Abstract
We present a protocol for determining the relative orientation and dynamics of A-form helices in 13C/15N isotopically enriched RNA samples using NMR residual dipolar couplings (RDCs). Non-terminal Watson–Crick base pairs in helical stems are experimentally identified using NOE and trans-hydrogen bond connectivity and modeled using the idealized A-form helix geometry. RDCs measured in the partially aligned RNA are used to compute order tensors describing average alignment of each helix relative to the applied magnetic field. The order tensors are translated into Euler angles defining the average relative orientation of helices and order parameters describing the amplitude and asymmetry of interhelix motions. The protocol does not require complete resonance assignments and therefore can be implemented rapidly to RNAs much larger than those for which complete high-resolution NMR structure determination is feasible. The protocol is particularly valuable for exploring adaptive changes in RNA conformation that occur in response to biologically relevant signals. Following resonance assignments, the procedure is expected to take no more than 2 weeks of acquisition and data analysis time.
INTRODUCTION
The functions of many regulatory RNAs involve large changes in conformation that occur in response to a range of cellular signals, including recognition of proteins and ligands, metal binding, changes in temperature and RNA synthesis itself1–4. Such conformational transitions allow one RNA molecule to carry out many biochemical transactions. For example, the RNA conformation required for the assembly of a complex ribonucleoprotein may differ from that required for executing the ribonucleoprotein function5,6. Conformational changes also provide a basis for sensing signals and transmitting regulatory responses. For example, a large class of mRNA riboswitches regulate gene expression by changing conformation in response to recognition of small metabolite molecules or changes in temperature7,8.
These and many other examples (see refs. 3,4,6 and references therein) illustrate how a broad structural landscape—rather than a single conformation—often needs to be characterized in order to fully understand how regulatory RNAs function at the atomic level. This structural landscape is vast when one considers the potentially innumerable RNA conformations that could be targeted by small molecules in order to combat infectious diseases9–11. Characterizing such a large number of RNA conformations at atomic resolution is a challenge that cannot be met by current techniques. Despite significant advances, RNA structure determination by X-ray crystallography and NMR spectroscopy still requires several months. X-ray crystallography is limited to conditions that yield well-diffracting crystals and this can preclude insight into less-ordered conformers of the RNA structural landscape. While NMR spectroscopy can be applied under a variety of conditions, it is currently limited to RNA molecules of the size of ~100 nt (ref. 12). Moreover, until recently, heavy reliance on short-range distance constraints made it difficult to reliably define global aspects of RNA architecture13–15.
Herein, we describe an NMR protocol that can be used to rapidly characterize the relative orientation and dynamics of RNA A-form helices that relies on the measurement of RDCs in partially aligned RNAs (Fig. 1) (see refs. 16,17). The relative orientation and dynamics of A-form helices is a feature of RNA conformation that goes through important changes during the course of folding, recognition and catalysis2,3,18,19. It is also a feature of RNA architecture that is particularly susceptible to artificial distortions owing to crystal packing forces20. The protocol, first described in refs. 21–23, combines long-range orientational constraints derived from RDCs with the ability to accurately model A-form helical domains using a standard idealized geometry15,24. Compared to other techniques used to characterize interhelical bending, including gel mobility measurements and transient electric birefringence25,26, the current protocol allows one to directly distinguish between fixed bends and interhelical flexibility and provides three-dimensional information regarding interhelical bend and twist angles. The orientational information obtained is highly complementary to the distance information obtained from other techniques such as fluorescent resonance energy transfer, providing ample opportunities for synergistic integration.
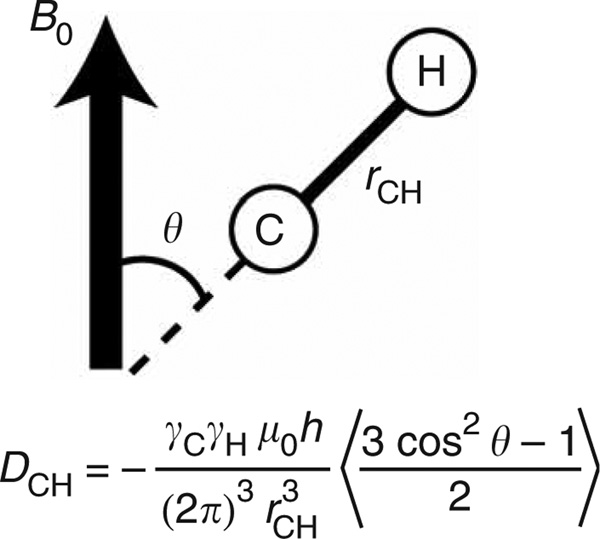
Figure 1.
RDCs (Dij) between spins i and j provide long-range constraints on the average orientation (θ) of the internuclear bond vector relative to the applied magnetic field.
The protocol trades structural resolution for greater efficiency of application and potentially broader applicability to larger RNA systems. Thus, while it does not yield complete high-resolution structures, it provides a basis for exploring the relative orientation and dynamics of helical domains under a variety of conditions of interest in large molecular systems. This high efficiency and broad applicability arises for a number of reasons. First, the protocol bypasses the rate- and size-limiting requirement for comprehensive assignments of resonances and NOEs. Rather, assignments are required only for a subset of resonances in A-form helices (Fig. 2a). Not only are these assignments the easiest to establish in nucleic acids using conventional methods27–29, there are possibilities for integrating RDC measurements into the assignment process thereby enhancing the efficiency and robustness of application even further30,31. Second, the conformation of Watson–Crick (WC) base pairs in helical stems does not need to be determined— rather it is modeled a priori using the idealized A-form geometry32,33 taking into account parameterized structural deviations23. Third, computational methods are available for interpreting RDCs in a semi-automated manner23,31,34–36 (Fig. 2b). The protocol in its current form yields 4n−1 degenerate solutions for orienting n helices and this degeneracy has to be lifted with the use of other experimental or geometrical restraints. Note that many variants of this protocol, some of which allow simultaneous local structure determination of molecular fragments, have also been described34–38.
