Abstract
One of the early stages of Klebsiella pneumoniae airway infections may involve biofilm formation. Bacterial biofilm formation is frequently investigated using in vitro techniques that facilitate identification and analysis of individual genes. We investigated the correlation between K. pneumoniae biofilm formation in vitro and ability to cause infection in vivo following construction of a bank of mini-Tn5 mutants.
The role of biofilm formation and development by bacteria has been suggested to be an important stage in the pathogenesis of numerous bacterial species (1, 3, 19). The establishment of biofilms by pathogenic bacteria on the tissues of susceptible hosts is believed to inhibit the effectiveness of antibiotic treatment, protect against host defense mechanisms, and facilitate bacterial communication leading to expression of virulence determinants. In order to produce a biofilm on appropriate surfaces, several investigations have established that multiple genetic systems are involved and biofilm development can be inhibited or altered in a broad range of mutants (2, 5, 10, 12, 20, 21). We have previously demonstrated that growth of Klebsiella pneumoniae on abiotic surfaces is facilitated, in part, by the MrkA type 3 fimbrial protein, whereas growth on surfaces coated with a human extracellular matrix (HECM) requires the presence of the type 3 fimbrial adhesin MrkD (6, 7). In order to investigate in vitro the genetics of biofilm formation by K. pneumoniae on surfaces, we constructed a bank of insertion mutants in strain 43816, a K2-positive isolate that is also virulent in a murine model of respiratory infection. Mutants that were altered in biofilm formation in vitro were also examined for their ability to infect animals in vivo.
Identification of biofilm-defective mutants.
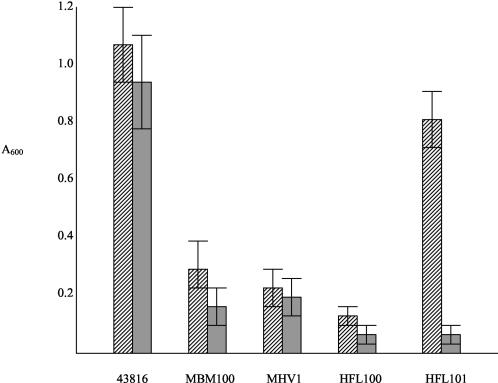
Following construction of a bank of Tn5 insertion mutants in K. pneumoniae 43816 as previously described by our group (7), a total of 2,000 mutants were examined for their ability to grow on either abiotic plastic surfaces or on HECM-coated microtiter plates. The development of a biofilm in vitro was detected using the procedure of O'Toole and Kolter (11), and the method of coating microtiter plates with HECM has been described in detail by us elsewhere (14). Four mutants consistently demonstrated a reduced ability to grow on at least one of the two surfaces. One of these mutants, K. pneumoniae MBM100, was unable to express type 3 fimbriae and has previously been described by our group (7). The remaining three mutants were fully fimbriate and, therefore, the decrease in their ability to form biofilms in vitro was not due to the lack of type 3 fimbriae on their surfaces. The biofilm phenotype of each mutant is shown in Fig. 1. Three mutants exhibited significantly reduced growth on both abiotic and HECM-coated surfaces of microtiter plates, whereas one mutant, K. pneumoniae HFL101, demonstrated decreased growth only on HECM-coated surfaces (Fig. 1). In all cases the mutants exhibited similar growth rates in vitro when cultured in the media used in the biofilm assays. Therefore, the reduction in growth on the surfaces of the microtiter plates was not due to differential growth in the media during the course of the experiments.
FIG. 1.
Production of a biofilm on the surfaces of uncoated (hatched bars) and HECM-coated (shaded bars) microtiter plates by K. pneumoniae 43816 and mini-Tn5 insertion mutants. Bacteria were incubated in plates containing glycerol-Casamino Acids broth, and crystal violet was used to quantitate biofilm formation as previously described (7). The bars indicate means ± standard errors of the means from three experiments.
Site of mini-Tn5 insertion in biofilm-deficient mutants.
