Summary
Embryonic neocortical development depends upon balanced production of progenitors and neurons. Genetic mutations disrupting progenitor mitosis frequently impair neurogenesis, however the link between altered mitosis and cell fate remains poorly understood. Here we demonstrate that prolonged mitosis of radial glial progenitors directly alters neuronal fate specification and progeny viability. Live imaging of progenitors from a neurogenesis mutant, Magoh+/−, reveals mitotic delay significantly correlates with preferential production of neurons instead of progenitors, as well as apoptotic progeny. Independently, two pharmacological approaches reveal a causal relationship between mitotic delay and progeny fate. As mitotic duration increases, progenitors produce substantially more apoptotic or neurogenic progeny. We show apoptosis, but not differentiation, is p53-dependent, demonstrating these are distinct outcomes of mitotic delay. Together our findings reveal prolonged mitosis is sufficient to alter fates of radial glia progeny and define a new paradigm to understand how mitosis perturbations underlie brain size disorders such as microcephaly.
Graphical abstract
Introduction
During neurogenesis of the developing dorsal telencephalon, radial glial progenitors (RGCs) divide adjacent to the ventricle to generate new neurons and progenitors (Figure 1A) (Malatesta et al., 2000; Noctor et al., 2001). RGCs undergo symmetric proliferative divisions to self-renew, and asymmetric neurogenic divisions to generate a new RGC and either a neuron or intermediate progenitor (IP) (Franco and Müller, 2013). IPs undergo 1–2 self-renewal divisions before producing neurons (Kowalczyk et al., 2009). The precise balance of RGC divisions and resulting progeny ultimately influence the size and function of the mature neocortex. This is exemplified in human and mouse microcephaly models in which reduced brain size is associated with altered neurogenesis including common phenotypes of depleted progenitors, precocious neurons, and apoptosis (Asami et al., 2011; Gruber et al., 2011; Lizarraga et al., 2010; Marthiens et al., 2013; McIntyre et al., 2012; Silver et al., 2010; Xie et al., 2013; Yingling et al., 2008). As most human microcephaly-associated genes identified to date encode mitotic regulators, aberrant progenitor division has been invoked in the etiology of this disease (Hu et al., 2014). Yet how altered progenitor mitosis impacts cell fate changes associated with microcephaly remains unclear.
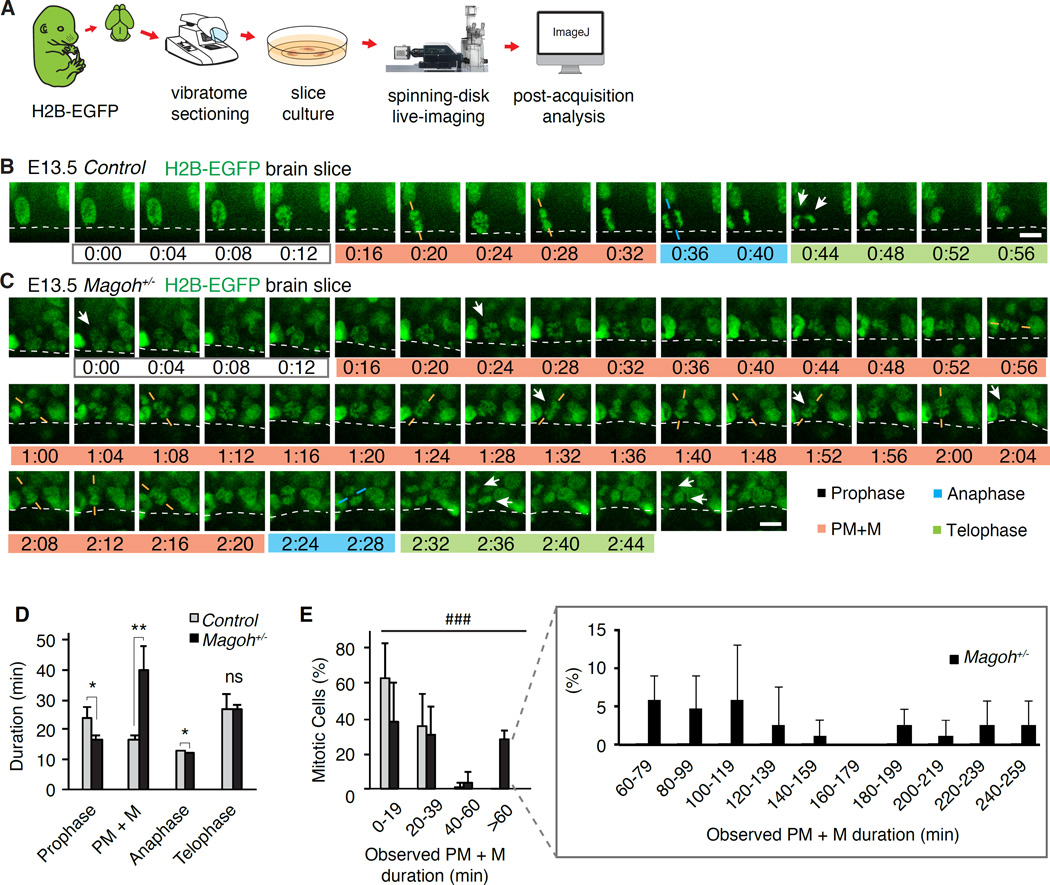
Figure 1. Cell cycle analysis of Magoh+/− radial glia progenitors shows a significant delay in mitosis.
(A) Cartoon depicting the potential fate of RGCs during the neurogenic period of corticogenesis. In this study RGCs were recognized as apically dividing cells, and/or Pax6+Tbr2−. (B) The question addressed by this article. (C–F) E13.5 sections from control (C,D) and Magoh+/− (E,F) cortices stained for EdU (red) and BrdU (green) (C, E), Tbr2 (magenta) and Pax6 (cyan) (D,F). (G) Duration of total cell cycle (Tc) and S phase (Ts) for indicated genotypes, calculated using cumulative BrdU/EdU labeling for RGCs (Pax6+Tbr2-). (H,I) E13.5 sections from control (H) and Magoh+/− (I) cortices stained for Ki67 (green) and Hoechst (white). Boxed insets: higher magnification views of mitotic RGCs (denoted by DNA condensation, arrowheads). (J) Proportion of Ki67+ control (grey) and Magoh+/− (black) mitotic RGCs. (K–N) E13.5 sections from control (K,L) and Magoh+/− (M,N) cortices stained for γ-tubulin (red) and Hoechst (green) (Dotted circle, mitotic RGC). (O) Proportion of RGCs in each mitosis phase for indicated genotypes. ns, not significant, * and #, P<0.05, *, t-test and #, Chi-square analysis. Scale bars: C-F: 50µm, H, I: 25µm, K-N: 10µm. Error bars, S.D.
Several aspects of progenitor mitosis have been implicated in neocortical fate specification. Spindle orientation is strongly linked to neuron production and survival, and generation of basally dividing cells (Asami et al., 2011; Konno et al., 2008; LaMonica et al., 2013; Lizarraga et al., 2010; Mora-Bermúdez et al., 2014; Xie et al., 2013; Yingling et al., 2008). Spindle size asymmetry is associated with asymmetric divisions (Delaunay et al., 2014). Mitotic progression defects can trigger mitotic catastrophe, in which progenitors die prior to division (Chen et al., 2014; Novorol et al., 2013) or massive apoptosis due to altered centrosome number (Insolera et al., 2014; Marthiens et al., 2013). However, the specific impact of prolonged mitosis on fate specification is poorly understood. An important unanswered question is whether there is a direct causal relationship between prolonged progenitor mitosis and pathogenic production of neurons and apoptotic progeny in the developing brain (Figure 1B).
Here we address this critical issue by demonstrating that prolonged M phase of RGCs directly alters progenitor and neuronal cell fates in the developing brain. First we employ live imaging of Magoh+/− brain slices and primary progenitors. This mutant exhibits microcephaly and neurogenesis phenotypes, which are attributable in part to altered levels of the microtubule protein Lis1 (Silver et al., 2010). Compared to non-delayed Magoh+/− RGCs, we find mitotically delayed Magoh+/− RGCs produce significantly fewer RGCs and more apoptotic progeny. Independently, we employ a reversible pharmacological paradigm using two distinct drugs to prolong mitosis both ex vivo and in vitro. We show prolonged RGC mitosis alone recapitulates altered differentiation and apoptotic fates, with only the latter occurring via a p53-dependent mechanism. Our results indicate prolonged mitosis of RGCs causes preferential generation of progeny with two mutually independent fates, differentiation and apoptosis.
