Abstract
Considerable evidence has suggested that the epigenetic regulation of N-methyl-D-aspartate (NMDA) glutamate receptors plays a crucial role in neuropsychiatric disorders. Previous exploratory studies have been primarily based on evidence from patients and have rarely sampled the general population. This exploratory study examined the relationship of single-nucleotide polymorphism (SNP) variations in the genes encoding the NMDA receptor (i.e., GRIN1, GRIN2A, GRIN2B, GRIN2C, and GRIN2D) with emotion and social behavior in adolescents. For this study, 832 tenth-grade Taiwanese volunteers were recruited, and their scores from the Beck Youth Inventories were used to evaluate their emotional and social impairments. Based on these scores, GRIN1 (rs4880213) was significantly associated with depression and disruptive behavior. In addition, GRIN2B (rs7301328) was significantly associated with disruptive behavior. Because emotional and social impairment greatly influence learning ability, the findings of this study provide important information for clinical treatment and the development of promising prevention and treatment strategies, especially in the area of psychological adjustment.
1. Introduction
Interest in the pathology of emotional disorders, such as depression and anxiety, has increased, primarily because the incidence of emotional disorders in adults, adolescents, and even children has dramatically increased over the past several decades [1]. Emotional disorders are often influenced by genetic and lifestyle factors [2, 3], and understanding the genetic etiologies of these diseases could provide valuable information for the development of effective therapies.
The neuronal N-methyl-D-aspartate receptor (NMDAR) has been postulated to play a key role in the pathophysiology of schizophrenia, bipolar disorder, and depression [4, 5]. The possible role of NMDAR signaling in the pathophysiology of emotional disorders has been supported by the following evidence: (a) bipolar disorder and major depression disorder are associated with altered levels of central excitation neurotransmitters [6, 7], (b) NMDAR expression, distribution, and function are decreased in patients with mood disorders [8], (c) the NMDAR modulator exerts a positive therapeutic effect on patients [9], and (d) antidepressants and mood stabilizers can improve NMDAR function [10, 11]. Therefore, genes involved in the NMDAR pathway might be important genetic regulators of human physiology that consequently influence mood diseases.
Recent studies have shown that many of the physiological and pharmacological properties of the NMDAR depend on the composition of its subunits [12]. NMDAR subtypes include at least one obligatory NR1 subunit in combination with different constellations of NR2 (i.e., GRIN2A, GRIN2B, GRIN2C, and GRIN2D) [13–15] and/or NR3 (i.e., GRIN3A and 3B) subunits. The alternative splicing of the GRIN1 gene yields eight different NR1 isoforms that form different combinations with NR2 and/or NR3. These isoforms display unique properties in the central nervous system that depend on their subunit composition (Figure 1). The NR1 subunit appears to be a common prerequisite for the expression of functional NMDARs, and the NR2 subunit is required for the efficient formation of functional NMDARs. Moreover, the NR2 subunits determine biophysiological ion channel properties, such as the mean conductance open time and sensitivity to an Mg2+ block [16]. The NR2 subunit assembly process may also be a critical factor in postsynaptic signaling pathways that direct synaptic plasticity [17]. The dysfunction of NMDA ligand-gated cation channels is an underling molecular mechanism for neurologic disorders, such as schizophrenia [14], psychiatric disorders [18], and neurodegenerative diseases [19]. Recent studies indicated that alterations in NR1 and NR2 transcript expression associated with the NMDAR stoichiometry in schizophrenia involved more complex cellular changes than previously assumed [20, 21]. The heteromeric nature of NMDARs provides a wide variety of receptor subtypes; thus, single-nucleotide polymorphisms (SNPs) in NMDAR subunits are likely responsible for creating various distinct NMDAR properties. Several SNPs in the NMDAR subunit genes have been shown to affect the role of NMDAR signaling in the pathophysiology of mood disorders by altering the expression, distribution, and function of the NMDARs [4, 22].
Figure 1.
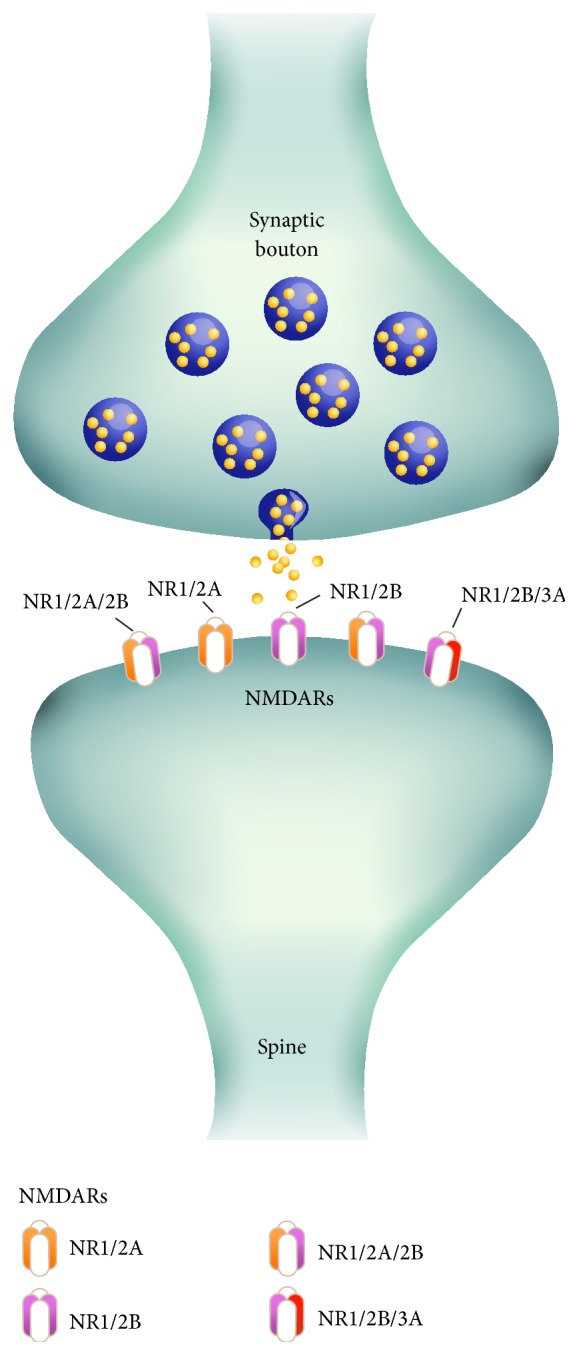
Different NMDA receptor stoichiometric subunits.