Either subcloning of Sau3A1 or NotI DNA fragments or arbitrary PCR analysis followed by DNA sequencing and comparison to genes and open reading frames deposited in databases was used to identify the sites of transposon insertion within the genome of K. pneumoniae 43816. As indicated previously, the mini-Tn5 of K. pneumoniae MBM100 had been located within the yadH gene (7). It is not clear why a mutation in yadH should result in a nonfimbrial phenotype, but restoration of type 3 fimbriation did restore the ability to form a biofilm (7). In Escherichia coli, yadH is a putative, membrane-bound component of an ABC transporter system.
For mutant K. pneumoniae MHV1, the site of Tn5 insertion is in the putative yciI gene, which is also found on the E. coli genome and the function of which is unknown. As in E. coli, the K. pneumoniae yciI gene is adjacent to tonB and, therefore, the mini-Tn5 insertion could have a polar effect on this gene. It has been demonstrated in E. coli that tonB mutants are attenuated and, as indicated below, K. pneumoniae MHV1 is avirulent in the mouse model of respiratory infection. Consequently, we cloned the K. pneumoniae tonB gene by PCR using the genome of strain 43816 as template. However, the recombinant plasmid possessing only the tonB gene did not complement K. pneumoniae MHV1 in restoration to virulence, although it partially increased the ability of transformants to form a biofilm in vitro compared to the mutant. However, the biofilm formed was still significantly less than that of the parental strain, K. pneumoniae 43816.
The third and fourth mutants isolated, K. pneumoniae HFL100 and HFL101, exhibited insertion of the mini-Tn5 at two distinct regions on the K. pneumoniae genome and were at sites most closely related to the yhdH and pduC genes of Salmonella enterica serovar Typhimurium, which encode a putative oxidoreductase and glycerol dehydratase, respectively (8). As for the mutants above, it cannot be concluded that these two genes specifically play a role in biofilm formation in vitro, since the insertion of the mini-Tn5 at these sites could have effects on adjacent genes. However, it is clear that these insertions give rise to reproducible and significant changes in the ability of the mutants to grow on solid surfaces in vitro (Fig. 1).
Virulence of mini-Tn5 insertion mutants.
As indicated in Table 1, K. pneumoniae 43816 is highly virulent in the murine model of airway infection. Intranasal inoculation of 103 organisms resulted in 100% lethality up to 5 days postinfection. Significant numbers of bacteria could be isolated from the lungs, liver, and spleen of surviving infected animals 48 h postinfection. As shown in Table 2, three of the mutants, K. pneumoniae MBM100, HFL100, and HFL101, that exhibited reduced biofilm formation in vitro were fully virulent in vivo, and all animals succumbed to infection, following intranasal inoculation, within 5 days. However, K. pneumoniae MHV1 was avirulent via the intranasal route of infection, and even after infection with relatively high doses (107 organisms) all animals remained healthy up to 28 days postinfection. In order to determine if strain MHV1 was reduced in virulence after direct inoculation without growth on the respiratory mucosa, individual groups (n = 5) of animals were injected intraperitoneally with either 103, 105, or 107 bacteria. Only at the high inoculum level (107 bacteria) were animals lethally infected (Table 2). These data suggest that yciI and the region around tonB in K. pneumoniae encode important virulence determinants of strain 43816. As mentioned above, infection of the urinary tract by E. coli is attenuated in tonB mutants (17), and this gene may facilitate the survival of bacteria both on mucosal surfaces and during systemic infection by facilitating effective uptake of iron from the host. However, as indicated above, the cloned tonB gene of K. pneumoniae did not restore virulence to strain MHV1. Therefore, we believe that both yciI and tonB play an important role in the virulence of K. pneumoniae 43816.
TABLE 1.
