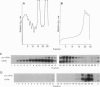
Abstract
Nonsense-mediated mRNA decay, the accelerated turnover of mRNAs transcribed from genes containing early nonsense mutations, is dependent on the product of the UPF1 gene in yeast. Mutations that inactivate UPF1 lead to the selective stabilization of mRNAs containing early nonsense mutations but have no effect on the half-lives of almost all other mRNAs. Since the transcripts of nonsense alleles are not typical cellular constituents, we sought to identify those RNAs that comprise normal substrates of the nonsense-mediated mRNA decay pathway. Many yeast pre-mRNAs contain early in-frame nonsense codons and we consider it possible that a role of this pathway is to accelerate the degradation of pre-mRNAs present in the cytoplasm. Consistent with this hypothesis, we find that, in a strain lacking UPF1 function, the CYH2, RP51B, and MER2 pre-mRNAs are stabilized 2- to 5-fold and are associated with ribosomes. We conclude that a major source of early nonsense codon-containing cytoplasmic transcripts in yeast is pre-mRNAs and that the UPF1 protein may be part of a cellular system that ensures that potentially deleterious nonsense fragments of polypeptides do not accumulate.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abovich N., Rosbash M. Two genes for ribosomal protein 51 of Saccharomyces cerevisiae complement and contribute to the ribosomes. Mol Cell Biol. 1984 Sep;4(9):1871–1879. doi: 10.1128/mcb.4.9.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baim S. B., Pietras D. F., Eustice D. C., Sherman F. A mutation allowing an mRNA secondary structure diminishes translation of Saccharomyces cerevisiae iso-1-cytochrome c. Mol Cell Biol. 1985 Aug;5(8):1839–1846. doi: 10.1128/mcb.5.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker G. F., Beemon K. Nonsense codons within the Rous sarcoma virus gag gene decrease the stability of unspliced viral RNA. Mol Cell Biol. 1991 May;11(5):2760–2768. doi: 10.1128/mcb.11.5.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann B., Potash M. J., Köhler G. Consequences of frameshift mutations at the immunoglobulin heavy chain locus of the mouse. EMBO J. 1985 Feb;4(2):351–359. doi: 10.1002/j.1460-2075.1985.tb03636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson M. R., Underbrink K. M., Fink G. R. Frameshift suppression Saccharomyces cerevisiae. II. Genetic properties of group II suppressors. Genetics. 1980 Aug;95(4):833–853. doi: 10.1093/genetics/95.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht J. A., Voelkel-Meiman K., Roeder G. S. Meiosis-specific RNA splicing in yeast. Cell. 1991 Sep 20;66(6):1257–1268. doi: 10.1016/0092-8674(91)90047-3. [DOI] [PubMed] [Google Scholar]
- Fink G. R. Pseudogenes in yeast? Cell. 1987 Apr 10;49(1):5–6. doi: 10.1016/0092-8674(87)90746-x. [DOI] [PubMed] [Google Scholar]
- Gaspar M. L., Meo T., Bourgarel P., Guenet J. L., Tosi M. A single base deletion in the Tfm androgen receptor gene creates a short-lived messenger RNA that directs internal translation initiation. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8606–8610. doi: 10.1073/pnas.88.19.8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goguel V., Rosbash M. Splice site choice and splicing efficiency are positively influenced by pre-mRNA intramolecular base pairing in yeast. Cell. 1993 Mar 26;72(6):893–901. doi: 10.1016/0092-8674(93)90578-e. [DOI] [PubMed] [Google Scholar]
- Gozalbo D., Hohmann S. Nonsense suppressors partially revert the decrease of the mRNA level of a nonsense mutant allele in yeast. Curr Genet. 1990 Jan;17(1):77–79. doi: 10.1007/BF00313252. [DOI] [PubMed] [Google Scholar]
- Herrick D., Parker R., Jacobson A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1990 May;10(5):2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käufer N. F., Fried H. M., Schwindinger W. F., Jasin M., Warner J. R. Cycloheximide resistance in yeast: the gene and its protein. Nucleic Acids Res. 1983 May 25;11(10):3123–3135. doi: 10.1093/nar/11.10.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin J. C., Thompson J. R., Woolford J. L., Jr Structure and expression of the Saccharomyces cerevisiae CRY1 gene: a highly conserved ribosomal protein gene. Mol Cell Biol. 1987 May;7(5):1764–1775. doi: 10.1128/mcb.7.5.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds P., Peltz S. W., Jacobson A., Culbertson M. R. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991 Dec;5(12A):2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- Losson R., Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrow R. E., Jacobson A. Identification and characterization of developmentally regulated mRNP proteins of Dictyostelium discoideum. Dev Biol. 1986 Jul;116(1):213–227. doi: 10.1016/0012-1606(86)90058-8. [DOI] [PubMed] [Google Scholar]
- Maquat L. E., Kinniburgh A. J., Rachmilewitz E. A., Ross J. Unstable beta-globin mRNA in mRNA-deficient beta o thalassemia. Cell. 1981 Dec;27(3 Pt 2):543–553. doi: 10.1016/0092-8674(81)90396-2. [DOI] [PubMed] [Google Scholar]
- Morse D. E., Yanofsky C. Polarity and the degradation of mRNA. Nature. 1969 Oct 25;224(5217):329–331. doi: 10.1038/224329a0. [DOI] [PubMed] [Google Scholar]
- Nilsson G., Belasco J. G., Cohen S. N., von Gabain A. Effect of premature termination of translation on mRNA stability depends on the site of ribosome release. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4890–4894. doi: 10.1073/pnas.84.14.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M., Scafe C., Sexton J., Young R. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol. 1987 May;7(5):1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen S. Escherichia coli ribosomes translate in vivo with variable rate. EMBO J. 1984 Dec 1;3(12):2895–2898. doi: 10.1002/j.1460-2075.1984.tb02227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltz S. W., Brewer G., Bernstein P., Hart P. A., Ross J. Regulation of mRNA turnover in eukaryotic cells. Crit Rev Eukaryot Gene Expr. 1991;1(2):99–126. [PubMed] [Google Scholar]
- Pikielny C. W., Rosbash M. mRNA splicing efficiency in yeast and the contribution of nonconserved sequences. Cell. 1985 May;41(1):119–126. doi: 10.1016/0092-8674(85)90066-2. [DOI] [PubMed] [Google Scholar]
- Steel L. F., Jacobson A. Translational control of ribosomal protein synthesis during early Dictyostelium discoideum development. Mol Cell Biol. 1987 Mar;7(3):965–972. doi: 10.1128/mcb.7.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swida U., Thüroff E., Käufer N. F. Intron mutations that affect the splicing efficiency of the CYH2 gene of Saccharomyces cerevisiae. Mol Gen Genet. 1986 May;203(2):300–304. doi: 10.1007/BF00333970. [DOI] [PubMed] [Google Scholar]
- Teem J. L., Rosbash M. Expression of a beta-galactosidase gene containing the ribosomal protein 51 intron is sensitive to the rna2 mutation of yeast. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4403–4407. doi: 10.1073/pnas.80.14.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]