (7, 27, 35). Specifically, the LRR proteins of the TpLRR family have been associated with bacterial cell surfaces, and these were identified as TpLRR from Treponema pallidum (44), BspA from Tannerella forsythensis (formerly Bacteroides forsythus) (43), and PcpA from Streptococcus pneumoniae (42).
T. pallidum belongs to the same genus as T. denticola (3), and Tannerella forsythensis is known as a putative periodontal pathogen like T. denticola (15, 16). It has been suggested that TpLRR may facilitate interactions between components of the Treponema cell envelope (44) and BspA may play roles in adherence to oral tissues and triggering of the host immune responses (17, 43). However, the exact function of these cell surface-associated proteins of the TpLRR family is unknown.
In the present study we identified the leucine-rich repeat gene lrrA from T. denticola and constructed a homologous lrrA-inactivated mutant. In addition, the lrrA gene was expressed in another strain of T. denticola, ATCC 33520, which was shown to lack this gene. Examination of these constructs suggested several functions of the T. denticola LrrA which might be important in the virulence of these organisms.
MATERIALS AND METHODS
Bacterial strains and cultivation.
T. denticola ATCC 35405 and ATCC 33520 were grown at 37°C under anaerobic conditions (85% N2, 10% H2, 5% CO2) in TYGVS medium (37). Tannerella forsythensis ATCC 43037 and its BspA-defective mutant, BFM-571 (17), were grown in BF broth under anaerobic conditions (43). Porphyromonas gingivalis strain 381 and Fusobacterium nucleatum NCTC 11326 were grown in tryptic soy broth (Becton Dickinson and Co., Franklin Lakes, N.J.) supplemented with hemin (10 μg/ml) and menadione (5 μg/ml) under anaerobic conditions. Escherichia coli JM109 was used as a host strain for plasmid construction and expression of fusion proteins. Ampicillin was added to Luria broth (LB) (Becton Dickinson and Co.) as indicated.
Cloning and sequencing of the lrrA gene of T. denticola.
The lrrA gene of T. denticola ATCC 35405 was amplified with oligonucleotide primers plrrAf (5′-AATCTTTAAGGAGGTTTGAC-3′) and plrrAr (5′-TATTTTGGGAAACGGCAG-3′) on the basis of the genome database of T. denticola from the Institute for Genomic Research (http://www.tigr.org). The PCR product was treated with Exo/SAP (Amersham Biosciences, Piscataway, N.J.) and sequenced to confirm that its sequence was identical to that in the genome database of T. denticola ATCC 35405.
Construction of the T. denticola lrrA mutant.
The lrrA gene-defective mutant of T. denticola ATCC 35405 was isolated by a strategy commonly used for construction of T. denticola mutants in this laboratory (31). A portion of the lrrA gene was amplified with oligonucleotide primers plrrAfm (5′-AATAGGCTACAAGTGTGC-3′) and plrrArm (5′-TATTTTGGGAAACGGCAG-3′). The amplified fragment was cloned into the pCR2.1 vector (Invitrogen Corp., Carlsbad, Calif.). The resulting plasmid, pCRlrrA, was cut with AccI within the lrrA fragment, and an ermF-ermAM cassette from pVA2198 was inserted into the AccI site of pCRlrrA to construct plasmid plrrAerm. Plasmid plrrAerm was then linearized following EcoRV digestion, and 10 μg of linearized plasmid was used to transform T. denticola ATCC 35405 by electroporation as described previously (31). The erythromycin-resistant transformants were isolated and inoculated into TYGVS medium containing erythromycin (40 μg/ml).
Southern blot analysis.
Chromosomal DNA of T. denticola was isolated and digested with PvuII. The digested DNA was separated on 1.0% agarose gels and transferred to Hybond-N+ membranes (Amersham Biosciences) by capillary transfer. The digoxigenin-labeled probe was the 0.31-kb lrrA fragment amplified by PCR with two synthetic oligonucleotide primers, pA210f (5′-AATAGGCTACAAGTGTGC-3′) and pA520r (5′-AAGCATACTCGCCTATTG-3′) from strain ATCC 35405. Digoxigenin labeling, hybridization, and detection were performed with a DIG DNA detection system (Roche Diagnostics Corporation, Indianapolis, Ind.) according to the manufacturer's protocol.
Expression of the lrrA gene in T. denticola.
The lrrA expression transformant of T. denticola ATCC 33520 was constructed with a T. denticola shuttle vector system (4). The lrrA gene containing an upstream region including the Shine-Dalgarno sequence was amplified by PCR with primers pKMAf (5′-TTTTTgtcgacAATCTTTAAGGAGGTTTGACATG-3′) and pKMAr (5′-TTTTTagatctCTAGTTTTTCTTCCAGTATTTATC-3′). The amplified fragment, to which SalI and BglII recognition sites were added (shown in lowercase letters), was inserted into the SalI and BglII sites of shuttle vector pKMR4PE. The resulting plasmid, pKMlrrA, was transformed into T. denticola ATCC 33520. The transformant was examined for expression of the LrrA protein by Western blot analysis.
Overexpression and purification of the LrrA protein.
The different regions of the PCR-amplified lrrA gene to which BamHI and PstI recognition sites had been added were cloned between the BamHI and PstI sites of a high-level expression hexahistidine-tagged recombinant protein vector pQE80 (Qiagen Inc., Valencia, Calif.). After verification of the resultant nucleotide sequences, the plasmids encoding the different regions of hexahistidine-tagged modified LrrA proteins were used for expression of the LrrA proteins (His-LrrA, His-LrrAΔC, His-LrrAΔN, and His-LrrAΔNC). Cells of Escherichia coli transformants carrying these plasmids were grown in LB medium. Expression of the chimeric genes was induced by addition of 1 mM isopropylthiogalactopyranoside (IPTG), and the recombinant proteins were purified by Ni2+-nitrilotriacetic acid spin affinity column chromatography (Qiagen Inc.) and HiTrap chelating HP column chromatography from the HisTrap kit (Amersham Biosciences). Proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (15% PAGE) and visualized by staining with Coomassie brilliant blue R.
Preparation of anti-LrrA antibody.
The recombinant protein His-LrrAΔN was separated by SDS-12% PAGE and visualized by staining with Coomassie brilliant blue R. A gel strip containing the His-LrrAΔN protein band was used as an antigen. Antiserum containing polyclonal antibodies against His-LrrAΔN (anti-LrrA serum) was obtained by injecting the antigen into chickens with custom antibody services (ProSci Inc., Poway, Calif.).
Western blot analysis of T. denticola with anti-LrrA antiserum.