Infectivity and virulence of K. pneumoniae 43816 in the murine model of infection
| Inoculum size (CFU) | Route of inoculationa | % Lethality (no. of animals) | Mean time (days) to death | No. of bacteria/g of tissue
|
||
|---|---|---|---|---|---|---|
| Lung | Liver | Spleen | ||||
| 103 | IN | 100 (14) | 4.1 ± 1.1 | 1.60 × 106 | 4.62 × 104 | 3.61 × 105 |
| 104 | IN | 100 (5) | 4.0 ± 1.5 | NDb | ND | ND |
| 107 | IN | 100 (8) | 2.5 ± 1.5 | 4.45 × 1010 | 3.49 × 108 | 5.39 × 108 |
| 108 | IN | 100 (14) | 2.7 ± 2.0 | ND | ND | ND |
| 103 | IP | 100 (10) | 3.0 ± 2.0 | ND | ND | ND |
IN, intranasal; IP, intraperitoneal.
ND, not determined.
TABLE 2.
Biofilm phenotype and mouse virulence of K. pneumoniae strains
| Strain | Site of Tn insertion | Biofilm in vitroa | Virulence in vivob |
|---|---|---|---|
| 43816 | + | + | |
| MBM100 | yadH | − | + |
| MHV1 | yciI | − | − |
| HFL100 | yhdH | − | + |
| HFL101 | pduA | − (+)c | + |
Biofilm formation determined using the microtiter plate assay after incubation for 18 h at 37°C.
Virulence measured as 100% lethality following intranasal inoculation with 103 CFU.
Strain HFL101 exhibited reduced biofilm growth on HECM-coated surfaces but not on untreated plates.
Since it has been suggested that biofilm formation by pathogenic bacteria is an important stage in the infective process for many different species, we decided to investigate the correlation between biofilm formation in vitro and virulence in vivo for K. pneumoniae. In our studies, three of four independent biofilm-defective mutants of K. pneumoniae 43816 exhibited virulence properties identical to that of the parental strain, suggesting that the inability, or reduced ability, to grow on solid surfaces in vitro does not always correlate with a reduced ability to grow in vivo. In the murine model of Klebsiella airway infection, the bacteria can colonize the respiratory tract and subsequently invade through this organ system. Consequently, growth on the respiratory mucosa would represent an important stage of infection. However, it is difficult to mimic the conditions encountered in vivo by growth in vitro. Clearly, our results indicate that some of the genes responsible for the development and growth of K. pneumoniae 43816 on the surfaces of microtiter plates are different from those required for growth in the murine lung. In some cases, however, in vitro screening for determinants that also play a role in vivo can reveal important virulence factors. K. pneumoniae MHV1 is a mutant that is severely attenuated for growth in vivo and was originally isolated as a strain with a reduced ability to form a biofilm in vitro. Therefore, for K. pneumoniae pathogenesis, the in vitro biofilm assay has the potential to identify putative virulence genes, but individual determinants have to be assessed in vivo to confirm their role.
Another consideration, however, when investigating K. pneumoniae airway infections is the relevance of the animal model system to mimic human infections. For example, although K. pneumoniae 43816 is virulent for mice, many isolates implicated in human airway infections are avirulent in the murine model, even though they belong to the same capsular serotype (Table 3). Many epidemiologic studies have indicated that human airway infections are most frequently associated with the K2 capsular serotype of K. pneumoniae (4, 9, 15). However, as indicated in Table 3, K2-positive isolates implicated in human infections do not consistently cause infections in the mouse model. Also, infection of mice by strain 43816 can occur in healthy animals, whereas the majority of human infections by K. pneumoniae occur in compromised individuals (13, 16, 18). Consequently, we tested several clinical isolates of K. pneumoniae, representing different capsular serotypes and isolated from either blood or tracheal aspirates of infected individuals, for their virulence in the murine model (Table 3). Most of these isolates did not cause lethal infections following intranasal inoculation of animals, unlike strain 43816. Our group has recently developed an in vitro system to investigate growth of human isolates of K. pneumoniae on human extracellular matrices in a continuous-culture flow cell (6). We believe that the use of this system in conjunction with the animal model of infection provides complementary approaches to examine the stages of infection by K. pneumoniae. Our results highlight the importance of using both in vivo and in vitro approaches to investigate the pathogenesis of an organism that causes human airway infections.