Two SNPs of GRIN1 merit special attention: rs11146020 (G1001C) and rs4880213 (GRIN1 5′-upstream region). The GRIN1 (rs11146020) gene is located at 9q34, a locus in the promoter region of the NMDAR subunit that has been linked to schizophrenia [23, 24]. The GRIN1 (rs11146020) gene product plays a fundamental role in many brain functions, and its involvement in the pathogenesis of schizophrenia has been widely investigated [25–27]. Only a few studies have focused on GRIN1 rs4880213 and the pathogenesis of schizophrenia or mood disorders. Two studies showed that individuals with the GRIN1 rs4880213 C/C variation displayed more severe disability and reduced NMDAR-mediated cortical excitability than individuals with the C/T or T/T variation [28, 29].
Further studies revealed that variations in the NMDAR 2B subunit gene (GRIN2B) were associated with schizophrenia, psychiatric disorders, and brain plasticity. For example, in human attention performance studies, subjects that were homozygous for the frequent C allele of the GRIN2B rs1806201 variation displayed more altered network scores than subjects in the other two genotype groups (C/T and T/T) [30]. The other 2B subunits of NMDAR have promising candidate SNP variants (i.e., rs1805476, rs1805477, rs1805501, and rs1805502) that affect the genetic susceptibility to obsessive-compulsive disorder [31]. In a functional study of protein expression, the GRIN2B rs1805502 (T5988C) C allele was associated with reduced GRIN1 mRNA and protein expression in schizophrenic patients [5]. In pharmacological interventions, the same variant of NMDAR (GRIN2B) has been proposed to act as a genetic predictor of treatment-resistant depression (TRD) in patients with major depressive disorder (MDD) [32].
Understanding the involvement of GRIN genes in neurocognitive deficits will shed light on the importance of links between subunit involvement in plasticity paradigms and behavior. However, most previous studies of SNP associations with schizophrenia only investigated one or two genes in NMDARs, particularly GRIN1, GRIN2A, and GRIN2B [5, 24, 33]. Therefore, in this study, we used more extensive systemic sequencing to identify SNPs that may affect multiple genes related to NMDARs. Our results further elucidate the role of NMDAR signaling in emotion and social behavior.
This exploratory study examined the relationship of SNP variations in NMDAR genes with emotion and social behavior in adolescents. Data from randomized controlled trials could provide convincing evidence on the mechanisms of genetic and epigenetic effects on emotion [34]. However, previous exploratory studies have been primarily based on evidence from patients and have rarely sampled the general population. In addition, previous studies of schizophrenia revealed that the total number of NMDARs remains normal as the patient ages, but the receptor stoichiometry appears to change with age [35]. Therefore, an investigation of the impact of epigenetic changes on emotion and behavior in different cross sections of the population could be important for understanding the underlying mechanisms of mood disorders. To the best of our knowledge, the association between genetic SNPs and emotion in adolescents has not yet been investigated. This study aimed to fill this gap. Furthermore, epidemiological studies have linked mood disorders to gender differences. Specifically, women are more likely than men to be diagnosed with depression [36, 37]. Therefore, the current study aimed to control for the influence of gender on the effect of genetic polymorphisms on emotion using statistics.
2. Materials and Methods
2.1. Participants
Three public senior high schools were selected (one in southern Taiwan, one in central Taiwan, and one in northern Taiwan). A total of 832 tenth-grade volunteers, 269 of whom were male, were recruited for this study. The mean age of the subjects was 16.3 years (SD: 0.5; age range: 16-17 years). The volunteers and their parents were explicitly informed about the plan, protocol, and procedure for the study, and written consent was obtained prior to the start of the study. This study was approved by the institutional review board of the National Taiwan University Hospital and the ClinicalTrials.gov registry of clinical trials (ClinicalTrials.gov identifier: NCT00713570).