For detection of the expression of T. denticola LrrA proteins with anti-LrrA serum, proteins (10 μg) were treated by boiling for 3 min in Laemmli sample buffer (Sigma Chemical Co., St. Louis, Mo.). These samples were separated by SDS-12% PAGE and blotted onto a polyvinylidene difluoride membrane (Bio-Rad Laboratories, Hercules, Calif.) with a protein transfer system (Bio-Rad Laboratories). The membrane was immunostained with a 1:2,000-diluted anti-LrrA serum for 2 h at room temperature, followed by 1:10,000-diluted horseradish peroxidase-conjugated goat anti-chicken immunoglobulin Y (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.) for 1 h at room temperature and detected by a horseradish peroxidase assay kit (Kirkegaard & Perry Laboratories Inc., Gaithersburg, Md.).
Immunogold labeling and observations by transmission electron microscopy.
Immunogold labeling was performed by the modified method of Ishihara et al. (18). Briefly, 3-day-cultured T. denticola cells were harvested by centrifuging at 3,000 × g for 10 min and washed with phosphate-buffered saline (PBS) twice. Bacterial cells were then adjusted to an optical density at 600 nm of 0.5. Bacterial cell suspensions were next reacted with 10-fold-diluted anti-LrrA serum on 200-mesh collodion membranes (Nisshin EM Co. Ltd., Tokyo, Japan) at 37°C for 30 min. After primary antibody reactions, membranes were washed with PBS twice, and the membranes were reacted with anti-chicken IgG rabbit serum conjugated to 10-fold-diluted gold particles (10 nm) (BB International, Cardiff, United Kingdom) at 37°C for 30 min. After reaction, the membranes were washed twice with PBS. The membranes were then negatively stained with 4% uranyl acetate solution. Four samples were made for each specimen. Observations were made with an AEM H-7100 transmission electron microscope (Hitachi Co, Ltd., Tokyo, Japan).
Attachment of T. denticola to HEp-2 epithelial cells.
HEp-2 epithelial cells were incubated in Dulbecco's modified Eagle's medium (DMEM; Invitrogen Corp.) supplemented with fetal bovine serum (Invitrogen Corp.) under 5% CO2 at 37°C. After the HEp-2 cells formed a tissue layer, the medium was changed to unsupplemented DMEM for the experiments. T. denticola strains were harvested and resuspended in unsupplemented DMEM to a density of 5 × 108 cells/ml. The bacterial suspensions (108 cells) were added to the HEp-2 tissue layers and incubated for 1 h at 37°C. After incubation, the tissue layers were washed twice with PBS containing 0.5% Tween 20 (PBST). The attached T. denticola cells were detected by measuring dentilisin activity (18) with a Chymotrypsin substrate II fluorogenic substrate (Calbiochem, La Jolla, Calif.). The numbers of attached T. denticola cells were calculated as the chemotrypsin activity of each strain per 106 cells. Attachment was calculated relative to that of strain ATCC 35405, which was adjusted to 100%.
Coaggregation assays.
Coaggregation activity between T. denticola and heterologous bacteria was determined with cells washed and suspended in PBS. The suspensions of each bacteria were adjusted to an optical density of 1.0 at 600 nm. Equal volumes of each suspension were mixed, and the optical density at 600 nm was determined over time at room temperature. The percent coaggregation was calculated by the following equation: coaggregation = [(preincubation value [OD600] − sample value [OD600])/(preincubation value [OD600])] × 100.
Binding assays of the LrrA derivatives to the recombinant Bsp70 protein.
The wells of 96-well microtiter plates (Invitrogen Corp.) were coated with 2 μg of recombinant Bsp70 (amino acid residues 17 to 724 of BspA) (43) dissolved in PBS following overnight incubation at 4°C. The protein-coated wells were then blocked with 0.5% bovine serum albumin in PBS for 1 h at room temperature. Various concentrations (1 and 10 pmol) of the His-tagged LrrA derivatives in 0.2% bovine serum albumin-PBS were prepared for binding assays. The proteins were added to the pretreated wells and incubated for 1 h at 37°C, followed by washing three times with PBST. The bound proteins were detected by QIAexpress Assay of 6×His-tagged proteins with anti-His horseradish peroxidase conjugate (Qiagen Inc.) according to the supplier's instructions. The anti-His horseradish peroxidase conjugate was diluted to 1:1,000 and developed with Tris-maleate buffer (Kirkegaard & Perry Laboratories, Gaithersburg, Md.).
Swarming assay.
T. denticola was incubated in TYGVS medium swarm plates containing 0.8% SeaPlaque agarose (FMC BioProducts, Rockland, Maine) at 37°C anaerobically, and swarming was monitored visually.
Tissue penetration assay.
Tissue penetration was determined by a modification of the penetration assay of Lux et al. (33). Tight-junctioned HEp-2 (now recognized as HeLa contaminants) tissue layers were incubated in supplemented DMEM in 3.0-μm Falcon cell culture inserts (Becton Dickinson Co.) under the culture conditions used for the attachment assays. T. denticola strains were harvested and resuspended in unsupplemented DMEM to a density of 5 × 108 cells/ml. The suspensions (2.5 × 108 cells) were added to the tissue layers and coincubated for 8 h under anaerobic conditions. Penetration rates were determined by counting cell numbers with a counting chamber (Hausser Scientific Partnership, Horsham, Pa.). The resistance of tissue layers was measured directly before and after each experiment to be sure that the electric resistance of tissues remained between 12 and 14 Ω. To monitor the viability of the tissue layers, the cytotoxicity of the HEp-2 tissue layers was measured with an in vitro toxicology assay kit based on lactic dehydrogenase (Sigma Chemical Co.).
Nucleotide sequence accession number.
The nucleotide sequence of the lrrA gene has been deposited in the GenBank database and assigned accession number AY545217.
RESULTS
Cloning and sequencing of the lrrA gene.
The LRR sequence in T. denticola was identified in the genome database of T. denticola ATCC 35405 from TIGR with the BspA sequence. As a result, a gene encoding a highly conserved LRR sequence was found in the database in addition to several other potential LRR protein genes. One of these genes was then isolated to confirm its nucleotide sequence (Fig. 1). After verification of the nucleotide sequence, the full-length gene, consisting of 1,068 bp, was named lrrA (leucine-rich repeat A). The lrrA gene was found to encode a putative protein consisting of 355 amino acid residues with a predicted molecular mass of 37,231 Da. A putative Shine-Dalgarno sequence was also identified in the sequence. Although the TpLRR protein from T. pallidum may not be a true lipoprotein, a potential signal peptidase cleavage site was identified in LrrA along with a consensus spirochetal lipoprotein box (13). This suggested that the LrrA protein may be a lipoprotein with a predicted molecular mass of 32,829 Da for the mature protein.
FIG. 1.
Nucleotide and deduced amino acid sequences of the lrrA gene of T. denticola ATCC 35405. The amino acid sequences of the LRR regions are shown in boldface letters and underlined. The potential Shine-Dalgarno sequence (SD) and spirochetal lipobox are underlined. A putative signal peptidase II cleavage site (SP) is indicated by an arrow.