Results
In vivo cell cycle analysis reveals Magoh-deficient radial glia exhibit a significant mitotic delay
We previously discovered that mice haploinsufficient for Magoh exhibit microcephaly and severe neurogenesis defects, including depleted IPs, ectopic neurons, and apoptosis (McMahon et al., 2014; Silver et al., 2010). Although Magoh is required for proper mitosis of cultured cells and melanoblasts (Silver et al., 2010; 2013), it remains unclear if it regulates mitosis duration of neural progenitors and if this is relevant for altered cell fate during corticogenesis. Moreover G1 and S phase durations are linked to RGC proliferative behavior (Arai et al., 2011; Lange et al., 2009; Pilaz et al., 2009), but the role of M phase in RGC function is poorly understood. We assessed population level cell-cycle using a cumulative BrdU/EdU labeling paradigm at E13.5 (Quinn et al., 2007), when neurogenesis is markedly disrupted in Magoh+/− (Silver et al., 2010) (Figure 1C–G). Relative mitotic index was calculated as the fraction of mitotic cells amongst Ki67+ cycling RGCs (Figure 1H–J). Although total cell-cycle (Tc) and S-Phase (Ts) were similar between control and Magoh+/− RGCs (Figure 1G), mitotic index was increased in Magoh+/− RGCs (Figure 1J, P<0.05). These results suggest that within the normal Tc-Ts fraction, RGC mitosis is lengthened. Using γ-Tubulin and Hoechst staining to mark centrosomes, we quantified a significantly higher fraction of Magoh+/− RGCs in prometaphase (PM) and metaphase (M) (Figures 1K–O). These findings were corroborated by fixed and live analyses of Magoh siRNA-treated HeLa cells, which displayed increased mitotic duration (Figure S1A–C, Movies S5). Overall these data indicate Magoh+/− RGCs exhibit significant mitotic delay.
In Magoh+/− brains, mitotic delay is concomitant with increased neuronal apoptosis (Silver et al., 2010). To examine if apoptosis is due to a requirement of Magoh in postmitotic neurons we used a conditional MagohLox/Lox allele. Magoh depletion from neural progenitors using Emx1-Cre causes severe microcephaly and apoptosis (McMahon et al., 2014)(Figure S1D, E). In contrast, Magoh loss from post-mitotic neurons using CamKIlα-Cre caused no discernable apoptosis or altered neuron number, suggesting Magoh is not required for post-mitotic neuron viability (Figure S1F–H). These experiments indicate Magoh deficiency in progenitors is a likely cause of apoptosis. Altogether these data prompted us to further examine M phase to assess its relevance for the Magoh+/− phenotype.
Live imaging reveals Magoh-deficient radial glia exhibit prolonged prometaphase
We evaluated mitosis progression live using time-lapse imaging of dividing RGCs in organotypic brain slices prepared from E13.5 Tg-H2B-EGFP (control) and Tg-H2B-EGFP;Magoh+/− embryos (Figure 2A–C, Movies S1,S2) (Pilaz and Silver, 2014). Control RGCs completed mitosis on average in 79.9 minutes (Figure S2A), similar to previous measurements of mitosis duration (Haydar et al., 2003). In contrast Magoh+/− RGCs were markedly delayed in mitosis, averaging 94.7 minutes. RGC mitotic delay was also evident in Syto11 -stained Magoh+/− brains (Figure S2B, Movie S3). Live imaging of Magoh+/− brains electroporated with mCherry-α-Tubulin demonstrated RGCs contained mitotic spindles, confirming they were mitotic (Figure S2C, D, Movie S4). Hence both fixed and live analyses show mitosis is prolonged in Magoh+/− RGCs.
Figure 2. Live imaging of embryonic brain slices reveals Magoh+/− radial glia progenitors exhibiting prolonged mitosis.
(A) Protocol utilized to visualize mitosis progression in live brain slices. (B, C) Time-lapse images of dividing progenitors in E13.5 H2B-EGFP control (B) and Magoh+/− (C) brain slices. (Dotted lines, ventricular border; arrows, progenitor and progeny; orange lines, metaphase plate orientation; blue lines, cleavage plane orientation). time=h:min, and mitotic stages are indicated beneath each panel. Time-lapse panels correspond to Movies S1 and S2. (D) Duration of mitosis stages and (E) distribution of prometaphase (PM)+metaphase (M) duration of E13.5 progenitors in indicated genotypes. *, P<0.05, **, P<0.01, ###, P<0.001. *, t-test and #, Chi-square analysis. Scale bars: B, C: 10µm. Error bars, S.D.
Live imaging uncovered a significant delay in prometaphase/metaphase (PM+M). On average PM+M was 2.4-fold longer for Magoh+/− RGCs than control (compare 39.7 to 16.8 minutes) (Figure 2D). Magoh+/− RGCs exhibited a broad range of PM+M durations, with 66% dividing normally (<60 minutes) and 34% significantly delayed (≥60 minutes, P<0.001) (Figure 2E). Amongst those Magoh+/− delayed progenitors, 62% eventually divided (Figure S2E). During the 5-hr imaging session we never observed mitotic catastrophe, suggesting most delayed Magoh+/− RGCs eventually divide. Together these data indicate a significant fraction of Magoh+/− RGCs progress slower through PM+M.
We sought to further define the cellular mechanism by which Magoh levels affect mitotic progression. Defective centrosome number and maturation are associated with microcephaly and mitosis defects (Gruber et al., 2011; Insolera et al., 2014; Marthiens et al., 2013). However Magoh deficient RGCs showed normal centrosome distance and number as demonstrated by γ-Tubulin staining (Figure 1K–N, Figure S3A). Magoh depleted HeLa cells also showed normal centriole number and centrosome maturation as assessed with ODF2 staining (Figure S3K–Q). Analysis of Ndc80/Hec1, Mad1, and CENP-A staining in Magoh siRNA-treated HeLa cells indicates intact kinetochore function and microtubule attachments, respectively (Figure S3R–QQ). Together these data reveal Magoh does not impact centrosome maturation or duplication, or chromosome attachments.
Both Magoh+/− RGCs and Magoh depleted HeLa cells showed evidence of altered mitotic microtubules. Staining for EB1, a microtubule plus-tip binding protein, revealed aberrant spindle microtubules in Magoh knockdown cells (Figure S3RR, SS). Acetylated Tubulin staining of Magoh+/− RGCs also revealed altered mitotic spindle morphology (Figure S3F–I). Magoh+/− brains showed reduced protein levels of Eg5 (Kinesin5/Kif11)(Skoufias et al., 2006) and Numa(Gaglio et al., 1995), both required for mitotic microtubules (Figure S3B–J). Of note, mutations in KIF11 cause human microcephaly (Ostergaard et al., 2012). Together with the previous finding that Magoh controls Lis1 protein levels (Silver et al., 2010), these analyses indicate altered microtubule regulation, rather than centrosome dysfunction, is associated with Magoh+/− mitotic defects (Figure S3TT).
We next assessed if prometaphase delay was associated with aneuploidy. Control and Magoh+/− E13.5 RGCs showed no evidence of aneuploidy, as evaluated by DNA FISH (Figure S4A–D). This result was not due to death of aneuploid cells, as evidenced by normal ploidy in a p53−/− background, which prevents apoptosis (Figure S4E). Thus, in contrast to some microcephaly models (Marthiens et al., 2013), aneuploidy is not associated with Magoh+/− delayed RGCs.
As spindle orientation is strongly implicated in neurogenesis and microcephaly phenotypes (Asami et al., 2011; Xie et al., 2013; Yingling et al., 2008), it is possible mitotic delay causes spindle mis-orientation, or alternatively these could be independent phenotypes. To distinguish between these scenarios, we analyzed spindle orientation and rotation in our 3D time-lapse imaging of E13.5 mitotic RGCs (Figure 2B, C, Movies S1–S4). Magoh+/− PM+M spindles underwent 1.6-fold more rotation than control (Figure S4G–J)(Haydar et al., 2003). In control anaphase progenitors, most cleavage planes were oriented between 60–90°, as previously reported (Konno et al., 2008; Xie et al., 2013) (Figure S4K, L). In comparison, Magoh+/− anaphase progenitors exhibited a significantly broader range of cleavage plane orientations spanning from 0–90° (Figure S4L, M). This corroborates previous analyses of fixed Magoh+/− E11.5 and E12.5 brains (Silver et al., 2010). Importantly, we observed no significant correlation between prometaphase duration and altered spindle rotation or anaphase cleavage plane orientation (Figure S4J, M). Thus in E13.5 Magoh+/− brains, prolonged mitosis and altered spindle orientation can occur in independent progenitors.