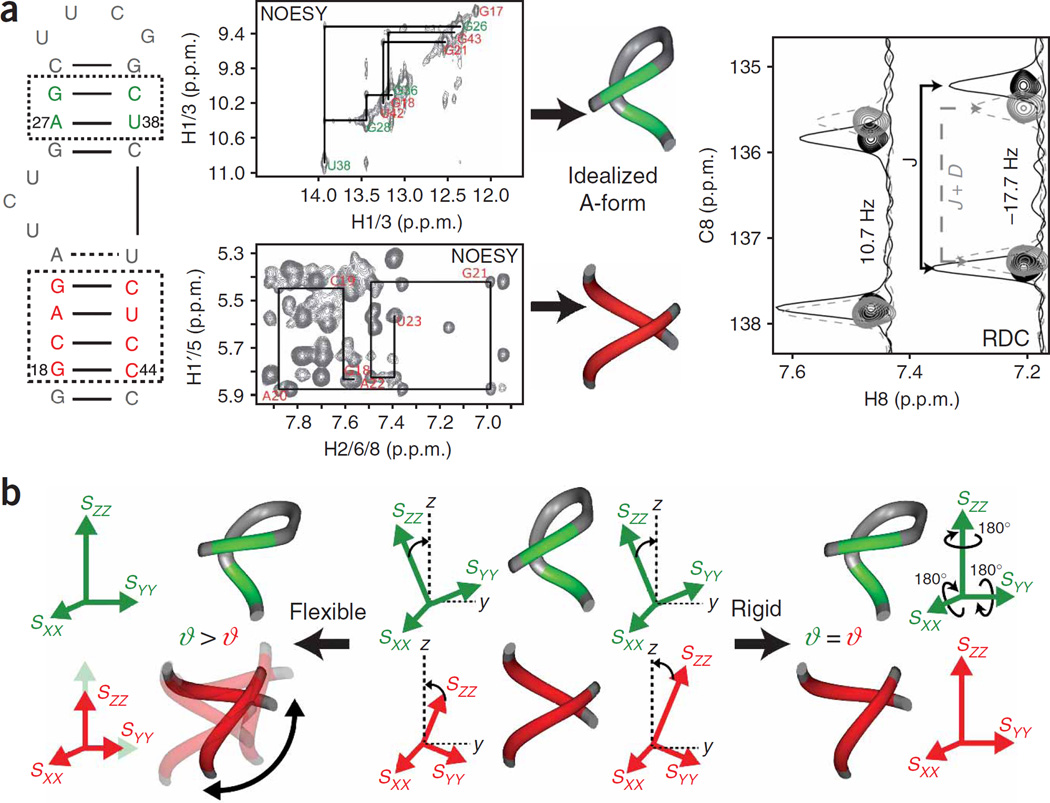
Figure 2.
Determining the relative orientation and dynamics of A-form helices using an order tensor analysis of RDCs46, 99. (a) Watson-Crick (WC) base pairs that are flanked by other WC base pairs are identified based on the predicted RNA secondary structural model. Resonance assignments in WC base pairs are established using NOESY connectivity (shown in the figure) or using through bond correlation experiments. The local structure of the experimentally verified WC pairs is modeled using the idealized A-form geometry23. Next, NMR experiments (Table 1) are used to measure splittings between various nuclei under aligned (J + D) (Table 2) and unaligned (J) conditions. Note that differences in the chemical shifts (center of doublet) between aligned and unaligned conditions arise owing to a combination of RCSA contributions and different lock frequencies as a result of quadrupolar splitting of the D2O signal in the aligned state. RDCs are computed from the differences in these values and, together with the idealized A-form PDBs, are used to compute order tensors for each helix. (b) The helix order tensor frames (Szz, Syy, Sxx) are superimposed to yield the relative orientation of helices subject to a 4n−1-fold degeneracy arising owing to allowed 180° inversions about the principal Szz, Syy and Sxx directions78. Information about interhelix motions is obtained from the relative ratios of the generalized degree of order (ϑ) obtained for each helix (ϑi /ϑj = ϑint; ϑi <ϑj). The ϑint value ranges between 0 for maximum interhelix motions and 1 for interhelix rigidity. Owing to motional couplings, the ϑint value will generally underestimate the real amplitude of interhelical motions.
Experimental design
Long-range orientational information from RDCs
RDCs arise owing to incomplete averaging of the dipolar interaction in partially aligned molecules16,17,39. They report on the orientation of bond vectors relative to the applied magnetic field, and specifically the time-averaged function , where θ is the angle between the bond vector and the magnetic field (Fig. 1). RDCs provide a straightforward approach for obtaining information regarding the relative orientation and dynamics of molecular fragments particularly when their local conformation and specifically the orientation of RDC-targeted bond vectors are known a priori (Fig. 2a) (see refs. 13,40,41).
In this protocol, RNA fragments consist of two or more nonterminal contiguous hydrogen-bonded WC base pairs (Fig. 2a).
Recently, we conducted a statistical survey23 of 421 such WC base pairs derived from 40 unbound and bound RNA X-ray structures (solved with <3 Å resolution) and the 2.4 A° X-ray structure of the ribosome42. The results showed that the local conformation of such WC base pairs can be modeled a priori using a standard idealized A-form helix geometry for analyzing RDC accu-rately32,33 (Fig. 2a). These WC base pairs can be experimentally identified/verified using NOESY connectivity and trans-hydrogen bond JNN-COSY type NMR experiments for directly detecting N–H–N hydrogen bonds43,44 (Fig. 2a). WC base pairs that are flanked by G-U pairs or non-canonical motifs can also be used although higher levels of structural noise need to be considered in the analysis (C.M., K.G. & H.M.A., unpublished findings).
With the local conformation of WC base pairs in hand, the measurement of more than five independent RDCs per helix allows determination of five elements of an order or alignment tensor16,45. The order tensor describes the average alignment of each helix relative to the applied magnetic field. Three Euler angles specify a helix-fixed order tensor frame (Sxx, Syy, Szz) that describes the average orientation of helices relative to the applied magnetic field. The average orientation of fragments—one relative to the other—can be obtained by superimposing their order tensor frames (Fig. 2b) (see refs. 24,46,47). The latter statement amounts to insisting that helical fragments share, on average, a common view of the magnetic field direction when assembled into a proper structure—similar to how countries in a properly assembled map report a common compass bearing. Two additional principal order tensor parameters describe the degree
and asymmetry
of helix alignment24,47. These parameters can be compared for various helices to obtain information about relative helix motions over sub-millisecond timescales47 (Fig. 2b). While helices will report identical parameters when they are held rigid relative to one another, interhelix motions can lead to differences. Specifically, the ϑ value for a given helix will be attenuated relative to the value observed for a helix that more strongly dominates total alignment, with the degree of attenuation generally increasing with motional amplitudes. Although often difficult to determine reliably, the asymmetry parameter (η) can provide insight into the directionality of interhelix motions with spatially isotropic (directionless) motions having a smaller effect on the relative helix η values compared to anisotropic (directional) motions24,47.