2.2. Gene Screening, Variation Analysis, and Bioinformatics
We investigated the association of 59 SNPs in the NMDAR genes (i.e., GRIN1, GRIN2A, GRIN2B, GRIN2C, and GRIN2D) with emotion and behavior in 832 Han Chinese subjects. Fifty-nine SNPs from the entire set of candidate genes associated with NMDARs were identified based on information available in the Entrez Gene (http://www.ncbi.nlm.nih.gov/gene), HapMap (http://www.hapmap.org), and Ensembl (http://www.ensembl.org/Homo_sapiens) databases. In a pilot study, we analyzed genotypes by sequencing DNA from 20 subjects, and we assumed that some genetic polymorphisms involved in functional NMDAR subunits might contribute to individual differences in emotion and social behavior (GSJUNIOR_A16, Roche Union ClinBio Co. Ltd.). SNPs with a minor allele frequency greater than 5% in the pilot study that were identified in the analyzed samples were considered the most promising candidates, and representative common variants were selected. From the pilot study, 8 SNPs from the GRIN1 and GRIN2 genes were identified as the most promising candidates. The samples were genotyped by DNA sequencing of the relevant PCR products using an ABI Prism_ BigDye Terminator v3.1 Cycle Sequencing Ready Reaction kit and an ABI Prism_3730 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). All participants were screened for the 8 selected SNPs using a custom/commercial TaqMan SNP genotyping assay according to the manufacturer's instructions (Life Technologies, Carlsbad, CA, USA) and a ViiA 7 real-time PCR system (Life Technologies).
2.3. Emotion Assessment
The subjects' emotional behaviors were assessed via the second edition of the complete Beck Youth Inventories (BYI-II) [38], which is a commonly used and thoroughly standardized test that suitably assesses the diversity of emotion and behavior in adolescents. The BYI-II consists of five inventories: anxiety, depression, anger, disruptive behavior, and self-concept, and each of these inventories contains 20 items that are assessed based on five self-reported scales. Higher scores for the inventories of anxiety, depression, anger, or disruptive behavior indicate a tendency towards emotional instability. In contrast, a higher score in the self-concept inventory reveals that the student has a positive sense of self.
2.4. Statistical Analysis
Two-tailed t-tests were used to assess the significance of differences in the BYI-II subtask scores between males and females. For each SNP, the participants were assigned to one of three groups based on their genotype, and deviation from the Hardy-Weinberg equilibrium was tested using a chi-squared test. An analysis of covariance (ANCOVA) was employed to determine the effect of each SNP genotype on behavior based on the BYI-II subtask scores. Previous studies revealed that gender is an important determinant of emotion. To assess the effect of genetic polymorphism on emotion, the gender of participants served as a covariate in this analysis to control for gender effects on BYI-II subtask scores. Post hoc analyses using pairwise comparisons were also used to identify trends in the data. The level of confidence was set at the 0.05 significance level. In consideration of type I errors, permutation tests (with N = 1,000 randomizations) [39] were performed to correct for multiple comparisons. The assumptions used for the ANCOVA and inferential statistical analyses were tested using SPSS version 22.0.
3. Results
NMDARs are tetramers that contain two GRIN1 subunits bound to two GRIN2 (A, B, C, or D) and/or GRIN3 (A or B) subunits. Many of the physiological and pharmacological properties of NMDARs depend on the specific GRIN2 and GRIN3 subunits. The observed distributions of the genotypes for each SNP appeared to follow Hardy-Weinberg equilibrium. The analyses of SNPs in NMDAR-related genes, genotypic distribution, chromosome localization, and genotype frequency are presented in Table 1.
Table 1.
The genotype distributions and chromosome locations of the SNPs.
| Gene/SNP ID | Allele/genotype | Subjects | Chromosome region | Genotype frequency | CHB population frequency∗ |
|---|---|---|---|---|---|
| GRIN1 | 9q34.3 | ||||
| rs4880213 | CC/CT/TT | 124/368/340 | 0.15/0.44/0.41 | 0.11/0.44/0.45 | |
| rs11146020 | CC/CG/GG | 565/239/28 | 0.68/0.29/0.03 | 0.72/0.24/0.04 | |
| GRIN2B | 12p12 | ||||
| rs7301328 | CC/CG/GG | 250/396/186 | 0.30/0.48/0.22 | 0.26/0.43/0.31 | |
| rs1806201 | AA/AG/GG | 223/438/171 | 0.27/0.53/0.20 | 0.22/0.55/0.22 | |
| rs1805247 | AA/AG/GG | 608/204/20 | 0.73/0.25/0.02 | 0.61/0.35/0.04 | |
| rs1805502 | AA/AG/GG | 608/203/20 | 0.73/0.24/0.03 | 0.61/0.35/0.04 | |
| rs3764028 | AA/AC/CC | 148/397/287 | 0.18/0.48/0.34 | 0.13/0.46/0.42 | |
| GRIN2C | 17q25 | ||||
| rs3744215 | AA/AC/CC | 143/423/266 | 0.17/0.51/0.32 | 0.15/0.47/0.38 |
∗Data from 1000 Genomes Project Phase 3; http://www.1000genomes.org/.
The demographic and BYI-II subtask scores of the participants are outlined in Table 2. Some differences between males and females were observed. Female subjects in this study tended to be more anxious, whereas male subjects exhibited more disruptive behaviors.
Table 2.