Live imaging clonal analysis reveals Magoh+/− radial glia exhibit mitotic delay, generate apoptotic progeny and undergo increased neurogenic divisions
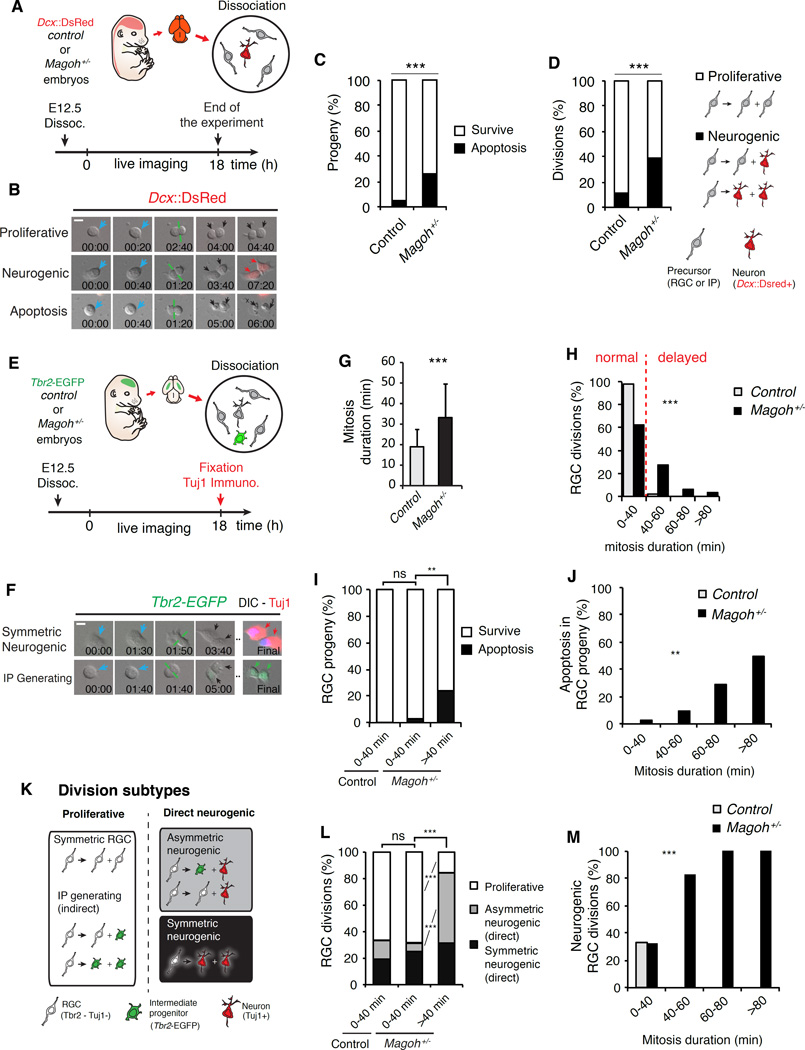
Altogether these data prompted us to examine if Magoh+/− progenitors exhibiting prolonged mitosis produce cells with altered fates. We employed live imaging of dividing progenitors coupled with clonal analysis of direct progeny (Shen et al., 2002). Dissociated progenitors were generated from control and Magoh+/− brains expressing Dcx::DsRed as a live reporter for new neurons (X. Wang et al., 2007). Analysis was at E12.5, when Magoh+/− brains contain ectopic neurons and apoptosis (evidenced by cleaved caspase 3 (CC3) staining) (Silver et al., 2010) (Figures 3A, B). As observed in vivo, Magoh+/− progenitors underwent 1.5-fold longer average mitoses than control (P<0.05, data not shown). Compared to control, this same cohort of Magoh+/− progenitors generated significantly more apoptotic progeny (at least 1 apoptotic daughter cell) (Figure 3C, Movie S6). In vivo, Magoh+/− apoptosis is accompanied by altered numbers of neurons and progenitors, prompting us to assess the fates of viable divisions. We defined proliferative divisions as production of two progenitors (RGCs and/or IPs, both DsRed-) and neurogenic divisions as generation of at least 1 neuron (DsRed+). In comparison to control, Magoh+/− E12.5 progenitors underwent significantly more neurogenic divisions (Figure 3D). Apoptosis and Dcx::DsRed expression were evident concurrently around 6 hours after division making it difficult to disentangle these two fates. Altogether, these findings indicate Magoh+/− progenitors exhibit prolonged mitosis, as well as altered proliferative, neurogenic, and apoptotic divisions.
Figure 3. Mitotically delayed Magoh+/ progenitors produce apoptotic progeny, more neurons and fewer progenitors.
(A) Overview of experiment for live imaging primary progenitors and visualizing cell fate using Dcx::DsRed (neurons, red). (B) Representative time-lapse images of dividing E12.5 progenitors as shown in Movie S6. (C) Proportion of surviving (white) and apoptotic (black) progeny from control and Magoh+/− progenitors. (D) Proportion of control and Magoh+/− progenitors undergoing proliferative (white) and neurogenic (black) divisions as indicated. (E) Overview of experiment for live imaging RGCs and visualizing cell fate using Tbr2-EGFP (IPs, green) and Tuj1 staining (neurons, red). (F) Representative time-lapse images of dividing E12.5 Tbr2-negative RGCs as shown in Movie S6. (G) Average mitosis duration for control (white) and Magoh+/− (black) RGCs. (H) Distribution of mitosis durations of control (white) and Magoh+/− (black) RGCs. (I) Proportion of surviving (white) and apoptotic (black) progeny generated from normal and delayed RGCs divisions of indicated genotypes. (J) Apoptotic progeny relative to increasing mitosis duration for indicated genotypes. (K) Representation of different RGC division types. (L) Proportion of normal and delayed control and Magoh+/− RGC divisions generating proliferative and neurogenic progeny. (M) Neurogenic divisions relative to increasing mitosis duration for indicated genotypes. ns, not significant, **, P<0.01, ***P<0.001, Scale bars, B, F: 10 µm, Error bars, S.D.
We refined our live imaging analysis to specifically examine RGCs. Mutant mice were bred onto a Tbr2-EGFP background (Arnold et al., 2009). At E12.5, RGCs were identified as EGFP- (93% of dividing cells) and IPs were identified as EGFP+. We performed live imaging clonal analysis as before with two exceptions: only RGC divisions were analyzed, and progeny were assessed by immunostaining for neurons (Tuj1+), RGCs (Pax6+) and Tbr2-EGFP for IPs (Figures 3E, F). Consistent with all prior experiments described above, Magoh haploinsufficient RGCs displayed 1.8 fold longer mitoses relative to control (Figure 3G). All Magoh+/− RGCs produced significantly more apoptotic progeny than control RGCs, generating either 1 or 2 apoptotic progeny to equivalent degrees (Figure S5A, B). Additionally, all Magoh+/− RGCs underwent more neurogenic divisions than control (Figure S5C). Thus, two independent reporters (Dcx::DsRed and Tuj1 staining) reveal Magoh+/− progenitors/RGCs exhibit increased neuron-generating divisions.
Generation of aberrant progeny in mitotically delayed but not normally dividing Magoh+/− radial glia
These findings established that total progenitor and RGC populations exhibit mitotic delay concomitant with altered progeny cell fates. We hypothesized that amongst Magoh+/− RGCs, those delayed in mitosis preferentially generate neurons and apoptotic progeny. To assess this, we examined progeny derived from either delayed or non-delayed RGCs. Virtually all control RGCs (98%) completed mitosis within 40 minutes (Figures 3H, Figure S5D). Therefore, normal and delayed mitosis was classified as <40 or >40 minutes, respectively. In contrast, only 62% of Magoh+/− RGCs completed mitosis normally, and 38% were delayed (Figure 3H). This distribution of mitotic delay is remarkably similar to that observed in Magoh+/− slices (compare Figures 2E and 3H).
We then assessed production of apoptotic progeny in delayed versus non-delayed Magoh+/− RGCs. Both control and non-delayed Magoh+/− RGCs produced primarily viable progeny, with no significant difference between genotypes (Figure 3I). In sharp contrast, delayed Magoh+/− RGCs produced significantly more apoptotic progeny than non-delayed mutant RGCs (Figure 3I, P<0.01). Strikingly, as mitotic duration increased up to 80 minutes or greater, there was a significant correlative increase in production of apoptotic cells (Figure 3J, P<0.001). Consistent with this, apoptotic divisions were on average significantly longer than divisions generating viable progeny (Figure S5E). This suggests two important findings: that non-delayed Magoh+/− RGCs do not contribute significantly to apoptosis in Magoh+/− E12.5 brain, and that the probability of Magoh+/− RGCs generating non-viable progeny increases proportionally with mitotic duration.
We used the same strategy to assess neuron and progenitor production from mitotically delayed Magoh+/− RGCs. Relative to control and non-delayed Magoh+/− RGCs, mitotically delayed Magoh+/− RGCs exhibited striking and significant increases in neuron generation and proportionate decreases in RGC production (Figure 3K, P<0.001). Increased neurons were a result of more asymmetric neurogenic Magoh+/− divisions, and not increased symmetric neurogenic divisions or indirect (IP producing) divisions (Figures 3L, Figure S5G–I). Importantly, as seen for apoptosis, as Magoh+/− RGC mitosis duration lengthened, the probability of producing neurons significantly increased (Figures 3M, Figure S5F).
We also assessed the potential contribution of non-delayed Magoh+/− RGCs for neuron production. Distinguishing proliferative and neurogenic divisions into distinct subtypes, including RGC/IP generating divisions, revealed no significant differences between non-delayed control and Magoh+/− RGCs (Figures 3L, Figure S5G–I). Together with the prior data, this indicates non-delayed RGCs show no increased propensity for differentiation or apoptosis. As Magoh is a RNA binding protein, it is possible that delayed Magoh+/− RGCs carry other pro-neurogenic and pro-apoptotic influences or potentially accumulated damage. Regardless, live imaging analyses clearly demonstrate that Magoh+/− RGCs show a strong and highly significant correlation between mitotic delay and altered cell fate. Therefore, we sought to independently extend our findings to challenge the idea of a causal relationship between mitotic duration and cell fate of progeny.