The amino acid sequence of the LRR motifs of LrrA show high homology to other LRR sequences, including BspA of Tannerella forsythensis (57% identity), TpLRR of T. pallidum (35% identity), and PcpA of Streptococcus pneumoniae (28% identity) by BlastP 2.2.6. Analysis of the amino acid sequence of LrrA also revealed that LrrA contains eight tandem repeats of 23 amino acid residues with the consensus pattern LTSIxLPANLTTIGEYAFxGCTG (Fig. 1), where x is any residue, and is analogous to a consensus sequence pattern from the TpLRR family of LRR proteins, LxxLxLxxxLxxIgxxAFxxC/Nxx (22, 27). Therefore, LrrA shares a conserved pattern of LRRs similar to those of the TpLRR family.
Construction of the lrrA mutant and pKMlrrA transformant.
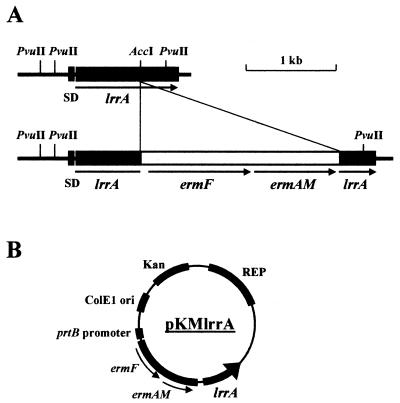
To determine the function of LrrA in T. denticola, an lrrA gene-defective mutant was constructed by allelic mutagenesis. A 905-bp portion of the lrrA gene was cloned into vector pCR2.1, and a 2.4-kb ermF-ermAM cassette was inserted into the AccI site of the lrrA gene to construct plasmid plrrAerm. Plasmid plrrAerm was then linearized with EcoRV and electroporated into T. denticola ATCC 35405. An insertion of the ermF-ermAM cassette into the lrrA gene on the chromosome by double crossover results in erythromycin-resistant (Ermr) transformants (Fig. 2A). The gene insertion in the transformants was confirmed by Southern blot analysis with a 311-bp fragment of the lrrA gene as a probe (Fig. 3). A 1.2-kb band was observed in strain ATCC 35405 (lane 2), while a lager 3.6-kb band was observed in the Ermr transformant AK-1 (lane 3). By contrast, no positive band was detected for strain ATCC 33520 (lane 1), indicating that a gene highly homologous to lrrA is absent in this strain. Therefore, to further analyze the function of LrrA in T. denticola, an lrrA gene expression plasmid was introduced into strain ATCC 33520 with a T. denticola shuttle vector system (4). The lrrA gene containing its Shine-Dalgarno sequence was cloned into T. denticola shuttle vector pKMR4PE (Fig. 2B). The resulting plasmid, pKMlrrA, was then transformed into strain ATCC 33520. The resulting transformant was confirmed for expression of LrrA by Western blot analysis with anti-LrrA serum (see Fig. 5).
FIG. 2.
Construction of the lrrA gene-inactivated mutant (AK-1) of strain ATCC 35405 and the lrrA gene expression plasmid (pKMlrrA). (A) The erythromycin resistance cassette (ermF-ermAM) used to inactivate the lrrA gene was inserted at the AccI site into the LRR region. (B) The lrrA gene containing a Shine-Dalgarno sequence (indicated by the arrow) was cloned into T. denticola shuttle vector pKMR4PE to construct lrrA gene expression plasmid pKMlrrA.
FIG. 3.
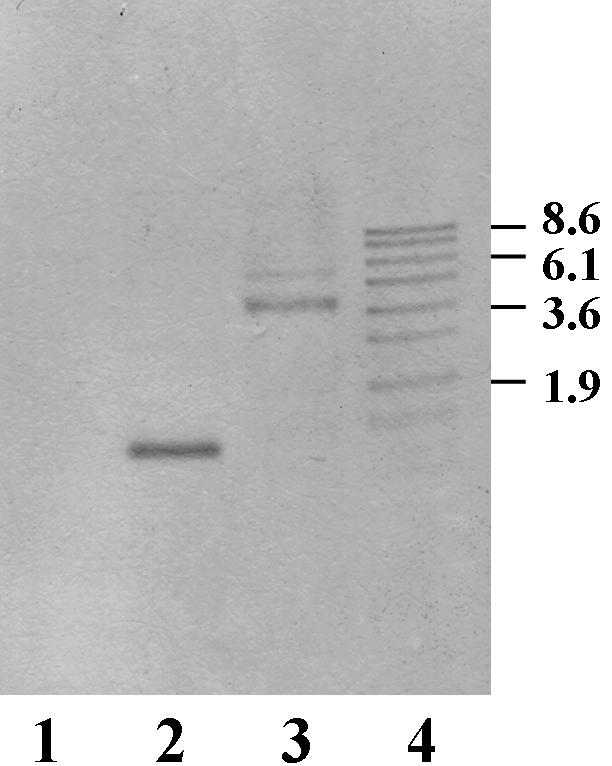
Southern blot analysis of T. denticola ATCC 35405, the lrrA::erm mutant AK-1, and strain ATCC 33520. Chromosomal DNAs from strain ATCC 33520 (lane 1), strain ATCC 35405 (lane 2), and mutant AK-1 (lane 3) were digested with PvuII. The digoxigenin-labeled probe was the 0.31-kb lrrA fragment from strain ATCC 35405. Digoxigenin-labeled DNA molecular size markers (in kilobases) are shown in lane 4.
FIG. 5.
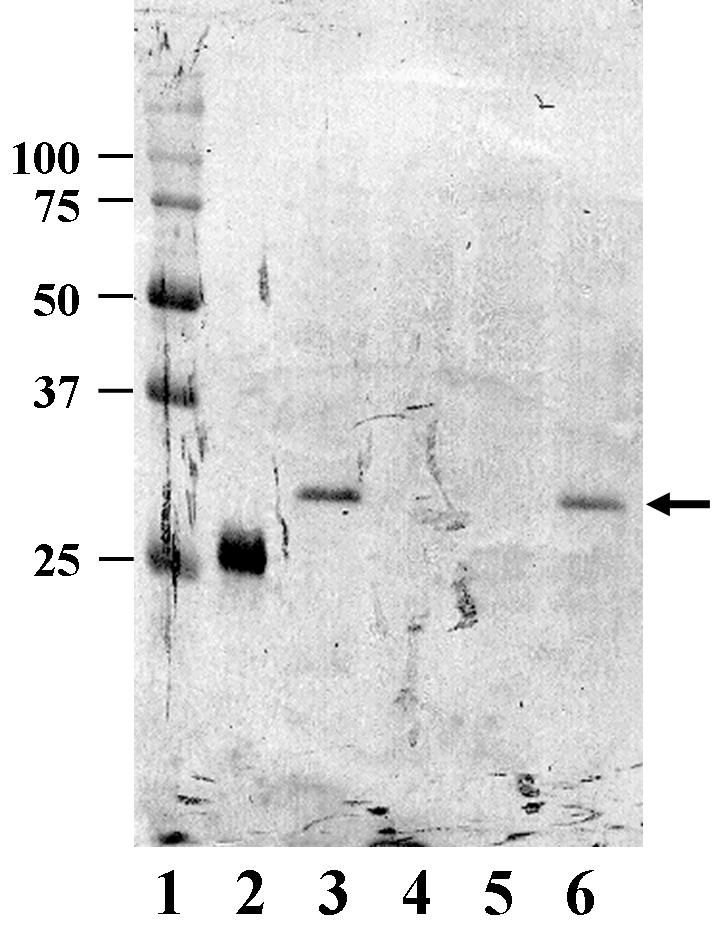
Western blot analysis of expression of the LrrA protein in different T. denticola strains. Proteins were separated by SDS-12% PAGE and transferred onto a polyvinylidene difluoride membrane. Lanes: 1, molecular mass markers; 2, purified His-LrrAΔN (positive control); 3, strain ATCC 35405; 4, mutant AK-1; 5, strain ATCC 33520; 6, shuttle vector pKMlrrA expressed in ATCC 33520. An arrowhead indicates the bands which reacted with anti-LrrA serum. The molecular mass sizes (in kilodaltons) are shown on the left.