Although an idealized A-form geometry is assumed for the WC base pairs, structural deviations can arise and this uncertainty (referred to as “structural noise”48) must be propagated into the RDC-derived order tensor parameters and ultimately the relative orientation and dynamics of helices. To this end, the statistical survey of RNA X-ray structures was used to parameterize standard angular deviations in base pair and base pair step angles (buckle (κ), propeller (ω), opening (σ), incline (η), (tip (θ), twist (Ω) and sugar torsions (υ0–υ4)) relevant for the analysis of one bond C–H and N–H RDCs (Fig. 2a) (ref. 23). The effects of A-form structural noise as well as RDC measurement uncertainty are taken into account in the determination of order tensors in the program AFORM-RDC available from the corresponding author23. Other more general approaches for dealing with structural noise in the determination of alignment tensors have also been described48.
Limits of applicability and practical considerations
There are three main considerations in implementing the presented protocol.
First, how many RDCs are needed per helical fragment to carry out the order tensor analysis? A minimum of five spatially independent (i.e., non-parallel) RDCs are strictly needed to solve for the five order tensor parameters. However, in practice, satisfying this condition requires the measurement of a larger number of RDCs–at least 8 one-bond C–H and N–H RDCs—in both sugar and base moieties—will ideally yield a spatial distribution defined by a condition number (CN) <5 (refs. 21,47). For ≥11 RDCs with CN<5, A-form structural noise and typical RDC uncertainty (~1.5 Hz) is expected to lead to average errors in the magnitude and orientation of the principal axis of order that are <9% and <4°, respectively23. The errors decrease to <5% and <4° for ≥17 RDCs (ref. 23). The choice of RDCs to be measured is guided by the desire to maximize the magnitude: precision of measurement ratio and spatial distribution of the targeted vectors. The most optimum and commonly targeted RDCs are those between directly bonded C–H and N–H nuclei, which yield the largest RDC magnitudes (Fig. 3a). Additional one-, two- and three-bond RDCs can also be measured (Fig. 3b) using some of the pulse sequences listed in Table 1. The latter RDCs are smaller and may prove difficult to measure in larger RNAs (>60 nt). As long as more than eight RDCs have been measured with CN<5, the protocol implemented in AFORM-RDC will yield faithful estimates of the order tensor error due to both structural noise and RDC uncertainty23. Additionally, although not discussed here, it should also be possible to include nucleobase residual chemical shift anisotropies (RCSAs), which can be measured abundantly in larger RNAs owing to favorable TROSY effects49–52.
Figure 3.
Typical RDCs measured in base and sugar moieties of RNA using the pulse sequences listed in Table 1. (a) One-bond C–H and N–H RDCs are the most commonly targeted interactions owing to their favorable size but smaller one-bond C–C and C–N as well as (b) two- and three-bond RDCs can be measured. The motionally non-averaged C–H and N–H bond lengths used in the order tensor analysis are N1/3–H1/3 = 1.01 Å, C–Hbase = 1.08 Å, C–Hribose = 1.09 Å (ref. 100). All other bond lengths can be obtained from ref. 101.
TABLE 1.
Pulse sequences for measuring RDCs in nucleic acids.
| Pulse sequence | Reference | Type of RDCs | Comments |
|---|---|---|---|
| HC[C](C) hd-TROSY-E.COSY | 61 |
1DC2H2, 1DC5H5, 1DC6H6, 1DC8H8, 1DC4C5, 1DC5C6, 2DC5H6, 2DC6H5 and 2DC4H5 |
Pseudo-3D experiments for homonuclear decoupling employing TROSY and E.COSY elements. Demonstrated on a 24-nt RNA at 25 °C |
| CH2-S3E HSQC | 104 |
1D(C5′H5′+ c5′H5″) and 2D(H5′H5″) (in DNA only 1D(C2′H2″+C2′H2″) and 2D(H2′H2″) |
2D experiments with spin-state selection for detection of up- or downfield carbon components of CH2 spin states. Demonstrated on a 24-nt RNA at 25 °C |
| 3D S3CT E.COSY | 105 |
1DC4′H4′, 2DC5′H4′1D(C5′H5′+C5′H5″), (1D C5′H5′[′]−2DH5′H5″), 2DC4′H5′+C4′H5″ and 3DH4′H5′[′] |
3D experiments for measuring RDCs in methine-methylene C–H pairs. One experiment yields eight splittings. Demon- strated on 24-nt RNA at 25 °C |
| H1C1C2 E.COSY | 106 |
1DC1′H1′, 1DC2′H2′, 2DC1′H2′, 2DC2′H1′ and 1DH1′H2′ |
3D experiment utilizing E.COSY for measuring five splittings in one experiment. Demonstrated on a 24-nt RNA at 25 °C |
| IPAP HN-HSQC, IPAP H(N)C-HSQC |
107 |
1DN1H1, 1DN3H3, 2DH1C2, 2DH1C6, 2DH3C2 and 2DH3C4 |
2D experiments yielding 1–2 couplings per experiment. Demonstrated on ubiquitin. |
| 3D IPAP-HCcH-COSY 3D relay-HCcH-COSY |
108 | 1DC2′H2′ and 1DC3′H3′ | Uses C1′H1′to alleviate spectral overcrowding in the C2′H2′and C3′H3′region. Demonstrated on a 42-nt RNA at 25 °C |
| MQ-HCN | 109 |
1DC1′H1′, 1DC1′N1/N9, 1DC1′C2′, 2DH1′N1/9, 2DH1′C2′, 2DH1′N1/9, 1DC6H6, 1DC6N1, 1DC6C5, 1DC8H8, 1DC8N9, 2DH8N9, 2DH6N1 and 2DH6C5 |
Suite of six MQ based 3D experiments. One to two splittings per experiment. Demonstrated on a 36-nt DNA in a 47 kDa complex |
| S3E IS[T] | 110 | 1D and 2D | 2D experiments for measuring most of the one- and two- bond splittings. Demonstrated on 24-nt DNA at 15 °C |
| 13C-1H TROSY | 111 | 1DC2H2, 1DC5H5, 1DC6H6 and 1DC8H8 | Sensitivity enhanced using TROSY and native 13C magne- tization. Demonstrated on 15% randomly 13C-labeled 33-nt RNA at 25 °C |
| 3D MQ/TROSY-HCN-QJ | 112 |
1DC1′N9, 1DC8N9, 1DC4N9, 1DC1′N1, 1DC6N1 and 1DC2N1 |
3D quantitative J-modulated experiments for measuring one-bond C–N splittings. Demonstrated on a 24-nt RNA at 8 °C |
Second, the order tensor analysis of RDCs assumes that one fragment dominates overall molecular alignment of the RNA24,47,53,54. This “decoupling limit” (i.e., when individual helix motions are decoupled from global motions) is readily satisfied when one helix dominates alignment or when helices are held rigid relative to one another. In general, the alignment of polyanionic nucleic acids in neutral or negatively charged ordering media will be governed by the anisotropy of the overall molecular shape. For A-form helical fragments longer than approximately 7 bp, in which the helix length exceeds the diameter, the structural anisotropy and thus the degree of order are expected to increase with helix length. Thus, longer helical fragments can be expected to dominate overall alignment.