Distribution of selected characteristics of the participants.
| Male | Female | P | |
|---|---|---|---|
| Age (yrs) | 16.8 (0.32)∗ | 16.8 (0.30) | |
| BYI-II | |||
| Anxiety | 16.2 (8.9) | 18.5 (8.8) | <0.01 |
| Depression | 13.6 (9.4) | 15.1 (9.3) | |
| Anger | 11.7 (8.9) | 12.4 (7.8) | |
| Disruptive behavior | 8.2 (7.2) | 6.6 (4.9) | <0.01 |
| Self-concept | 36.0 (9.6) | 36.5 (7.9) |
∗Mean (SD).
As shown in Table 3, the emotional instability significantly differed in individuals carrying the GRIN1 (rs4880213) and GRIN2B (rs7301328) polymorphisms. After the multiple comparison correction by 1,000 permutation tests, GRIN1 (rs4880213) was significantly associated with depression (P = 0.04) and disruptive behavior (P = 0.04), and GRIN2B (rs7301328) was significantly associated with disruptive behavior (P = 0.05). The other six SNPs were not significantly associated with behavior based on the BYI-II analysis.
Table 3.
Genotype distributions and comparisons of subtask scores and genotype groupings.
| C/C | C/T | T/T | F | P cor | |
|---|---|---|---|---|---|
| GRIN1 rs4880213 | |||||
| Anxiety | 19.1 ± 9.5∗ | 18.2 ± 9.0 | 17.0 ± 8.5 | 2.6 | >0.05 |
| Depression | 16.5 ± 9.8 | 15.0 ± 9.5 | 13.5 ± 8.8 | 4.6 | 0.04 |
| Anger | 12.3 ± 7.8 | 12.8 ± 8.3 | 11.6 ± 8.0 | 1.4 | >0.05 |
| Disruptive behavior | 7.2 ± 5.6 | 7.7 ± 6.1 | 6.4 ± 5.1 | 4.1 | 0.04 |
| Self-concept | 36.0 ± 7.9 | 36.2 ± 9.2 | 36.7 ± 8.0 | 0.4 | >0.05 |
|
| |||||
| C/C | C/G | G/G | |||
|
| |||||
| GRIN2B rs7301328 | |||||
| Anxiety | 17.5 ± 8.6 | 17.8 ± 9.0 | 18.6 ± 9.3 | 0.8 | >0.05 |
| Depression | 14.1 ± 8.9 | 14.6 ± 9.1 | 15.5 ± 10.5 | 1.1 | >0.05 |
| Anger | 11.6 ± 7.8 | 12.3 ± 7.8 | 13.1 ± 9.2 | 1.5 | >0.05 |
| Disruptive behavior | 6.3 ± 5.6 | 7.2 ± 5.6 | 7.9 ± 5.8 | 3.7 | 0.05 |
| Self-concept | 36.2 ± 8.9 | 36.9 ± 7.6 | 35.7 ± 9.5 | 1.2 | >0.05 |
∗Mean values ± standard deviations.
P cor: multiple comparison correction by 1,000 permutation tests.
The ANCOVA of the depression subtask scores revealed a significant main effect for the three genotype groups of GRIN1 (rs4880213; Table 4, F[2,829] = 4.5, P < 0.05). The pairwise test revealed that the depression subtask scores of the T/T genotype group were significantly lower than those of the C/C and C/T genotype groups (Table 5, C/C > C/T, P = 0.005; C/T > T/T, P = 0.04). The GRIN1 (rs4880213) polymorphisms also influenced the disruptive behavior scores (Table 6, F[2,829] = 4.0, P < 0.05). Subjects with the T/T genotype group tended to score lower on the disruptive behavior subtask (Table 7, C/T > T/T, P = 0.005). Furthermore, the disruptive behavior scores were significantly associated with the GRIN2B rs7301328 polymorphism (Table 8, F[2,829] = 4.2, P = 0.05). As shown in Table 9, a pairwise comparison analysis revealed that the disruptive behavior subtask scores of the G/G genotype group were significantly lower than those of the C/C genotype group (P = 0.004).
Table 4.
ANCOVA summary for the depression subtask scores.
| Source of variance | Df | SS | MS | F | P |
|---|---|---|---|---|---|
| GRIN1 rs4880213 | 2 | 787.6 | 393.8 | 4.5 | 0.04∗ |
| Gender | 1 | 278.3 | 278.3 | 3.2 | 0.07 |
| Residual | 828 | 21794 | 26.3 |
∗Multiple comparison correction by 1000 permutation tests.
Table 5.
Pairwise comparisons of three GRIN1 rs4880213 genotype groups in terms of adjustments of depression subtask scores.
| GRIN1 rs4880213 | Madj | SE | Pairwise comparison |
|---|---|---|---|
| C/C | 16.5 | 0.9 | C/C > T/T (P = 0.005) |
| C/T | 15.1 | 0.5 | C/T > T/T (P = 0.04) |
| T/T | 13.5 | 0.5 |
Table 6.
ANCOVA summary for the disruptive behavior subtask scores.
| Source of variance | Df | SS | MS | F | P |
|---|---|---|---|---|---|
| GRIN1 rs4880213 | 2 | 254.3 | 127.1 | 4.0 | 0.04∗ |
| Gender | 1 | 335.2 | 335.2 | 10.5 | 0.001 |
| Residual | 828 | 21794 | 26.5 |
∗Multiple comparison correction by 1,000 permutation tests.
Table 7.