Overexpression and purification of LrrA.
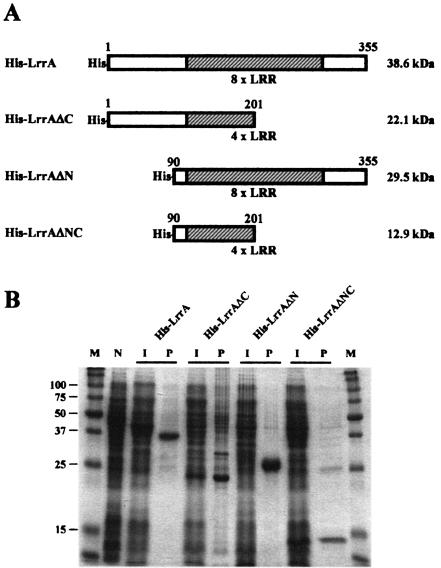
In order to prepare anti-LrrA antibody and identify the functional motifs of LrrA, various fragments of LrrA were expressed as fusion proteins with His tags in E. coli with the high-level expression system vector pQE80 (Fig. 4A). The fusion proteins included a full-length LrrA containing eight LRRs from residues 1 to 355 (His-LrrA), a C-terminal deletion LrrA containing four LRRs from residues 1 to 201 (His-LrrAΔC), an N-terminal deletion LrrA containing eight LRRs from residues 91 to 355 (His-LrrAΔN), and both N- and C-terminally deleted LrrA containing four LRRs from residues 91 to 201 (His-LrrAΔNC). These recombinant proteins were purified by affinity column chromatography and analyzed by SDS-15% PAGE. The results of SDS-PAGE analysis indicated that the recombinant proteins were expressed in sizes similar to the predicted molecular masses (His-LrrA, 38.6 kDa; His-LrrAΔC, 22.1 kDa; His-LrrAΔN, 29.5 kDa; and His-LrrAΔNC, 12.9 kDa). The recombinant proteins were purified as described earlier, and His-LrrAΔN was used for preparation of the anti-LrrA antibody.
FIG. 4.
Overexpression and purification of the various LrrA fragments. (A) Schematic representation of the various LrrA fragments. (B) Purification of the His-tagged LrrA derivatives. Fusion proteins (His-LrrA, His-LrrAΔC, His-LrrAΔN, and His-LrrAΔNC) were expressed in E. coli and purified on Ni-nitrilotriacetic acid columns. N, noninduced cells; I, cells induced with IPTG; P, purified protein; M, size markers. Proteins were separated by SDS-15% PAGE and visualized by staining with Coomassie brilliant blue R.
Expression of T. denticola LrrA protein in T. denticola.
Proteins were extracted from various T. denticola strains and fractionated by SDS-12% PAGE. Positive bands reacting with anti-LrrA serum were detected for strain ATCC 35405, the pKMlrrA transformant of strain ATCC 33520 (ATCC 33520/pKMlrrA) and His-LrrAΔN (positive control) (Fig. 5). These bands in strains ATCC 35405 and ATCC 33520/pKMlrrA each corresponded to a mass of approximately 32 kDa, consistent with the predicted molecular mass of LrrA. This result indicated that LrrA was expressed in strains ATCC 35405 and ATCC 33520/pKMlrrA; however, mutant AK-1 as well as strain ATCC 33520 were defective in LrrA expression.
LrrA is associated with the cell surface of T. denticola.
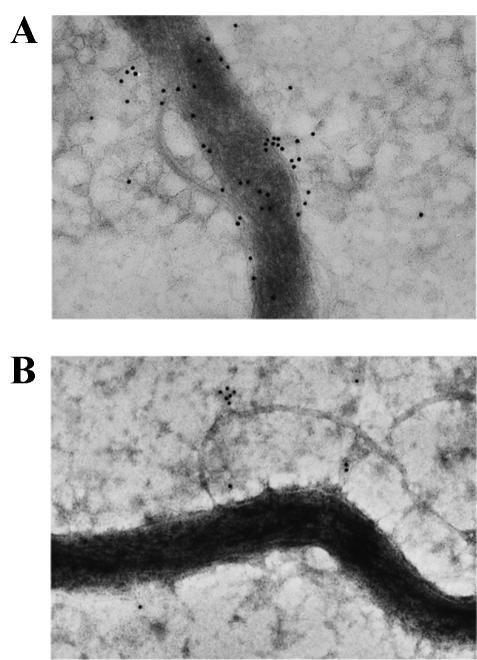
Since the sequencing data suggested that LrrA may be a lipoprotein associated with the extracytoplasmic fraction of strain ATCC 35405, immunoelectron microscopy was performed to localize LrrA in strain ATCC 35405. As shown in Fig. 6A, cells of strain ATCC 35405 incubated with anti-LrrA serum displayed gold particles associated with the cell surface of the organism. However, labeling was not observed in the lrrA mutant AK-1 (Fig. 6B). These results demonstrated that the LrrA protein is a component of the extracytoplasmic cell fraction of T. denticola, consistent with its identity as a lipoprotein.
FIG. 6.
Immunoelectron microscopy of T. denticola ATCC 35405 (A) and LrrA-deficient mutant AK-1 (B) cells with anti-LrrA serum. The size of the gold particles is 10 nm.
Attachment of T. denticola to HEp-2 cell tissue layers.
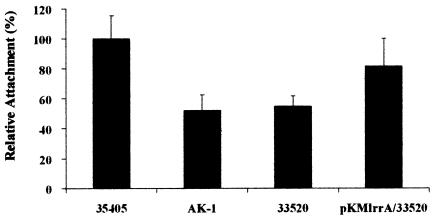
To investigate the attachment of T. denticola to epithelial cells, attachment assays were performed with HEp-2 cell cultures. Attachment to HEp-2 cell tissue layers was measured for four different T. denticola strains, ATCC 35405, AK-1, ATCC 33520, and ATCC 33520/pKMlrrA. The chymotrypsin specific activities of these strains as a percentage of that of strain ATCC 35405 were 79.8 ± 6.0% for mutant AK-1, 12.6 ± 0.7% for strain ATCC 33520, and 16.7 ± 3.5% for ATCC 33520/pKMlrrA.