Pharmacologically induced mitotic delay causes generation of fewer progenitors, more neurons, and apoptotic cells
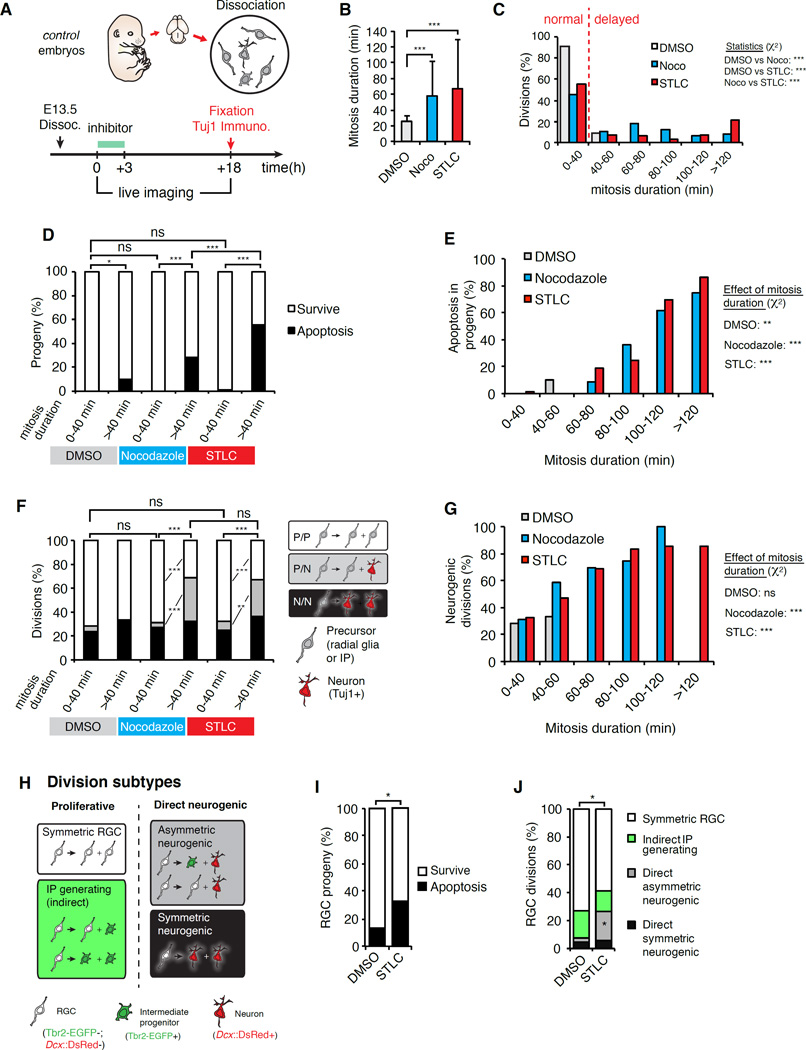
To test the hypothesis that prolonged mitosis alone can directly impact progeny fate, we established an experimental approach to reversibly delay control progenitors in prometaphase. We used two traditional mitotic inhibitors, S-trityl-L-cysteine (STLC), which inhibits Eg5-mediated centrosome separation (Skoufias et al., 2006) and nocodazole, which depolymerizes microtubules (Bazzi and Anderson, 2014; Uetake and Sluder, 2010). The use of pharmacology in brain slices is well established for studying neural progenitor mitosis (Mora-Bermúdez et al., 2014; Ostrem et al., 2014; Tsai et al., 2007). E13.5 brain slices were cultured in media containing either STLC, nocodazole or DMSO (control) for 3 hours, and subsequently cultured in drug-free media for up to 14 hours (Figure 4A–G). Compared to control, both STLC and nocodazole treatment significantly increased the number of mitotic cells (Figures 4A–F, H). Upon inhibitor washout, prometaphase-arrested progenitors proceeded to anaphase within 2–3 hours in STLC treated slices, and within 1 hour in nocodazole treated slices (Figures 4A–F, H). Neither drug significantly impacted anaphase cleavage plane orientation (Figures 4I, J). Moreover there was no increased aneuploidy in STLC treated brain slices (Figures 4K–N). Hence both pharmacological treatments reversibly prolong prometaphase of neural progenitors without perturbing ploidy or cleavage plane orientation.We next coupled EdU pulse-chase with the pharmacological paradigm to assess progeny fate at a population level. We pulsed slices with EdU for 30 minutes to indelibly label a cohort of S phase progenitors, and subsequently applied either DMSO or a mitotic inhibitor for 3 additional hours (Figures 5A–C, Figure S6A, B). Prior to drug treatment, the majority of EdU+ cells were Pax6+ RGCs, and 16% were Tbr2+ IPs (Figure S6C). The 1-hour delay between EdU pulse and drug-treatment corresponds to the time required for EdU+ progenitors to enter mitosis (Takahashi et al., 1995). Over the 3-hour drug treatment, EdU+ progenitors continued to enter mitosis, but were stalled at prometaphase (Figure 5C). Both drug treatments led to significantly more mitotically delayed progenitors within the EdU+ population compared to control (Figure 5B). At 6-hours post-washout, the localization of EdU+ cells in STLC treated slices was similar to control-treated slices 3 hours earlier, consistent with mitotic delay (Figure S6B). By 14-hours post-washout, EdU+ cells were located in the basal VZ and SVZ/CP layers (Figures 5D–O, Figure S6B). Given cell cycle and migration kinetics this population corresponds to progeny of RGCs and IPs (Takahashi et al., 1995). Based on published cell cycle measurements of progenitors (Arai et al., 2011; Takahashi et al., 1995) we can infer that by 14-hours post-washout, new EdU+ progeny do not have time to complete a second round of mitosis. Thus a significant proportion of EdU+ cells are direct progeny of mitotically delayed progenitors.
Figure 4. Pharmacological inhibitors reversibly prolong progenitor prometaphase.
(A–F) Hoechst-stained sections from slices treated with DMSO, STLC or nocodazole (for 3 hours) fixed at +0 hours (A–C), or +3 hour washout (D–F). At 0 hours STLC and nocodazole treated progenitors accumulate in prometaphase (brown arrows) and by 3 hours anaphase cells are evident (blue arrowheads). (G) Timeline of slice culture experiment. (H) Quantification of mitoses in different conditions. Stars indicate significant differences between DMSO and STLC (red) and DMSO and nocodazole (blue) conditions at each time point. Absence of a star indicates ns. (I,J) Average spindle orientation (I) and distribution of angles (J) in DMSO treated (grey), STLC treated (black), and nocodazole treated (blue) brain slices after 3-hour (STLC) and 1-hour washout (nocodazole). (K–N) DNA FISH analysis of STLC treated brain slice (K) with higher magnification images (L,M). (N) Graph depicting lack of significant aneuploidy in STLC treated slices. ns, not significant, *P<0.05, **, P<0.01, ***, P<0.001. Scale bars: A-F: 10µm, K: 20µm, L, M: 5µm. Error bars, S.D.
Figure 5. Prometaphase-delay of progenitors causes ectopic neurons, reduced progenitors, and apoptosis in embryonic brain slices.
(A) Timeline of the slice culture experiment. (B) Quantification of the fraction of EdU+ mitotic cells in different conditions. Stars indicate significant differences between DMSO and STLC (red) and DMSO and nocodazole (blue) conditions, at each time point. (C) Paradigm for EdU pulse-chase of E13.5 brain slices. (D,J) Sections from E13.5 brain slices 14 hours after washout, using paradigm outlined in (A,C), and stained for EdU (red), CC3 (blue) and Tuj1 (green). (E-H, K-N) High magnification images of (D, J) stained for EdU (red), CC3 (white or blue), and Tuj1 (green). (I, O) High magnification images of (H, N) depicting more EdU+Tuj1+ cells (yellow) in the STLC treated brain slices versus control. (P, Q, W, X) Percentage of EdU+ cells in DMSO (grey), nocodazole (blue) and STLC (red) treated slices that are CC3+ (P), Tuj1+ (Q), Tbr2+ (W), Pax6+ (X) after 14 hour washout. (R) Percentage of CC3+EdU+ cells in each condition that are Tuj1+ neurons (black) or TuJ1- (white). (S–V) Section from E13.5 brain slices 14 hours after washout, stained for EdU (red), Pax6 (blue) and Tbr2 (green). ns, not significant, *, P<0.05, **, P<0.01, ***, P<0.001. Scale bars, D, J: 500 µm, E–I, K–O: 20 µm, S–V: 10µm. Error bars, S.D.
We next assessed apoptosis of EdU+ cells 14 hours post-washout following progenitor mitotic delay. STLC and nocodazole treated slices had 21-fold and 8-fold higher respective fractions of apoptotic (CC3+) EdU+ cells compared to control (Figure 5D–P). Apoptotic cells were not detectable until 6 hours post-washout, well after the drug was absent from the media and after progenitors had exited mitosis, arguing against global cytotoxicity (Figure S6G). Consistent with the specificity of apoptosis, the majority of CC3+ cells were EdU+ in both STLC and nocodazole treated slices (91% and 85%, respectively) (Figure S6E). Altogether these pulse-chase experiments demonstrate that prolonged prometaphase is associated with production of more apoptotic cells, primarily in progeny of mitotically delayed progenitors.
We next assessed neuronal and precursor fates following mitotic delay. We quantified the proportion of EdU-positive cells at 14-hours post-washout as neurons, RGCs, or IPs using TuJ1, Pax6, and Tbr2 staining, respectively (Figures 5D–O, Q, 5S–X, S6D, F). Strikingly, compared to control, prometaphase delay resulted in 1.5-fold more neurons, evident with either STLC or nocodazole treatment (Figure 5Q). Concomitant with the increase of neurons, we observed a significant 25% reduction in IPs (EdU+Tbr2+) with either STLC or nocodazole (Figure 5W). RGCs (EdU+Pax6+Tbr2+) were significantly reduced in STLC, but not in nocodazole (Figure 5X). Compared to STLC, nocodazole induces shorter mitotic arrest, which could explain the reduced impact on RGCs. It is possible these population changes could be due to preferential progenitor apoptosis. However, apoptosis was evident in equivalent fractions of both EdU+ neurons and EdU+ progenitors, suggesting it impacts both populations to a similar extent (Figure 5R). In sum these data show prometaphase delay produces imbalanced numbers of neurons and progenitors.