Pairwise comparisons of three GRIN1 rs4880213 genotype groups in terms of adjustments of disruptive behavior subtask scores.
| GRIN1 rs4880213 | Madj | SE | Pairwise comparison |
|---|---|---|---|
| C/C | 7.3 | 0.5 | C/T > T/T (P = 0.005) |
| C/T | 7.7 | 0.3 | |
| T/T | 6.4 | 0.3 |
Table 8.
ANCOVA summary for the disruptive behavior subtask scores.
| Source of variance | Df | SS | MS | F | P |
|---|---|---|---|---|---|
| GRIN2B rs7301328 | 2 | 269.2 | 269.2 | 4.2 | 0.05∗ |
| Gender | 1 | 386.3 | 386.3 | 12.1 | 0.001 |
| Residual | 828 | 21779 | 26.3 |
∗Multiple comparison correction by 1,000 permutation tests.
Table 9.
Pairwise comparisons of three GRIN2B rs7301328 genotype groups in terms of adjustments of disruptive behavior subtask scores.
| GRIN2B rs7301328 | Madj | SE | Pairwise comparison |
|---|---|---|---|
| C/C | 6.2 | 0.5 | C/C > G/G (P = 0.004) |
| C/G | 7.2 | 0.3 | |
| G/G | 8.0 | 0.3 |
4. Discussion
All NMDARs appear to function as heteromeric assemblies that consist of multiple NR1 subunits and at least one type of NR2. The NR3 subunit does not form functional receptors alone but can coassemble with NR1/NR2 complexes. The temporal and spatial expression patterns of the NR2 and NR3 subunits were recently shown to differ in the brain, and the expression of NMDAR subtypes also vary by cell type and subcellular localization [40, 41]. In situ hybridization studies have shown that the mRNAs for NMDAR subunits are differentially distributed throughout the brain, and the expression patterns of these mRNAs change strikingly during development [42]. This study aimed to determine whether any other SNPs are responsible for the distinct properties of the NMDAR that are associated with emotion and behavior, especially in adolescents. Previous studies show that GRIN1 (rs11146020) was strongly associated with bipolar disorder [4] and depressive symptoms [43]. The incidences of depression and disruptive behavior were significantly lower in participants that carried the GRIN1 T/T genotype (SNP rs4880213) than in members of the two other groups; this result was consistent with Georgi's study, which showed that the T allele was less frequent in schizophrenia patients with a lifetime history of depression than in control. Francesco et al. [44] suggested that the T/T genotype of the GRIN1 rs4880213 SNP is associated with reduced intracortical inhibition, enhanced glutamatergic excitation, and enhanced glutamate NMDAR function. Francesco's study also revealed that the GRIN1 and GRIN2B subunits of NMDARs are involved in regulating cortical excitability and plasticity in the human cortex. Rossi et al. [28] also indicated that the C allele of rs4880213 is associated with reduced NMDAR-mediated cortical excitability. Notably, a number of studies have indicated that NMDAR dysfunction is associated with depression syndrome [10]. Thus, the homozygosity of the GRIN1 rs4880213 T allele might increase glutamate NMDAR function and stabilize the mood status compared with other alleles. However, because the effect of the GRIN1 rs4880213 SNP on NMDAR function has not yet been analyzed at the molecular or metalevel, the mechanism by which the GRIN1 rs4880213 SNP affects NMDAR function needs to be investigated.
Our results also showed that the homozygosity of the C allele of the GRIN2B (rs7301328) is associated with increases in disruptive behavior; this finding is consistent with a report by Ohtsuki et al. [45], which showed that the G allele of the 366C/G (rs7301328) polymorphism is more common in patients than in population-based controls. The GRIN2B SNP (rs7301328) has been linked to bipolar disorder [24], schizophrenia, and other neuropsychiatric disorders; however, to the best of our knowledge, few studies have investigated the influence of the GRIN2B SNP (rs7301328) at the molecular level, including its effect on NMDAR function or on the level of GRIN1 or GRIN2 expression. In addition, the currently available studies do not conclusively show that unique GRIN2 subunits selectively mediate directions of plasticity, especially regarding long-term depression (LTD). In some studies, behavioral impairment can be related to a selective deficit in one direction of plasticity. Moreover, a GRIN2B-dependent LTD-like process has also often been implicated in mechanisms that support reversal learning [4, 46]. A rigorous testing of this hypothesis based on an analysis of more complete information concerning the effect of the GRIN2B SNP (rs7301328) would be interesting.
This study did not identify significant associations between the other polymorphisms and behavioral indices. Previous exploratory studies have indicated that GRIN1 (rs11146020) is a good candidate for susceptibility to schizophrenia [24, 25, 33], and other studies have suggested that the combined effects of GRIN1 and GRIN2B gene polymorphisms, including rs1805247 and rs1805502, might be involved in the etiology of schizophrenia. However, we did not identify such an association. Notably, the students in this study were physically and psychologically healthy, and identifying significant differences in emotional performance among different genotype groups is more difficult in a healthy population than in patients who are suffering from single-gene diseases or psychiatric disorders.