The attachment of mutant AK-1 to HEp-2 cell tissue layers was reduced by 47.9% compared to that of parental strain ATCC 35405 (Fig. 7). Strain ATCC 33520 demonstrated weaker attachment (54.6%) in comparison with the LrrA-bearing strain ATCC 35405. Moreover, the attachment of ATCC 33520/pKMlrrA was slightly enhanced (81.6%) in comparison to its parental strain ATCC 33520. These results suggested that LrrA plays a role in the attachment of T. denticola to epithelial cells.
FIG. 7.
Attachment of T. denticola strains to HEp-2 cells. T. denticola strains ATCC 35405, AK-1, ATCC 33520, and ATCC 33520/pKMlrrA) were added to HEp-2 cell tissue layers and incubated for 1 h at 37°C. Attached cells were detected by chymotrypsin activity with a fluorogenic substrate. Attachment values are indicated relative to that of ATCC 35405, which are set at 100%. Values are means and standard deviations (error bars) for three separate experiments performed in triplicate.
Coaggregation activity between T. denticola and heterologous bacteria.
To analyze the role of LrrA in the coaggregation between T. denticola and heterologous oral bacteria, coaggregation activities were determined with T. denticola strains ATCC 35405, AK-1, ATCC 33520, and ATCC 33520/pKMlrrA.
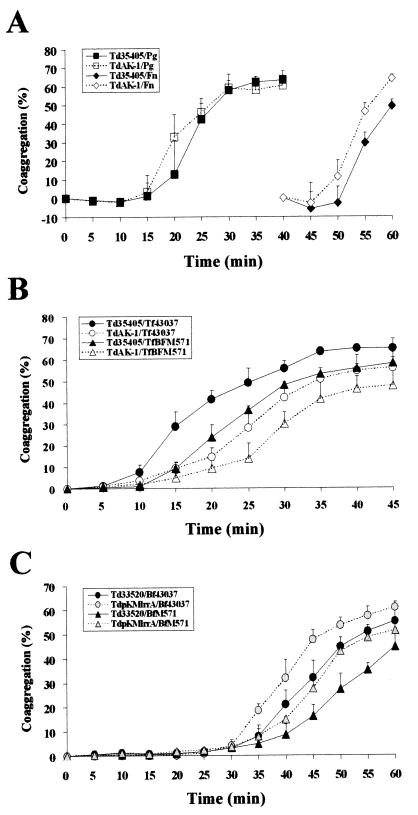
Coaggregations between T. denticola mutant AK-1 and Porphyromonas gingivalis or Fusarium nucleatum were not significantly different compared to that of parental strain ATCC 35405 (Fig. 8A). By contrast, mutant AK-1 was attenuated in coaggregation with Tannerella forsythensis (Fig. 8B). In addition, a reduction in coaggregation was likewise observed between T. denticola ATCC 35405 and the Tannerella forsythensis BspA mutant BFM-571 (Fig. 8B). Interestingly, strain ATCC 33520 appeared to aggregate with Tannerella forsythensis at a slower rate than strain ATCC 35405. Moreover, ATCC 33520/pKMlrrA was increased in coaggregation with Tannerella forsythensis relative to its parental strain ATCC 33520 (Fig. 8C). These results, taken together, suggested that the coaggregation between T. denticola and Tannerella forsythensis is mediated, in part, by interaction of the LrrA protein with the corresponding LRR protein, BspA, of Tannerella forsythensis. Neither T. denticola nor Tannerella forsythensis exhibited autoaggregation under these conditions.
FIG. 8.
Coaggregation between T. denticola and heterologous bacteria. (A) Coaggregation between T. denticola and heterologous bacteria. The percent coaggregation was calculated as described in the text. The data are the averaged values from three separate experiments. Td35405, T. denticola ATCC 35405; TdAK-1, T. denticola LrrA-deficient mutant AK-1; Tf, Tannerella forsythensis 43037; Pg, P. gingivalis 381; Fn, F. nucleatum 11326. (B) Effects of LRR proteins (LrrA and BspA) on coaggregation between T. denticola and Tannerella forsythensis. TfBFM571, Tannerella forsythensis BspA-deficient mutant BFM-571. (C) Effects of LrrA expression on coaggregation between T. denticola and Tannerella forsythensis. Td33520, T. denticola ATCC 33520; ATCC 33520/pKMlrrA, LrrA expression transformant of ATCC 33520.
Binding assays of LrrA derivatives to recombinant Bsp70.
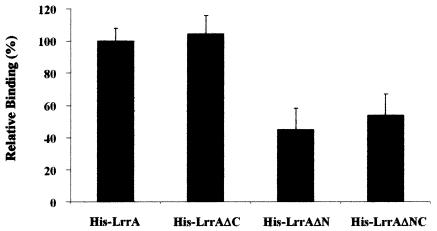
Since mutant analysis suggested that T. denticola-Tannerella forsythensis interactions may be mediated by LrrA-BspA binding, the interaction of the LrrA derivatives with BspA was analyzed directly. The binding assays demonstrated that the specific binding activity of the LrrA derivatives to recombinant Bsp70 was 104.2 ± 11.4% for His-LrrAΔC, 45.2 ± 13.2% for His-LrrAΔN, and 53.6 ± 13.4% for His-LrrAΔNC compared with His-LrrA (Fig. 9). These results suggested that the specific binding of LrrA to recombinant Bsp70 is mediated through the N-terminal region of LrrA, which is devoid of the LRR sequences.
FIG. 9.
Binding of the LrrA derivatives to BspA (recombinant Bsp70). Binding activities of the various LrrA derivatives were determined by enzyme-linked immunosorbent assay. Microtiter plates coated with recombinant Bsp70 were incubated with the LrrA derivatives His-LrrA, His-LrpAΔC, His-LrpAΔN, and His-LrpAΔNC for 1 h at room temperature. Bound proteins were incubated with 1:1,000-diluted anti-His-horseradish peroxidase conjugate and developed with TMB. Binding values are indicated relative to binding of His-LrrA, which was set at 100%. Values are means and standard deviations (error bars) for three separate experiments performed in duplicate.
Swarming activity of mutant AK-1 and transformant ATCC 33520/pKMlrrA.
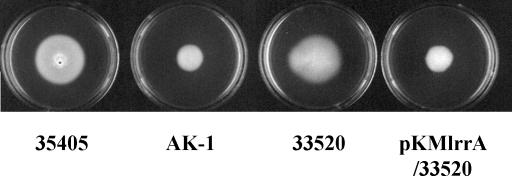
The growth rates of mutant AK-1 and ATCC 33520/pKMlrrA were similar to that of the parental strains in TYGVS medium (data not shown). These strains and the parental strains also appeared to exhibit similar motility in liquid culture when observed by dark-field microscopy. However, mutant AK-1 was shown to be deficient in swarming on TYGVS swarm plates (containing 0.8% SeaPlaque agarose) (Fig. 10). Surprisingly, the ATCC 33520/pKMlrrA construct was also decreased in swarming compared to its parental strain ATCC 33520 without pKMlrrA or with the shuttle vector plasmid pKMR4PE alone. These results indicated that mutant AK-1 and ATCC 33520/pKMlrrA were reduced in swarming activities in high-viscosity medium. These data suggest that the swarming activity of T. denticola can be influenced by the presence or absence of LrrA in the spirochetes.