Additionally, two other regimes can be identified when helices are flexible. In the extreme coupling limit, helices have similar size and shape and contribute equally to total alignment. If motions of the helices—one relative to the other—result in equivalent changes in total alignment, then similar degrees of helix order will be observed regardless of interhelical motions54. In such cases, observation of ϑint values equal to one does not rule out the presence of interhelix motions. Note, however, that depending on the interhelix motional trajectory, inequivalent ϑ’s can be observed for helices even if they have equivalent size and shape. For example, twisting around the axis of a given helix will result in a reduction of its ϑ without affecting the ϑ value observed in another helix. In the intermediate coupling limit, which is the most common scenario, one helix partially (but not completely) dominates total alignment. Here, the derived motional amplitudes will underestimate the real motional amplitudes24. Simulations (data not shown) as well as experimental results show that differences on the order of three base pairs can be sufficient to take an RNA system outside the extreme coupling limit and into the intermediate regime21,22,24,38,54,55. Extensive elongation of helices using isotopic labeling strategies that render elongation residues invisible can also be used to bring the RNA system closer to within the decoupling limit56.
Finally, note that the order tensor analysis assumes that local fluctuations are similar in magnitude for the WC base pairs in various helical segments. A survey of NMR relaxation studies and analysis of a molecular dynamics simulation support this assumption for WC base pairs flanking other WC base pairs23. Nevertheless, it will be important to independently establish the structural stability of WC base pairs, as noted earlier, by using NOESY connectivity and JNN-COSY experiments for detecting hydrogen bond alignments43,44 (Fig. 2a). Note that a greater degree of local motions may arise in WC base pairs flanking non-canonical motifs23.
MATERIALS
REAGENT SETUP
13C/15N-labeled RNA sample (typically >0.2 mM) in NMR buffer
The protocol assumes that 1H, 13C, 15N resonances in A-form helices have been assigned using established methods27–29. The experiments are carried out on uniformly (or residue-specific) 13C/15N-labeled RNA samples (typically >0.2 mM).
Ordering medium
All of the ordering media listed in Table 2 have been used to align nucleic acids. The most popular and commonly used ordering medium is Pf1 phage57. Relative to other media, Pf1 phage is tolerant to the high salt concentrations used in nucleic acid samples and is negatively charged, thus reducing the possibility of adverse intermolecular interactions. Pf1 phage is available commercially or can be prepared using the methods described in ref. 57. Typically, a Pf1 solution is exchanged into the NMR buffer by repeated (at least three) rounds of ultracentrifugation (1 h at 4 °C in a Beckman TLA-100.3 rotor at 475,000g, or 3–6 h in a VTi50 rotor at 200,000g) followed by resuspension of pellet into the NMR buffer. Alternatively, one can dialyze Pf1 phage into the desired buffer. After completing the aligned experiments, the same ultracentrifugation procedure can in principle be used to recover the nucleic acid (supernatant) from the phage solution (pellet). Note that it will generally be difficult to achieve perfect separation of the nucleic acid sample from the phage medium.
TABLE 2.
Alignment media used in studies of nucleic acids.
| Medium | Reference | Temp. range (°C) | Features and limitations |
|---|---|---|---|
| DMPC:DHPC (“Bicelles”) | 113,114 | 27–45 | Perpendicular alignment disc-like shape. Sensitive to ionic conditions |
| Rod-shaped viruses (Pf1 phage and TMV) |
57,63,64 | 5–60 | Parallel alignment rod-like shape. Negatively charged. Most widely used |
| Purple membrane | 115,116 | − 269 to 69 | Parallel alignment disc-like shape. Stable in the pH range 2.5–10, and salt concentrations up to 5 M |
| Polyacrylamide gels | 117,118 | 5–45 | Mechanical gel. Very stable and inert |
|
n-Alkyl-ply(ethylene glycol)/n-alkyl alcohol or glucopone/n-hexanol (PEG) |
119,120 | 0–40 | Perpendicular alignment lamellar shape. Insensitive to pH, and moderately sensitive to salt concentrations |
NMR buffer
Sample conditions, including pH, temperature, monovalent and divalent ion concentrations as well as the RNA refolding procedure may have to be optimized at the onset of an investigation to ensure proper folding of the RNA. Imino proton NMR spectra are a convenient way to probe hydrogen bond alignments and thus the RNA secondary structure. Typically, one expects a single imino proton resonance for every guanine and uridine base involved in N–H— X hydrogen bonding. In general, fast cooling of dilute (~10 µM) RNA samples in low salt (desalted buffer) favors hairpin structures, whereas slow cooling of more concentrated NMR samples in high salt favors formation of duplexes29. The standard buffer used in our laboratory consists of 15 mM sodium phosphate, pH ~6.4, 0.1 mM EDTA and 25 mM sodium chloride.
EQUIPMENT SETUP
NMR spectrometer equipped with a triple resonance probehead. Computer programs used for the analysis of RDCs (A-form-RDC and RAMAH) and calculation of interhelical angles (Euler-RNA(A-form)) can be obtained by emailing H.M.Al-Hashimi (hashimi@umich.edu) or by downloading them from the website http://www-personal.umich.edu/~hashimi/.
PROCEDURE
Experimental validation of predicted WC base pairs
-
1|
Divide the RNA secondary structure into constituent helical stems and identify contiguous WC base pairs that are flanked by other WC base pairs (Fig. 2a).
-
2|
Use imino proton line widths, NOE and trans-hydrogen bond (e.g., JNN-COSY)43,44 connectivity to validate hydrogen bonding in the target WC base pairs (Fig. 2a).