Prometaphase-delayed progenitors generate significantly more neurons, fewer progenitors and apoptotic progeny
With the observation that mitotic delay induces population level changes, we next used live imaging to directly follow individual progenitor divisions and fate of their progeny. A similar paradigm was used as described in Figure 3A, except E13.5 progenitors were treated with DMSO, STLC, or nocodazole for the first 3 hours of imaging (Figure 6A). Importantly, with this paradigm we captured co-cultured progenitors with different mitosis durations, depending upon their cell cycle state at the beginning of the drug treatment. We noted no significant apoptosis of interphase progenitors treated with either STLC or nocodazole (Figure S7A). This suggests neither drug impacts overall cell viability, as seen in slice culture experiments. On average, control E13.5 progenitors remained in mitosis about 22 minutes (Figures 6B). In contrast, mitoses of nocodazole and STLC treated progenitors were on average 54 and 63 minutes, respectively (Figure 6B). 91% of control progenitors completed mitosis within 40 minutes, whereas only 45% of nocodazole and 52% of STLC treated progenitors exhibited normal mitosis duration (Figure 6C, Figure S7B). These data indicate nocodazole and STLC treatments can be used to prolong mitosis and evaluate its direct impact on progeny cell fate.
Figure 6. Prolonged mitosis of progenitors directly causes production of neurons and progenitors and apoptotic progeny.
(A) Overview of experiment for live imaging progenitors and visualizing cell fate as exemplified in Movies S6 and S7. (B) Average mitosis duration for DMSO (grey), nocodazole (noco, blue) and STLC (red) treated progenitors. (C) Distribution of mitosis durations of DMSO (grey), nocodazole (blue) and STLC (red) progenitors. (D) Proportion of surviving (white) and apoptotic (black) progeny generated from normal and delayed RGCs divisions for indicated treatments. (E) Apoptotic progeny observed relative to increasing mitosis duration for indicated treatments. (F) Proportion of normal and delayed progenitors undergoing P/P (white), P/N (grey), or N/N (black) divisions for indicated treatments. (G) Neurogenic divisions relative to increasing mitosis duration for indicated treatments. (H) Different subtypes of RGC divisions. (I) Proportion of surviving (white) and apoptotic (black) progeny from DMSO and STLC treated RGCs. (J) Proportion of different divisions from DMSO and STLC treated RGCs. ns, not significant, *, P<0.05, ***, P<0.001. Error bars, S.D.
With this paradigm we examined apoptosis in progeny derived from delayed and non-delayed progenitors. Strikingly, we discovered causal relationships between mitotic delay and subsequent apoptosis for all three conditions: control, STLC and nocodazole treated progenitors. The small fraction of control E13.5 progenitors displaying prolonged mitosis generated significantly more apoptotic progeny, an outcome also manifest by mitotically delayed E14.5 progenitors (Figures 6D, Figure S7H, I). Analysis of the drug treated progenitors showed a dramatic trend. Delayed progenitors treated with either inhibitor produced significantly more apoptotic progeny than non-delayed counterparts. Approximately 70–80% of apoptotic divisions gave rise to 2 dying cells (Figures 6D, Figure S7C). Most strikingly, as mitosis duration lengthened in drug treated progenitors, the probability of producing non-viable progeny significantly increased (Figure 6E). For example, STLC treated progenitors delayed for 60 minutes produced viable progeny in 80% of their divisions, whereas those delayed beyond 120 minutes underwent only 14% viable divisions. This relationship between mitosis duration and increased production of apoptotic progeny parallels results obtained with Magoh+/− delayed RGCs (compare to Figure 3J). For all conditions, non-delayed progenitors divisions produced almost 100% viable progeny, consistent with the notion that exposure to inhibitors alone is insufficient to induce cell death (Figures 6D, E).
We then assessed the causal relationship between prolonged mitosis and neuron and progenitor production. We quantified divisions as follows: proliferative P/P (both Tuj1- progenitors, neurogenic P/N (1 Tuj1+), and neurogenic N/N (both Tuj1+) (Figure 6F, Figure S7D). Comparison of non-delayed DMSO or drug treated progenitors showed no significant difference in differentiation or proliferation, indicating pharmacological treatment alone in cell culture is insufficient to induce differentiation (Figure 6F). Strikingly, only those precursors with prolonged mitosis showed altered proliferative and neurogenic divisions, evident to a similar extent with either STLC or nocodazole (Figures 6F, G, S7D). Moreover, as mitosis duration was prolonged using either drug, there was a concomitant increase in neurogenic divisions, as evident in Magoh+/− delayed progenitors (Figure 6G, P<0.001, both). Although control delayed E13.5 progenitors did not show altered neuron production, control E14.5 progenitors, which have higher neurogenic potency, did show a correlation between prolonged mitosis and increased neurogenic divisions (Figure S7J).
Finally we queried if these phenotypes were evident specifically in E13.5 RGCs (Figure 6H). The same causal relationship emerged as in the previous experiment. Prometaphase-delayed RGCs (DsRed-Tbr2-) generated significantly more apoptotic progeny compared to control, and underwent significantly more direct neurogenic divisions (Figures 6I, J). We noted no notable increase in indirect IP generating divisions (Figures 6J, Figure S7E–G). Thus both Magoh+/− and STLC treated RGCs produce more neurons specifically via asymmetric direct neurogenic divisions. Taken together, these pharmacology results demonstrate prolonged mitosis alters the balance of progenitor division subtypes, directly leading to increased neuron production, decreased RGC production, and increased apoptotic progeny.
p53 signaling distinguishes mitotic mechanisms of differentiation and apoptosis
These findings indicate abnormally long mitosis induces both differentiation and cell death in progeny. However, it remains unclear if these fates are caused by distinct molecular events and how mitotically delayed progenitors direct progeny to die or differentiate. To begin to address these questions, we examined STLC delayed progenitors to identify candidate pathways linking prolonged mitosis to apoptosis and differentiation. In mitotically delayed immortalized cells, DNA damage response (DDR) can be increased (Ganem and Pellman, 2012). Indeed, we noted significantly higher DDR in mitotically delayed but not non-delayed progenitors, as assessed by γH2AX staining (Figures 7A–E). Interestingly, depletion of Magoh either genetically or in immortalized cells also triggers increased DDR (Silver et al., 2010). p53 is induced following DDR and is also upregulated following PM-arrest (Bazzi and Anderson, 2014; Uetake and Sluder, 2010), suggesting it could be an important pathway downstream of damage and mitotic delay. In addition, both p53 signaling and DDR are implicated in apoptosis and differentiation of various stem cell populations, including adult neural stem cells (Gil-Perotin et al., 2006; Inomata et al., 2009; J. Wang et al., 2012).
Figure 7. p53 signaling distinguishes apoptosis and differentiation fates of mitotic progenitors in embryonic brain slices.
(A–D) γH2AX staining of non-delayed and STLC delayed progenitors (dotted lines). (E) Graph depicting relative γH2AX staining intensity in delayed versus non-delayed progenitors. (F–I) Sections from E13.5 brain slices treated with either DMSO (F, G) or STLC (H, I) after 0h (F, H) or 3h washout (G, I) and stained for P53 (green). (J) Quantification of nuclei with high P53 signal. (K) Timeline of the slice culture experiment. (L–O) Sections from E13.5 control (L, N) and p53−/− (M, O) brain slices 14h after washout from either DMSO (L, M) or STLC treatment (N,O) and stained for CC3 (green) and Hoechst (blue). (P, Q) Fraction of EdU+ cells in DMSO (grey) and STLC (red) treated E13.5 p53−/− slices that are CC3+ (P) or Tuj1+ (Q) after 14h washout. (R) Fold changes in the fraction of neurons (Tuj1+), IPs (Tbr2+), and RGCs (Pax6+ Tbr2-) in STLC/DMSO treated slices from WT (grey) vs p53−/− (black) embryonic brains. ns, not significant, *, P<0.05, **, P<0.01, *** P<0.001. Scale bars, A–D: 10 µm, F–I: 50µm, L–O: 75µm. Error bars, S.D.
We hypothesized that p53 induces both apoptosis and neuronal differentiation following mitotic delay. We first assessed accumulation of p53 in nuclei of STLC treated slices, as a proxy for pathway activation (Insolera et al., 2014). By 3 hours following pharmacological treatment, we observed a significant 50-fold increase in p53+EdU+ cells (Figures 7F–J, Figure S8A, B). To assess the role of p53 signaling in apoptosis and differentiation, we repeated the EdU pulse-chase experiments described above in a p53−/− background, using STLC to reversibly delay progenitors in prometaphase (Figure 7K). 14 hours after washout, we observed a complete rescue of apoptosis in STLC treated p53-null slices (Figures 7L–P). If p53 were also required for differentiation, then we expected equivalent rescue of neuron, RGC, and IP number. However, this was not the case. Compared to DMSO treated p53−/− slices, significantly more neurons (Tuj1+EdU+) were present in STLC treated p53−/− slices (Figures 7Q). This indicates aberrant neuronal differentiation occurs in the absence of p53. We noted similar fold changes in the fraction of EdU+ neurons and RGCs in both the p53 wild-type and null backgrounds (Figure 7R). In contrast, IP depletion was rescued in the p53−/− background. This result could be due to either a role of p53 in IP apoptosis or IP generation. However if p53 were required for IP generation, one would expect reciprocal rescue of Pax6 and Tuj1 populations, which was not evident. Along with the data in control slices (Figure 5R) these findings indicate the following outcomes of mitotic delay: RGCs are depleted due to both apoptosis and fewer RGC producing divisions; IPs are depleted primarily due to apoptosis; neurons are increased due to more neurogenic divisions. These data are consistent with clonal analysis of drug-treated and Magoh+/− progenitors, in which mitotic delay did not dramatically impact IP generating divisions.