FIG. 10.
Swarming activity of T. denticola. T. denticola strains ATCC 35405, AK-1, ATCC 33520, and ATCC 33520/pKMlrrA cultured in TYGVS medium swarm plates (containing 0.8% agarose) were monitored visually after 7 days.
Tissue penetration by T. denticola strains.
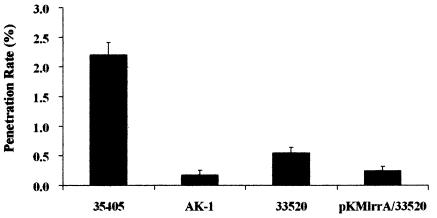
Motility and chemotaxis were reported to be important in tissue penetration (33, 34); therefore, poor swarming may influence tissue penetration. Tissue penetration ability was estimated with an oral epithelial cell line-based tissue penetration assay basically according to the method of Lux et al. (33). From microscopic observations, the human cell line HEp-2 was demonstrated to form tight-junctioned tissue layers with an electric resistance of >10 Ω and maintained viability under anaerobic conditions for at least 8 h. Cytotoxicity was not detected in the HEp-2 cell cultures under these conditions. Penetration rates were determined as 2.20 ± 0.22% for strain ATCC 35405, 0.17 ± 0.08% for mutant AK-1, 0.55 ± 0.09% for strain ATCC 33520, and 0.25 ± 0.07% for ATCC 33520/pKMlrrA (Fig. 11). The weaker swarming strains, mutant AK-1 and ATCC 33520/pKMlrrA, showed significantly reduced penetration compared to their respective parental strains. The penetration of mutant AK-1 was reduced by approximately 90% compared to that of strain ATCC 35405. In addition, the ATCC 33520/pKMlrrA construct showed an approximately 50% reduction in penetration relative to strain ATCC 33520. These results suggested that tissue penetration ability was correlated with swarming activity. In addition, the weaker attachment of strain ATCC 33520 to HEp-2 cells compared to strain ATCC 35405 (Fig. 6) can also affect relative tissue penetration rates.
FIG. 11.
Tissue penetration of T. denticola. Penetration rates were determined by counting cell numbers after 8 h of incubation of T. denticola strains ATCC 35405, AK-1, ATCC 33520, and ATCC 33520/pKMlrrA with HEp-2 cell tissue layers. Values are means and standard deviations for three separate experiments performed in duplicate. The resistances of the tissue layers were between 12 and 14 Ω for the experiments.
DISCUSSION
LRRs have been reported to be generally useful as protein-binding motifs (20-22, 26, 27). The major function of LRRs may be to provide a structural framework for the formation of protein-protein interactions (27). In this study, an lrrA gene identified in the genome database of T. denticola ATCC 35405 was used to analyze the function of LRR proteins in T. denticola. The lrrA gene was characterized as encoding a polypeptide of 355 amino acid residues that contains eight tandem repeats of 23 amino acid residues and exhibits high homology in the LRR region with other LRR proteins, such as BspA and TpLRR. Recently, the TpLRR family has been proposed as one of the subfamilies of LRR proteins (22). BspA and TpLRR belong to the TpLRR family, exhibiting consensus pattern LxxLxLxxxLxxIgxxAFxxC/Nxx. The consensus pattern of the LRR in LrrA was identical to that of the TpLRR family. Analysis of the functions of the TpLRR family has suggested that TpLRR proteins may facilitate interactions of components of the cell envelope with the environment (44). Furthermore, BspA may play roles in adherence to oral tissues and triggering of host immune responses (17, 43). However, only two genes of the TpLRR family have been analyzed until now, and it was not clear what role LRR proteins had in T. denticola.
Southern blot analysis demonstrated that T. denticola ATCC 35405 contains an lrrA gene, while strain ATCC 33520 does not. Expression of the LrrA protein in strain ATCC 35405 as well as in strain ATCC 33520 transformed with the lrrA gene was confirmed by Western blot analysis with anti-LrrA serum. Furthermore, the data from immunoelectron microscopy suggested that LrrA is associated with the cell surface of T. denticola, although the precise location has not yet been determined. LRR proteins of the TpLRR family have also been reported to be cell surface associated (22). Specifically, it has been proposed that BspA may be concerned with cellular adhesion by Tannerella forsythensis (17, 43). The association of LrrA with the surface of T. denticola is also consistent with the observation from this study that LrrA plays a role in interaction of this spirochete with its oral environment.
The ability to attach to extracellular matrix molecules as well as bacterial partners may play an important role in the initiation of bacterial infection. T. denticola has been reported to attach to epithelial cells and extracellular matrix components (1, 6, 14, 23-25, 38, 39, 48). The present results regarding the attachment of T. denticola to HEp-2 cell tissue layers indicated that the LrrA protein plays a role in these interactions. Therefore, LrrA may be involved in the attachment of the spirochete to the epithelial cell layer lining the gingival crevice.
Coaggregation is a process by which bacteria attach to one another via specific cell surface molecules (40). These interactions are believed to contribute to the development of multispecies biofilms (40); therefore, coaggregation has important implications for the bacteria isolated from dental plaque biofilms. Many different coaggregation interactions have now been identified between oral bacteria (29, 30), and that of T. denticola has been reported to be influenced by P. gingivalis and F. nucleatum (12, 28). Some of the coaggregation adhesion proteins are multifunctional and can often interact with host oral surfaces as well as host proteins (40). Therefore, it is likely that LrrA may also be multifunctional and play a role in multiple interactions. The influence of LrrA on the coaggregation between T. denticola and heterologous bacteria indicated that LrrA did not affect coaggregations with P. gingivalis and F. nucleatum. However, coaggregation between T. denticola and Tannerella forsythensis was influenced by the LRR proteins LrrA and BspA. These results suggested that the coaggregation between T. denticola and Tannerella forsythensis is mediated, in part, by interaction of LrrA with BspA.
In vitro binding assays of the LrrA derivatives suggested that specific binding of LrrA to recombinant Bsp70 is not mediated by the interaction of the LRR domains but is mainly mediated through the N-terminal region of LrrA. Most LRR proteins have been previously reported to contain flanking regions that are an integral part of the LRR domain (2, 27). The LRR-associated motif LRR cap was identified in flanking regions and proposed to play a role in shielding the hydrophobic core of the LRRs from solvents (2, 27). In the case of LrrA, the LRR cap motif was not identified; however, a functional motif similar to an LRR cap may exist in the N-terminal region and could interact with the LRR domain of BspA. More detailed investigations will be necessary to identify the motifs involved in LrrA-BspA interactions.