Δ CRITICAL STEP Although not expected, WC base pairs exhibiting severely exchange-broadened imino proton signals and/or unusually weak NOEs and trans-hydrogen bond connectivity should be excluded from analysis even if they meet the criteria of being flanked by other WC base pairs.
Measurement of RDCs
-
3|
Record NMR experiments for measuring splittings (Fig. 3) in the unaligned RNA sample using the pulse sequences listed in Table 1.
-
4|
Process NMR spectra and measure splittings.
! CAUTION In frequency domain experiments, phase distortions owing to improper calibration of timing delays and/or shaped pulses can yield splitting measurement errors that are larger than theoretical limits (approximately given by 0.7*Linewidth* (1/signal:noise)) (see ref. 58).
Δ CRITICAL STEP The unaligned splittings correspond to isotropic scalar couplings (J) plus contributions (typically <1–2 Hz for C–H bonds in RNAs <40 nt at 600 MHz) from magnetic field-induced RDCs (Dfield) and dynamic frequency shifts (DFS). The value of Dfield increases quadratically with the magnetic field strength (i.e., with B2). DFS are contributions to chemical shifts and scalar couplings arising from the imaginary component of the spectral density function for cross-correlation between dipolar and CSA relaxation mechanisms59. At magnetic fields above 400 MHz, the DFS contribution to the apparent splittings is nearly constant (within 0.1 Hz) (see ref. 60). At 600 MHz, it is computed to be −1.2 and 0.6 Hz for directly bonded C–H and N–H bonds in the nucleobases of an RNA molecule tumbling with an overall correlation time of 6 ns. These DFS contributions will nearly perfectly cancel out when computing RDCs from the difference in splittings observed in the aligned and unaligned states particularly when using the same magnetic field to measure aligned and unaligned samples (>400 MHz). Typical values for isotropic splittings are provided in ref. 61.
-
5|
Align RNA sample by dissolution into an appropriate ordering medium62 (Table 2). Pf1 phage is the most popular commercially available medium for aligning nucleic acids57,63.
For small to moderately sized RNAs (25–50 nt), 18–25 mg ml−1 phage will generally yield an optimal level of alignment (~20 Hz maximum N–H) (ref. 64). Typically, we add a preconcentrated RNA solution (~0.5–1.5 mM) in NMR buffer to a desired volume of Pf1 phage (50 mg ml−1) in NMR buffer in an Eppendorf tube and then gently transfer the phage/RNA solution into the NMR tube avoiding bubble formation. Other media that have been used to align nucleic acids are shown in Table 2.
It is important to verify that the ordering medium does not interfere with the RNA conformation by comparing chemical shifts obtained in the unaligned and aligned samples. Note that small variations in the chemical shifts of nucleobase carbons and nitrogens are expected between unaligned and aligned samples owing to incomplete averaging of RCSAs50,51,65. These RCSA contributions scale linearly with the magnetic field and degree of order. Typical values are shown in Table 3.
RNA molecules can also be aligned spontaneously (magnetic field-induced alignment) owing to their large magnetic susceptibility (χ) tensor arising from constructive summation of the individual nucleobase χ-tensors66–68. The degree of alignment scales quadratically with the magnetic field strength for typical RNA samples (~40 nt)—at 900 MHz, the degree of alignment is ~3- to 6-fold smaller than the optimum level obtained by using ordering media.
! CAUTION The concentration of ordering medium needed for optimum alignment may be significantly smaller for much larger RNAs (» 50 nt). If a model structure for the RNA is available, programs for predicting steric alignment69 can be used to assess relative levels of order and the ordering medium concentration adjusted accordingly. In addition, the RNA concentration suitable for a given ordering medium may be substantially smaller for larger RNAs.
? TROUBLESHOOTING
-
6|
Use the same NMR experiments to measure splittings in the aligned sample. Subtract the value of splittings measured in aligned (J + D) and unaligned (J) samples to compute RDCs (Fig. 2a).
! CAUTION To avoid differential contributions from magnetic field-induced RDCs and relaxation interference effects, splittings in unaligned and aligned samples should be measured at the same magnetic field strength.
Δ CRITICAL STEP It is advisable to estimate the experimental RDC uncertainty from the standard deviation in duplicate measurements. Resonances exhibiting significant differences (>3σ) as a result of considerable broadening, overlap, presence/ absence of unresolved multiplets should be discarded unless a weighted fit is carried out in the order tensor determination. Depending on the RNA sample, we typically obtain σ values ranging between 0.5 and 3 Hz for one-bond C–H RDCs.
TABLE 3.
Changes in the chemical shifts of protonated carbons and nitrogens upon alignment owing to RCSA contributions assuming 1.0 × 10−3 degree of order. Corresponding shifts for attached protons are ±8 ppb.
| Atom | Base (ppb) | Sugar (ppb) | ||||
|---|---|---|---|---|---|---|
| 2/5/6/8 | 1′ | 2′ | 3′ | 4′ | 5′ | |
| 13C | ±170 | ±23 | ±20 | ±67 | ±67 | ±40 |
| 15N | ±100 | NA | NA | NA | NA | NA |
Order tensor analysis
-
7|
Build idealized A-form helices (PDB files) corresponding to the sequence of the targeted WC base pairs in each helix (Fig. 2a). For example, to build an A-form helix using the Biopolymer module of Insight II 2000.1 (Molecular Simulations Inc.), click on the module icon in the upper left corner and select append from the nucleotide menu. In the pop-up box, select “A_RNA_Duplex”. Input a name for the molecule into the text field. Next, select the appropriate WC base pair in the Nucleotide text field. Continue to append base pairs—following along in sequence from 5’ to 3’—until you have completed building the desired helix. Click cancel and then select the File menu and choose the desired export option for the helix coordinates.
! CAUTION The idealized A-form helices should conform to published parameters23,32,33. If building helices using INSIGHT II 2000.1 (Molecular Simulations Inc.), care needs to be taken to correct the propeller twist angles to the proper value of −14.5°. Programs such as Curves 5.1 (ref. 70), FreeHelix98 (ref. 71), 3DNA (refs. 32,72), SCHNAaP73, NUPARM and NUCGEN74 can be used to compute relevant helix parameters.
-
8|
Compute five-order tensor elements for each A-form helix by fitting the RDCs in each case to the A-form PDB coordinates.
Δ CRITICAL STEP Several programs are available to carry out such calculations, including ORDERTEN-SVD46, REDCAT75, PALES69, iDC76, CONFORMIST77 and RAMAH51.
! CAUTION Non-ideal WC base pairs as identified in Step 2 are excluded from this analysis.