TABLE 3.
Virulence of clinical isolates of K. pneumoniae in the murine model of airway infection following intranasal inoculation
| Strain | Serotype | Inoculum size (CFU) | No. of animals that died/no. inoculated |
|---|---|---|---|
| 43816 | K2 | 103 | 14/14 |
| i220-94 | K2 | 103 | 0/10 |
| 108 | 1/9 | ||
| i222-94 | K2 | 108 | 0/10 |
| IA565 | NDa | 108 | 0/10 |
| THK12133 | K57 | 108 | 0/5 |
| THK12127 | K26 | 103 | 0/7 |
| 108 | 3/7 | ||
| UIR908 | ND | 108 | 0/5 |
ND, not determined.
Editor: V. J. DiRita
REFERENCES
- 1.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 2.De Kievit, T. R., and B. H. Iglewski. 1999. Quorum sensing, gene expression, and Pseudomonas biofilms. Methods Enzymol. 310:117-128. [DOI] [PubMed] [Google Scholar]
- 3.Donlan, R. M. 2001. Biofilm formation: a clinically relevant microbiological process. Clin. Infect. Dis. 33:1387-1392. [DOI] [PubMed] [Google Scholar]
- 4.Edwards, P. R., and M. A. Fife. 1952. Capsule types of Klebsiella. J. Infect. Dis. 91:92-104. [DOI] [PubMed] [Google Scholar]
- 5.Haugo, A. J., and P. I. Watnick. 2002. Vibrio cholerae CytR is a repressor of biofilm development. Mol. Microbiol. 45:471-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jagnow, J., and S. Clegg. 2003. Klebsiella pneumoniae MrkD-mediated biofilm formation on extracellular matrix- and collagen-coated surfaces. Microbiology 149:2397-2405. [DOI] [PubMed] [Google Scholar]
- 7.Langstraat, J., M. Bohse, and S. Clegg. 2001. Type 3 fimbrial shaft (MrkA) of Klebsiella pneumoniae, but not the fimbrial adhesin (MrkD), facilitates biofilm formation. Infect. Immun. 69:5805-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 9.Nassif, X., and P. J. Sansonetti. 1986. Correlation of the virulence of Klebsiella pneumoniae K1 and K2 with the presence of a plasmid encoding aerobactin. Infect. Immun. 54:603-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 11.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 12.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis, and type 1 pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 13.Pugliese, G., and D. A. Lichtenberg. 1987. Nosocomial bacterial pneumonia: an overview. Am. J. Infect. Control 15:249-265. [DOI] [PubMed] [Google Scholar]
- 14.Schurtz Sebghati, T. A., T. K. Korhonen, D. B. Hornick, and S. Clegg. 1998. Characterization of the type 3 fimbrial adhesins of Klebsiella strains. Infect. Immun. 66:2887-2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tarkkanen, A. M., B. L. Allen, P. H. Williams, M. Kauppi, K. Haahtela, A. Siitonen, I. Orskov, F. Orskov, S. Clegg, and T. K. Korhonen. 1992. Fimbriation, capsulation, and iron-scavenging systems of Klebsiella strains associated with human urinary tract infection. Infect. Immun. 60:1187-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tillotson, J. R., and A. M. Lerner. 1966. Pneumonias caused by gram-negative bacilli. Medicine 45:65-76. [DOI] [PubMed] [Google Scholar]
- 17.Torres, A. G., P. Redford, R. A. Welch, and S. M. Payne. 2001. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect. Immun. 69:6179-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Alphen, L., H. M. Jansen, and J. Dankert. 1995. Virulence factors in the colonization and persistence of bacteria in the airways. Am. J. Respir. Crit. Care Med. 151:2094-2100. [DOI] [PubMed] [Google Scholar]
- 19.Watnick, P., and R. Kolter. 2000. Biofilm, city of microbes. J. Bacteriol. 182:2675-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watnik, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae biofilm. Mol. Microbiol. 34:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]