The above data indicate that following mitotic delay at the population level, p53 signaling is required for apoptosis, but not for neuronal differentiation. We sought to directly assess the requirement of p53 signaling in these progeny fates using clonal live imaging (Figure 8A). Similar to wild-type, p53-depleted progenitors exhibited mitotic delay following STLC treatment (compare Figures 6B, C to 8B, C). Mitotically delayed p53−/− progenitors produced negligible apoptotic progeny, similar to slice experiments (Figures 8D, S8C, D). Also reinforcing the slice findings, mitotic-delay induced differentiation was p53-independent. Compared to non-delayed p53−/− progenitors, delayed p53−/− progenitors underwent significantly more neurogenic but fewer proliferative divisions (Figure 8E). Notably, both p53 null and wild-type progenitors showed similar fractions of proliferative and neurogenic divisions (Figures 8E). Thus, in the absence of apoptosis, there is no significant shift in the balance of proliferative and neurogenic divisions. These data demonstrate that prolonged mitosis of progenitors induces apoptosis in a p53-dependent fashion. In contrast p53 signaling is not involved in the induction of differentiation following prolonged mitosis (Figure 8F–H). Altogether these analyses reveal that apoptosis and differentiation are mutually exclusive outcomes of mitotic delay.
Figure 8. Mitotically delayed progenitors directly produce p53-dependent apoptotic progeny and p53-independent neuronal progeny.
(A) Overview of experiment for live imaging progenitors and visualizing cell fate. (B, C) Average mitosis duration (B) and distribution of mitosis durations (C) for DMSO (white) and STLC (red) treated p53−/− progenitors. (D) Absence of apoptosis (black) in both DMSO and STLC treated p53−/− progenitors. (E) Proportion of P/P (white), P/N (grey), and N/N (black) divisions of delayed and non-delayed progenitors from indicated treatments. (F–I) Models depicting how prolonged neural progenitor mitosis influences the survival and fate of direct progeny. RGCs with prolonged mitosis directly produce neurons at the expense of new RGCs in a p53-independent manner (F, H). With excessively longer mitosis, RGCs are more likely to generate apoptotic progeny, via p53-dependent mechanisms (F, I). ns, not significant, *, P<0.05, ***, P<0.001, ns, not significant.
Discussion
Control of cell cycle progression has been linked to regulation of cortical development (Arai et al., 2011; Lange et al., 2009; Pilaz et al., 2009; Takahashi et al., 1995), yet the specific impact of prolonged M phase in specification of neural cell fates has remained enigmatic. Using genetic models and 2 independent pharmacological approaches we demonstrate that prolonged mitosis directly alters balanced divisions of radial glial progenitors, generating more neurons and apoptotic progeny. Our findings indicate that prolonged mitosis in the Magoh+/− mutant contributes to aberrant neurogenesis and is likely a major contributor to microcephaly of these animals. Taken together our study has implications for understanding the behavior of neural stem cell populations and for the etiology of microcephaly.
Prometaphase delay produces common microcephaly phenotypes
Mitosis defects are proposed to underlie microcephaly and associated neurogenesis phenotypes. Several specific mitotic mechanisms have been invoked to explain this. Altered centrosome number and spindle orientation cause apoptosis, premature differentiation, and mislocalized progenitors (Insolera et al., 2014; Marthiens et al., 2013). Progenitor depletion and apoptosis can also arise from mitotic catastrophe in which progenitors die prior to division (Chen et al., 2014; Novorol et al., 2013). Our study highlights prolonged prometaphase as an additional mechanism for influencing cell fate, which can occur independent of aneuploidy, altered cleavage plane orientation, and centrosome integrity. Many mouse microcephaly mutants are reported to increase mitotic index, but few if any studies have directly analyzed mitotic duration using live imaging. Thus it will be of interest to examine if other microcephaly-associated genes also operate via prolonged mitosis. Moreover, we propose prolonged metaphase duration is a valuable parameter to examine in human organoid models of microcephaly (Lancaster et al., 2013).
Exposing mechanisms by which Magoh influences neurogenesis
We previously demonstrated Magoh haploinsufficient mice exhibit microcephaly due to apoptosis, precocious neurons and reduced IPs, however the cellular cause of these phenotypes was unclear (Silver et al., 2010). We can now pinpoint altered balance of neurogenic, proliferative, and viable RGC divisions as a major contributing mechanism. At E12.5 delayed Magoh+/− RGCs directly produce fewer RGCs, more neurons, and more apoptotic progeny. As mitosis duration of Magoh deficient RGCs lengthens, so does the probability of producing fewer progenitors and non-viable cells. The net result is a severe depletion of the progenitor pool, which is predicted to ultimately reduce neurons, as is evident in E18.5 Magoh+/− brains (Silver et al., 2010). Interestingly, RGCs are not depleted at the population level in microcephalic Magoh+/− cortical columns (Silver et al., 2010), suggesting Magoh regulation of RGCs is especially relevant in the tangential dimension. Magoh is part of the exon junction RNA binding complex, components of which are associated with human neurodevelopmental phenotypes including microcephaly (Mao et al., 2015; Pilaz and Silver, 2015). Our findings may have broad relevance for understanding roles of this complex in human brain development.
It is striking that only mitotically delayed Magoh+/− RGCs produce altered progeny. However, Magoh+/− delayed RGCs are likely to carry additional factors that influence cell fate. In support of this, for identical mitotic durations of 60–80 minutes, Magoh+/− E12.5 progenitors produce substantially more apoptotic progeny than STLC delayed E13.5 progenitors. What could these contributing factors be? Magoh+/− brains have increased DNA damage (Silver et al., 2010), and it is plausible that mitotic delay is a consequence of earlier damage, which could also influence progeny fate. It is also possible that mitotically delayed Magoh+/− progenitors express a slightly different repertoire of Magoh-dependent pro-differentiation determinants, such as Ngn2, Tbr2, and Lis1 protein (Silver et al., 2010). In considering these possibilities, it will be of interest to understand why only a sub-population of RGCs are affected by Magoh haploinsufficiency. One potential explanation is stochastic Magoh levels but another intriguing idea is that delayed progenitors are fundamentally distinct. Indeed heterogeneous populations of RGCs have been reported (Franco and Müller, 2013; Tyler et al., 2015). Defining transcriptomic and proteomic differences in Magoh+/− progenitors with normal and prolonged mitoses will help address these fascinating questions.
How does Magoh influence mitosis? The most striking defect in Magoh depleted mitotic cells was in microtubule organization, leading us to posit Magoh regulates mitotic progression by controlling spindle dynamics and anaphase onset. Consistent with this notion, mago nashi, the Magoh ortholog, is required for microtubule dynamics in Drosophila oocytes (Micklem et al., 1997). Our data indicate Magoh can influence microtubules indirectly, by modulating expression of key microtubule/mitosis regulatory proteins. However, Magoh may also directly influence microtubule organization; it localizes to the mitotic spindle (Ishigaki et al., 2013), was identified in proteomic studies as a mitotic spindle component (Bonner et al., 2011; Sauer, 2004), and physically associates with the microtubule-associated protein MAP1B (Tretyakova, 2005). It will be interesting to assess if Magoh’s RNA binding function is necessary for microtubule regulation and if so to identify its RNA cargos specifically at M phase. Future studies aimed at understanding how Magoh influences microtubules will help illuminate the root causes of the mitosis delay.
Potential mechanisms linking prometaphase to altered cell fate
We show mitotically delayed progenitors have increased propensity for generating apoptotic progeny and differentiated neurons. Our data indicate that as mitosis lengthens, the probability for producing apoptotic progeny increases. In Magoh+/−, STLC and nocodazole treated progenitors, extremely long mitoses generate primarily apoptotic fates, whereas increased neurogenic divisions are first evident in shorter delays. We speculate that neural progenitors have an internal clock, which measures mitosis duration. This model has been invoked to explain why mitotically delayed immortalized cells produce G1-arrested progeny (Uetake and Sluder, 2010). Progenitors with longer mitoses may be deemed damaged or problematic, forcing their progeny to die or exit the cell cycle. In support of this model, our genetic and pharmacology experiments indicate delayed cells generate aberrant neurons via direct neurogenic rather than indirect IP divisions. Thus mitotic delay causes progenitors to preferentially produce post-mitotic or apoptotic progeny rather than generating new progenitors.
How does longer mitosis induce cell fate changes in progeny? In this study we examined several potential mechanisms. In drug-treated progenitors neither increased aneuploidy nor misoriented spindles explain observed cell fate changes. In contrast, mitotically-delayed progenitors showed increased γH2AX staining prior to completion of mitosis. Subsequent progeny exhibited p53 activation, which induced apoptosis. As p53 loss rescued apoptosis but not differentiation, it is intriguing to consider if other DDR-dependent signaling pathways promote differentiation. Notably, DDR can trigger differentiation of stem cell populations including adult neural stem cells, making it an interesting avenue for future studies (Gil-Perotin et al., 2006; Inomata et al., 2009; Sherman et al., 2011; J. Wang et al., 2012). Identifying the molecular mechanisms driving delay-induced differentiation is a high priority for future studies.