-
9|
Examine the correlation between measured and back-calculated RDCs. Major outliers should be interrogated for possible measurement errors.
! CAUTION For a small number of RDCs (<11), the major outliers may not necessarily correspond to the ‘bad’ data (but could instead be data points that exhibit a good fit). This underscores the need to independently identify poor RDC measurements early on in the analysis.
-
10|
Use AFORM-RDC23 or other approaches48 to estimate the order tensor error owing to structural noise and RDC measurement uncertainty.
Determining the average interhelix alignment
-
11|
Superimpose the best-fit order tensor frames determined for the various helices by rotating each helix into the principal axis system (PAS) of the computed best-fit order tensor. In the PAS frame, the principal Szz, Syy and Sxx directions are oriented along the z, y and x axes, respectively, of the molecular frame (Fig. 2b).
Δ CRITICAL STEP There are four ways for rotating helices into their order tensor PAS involving 180° rotations about the principal Sxx, Syy and Szz directions. Owing to this fourfold degeneracy, n helices can be assembled in 4n−1 distinct structures that satisfy measured RDCs78 (Fig. 2b). Half of these solutions can generally be eliminated because they do not satisfy the RNA secondary structure. The remaining twofold degeneracy typically involves rotations about the long axis of the RNA and must be eliminated based on bond connectivity considerations, other experimental restraints and/or by measuring RDCs in a second non-collinear alignment medium78.
Δ CRITICAL STEP Extended RNA conformations (e.g., involving coaxially stacked helices) will often have nearly axially symmetric (η~0) order/alignment tensors. In these cases, rotations of helices around the effective long axis (Szz) of the RNA molecule will be ill-defined. As a result, interhelix bend angles tend to be much better defined than corresponding interhelix twist angles.
? TROUBLESHOOTING
-
12|
Assemble the rotated helices into an overall RNA structure. Here, a molecular visualization program can be used to open and view the helices in their respective order tensor PAS frames. The helices are translated taking care not to rotate them away from their PAS frames and connected in such a way that satisfies experimental or geometrical restraints. For the example shown in Figure 4, the helices were translated to satisfy a direct phosphodiester linkage between the two helices (distance between O3’ atom of residue C39 in helix II and P atom of residue U40 in helix I is set to 1.58 Å).
-
13|
Translate the uncertainty in the order tensor PAS derived by the program AFORM-RDC23 or other approaches into uncertainty in the relative orientation of RNA helices48.
Δ CRITICAL STEP For the above, it is useful to use a convention for specifying the relative orientation of helices. Starting with a coaxial alignment for helices i and j with the helix axis oriented along the molecular z direction, the orientation of helix i relative to a reference helix j can be specified using three Euler angles (αh, θh, γh) that transform (through the rotation matrix Rij(αh, θh, γh)) helix i from an orientation that is perfectly coaxial with helix j to that observed in the RDC-derived interhelix structure. The angle αh defines a twist around the helix axis j, θh defines the interhelix bend and γh defines a twist around the helix axis i. The sum of αh + γh yields the interhelix twist angle (ξ). In this work, the 3′ helices are used as the reference and positive angles refer to anticlockwise rotation of the molecular frame (or clockwise rotation of the object). Thus, positive and negative interhelix twist angles (ξ) correspond to over- and undertwisting, respectively. Note R(αh, θh, γh) = R(αh ± 180°, – θh, γh ± 180) = R(αh ± 180° –θh γh ± 180°) and Rji = Rij−1 = R (−γh – θh −αh). The rotation matrix Rij can be expressed in terms of the rotation matrices () that diagonalize the helix i and j order tensor, respectively, obtained when using the coaxial helix molecular frame. The relation, , can be used to propagate the experimental error in estimated using A-form-RDC into an error in the structural parameters defined by Rij.
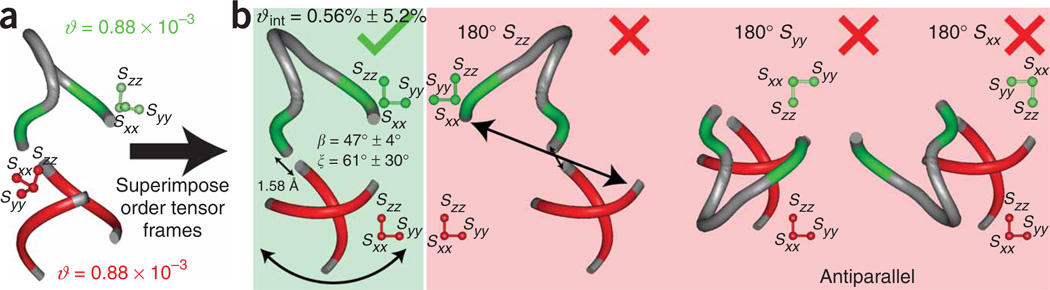
Figure 4.
Example illustrating application of protocol in the determination of the relative orientation and dynamics of two helices in the free state of HIV-1 TAR RNA. (a) Idealized A-form helices are used to determine order tensors for each helix in TAR RNA, with 18 (12 base, 6 sugar) and 22 (13 base, 9 sugar) one-bond C–H RDCs used in the analysis of helices I and II, respectively. (b) Superposition of the experimentally determined order tensor frames yields one of four solutions for the relative orientation of helices. Three additional degenerate solutions (180° Szz, 180° Syy, 180° Sxx) are generated by subsequent rotation of a given helix (in this case helix II) by 180° about each of the three helix II principal axes Szz, Syy and Sxx, respectively. In each case, the helices are translated/assembled by setting the distance between 03′ of residue 39 and P of residue 40 equal to 1.58 Å. The 180° Szz solution is discarded because it yields a distance between 03′ of residue 22 and P of residue 40 that is long to be satisfactorily connected by a trinucleotide bulge, whereas solutions 180° Syy and 180° Sxx are discarded because they lead to an antiparallel helix alignment that is inconsistent with the TAR secondary structure.
Characterizing interhelix motions
-
14|
The amplitude of motions between helices i and j can be obtained from the ratios of their respective ϑs obtained from the order tensor analysis, ϑint = ϑ (i) / ϑ (j), (ϑ (i)<ϑ (j)) (ref. 47) (Fig. 2b). The ϑint value ranges between 1 for interhelix rigidity and 0 for maximum interhelix motions.
! CAUTION A ϑint<1 implies interhelix motions but the motional amplitudes are likely to be underestimated by couplings between helix motions and overall alignment24,54. In contrast, ϑint~1 implies either interhelix rigidity or that interhelix motions evade detection owing to motional couplings. Helix elongation can be used to resolve such ambiguities56.