Here, we report an exploratory study of SNPs in the NMDAR GRIN1 and GRIN2 subunit genes in a healthy adolescent population. Specifically, we verified that the two SNPs (rs4880213 and rs7301328) influenced emotional performance in this adolescent population. Nevertheless, this study is subject to some limitations. First, most of the participants were relatively physically healthy. Nevertheless, the effect size for the association between the examined SNPs and behavior was narrow. However, because observing significant differences within a normal population is difficult, future studies may rely on the data presented herein to explore mechanisms relevant to this association, both at the clinical and molecular levels. The other limitation of the present study is differences in the socioeconomic levels of students which may have influenced the emotional behavior assessments. All volunteers in this study lived in metropolitan areas of Taiwan. Studies showed students socioeconomic levels were likely approximately equivalent in metropolitan areas of Taiwan [47]. Therefore, the impact of social economy on emotion in this study might be less significant. However, because the current study did not measure the effects of socioeconomic status on emotional behavior, we cannot conclusively rule out such an effect. Subsequent studies should rigorously examine this effect.
Educational researchers, teachers, and counselors might be interested in the implications of the effects of genetic polymorphisms identified herein. Specifically, students who do not perform as well as others academically due to poor cognitive abilities or emotional self-control show a decreased willingness to learn. Genotyping these students could provide educators with a strategic understanding of the potential innate emotional self-control of an individual student. Because human emotion is influenced by interactions between genetic variations and environmental factors, this knowledge could be used to provide an appropriate environment and monitor the emotions of a student during learning. These strategies could improve a student's interest in learning and achievement.
Supplementary Material
We investigated the association of 59 SNPs in the NMDAR genes (i.e., GRIN1, GRIN2A, GRIN2B, GRIN2C, and GRIN2D) with emotion and behavior in 832 Han Chinese subjects. Fifty-nine SNPs from the entire set of candidate genes associated with NMDARs were identified as shown supplementary material information in terms of their dbSNP IDs, genomic location, variation, and allele frequency.
Acknowledgments
The work in this study was supported by the National Science Council of Taiwan under Contracts NSC 102–2511-S-003-007-MY2/S-003-008-MY2, “Aim for the Top University Project NTNU,” and “International Research-Intensive Center of Excellence Program” (MOST 104–2911-I-003-301). The authors appreciate all the students and their families for giving their consent to participate in this project.
Conflict of Interests
None of the authors have any conflict of interests to report.
References
- 1.Merikangas K. R., Nakamura E. F., Kessler R. C. Epidemiology of mental disorders in children and adolescents. Dialogues in Clinical Neuroscience. 2009;11(1):7–20. doi: 10.31887/DCNS.2009.11.1/krmerikangas. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larson M. K., Walker E. F., Compton M. T. Early signs, diagnosis and therapeutics of the prodromal phase of schizophrenia and related psychotic disorders. Expert Review of Neurotherapeutics. 2010;10(8):1347–1359. doi: 10.1586/ern.10.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lencz T., Smith C. W., McLaughlin D., et al. Generalized and specific neurocognitive deficits in prodromal schizophrenia. Biological Psychiatry. 2006;59(9):863–871. doi: 10.1016/j.biopsych.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Mundo E., Tharmalingham S., Neves-Pereira M., et al. Evidence that the N-methyl-D-aspartate subunit 1 receptor gene (GRIN1) confers susceptibility to bipolar disorder. Molecular Psychiatry. 2003;8(2):241–245. doi: 10.1038/sj.mp.4001218. [DOI] [PubMed] [Google Scholar]
- 5.Weickert C. S., Fung S. J., Catts V. S., et al. Molecular evidence of N-methyl-D-aspartate receptor hypofunction in schizophrenia. Molecular Psychiatry. 2013;18(11):1185–1192. doi: 10.1038/mp.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coyle J. T. The glutamatergic dysfunction hypothesis for schizophrenia. Harvard Review of Psychiatry. 1996;3(5):241–253. doi: 10.3109/10673229609017192. [DOI] [PubMed] [Google Scholar]
- 7.Javitt D. C., Zukin S. R. Recent advances in the phencyclidine model of schizophrenia. The American Journal of Psychiatry. 1991;148(10):1301–1308. doi: 10.1176/ajp.148.10.1301. [DOI] [PubMed] [Google Scholar]