Mitotic regulation and the developing brain
Our study indicates prolonged mitosis can alter cell fates in a pathogenic state, but it is interesting to consider its potential role during normal development. Haydar et al. showed that over the course of corticogenesis as more neurons are being produced, neural progenitor metaphase duration increases (Haydar et al., 2003). We observed that prolonged mitosis could impact normal development as evidenced by significantly more apoptotic progeny from delayed control progenitors at E13.5 and E14.5. E14.5 control progenitors delayed in mitosis also underwent more neurogenic divisions. These outcomes were not evident at E12.5, perhaps because there are fewer delayed progenitors at this age. Thus, the relatively subtle relationships between duration and cell fate may be more pronounced at older stages when progenitor divisions are longer and more neurogenic, and/or when measured with less pro-proliferative culture conditions. Studies of older stages are also particularly relevant for assessing the role of mitotic duration in different progenitor populations, and in the generation of different neuronal and glial subtypes. Our findings provide rationale for future detailed analysis of the role of mitosis duration in influencing normal brain development.
Beyond normal mouse development, cell cycle differences are one of the most distinguishing traits differentiating the brains of mice, non-human primates and humans (Geschwind and Rakic, 2013). Such differences are postulated to influence brain size, shape and neuronal number. Thus in future studies we should consider not only the potential role that prolonged mitotic duration plays in the etiology of microcephaly, but also in defining species-specific differences in brain development.
Experimental Procedures
Statistical analyses and Quantitation
See Supplemental Table S1 for details regarding statistical analysis. All graphs depict average and SD values, and error bars denote biological variation. When possible all analyses were performed by more than person, blind to genotype. For this manuscript RGCs were denoted either by their division at the apical surface, expression of Pax6, or absence of Tbr2 expression at E12.5.
Live analyses
For clonal analysis, progenitors were cultured as previously described (Shen et al., 2002). For brain slice imaging, analysis was performed as previously described (Pilaz and Silver, 2014). Because pharmacology can potentially have off-target effects, in our study we rule out impacts of nocodazole and STLC upon interphase viability, cleavage plane orientation, and exclude aneuploidy as an outcome of STLC.
Mouse genetics
MagohMos2/+ (Magoh+/−)(Silver et al., 2010), MagohLoxP (McMahon et al., 2014), and Tbr2-EGFP (Arnold et al., 2009) mice were previously described. The following mouse strains were obtained from Jackson labs: B6.129S2-Emx1<tm1(cre)Krj/J, B6.Cg-Tg(Camk2a-cre)T29-1Stl/J, B6.129S2-Trp53tm1Tyj/J, B6.Cg-Tg(HIST1H2BB/EGFP)1Pa/J, and C57BL/6J-Tg(Dcx-DsREd)14Qlu/J. All experiments were approved by the Institutional Animal Care and Use Committee of Duke University.
Supplementary Material
Acknowledgements
We thank Dr. Hiro Matsunami, Cagla Eroglu, Dr. Terry Lechler, Dr. Beth Sullivan, Dr. Colette Dehay, Dr. Delphine Delaunay, Dr. Scott Soderling, Dr. Don Fox, and Silver lab members for careful reading of the manuscript and helpful discussions. We thank Autumn Rorrer, Eric Feeley, Andrew Muroyama, Hamid Hussaini, the Duke light microscopy and mouse transgenic core facilities for technical assistance. Tbr2-EGFP mice were a kind gift of Sebastian Arnold. This work was funded through grants to DLS (R01NS083897) and to Duke Cancer center (P30 CA014236).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
LJP, JM, and DS designed the study, analyzed all data and wrote the manuscript. LJP performed Figures 1,2,4,5,7, S2, S3, S4, S6. JM performed Figures 3,6,7,8, S1, S5, S7, S8. AS, TS, AL, and EM contributed to Figures S1, S3 and 4.
Please see Supplemental Experimental Procedures
References
- Arai Y, Pulvers JN, Haffner C, Schilling B, Nüsslein I, Calegari F, Huttner WB. Neural stem and progenitor cells shorten S-phase on commitment to neuron production. Nature Communications. 2011;2:154. doi: 10.1038/ncomms1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold SJ, Sugnaseelan J, Groszer M, Srinivas S, Robertson EJ. Generation and analysis of a mouse line harboring GFP in the Eomes/Tbr2 locus. Genesis. 2009;47:775–781. doi: 10.1002/dvg.20562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asami M, Pilz GA, Ninkovic J, Godinho L, Schroeder T, Huttner WB, Gotz M. The role of Pax6 in regulating the orientation and mode of cell division of progenitors in the mouse cerebral cortex. 2011;138:5067–5078. doi: 10.1242/dev.074591. [DOI] [PubMed] [Google Scholar]
- Bazzi H, Anderson KV. Acentriolar mitosis activates a p53-dependent apoptosis pathway in the mouse embryo. Proceedings of the National Academy of Sciences. 2014;111:E1491–E1500. doi: 10.1073/pnas.1400568111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner MK, Poole DS, Xu T, Sarkeshik A, Yates JR, III, Skop AR. Mitotic Spindle Proteomics in Chinese Hamster Ovary Cells. PLoS ONE. 2011;6:e20489. doi: 10.1371/journal.pone.0020489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-F, Zhang Y, Wilde J, Hansen KC, Lai F, Niswander L. Microcephaly disease gene Wdr62 regulates mitotic progression of embryonic neural stem cells and brain size. Nature Communications. 2014;5:3885. doi: 10.1038/ncomms4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco SJ, Müller U. Shaping Our Minds: Stem and Progenitor Cell Diversity in the Mammalian Neocortex. Neuron. 2013;77:19–34. doi: 10.1016/j.neuron.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio T, Saredi A, Compton DA. NuMA is required for the organization of microtubules into aster-like mitotic arrays. J Cell Biol. 1995;131:693–708. doi: 10.1083/jcb.131.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Pellman D. Linking abnormal mitosis to the acquisition of DNA damage. J Cell Biol. 2012;199:871–881. doi: 10.1083/jcb.201210040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, Rakic P. Perspective. Neuron. 2013;80:633–647. doi: 10.1016/j.neuron.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Perotin S, Marin-Husstege M, Li J, Soriano-Navarro M, Zindy F, Roussel MF, Garcia-Verdugo J-M, Casaccia-Bonnefil P. Loss of p53 induces changes in the behavior of subventricular zone cells: implication for the genesis of glial tumors. Journal of Neuroscience. 2006;26:1107–1116. doi: 10.1523/JNEUROSCI.3970-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber R, Zhou Z, Sukchev M, Joerss T, Frappart P-O, Wang Z-Q. MCPH1 regulates the neuroprogenitor division mode by coupling the centrosomal cycle with mitotic entry through the Chk1-Cdc25 pathway. Nat Cell Biol. 2011;13:1325–1334. doi: 10.1038/ncb2342. [DOI] [PubMed] [Google Scholar]
- Haydar TF, Ang E, Rakic P. Mitotic spindle rotation and mode of cell division in the developing telencephalon. Proc Natl Acad Sci USA. 2003;100:2890–2895. doi: 10.1073/pnas.0437969100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu WF, Chahrour MH, Walsh CA. The diverse genetic landscape of neurodevelopmental disorders. Annu Rev Genomics Hum Genet. 2014;15:195–213. doi: 10.1146/annurev-genom-090413-025600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inomata K, Aoto T, Binh NT, Okamoto N, Tanimura S, Wakayama T, Iseki S, Hara E, Masunaga T, Shimizu H, Nishimura EK. Genotoxic stress abrogates renewal of melanocyte stem cells by triggering their differentiation. Cell. 2009;137:1088–1099. doi: 10.1016/j.cell.2009.03.037. [DOI] [PubMed] [Google Scholar]
- Insolera R, Bazzi H, Shao W, Anderson KV, Shi S-H. Cortical neurogenesis in the absence of centrioles. Nat Neurosci. 2014;17:1528–1535. doi: 10.1038/nn.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki Y, Nakamura Y, Tatsuno T, Hashimoto M, Iwabuchi K, Tomosugi N. RNA-binding protein RBM8A (Y14) and MAGOH localize to centrosome in human A549 cells. Histochem Cell Biol. 2013 doi: 10.1007/s00418-013-1135-4. [DOI] [PubMed] [Google Scholar]
- Konno D, Shioi G, Shitamukai A, Mori A, Kiyonari H, Miyata T, Matsuzaki F. Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nat Cell Biol. 2008;10:93–101. doi: 10.1038/ncb1673. [DOI] [PubMed] [Google Scholar]
- Kowalczyk T, Pontious A, Englund C, Daza RAM, Bedogni F, Hodge R, Attardo A, Bell C, Huttner WB, Hevner RF. Intermediate neuronal progenitors (basal progenitors) produce pyramidal-projection neurons for all layers of cerebral cortex. Cereb Cortex. 2009;19:2439–2450. doi: 10.1093/cercor/bhn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMonica BE, Lui JH, Hansen DV, Kriegstein AR. Mitotic spindle orientation predicts outer radial glial cell generation in human neocortex. Nature Communications. 2013;4:1665. doi: 10.1038/ncomms2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin C-A, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C, Huttner WB, Calegari F. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell stem cell. 2009;5:320–331. doi: 10.1016/j.stem.2009.05.026. [DOI] [PubMed] [Google Scholar]