Analysis of swarming indicated that the affected LrrA expression strains, mutant AK-1 and ATCC 33520/pKMlrrA, exhibited decreased swarming on TYGVS swarm plates, even though each appeared to be as motile as the parental strains in liquid cultures. This change may also attenuate tissue penetration, as demonstrated in the present study, because previous studies reported that the tissue penetration of oral epithelial cell layers by T. denticola is likely dependent upon motility and chemotaxis (33, 34). The attenuated swarming by AK-1 suggested that the LrrA protein is involved in the swarming ability of strain ATCC 35405. However, strain ATCC 33520 swarms as well as strain ATCC 35405 without expression of an LrrA protein. This suggests that the expression of LrrA in strain ATCC 33520 “interferes” with the swarming mechanism of this strain. Perhaps the abnormal expression and localization of LrrA in strain ATCC 33520 interferes with the cell surface properties of this strain that are required for swarming. In addition, since dentilisin activity may itself play a role in the surface properties of T. denticola, it is possible that interactions between the protease and LrrA could be involved in some of the results demonstrated in this study. For example, the relatively low protease activity of strain ATCC 33520 relative to that of strain ATCC 35405 could influence such interactions and alter surface properties. Additional approaches are ongoing to determine the molecular basis for these novel observations.
In the present study, it was demonstrated that the functions of the cell surface-associated LRR protein LrrA in T. denticola are consistent with a significant role for this protein in virulence. The protein appears to play a role in both attachment to epithelial cells and coaggregation with other periodontal pathogenic bacteria such as Tannerella forsythensis. Both of these interactions may be important in the colonization of T. denticola into subgingival plaque. Furthermore, a role for LrrA in tissue penetration was demonstrated, and this property may also be an important virulence factor for oral spirochetes. Confirmation of these roles awaits the development of animal models involving T. denticola-induced periodontitis.
Acknowledgments
This study was supported in part by National Institutes of Health grants DE09821 and DE14749.
Editor: V. J. DiRita
REFERENCES
- 1.Carranza, N., Jr., G. R. Riviere, K. S. Smith, D. F. Adams, and T. Maier. 1997. Differential attachment of oral treponemes to monolayers of epithelial cells. J. Periodontol. 68:1010-1018. [DOI] [PubMed] [Google Scholar]
- 2.Ceulemans, H., M. D. Maeyer, W. Stalmans, and M. Bollen. 1999. A capping domain for LRR protein interaction modules. FEBS Lett. 456:349-351. [DOI] [PubMed] [Google Scholar]
- 3.Chan, E. C., and R. McLaughlin. 2000. Taxonomy and virulence of oral spirochetes. Oral Microbiol. Immunol. 15:1-9. [DOI] [PubMed] [Google Scholar]
- 4.Chi, B., S. Chauhan, and H. Kuramitsu. 1999. Development of a system for expressing heterologous genes in the oral spirochete Treponema denticola and its use in expression of the Treponema pallidum flaA gene. Infect. Immun. 67:3653-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dahle, U. R., L. Tronstad, and I. Olsen. 1993. Spirochaetes in oral infections. Endod. Dent. Traumatol. 9:87-94. [DOI] [PubMed] [Google Scholar]
- 6.Dawson, J. R., and R. P. Ellen. 1990. Tip-oriented adherence of Treponema denticola to fibronectin. Infect. Immun. 58:3924-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evdokimov, A. G., D. E. Anderson, K. M. Routzahn, and D. S. Waugh. 2001. Unusual molecular architecture of the Yersinia pestis cytotoxin YopM: a leucine-rich repeat protein with the shortest repeating unit. J. Mol. Biol. 312:807-821. [DOI] [PubMed] [Google Scholar]
- 8.Fenno, J. C., K. H. Muller, and B. C. McBride. 1996. Sequence analysis, expression, and binding activity of recombinant major outer sheath protein (Msp) of Treponema denticola. J. Bacteriol. 178:2489-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fenno, J. C., G. W. Wong, P. M. Hannam, K. H. Muller, W. K. Leung, and B. C. McBride. 1997. Conservation of msp, the gene encoding the major outer membrane protein of oral Treponema spp. J. Bacteriol. 179:1082-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fenno, J. C., P. M. Hannam, W. K. Leung, M. Tamura, V. J. Uitto, B. C. McBride. 1998. Cytopathic effects of the major surface protein and the chymotrypsinlike protease of Treponema denticola. Infect. Immun. 66:1869-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fenno, J. C., M. Tamura, P. M. Hannam, G. W. Wong, R. A. Chan, and B. C. McBride. 2000. Identification of a Treponema denticola OppA homologue that binds host proteins present in the subgingival environment. Infect. Immun. 68:1884-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grenier, D. 1992. Demonstration of a bimodal coaggregation reaction between Porphyromonas gingivalis and Treponema denticola. Oral Microbiol. Immunol. 7:280-284. [DOI] [PubMed] [Google Scholar]
- 13.Haake, D. A. 2000. Spirochaetal lipoproteins and pathogenesis. Microbiology 146:1491-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haapasalo, M., U. Singh, B. C. McBride, and V. J. Uitto. 1991. Sulfhydryl-dependent attachment of Treponema denticola to laminin and other proteins. Infect. Immun. 59:4230-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haffajee, A. D., M. A. Cugini, S. Dibart, C. Smith, R. L. Kent Jr., and S. S. Socransky. 1997. The effect of SRP on the clinical and microbiological parameters of periodontal diseases. J. Clin. Periodontol. 24:324-334. [DOI] [PubMed] [Google Scholar]
- 16.Haffajee, A. D., M. A. Cugini, S. Dibart, C. Smith, R. L. Kent Jr., and S. S. Socransky. 1997. Clinical and microbiological features of subjects with adult periodontitis who responded poorly to scaling and root planing. J. Clin. Periodontol. 24:767-776. [DOI] [PubMed] [Google Scholar]
- 17.Honma, K., H. K. Kuramitsu, R. J. Genco, and A. Sharma. 2001. Development of a gene inactivation system for Bacteroides forsythus: construction and characterization of a BspA mutant. Infect. Immun. 69:4686-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishihara, K., T. Miura, H. K. Kuramitsu, and K. Okuda. 1996. Characterization of the Treponema denticola prtP gene encoding a prolyl-phenylalanine-specific protease (dentilisin). Infect. Immun. 64:5178-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishihara, K., and K. Okuda. 1999. Molecular pathogenesis of the cell surface proteins and lipids from Treponema denticola. FEMS Microbiol. Lett. 181:199-204. [DOI] [PubMed] [Google Scholar]