- 8.Kristiansen L. V., Huerta I., Beneyto M., Meador-Woodruff J. H. NMDA receptors and schizophrenia. Current Opinion in Pharmacology. 2007;7(1):48–55. doi: 10.1016/j.coph.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 9.Ibrahim L., Diazgranados N., Jolkovsky L., et al. A randomized, placebo-controlled, crossover pilot trial of the oral selective NR2B antagonist MK-0657 in patients with treatment-resistant major depressive disorder. Journal of Clinical Psychopharmacology. 2012;32(4):551–557. doi: 10.1097/jcp.0b013e31825d70d6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berman R. M., Cappiello A., Anand A., et al. Antidepressant effects of ketamine in depressed patients. Biological Psychiatry. 2000;47(4):351–354. doi: 10.1016/s0006-3223(99)00230-9. [DOI] [PubMed] [Google Scholar]
- 11.Mathew S. J., Murrough J. W., aan het Rot M., Collins K. A., Reich D. L., Charney D. S. Riluzole for relapse prevention following intravenous ketamine in treatment-resistant depression: a pilot randomized, placebo-controlled continuation trial. International Journal of Neuropsychopharmacology. 2010;13(1):71–82. doi: 10.1017/s1461145709000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cull-Candy S. G., Leszkiewicz D. N. Role of distinct NMDA receptor subtypes at central synapses. Science's STKE. 2004;2004(255):p. re16. doi: 10.1126/stke.2552004re16. [DOI] [PubMed] [Google Scholar]
- 13.Erreger K., Geballe M. T., Kristensen A., et al. Subunit-specific agonist activity at NR2A-, NR2B-, NR2C-, and NR2D-containing N-methyl-D-aspartate glutamate receptors. Molecular Pharmacology. 2007;72(4):907–920. doi: 10.1124/mol.107.037333. [DOI] [PubMed] [Google Scholar]
- 14.Gielen M., Retchless B. S., Mony L., Johnson J. W., Paoletti P. Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature. 2009;459(7247):703–707. doi: 10.1038/nature07993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li R., Huang F.-S., Abbas A.-K., Wigström H. Role of NMDA receptor subtypes in different forms of NMDA-dependent synaptic plasticity. BMC Neuroscience. 2007;8, article 55 doi: 10.1186/1471-2202-8-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monyer H., Sprengel R., Schoepfer R., et al. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science. 1992;256(5060):1217–1221. doi: 10.1126/science.256.5060.1217. [DOI] [PubMed] [Google Scholar]
- 17.Liu L., Wong T. P., Pozza M. F., et al. Role of NMDA receptor subtypes in governing the direction of hippocampal synaptic plasticity. Science. 2004;304(5673):1021–1024. doi: 10.1126/science.1096615. [DOI] [PubMed] [Google Scholar]
- 18.Lerche H., Shah M., Beck H., Noebels J., Johnston D., Vincent A. Ion channels in genetic and acquired forms of epilepsy. Journal of Physiology. 2013;591(4):753–764. doi: 10.1113/jphysiol.2012.240606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Villmann C., Becker C.-M. On the hypes and falls in neuroprotection: targeting the NMDA receptor. The Neuroscientist. 2007;13(6):594–615. doi: 10.1177/1073858406296259. [DOI] [PubMed] [Google Scholar]
- 20.Cull-Candy S., Brickley S., Farrant M. NMDA receptor subunits: diversity, development and disease. Current Opinion in Neurobiology. 2001;11(3):327–335. doi: 10.1016/s0959-4388(00)00215-4. [DOI] [PubMed] [Google Scholar]
- 21.Kristiansen L. V., Beneyto M., Haroutunian V., Meador-Woodruff J. H. Changes in NMDA receptor subunits and interacting PSD proteins in dorsolateral prefrontal and anterior cingulate cortex indicate abnormal regional expression in schizophrenia. Molecular Psychiatry. 2006;11(8):737–747. doi: 10.1038/sj.mp.4001844. [DOI] [PubMed] [Google Scholar]
- 22.Dorval K. M., Burcescu I., Adams J., et al. Association study of N-methyl-D-aspartate glutamate receptor subunit genes and childhood-onset mood disorders. Psychiatric Genetics. 2009;19(3):156–157. doi: 10.1097/YPG.0b013e32832a5097. [DOI] [PubMed] [Google Scholar]
- 23.Kaufmann C. A., Suarez B., Malaspina D., et al. NIMH genetics initiative millennium schizophrenia consortium: linkage analysis of African-American pedigrees. American Journal of Medical Genetics. 1998;81(4):282–289. doi: 10.1002/(sici)1096-8628(19980710)81:460;282::aid-ajmg262;3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 24.Zhao X., Li H., Shi Y., et al. Significant association between the genetic variations in the 5′ end of the N-methyl-D-aspartate receptor subunit gene GRIN1 and schizophrenia. Biological Psychiatry. 2006;59(8):747–753. doi: 10.1016/j.biopsych.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 25.Begni S., Moraschi S., Bignotti S., et al. Association between the G1001C polymorphism in the GRIN1 gene promoter region and schizophrenia. Biological Psychiatry. 2003;53(7):617–619. doi: 10.1016/s0006-3223(02)01783-3. [DOI] [PubMed] [Google Scholar]
- 26.Chanasong R., Thanoi S., Watiktinkorn P., Reynolds G. P., Nudmamud-Thanoi S. Genetic variation of GRIN1 confers vulnerability to methamphetamine-dependent psychosis in a Thai population. Neuroscience Letters. 2013;551:58–61. doi: 10.1016/j.neulet.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 27.Galehdari H., Pooryasin A., Foroughmand A., Daneshmand S., Saadat M. Association between the G1001C polymorphism in the GRIN1 gene promoter and schizophrenia in the Iranian population. Journal of Molecular Neuroscience. 2009;38(2):178–181. doi: 10.1007/s12031-008-9148-5. [DOI] [PubMed] [Google Scholar]