• TIMING
Following resonance assignments, the procedure is expected to take no more than 2 weeks of acquisition and data analysis time.
? TROUBLESHOOTING
Step 5
If a precipitate forms, try to dilute your RNA concentration with a solution of phage at the desired concentration in your NMR buffer.
Step 11
It may be possible to eliminate a degenerate orientation based on an inability of each structure to correctly back-predict the total observed alignment tensor.
ANTICIPATED RESULTS
As an example, the protocol is applied to determine the relative orientation and dynamics of two helices in the free state of the HIV-1 transactivation response element (TAR). The two TAR helices are linked by a trinucleotide pyrimidine bulge24. A total of 18 (12 base, 6 sugar) and 22 (13 base, 9 sugar) one-bond C–H RDCs were measured in helices I and II, respectively, using ~22 mg ml−1 of Pf-1 phage ordering medium24. The RDCs were used to determine order tensors for each helix using the program RAMAH. The order tensor frames and degree of order for each helix are shown in Figure 4. The large difference between the helix ϑ’s implies the presence of interhelix motions. The error bars shown reflect a combination of RDC measurement uncertainty and A-form structural noise as implemented in the program A-form-RDC. The motional amplitudes are given by ϑint = ϑhelix I /ϑhelix II = 0.56 ± 5.2% (ref. 24). Superposition of the helix order tensor frames yields four possible solutions for the relative orientation of the two helices. Following an initial superposition, three additional solutions are generated by rotation of one helix (helix II) relative to the other 180° about the Sxx, Syy and Szz directions, respectively. The helices are translated to satisfy a direct phosphodiester linkage between the two helices (distance between 03’ atom of residue C39 in helix II and P atom of residue U40 in helix I is set to 1.58 Å). Two solutions (Sxx and Syy) are omitted as they lead to antiparallel helix orientations that are inconsistent with the TAR secondary structure. The third solution (Szz) is omitted because it leads to a distance between the 03’ ribose oxygen of residue 22 and the backbone phosphorous of residue 26 that cannot be satisfactorily linked by the trinucleotide bulge. The overall free TAR conformation is thus described by interhelix bend (β) and twist (ξ) angles of 47° ± 4° and 61° ± 30°, respectively, and a high degree of interhelix flexibility (ϑint = 0.56 ± 5.2%).
In general, the relative orientation and dynamics of RNA helices will depend on sequence/structural context, temperature and pH, as well as presence/absence of metals and bound protein/ligand molecules. Application of the presented protocol is beginning to illuminate salient trends and detailed relationships between interhelical bend, twist and flexibility, building on observations obtained previously by gel mobility measurements25,79,80 and transient electric birefringence25,26. In what follows, we summarize the results obtained thus far noting key trends and their possible explanation.
Figure 5a shows the relative orientation and dynamics of helices observed in three different RNA contexts in the absence and presence of Mg2+ ions. This includes HIV-1 TAR, the RNase P P4 helix containing a single pyrimidine bulge nucleotide21, and HIV-1 SL1 containing a purine-rich asymmetric four-nucleotide internal loop22. In Figure 5b, we show corresponding results for HIV-1 TAR bound to Mg2+ and four different small molecules containing a different number of cationic groups.
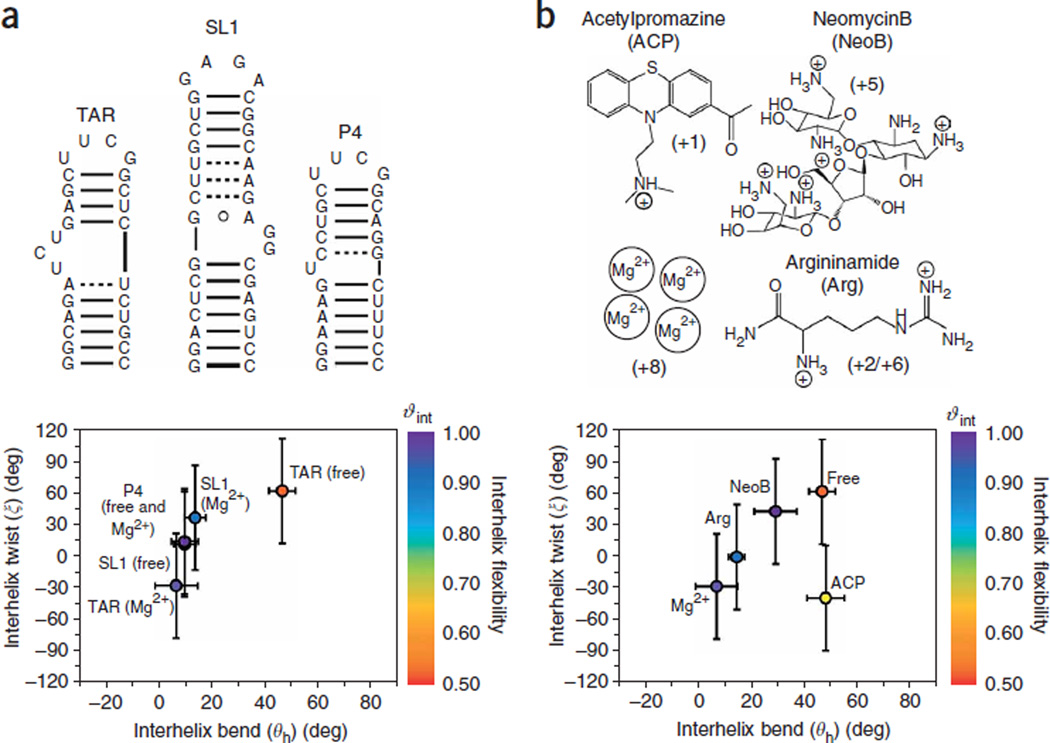
Figure 5.
The relative orientation (interhelix bend, θh, and twist angles, ξ) and dynamics (ϑint) of RNA helices obtained from order tensor analysis of RDCs under different contexts. (a) Three different RNA secondary structures (TAR, SL1, P4) in the presence and absence of Mg2+ (see refs. 21,22,87). (b) TAR RNA in four distinct ligand-bound forms87,96,97. The ϑint values are color-coded on each point. The large uncertainty in the interhelix twist angles (set at ± 50° (see ref. 102)) arises from near axial symmetry (η~0) of the order tensor. The net charge delivered by each small molecule upon binding is shown in parentheses. Although the ligand arginanamide (Arg) has a +2 charge, studies103 show that up to three molecules can bind to TAR contributing a total +6 charge. For Mg2+, the charge of +8 is shown based on the observation of four bound Ca2+ ions in the X-ray structure of TAR89.