- 20.Kajava, A. V. 1998. Structural diversity of leucine-rich repeat proteins. J. Mol. Biol. 277:519-527. [DOI] [PubMed] [Google Scholar]
- 21.Kajava, A. V. 2001. Proteins with repeated sequence—structural prediction and modeling. J. Struct. Biol. 134:132-144. [DOI] [PubMed] [Google Scholar]
- 22.Kajava, A. V., and B. Kobe. 2002. Assessment of the ability to model proteins with leucine-rich repeats in light of the latest structural information. Protein Sci. 11:1082-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keulers, R. A., J. C. Maltha, F. H. Mikx, and J. M. Wolters-Lutgerhorst. 1993. Attachment of Treponema denticola strains to monolayers of epithelial cells of different origin. Oral Microbiol. Immunol. 8:84-88. [DOI] [PubMed] [Google Scholar]
- 24.Keulers, R. A., J. C. Maltha, F. H. Mikx, and J. M. Wolters-Lutgerhorst. 1993. Attachment of T. denticola strains ATCC 33520, ATCC 35405, B11 and Ny541 to a morphologically distinct population of rat palatal epithelial cells. J. Periodontal. Res. 28:274-280. [DOI] [PubMed] [Google Scholar]
- 25.Keulers, R. A., J. C. Maltha, F. H. Mikx, and J. M. Wolters-Lutgerhorst. 1993. Involvement of treponemal surface-located protein and carbohydrate moieties in the attachment of Treponema denticola ATCC 33520 to cultured rat palatal epithelial cells. Oral Microbiol. Immunol. 8:236-241. [DOI] [PubMed] [Google Scholar]
- 26.Kobe, B., and J. Deisenhofer. 1994. The leucine-rich repeat: a versatile binding motif. Trends Biochem. Sci. 19:415-421. [DOI] [PubMed] [Google Scholar]
- 27.Kobe, B., and A. V. Kajava. 2001. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11:725-732. [DOI] [PubMed] [Google Scholar]
- 28.Kolenbrander, P. E., K. D. Parrish, R. N. Andersen, and E. P. Greenberg. 1995. Intergeneric coaggregation of oral Treponema spp. with Fusobacterium spp. and intrageneric coaggregation among Fusobacterium spp. Infect. Immun. 63:4584-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolenbrander, P. E. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54:413-437. [DOI] [PubMed] [Google Scholar]
- 30.Kolenbrander, P. E., R. N. Andersen, D. S. Blehert, P. G. Egland, J. S. Foster, and R. J. Palmer Jr. 2002. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 66:486-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, H., J. Ruby, N. Charon, and H. K. Kuramitsu. 1996. Gene inactivation in the oral spirochete Treponema denticola: construction of an flgE mutant. J. Bacteriol. 178:3664-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loesche, W. J. 1993. Bacterial mediators in periodontal disease. Clin. Infect. Dis. 16:S203-210. [DOI] [PubMed] [Google Scholar]
- 33.Lux, R., J. N. Miller, N. H. Park, and W. Shi. 2001. Motility and chemotaxis in tissue penetration of oral epithelial cell layers by Treponema denticola. Infect. Immun. 69:6276-6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lux, R., J. H. Sim, J. P. Tsai, and W. Shi. 2002. Construction and characterization of a cheA mutant of Treponema denticola. J. Bacteriol. 184:3130-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marino, M., L. Braun, P. Cossart, and P. Ghosh. 1999. Structure of InlB leucine-rich repeats, a domain that triggers host cell invasion by the bacterial pathogen L. monocytogenes. Mol. Cell 4:1063-1072. [DOI] [PubMed] [Google Scholar]
- 36.Mathers, D. A., W. K. Leung, J. C. Fenno, Y. Hong, and B. C. McBride. 1996. The major surface protein complex of Treponema denticola depolarizes and induces ion channels in HeLa cell membranes. Infect. Immun. 64:2904-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohta, K., K. K. Makinen, and W. J. Loesche. 1986. Purification and characterization of an enzyme produced by Treponema denticola capable of hydrolyzing synthetic trypsin substrates. Infect. Immun. 53:213-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olsen, I. 1984. Attachment of Treponema denticola to cultured human epithelial cells. Scand. J. Dent. Res. 92:55-63. [DOI] [PubMed] [Google Scholar]
- 39.Peters, S. R., M. Valdez, G. Riviere, and D. D. Thomas. 1999. Adherence to and penetration through endothelial cells by oral treponemes. Oral Microbiol. Immunol. 14:379-383. [DOI] [PubMed] [Google Scholar]
- 40.Rickard, A. H., P. Gilbert, N. J. High, P. E. Kolenbrander, and P. S. Handley. 2003. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 11:94-100. [DOI] [PubMed] [Google Scholar]
- 41.Riviere, G. R., K. S. Elliot, D. F. Adams, L. G. Simonson, L. B. Forgas, A. M. Nilius, and S. A. Lukehart. 1992. Relative proportions of pathogen-related oral spirochetes (PROS) and Treponema denticola in supragingival and subgingival plaque from patients with periodontitis. J. Periodontol. 63:131-136. [DOI] [PubMed] [Google Scholar]
- 42.Sanchez-Beato, A. R., R. Lopez, and J. L. Garcia. 1998. Molecular characterization of PcpA: a novel choline-binding protein of Streptococcus pneumoniae. FEMS Microbiol. Lett. 164:207-214. [DOI] [PubMed] [Google Scholar]
- 43.Sharma, A., H. T. Sojar, I. Glurich, K. Honma, H. K. Kuramitsu, and R. J. Genco. 1998. Cloning, expression, and sequencing of a cell surface antigen containing a leucine-rich repeat motif from Bacteroides forsythus ATCC 43037. Infect. Immun. 66:5703-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shevchenko, D. V., D. R. Akins, E. Robinson, M. Li, T. G. Popova, D. L. Cox, and J. D. Radolf. 1997. Molecular characterization and cellular localization of TpLRR, a processed leucine-rich repeat protein of Treponema pallidum, the syphilis spirochete. J. Bacteriol. 179:3188-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simonson, L. G., C. H. Goodman, J. J. Bial, and H. E. Morton. 1988. Quantitative relationship of Treponema denticola to severity of periodontal disease. Infect. Immun. 56:726-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Umemoto, T., Y. Nakatani, Y. Nakamura, and I. Namikawa. 1993. Fibronectin-binding proteins of a human oral spirochete Treponema denticola. Microbiol. Immunol. 37:75-78. [DOI] [PubMed] [Google Scholar]
- 47.Umemoto, T., M. Li, and I. Namikawa. 1997. Adherence of human oral spirochetes by collagen-binding proteins. Microbiol. Immunol. 41:917-923. [DOI] [PubMed] [Google Scholar]
- 48.Walker, S. G., J. L. Ebersole, and S. C. Holt. 1999. Studies on the binding of Treponema pectinovorum to HEp-2 epithelial cells. Oral Microbiol. Immunol. 14:165-171. [DOI] [PubMed] [Google Scholar]
- 49.Xu, X., S. C. Holt, and D. Kolodrubetz. 2001. Cloning and expression of two novel hemin binding protein genes from Treponema denticola. Infect. Immun. 69:4465-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]