- 28.Rossi S., Studer V., Moscatelli A., et al. Opposite roles of NMDA receptors in relapsing and primary progressive multiple sclerosis. PLoS ONE. 2013;8(6) doi: 10.1371/journal.pone.0067357.e67357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu S.-L., Wang W.-F., Shyu H.-Y., et al. Association analysis of GRIN1 and GRIN2B polymorphisms and Parkinson's disease in a hospital-based case-control study. Neuroscience Letters. 2010;478(2):61–65. doi: 10.1016/j.neulet.2010.04.063. [DOI] [PubMed] [Google Scholar]
- 30.Schulz S., Arning L., Pinnow M., Epplen J. T., Beste C. N-methyl-D-aspartate receptor 2B subunit (GRIN2B) gene variation is associated with alerting, but not with orienting and conflicting in the Attention Network Test. Neuropharmacology. 2012;63(2):259–265. doi: 10.1016/j.neuropharm.2012.02.024. [DOI] [PubMed] [Google Scholar]
- 31.Alonso P., Gratacós M., Segalàs C., et al. Association between the NMDA glutamate receptor GRIN2B gene and obsessive-compulsive disorder. Journal of Psychiatry & Neuroscience. 2012;37(4):273–281. doi: 10.1503/jpn.110109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang C., Li Z., Wu Z., et al. A study of N-methyl-D-aspartate receptor gene (GRIN2B) variants as predictors of treatment-resistant major depression. Psychopharmacology. 2014;231(4):685–693. doi: 10.1007/s00213-013-3297-0. [DOI] [PubMed] [Google Scholar]
- 33.Qin S., Zhao X., Pan Y., et al. An association study of the N-methyl-D-aspartate receptor NR1 subunit gene (GRIN1) and NR2B subunit gene (GRIN2B) in schizophrenia with universal DNA microarray. European Journal of Human Genetics. 2005;13(7):807–814. doi: 10.1038/sj.ejhg.5201418. [DOI] [PubMed] [Google Scholar]
- 34.Lester K. J., Eley T. C. Therapygenetics: using genetic markers to predict response to psychological treatment for mood and anxiety disorders. Biology of Mood & Anxiety Disorders. 2013;3(1, article 4) doi: 10.1186/2045-5380-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clinton S. M., Haroutunian V., Meador-Woodruff J. H. Up-regulation of NMDA receptor subunit and post-synaptic density protein expression in the thalamus of elderly patients with schizophrenia. Journal of Neurochemistry. 2006;98(4):1114–1125. doi: 10.1111/j.1471-4159.2006.03954.x. [DOI] [PubMed] [Google Scholar]
- 36.Holden C. Sex and the suffering brain. Science. 2005;308(5728):1574–1577. doi: 10.1126/science.308.5728.1574. [DOI] [PubMed] [Google Scholar]
- 37.Kessler R. C. Epidemiology of women and depression. Journal of Affective Disorders. 2003;74(1):5–13. doi: 10.1016/s0165-0327(02)00426-3. [DOI] [PubMed] [Google Scholar]
- 38.Steer R. A., Kumar G., Beck J. S., Beck A. T. Evidence for the construct validities of the beck youth inventories with child psychiatric outpatients. Psychological Reports. 2001;89(3):559–565. doi: 10.2466/pr0.2001.89.3.559. [DOI] [PubMed] [Google Scholar]
- 39.Edgington E. S. Randomization Tests. 3rd. New York, NY, USA: Marcel Dekker; 1995. [Google Scholar]
- 40.Paoletti P., Bellone C., Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nature Reviews Neuroscience. 2013;14(6):383–400. doi: 10.1038/nrn3504. [DOI] [PubMed] [Google Scholar]
- 41.Traynelis S. F., Wollmuth L. P., McBain C. J., et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacological Reviews. 2010;62(3):405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monyer H., Burnashev N., Laurie D. J., Sakmann B., Seeburg P. H. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 1994;12(3):529–540. doi: 10.1016/0896-6273(94)90210-0. [DOI] [PubMed] [Google Scholar]
- 43.Georgi A., Jamra R. A., Klein K., et al. Possible association between genetic variants at the GRIN1 gene and schizophrenia with lifetime history of depressive symptoms in a German sample. Psychiatric Genetics. 2007;17(5):308–310. doi: 10.1097/ypg.0b013e3280c1e5fb. [DOI] [PubMed] [Google Scholar]
- 44.Francesco M., Michele R., Hajime K., et al. Genetic variants of the NMDA receptor influence cortical excitability and plasticity in humans. Journal of Neurophysiology. 2011;106(4):1637–1643. doi: 10.1152/jn.00318.2011. [DOI] [PubMed] [Google Scholar]
- 45.Ohtsuki T., Sakurai K., Dou H., Toru M., Yamakawa-Kobayashi K., Arinami T. Mutation analysis of the NMDAR2B (GRIN2B) gene in schizophrenia. Molecular Psychiatry. 2001;6(2):211–216. doi: 10.1038/sj.mp.4000808. [DOI] [PubMed] [Google Scholar]
- 46.Dorval K. M., Wigg K. G., Crosbie J., et al. Association of the glutamate receptor subunit gene GRIN2B with attention-deficit/hyperactivity disorder. Genes, Brain and Behavior. 2007;6(5):444–452. doi: 10.1111/j.1601-183x.2006.00273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Y.-C., Liu T.-M. Educational attainment and urban/rural discrepancy: an analysis of spatial cluster. Journal of Population Studies. 2008;37:1–43. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
We investigated the association of 59 SNPs in the NMDAR genes (i.e., GRIN1, GRIN2A, GRIN2B, GRIN2C, and GRIN2D) with emotion and behavior in 832 Han Chinese subjects. Fifty-nine SNPs from the entire set of candidate genes associated with NMDARs were identified as shown supplementary material information in terms of their dbSNP IDs, genomic location, variation, and allele frequency.