- Lizarraga SB, Margossian SP, Harris MH, Campagna DR, Han A-P, Blevins S, Mudbhary R, Barker JE, Walsh CA, Fleming MD. Cdk5rap2 regulates centrosome function and chromosome segregation in neuronal progenitors. 2010;137:1907–1917. doi: 10.1242/dev.040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malatesta P, Hartfuss E, Gotz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. 2000;127:5253–5263. doi: 10.1242/dev.127.24.5253. [DOI] [PubMed] [Google Scholar]
- Mao H, Pilaz L-J, McMahon JJ, Golzio C, Wu D, Shi L, Katsanis N, Silver DL. Rbm8a haploinsufficiency disrupts embryonic cortical development resulting in microcephaly. Journal of Neuroscience. 2015;35:7003–7018. doi: 10.1523/JNEUROSCI.0018-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marthiens V, Rujano MA, Pennetier C, Tessier S, Paul-Gilloteaux P, Basto R. Centrosome amplification causes microcephaly. Nat Cell Biol. 2013;15:731–740. doi: 10.1038/ncb2746. [DOI] [PubMed] [Google Scholar]
- McIntyre RE, Lakshminarasimhan Chavali P, Ismail O, Carragher DM, Sanchez-Andrade G, Forment JV, Fu B, Del Castillo Velasco-Herrera M, Edwards A, van der Weyden L, Yang F, Sanger Mouse Genetics Project Ramirez-Solis R, Estabel J, Gallagher FA, Logan DW, Arends MJ, Tsang SH, Mahajan VB, Scudamore CL, White JK, Jackson SP, Gergely F, Adams DJ. Disruption of mouse Cenpj, a regulator of centriole biogenesis, phenocopies Seckel syndrome. PLoS Genet. 2012;8:e1003022. doi: 10.1371/journal.pgen.1003022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon JJ, Shi L, Silver DL. Generation of a Magoh conditional allele in mice. Genesis. 2014;52:752–758. doi: 10.1002/dvg.22788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micklem DR, Dasgupta R, Elliott H, Gergely F, Davidson C, Brand A, González-Reyes A, St Johnston D. The mago nashi gene is required for the polarisation of the oocyte and the formation of perpendicular axes in Drosophila. Curr Biol. 1997;7:468–478. doi: 10.1016/s0960-9822(06)00218-1. [DOI] [PubMed] [Google Scholar]
- Mora-Bermúdez F, Matsuzaki F, Huttner WB. Specific polar subpopulations of astral microtubules control spindle orientation and symmetric neural stem cell division. eLife. 2014 doi: 10.7554/eLife.02875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- Novorol C, Burkhardt J, Wood KJ, Iqbal A, Roque C, Coutts N, Almeida AD, He J, Wilkinson CJ, Harris WA. Microcephaly models in the developing zebrafish retinal neuroepithelium point to an underlying defect in metaphase progression. Open Biology. 2013;3:130065. doi: 10.1098/rsob.130065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostergaard P, Simpson MA, Mendola A, Vasudevan P, Connell FC, van Impel A, Moore AT, Loeys BL, Ghalamkarpour A, Onoufriadis A, Martinez-Corral I, Devery S, Leroy JG, van Laer L, Singer A, Bialer MG, McEntagart M, Quarrell O, Brice G, Trembath RC, Schulte-Merker S, Makinen T, Vikkula M, Mortimer PS, Mansour S, Jeffery S. Mutations in KIF11 cause autosomal-dominant microcephaly variably associated with congenital lymphedema and chorioretinopathy. Am J Hum Genet. 2012;90:356–362. doi: 10.1016/j.ajhg.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrem BEL, Lui JH, Gertz CC, Kriegstein AR. Control of Outer Radial Glial Stem Cell Mitosis in the Human Brain. CellReports. 2014;8:656–664. doi: 10.1016/j.celrep.2014.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilaz L-J, Patti D, Marcy G, Ollier E, Pfister S, Douglas RJ, Betizeau M, Gautier E, Cortay V, Doerflinger N, Kennedy H, Dehay C. Forced G1-phase reduction alters mode of division, neuron number, and laminar phenotype in the cerebral cortex. Proceedings of the National Academy of Sciences. 2009;106:21924–21929. doi: 10.1073/pnas.0909894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilaz L-J, Silver DL. Post-transcriptional regulation in corticogenesis: how RNA-binding proteins help build the brain. WIREs RNA. 2015;6:501–515. doi: 10.1002/wrna.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilaz L-J, Silver DL. Live imaging of mitosis in the developing mouse embryonic cortex. Journal of visualized experiments : JoVE. 2014:e51298–e51298. doi: 10.3791/51298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JC, Molinek M, Martynoga BS, Zaki PA, Faedo A, Bulfone A, Hevner RF, West JD, Price DJ. Pax6 controls cerebral cortical cell number by regulating exit from the cell cycle and specifies cortical cell identity by a cell autonomous mechanism. Dev Biol. 2007;302:50–65. doi: 10.1016/j.ydbio.2006.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer G. Proteome Analysis of the Human Mitotic Spindle. Molecular & Cellular Proteomics. 2004;4:35–43. doi: 10.1074/mcp.M400158-MCP200. [DOI] [PubMed] [Google Scholar]
- Shen Q, Zhong W, Jan Y, Temple S. Asymmetric Numb distribution is critical for asymmetric cell division of mouse cerebral cortical stem cells and neuroblasts. 2002;129:4843–4853. doi: 10.1242/dev.129.20.4843. [DOI] [PubMed] [Google Scholar]
- Sherman MH, Bassing CH, Teitell MA. Regulation of cell differentiation by the DNA damage response. Trends Cell Biol. 2011;21:312–319. doi: 10.1016/j.tcb.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DL, Leeds KE, Hwang H-W, Miller EE, Pavan WJ. The EJC component Magoh regulates proliferation and expansion of neural crest-derived melanocytes. Dev Biol. 2013;375:172–181. doi: 10.1016/j.ydbio.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DL, Watkins-Chow DE, Schreck KC, Pierfelice TJ, Larson DM, Burnetti AJ, Liaw H-J, Myung K, Walsh CA, Gaiano N, Pavan WJ. The exon junction complex component Magoh controls brain size by regulating neural stem cell division. Nat Neurosci. 2010;13:551–558. doi: 10.1038/nn.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoufias DA, DeBonis S, Saoudi Y, Lebeau L, Crevel I, Cross R, Wade RH, Hackney D, Kozielski F. S-trityl-L-cysteine is a reversible, tight binding inhibitor of the human kinesin Eg5 that specifically blocks mitotic progression. J Biol Chem. 2006;281:17559–17569. doi: 10.1074/jbc.M511735200. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Nowakowski RS, Caviness VS. The cell cycle of the pseudostratified ventricular epithelium of the embryonic murine cerebral wall. The Journal of Neuroscience : the official journal of the Society for Neuroscience. 1995;15:6046–6057. doi: 10.1523/JNEUROSCI.15-09-06046.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretyakova I. Nuclear Export Factor Family Protein Participates in Cytoplasmic mRNA Trafficking. J Biol Chem. 2005;280:31981–31990. doi: 10.1074/jbc.M502736200. [DOI] [PubMed] [Google Scholar]
- Tsai J-W, Bremner KH, Vallee RB. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nature Publishing Group. 2007;10:970–979. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- Tyler WA, Medalla M, Guillamon-Vivancos T, Luebke JI, Haydar TF. Neural Precursor Lineages Specify Distinct Neocortical Pyramidal Neuron Types. The Journal of Neuroscience : the official journal of the Society for Neuroscience. 2015;35:6142–6152. doi: 10.1523/JNEUROSCI.0335-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y, Sluder G. Prolonged Prometaphase Blocks Daughter Cell Proliferation Despite Normal Completion of Mitosis. Current Biology. 2010;20:1666–1671. doi: 10.1016/j.cub.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sun Q, Morita Y, Jiang H, Gross A, Lechel A, Hildner K, Guachalla LM, Gompf A, Hartmann D, Schambach A, Wuestefeld T, Dauch D, Schrezenmeier H, Hofmann W-K, Nakauchi H, Ju Z, Kestler HA, Zender L, Rudolph KL. A differentiation checkpoint limits hematopoietic stem cell self-renewal in response to DNA damage. Cell. 2012;148:1001–1014. doi: 10.1016/j.cell.2012.01.040. [DOI] [PubMed] [Google Scholar]
- Wang X, Qiu R, Tsark W, Lu Q. Rapid promoter analysis in developing mouse brain and genetic labeling of young neurons by doublecortin-DsRed-express. J Neurosci Res. 2007;85:3567–3573. doi: 10.1002/jnr.21440. [DOI] [PubMed] [Google Scholar]
- Xie Y, Jüschke C, Esk C, Hirotsune S, Knoblich JA. The phosphatase PP4c controls spindle orientation to maintain proliferative symmetric divisions in the developing neocortex. Neuron. 2013;79:254–265. doi: 10.1016/j.neuron.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yingling J, Youn YH, Darling D, Toyo-oka K, Pramparo T, Hirotsune S, Wynshaw-Boris A. Neuroepithelial Stem Cell Proliferation Requires LIS1 for Precise Spindle Orientation and Symmetric Division. Cell. 2008;132:474–486. doi: 10.1016/j.cell.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.