A general trend is observed between the degree of interhelical bending (θ), helical overtwisting (ξ) and interhelical flexibility (ϑint) (Fig. 5). The greater the bend angle, the greater the degree of overtwisting and interhelix flexibility. This behavior can be understood in terms of the stacking and electrostatic interactions at the bulge/internal loop that dictate the resulting orientation and dynamics of juxtaposed helices. Bulges and asymmetric internal loops induce interhelical bending and overtwisting for two main reasons25,79,80. First, by extending the bulge/internal loop conformation, interhelical bending alleviates electrostatic charge repulsion that would otherwise build up in coaxial structures owing to spatial confinement of bulge/internal loop phosphates. Second, interhelical bending accommodates looped in conformations allowing favorable bulge/internal loop stacking interactions. It is the stacking interactions of bulge/internal loop residues that account for the observed helical overtwisting. In the absence of tertiary contacts that stabilize the relative orientation of helices, the degree of interhelical bending, overtwisting and interhelix flexibility is expected to increase with the length of the bulge26,79,81–84, as is observed when comparing TAR and P4 both of which contain pyrimidine bulges (Fig. 5a). Unopposed asymmetric bulges are expected to give rise to greater bending/flexibility compared to opposed symmetric internal loops that may have compensating bending effects26,79,81–84, as observed when comparing TAR and SL1 (Fig. 5a).
By screening unfavorable electrostatic charge repulsion in and around the bulge/internal loops, divalent and monovalent ions (or small molecule containing positive groups) can help stabilize coaxial helical conformations25,26,85–88. However, this will often require the looping out of bulge/internal loop residues. Thus, the energetic gains owing to favorable coaxial helical stacking and metal binding have to offset the unfavorable loss of stacking interactions in bulge internal/loop residues. In the case of TAR, Mg2+ binding induces a large structural transition toward a rigid coaxial interhelical conformation (Fig. 5a) (see ref. 87). This transition is accompanied by looping out of the otherwise stacked nucleobases of pyrimidine bulge residues U23 and C24 (see refs. 87,89). In contrast to TAR, Mg2+ binding has an insignificant effect on the P4 conformation for which favorable coaxial helical stacking and looping out of the uridine bulge is already observed in the absence of Mg2+ (Fig. 5a) (ref. 21). Smaller conformational effects are also seen for SL1, which contains a purine-rich internal loop (Fig. 5a) (ref. 22).
Previous NMR studies have shown that TAR RNA undergoes conformational rearrangements upon binding to small molecule therapeutics bearing a different number and spatial arrangement of cationic groups90–95. Figure 5b shows these TAR conformational transitions as visualized through application of the presented RDC protocol24,87,96,97. Interestingly, one finds that molecules that contribute a larger number of cationic groups tend to stabilize more linear and rigid TAR conformations (Fig. 5b)—in analogy to the trend observed when adding Mg2+ metals (Fig. 5a) (see refs. 97,98).
The above illustrates the diversity of conformations that can populate the RNA structural landscape and the possibility for systematic characterization using the described protocol. In the future, we expect applications to more complex RNA contexts, including those involving long-range tertiary contacts and interhelical linkers composed of pseudoknots and junctions under a wide range of physiologically relevant conditions.
ACKNOWLEDGMENTS
We thank members of the Al-Hashimi laboratory for insightful comments and Dr. Alex Kurochkin for his expertise and for maintenance of the NMR instruments. H.M.A. acknowledges fruitful collaborations with the groups of Carol Fierke (The University of Michigan) and Ioan Andricioaei (The University of Michigan). We gratefully acknowledge the Michigan Economic Development Cooperation and the Michigan Technology Tri-Corridor for the support in the purchase 600 MHz spectrometer. This work was supported by funding from the NIH (RO1 AI066975-01).
Footnotes
COMPETING INTERESTS STATEMENT The authors declare no competing financial interests.
Reference
- 1.Williamson JR. Molecular biology—small subunit, big science. Nature. 2000;407:306–307. doi: 10.1038/35030271. [DOI] [PubMed] [Google Scholar]
- 2.Leulliot N, Varani G. Current topics in RNA-protein recognition: control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- 3.Al-Hashimi HM. Dynamics-based amplification of RNA function and its characterizationbyusing NMR spectroscopy. ChemBioChem. 2005;6:1506–1519. doi: 10.1002/cbic.200500002. [DOI] [PubMed] [Google Scholar]
- 4.Micura R, Hobartner C. On secondary structure rearrangements and equilibria of small RNAs. ChemBioChem. 2003;4:984–990. doi: 10.1002/cbic.200300664. [DOI] [PubMed] [Google Scholar]
- 5.Williamson JR. Assembly of the 30S ribosomal subunit. Q. Rev. Biophys. 2005;38:397–403. doi: 10.1017/S0033583506004264. [DOI] [PubMed] [Google Scholar]
- 6.Schroeder R, Barta A, Semrad K. Strategies for RNA folding and assembly. Nat. Rev. Mol. Cell Biol. 2004;5:908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- 7.Mandal M, Breaker RR. Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 8.Grundy FJ, Henkin TM. Regulation of gene expression by effectors that bind to RNA. Curr. Opin. Microbiol. 2004;7:126–131. doi: 10.1016/j.mib.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 9.Hermann T. Rational ligand design for RNA: the role of static structure and conformational flexibility in target recognition. Biochimie. 2002;84:869–875. doi: 10.1016/s0300-9084(02)01460-8. [DOI] [PubMed] [Google Scholar]
- 10.Tor Y. Targeting RNA with small molecules. ChemBioChem. 2003;4:998–1007. doi: 10.1002/cbic.200300680. [DOI] [PubMed] [Google Scholar]
- 11.Vicens Q, Westhof E. RNA as a drug target: the case of aminoglycosides. ChemBioChem. 2003;4:1018–1023. doi: 10.1002/cbic.200300684. [DOI] [PubMed] [Google Scholar]
- 12.D’Souza V, Dey A, Habib D, Summers MF. NMR structure of the 101-nucleotide core encapsidation signal of the Moloney murine leukemia virus. J. Mol. Biol. 2004;337:427–442. doi: 10.1016/j.jmb.2004.01.037. [DOI] [PubMed] [Google Scholar]
- 13.Bax A, Grishaev A. Weak alignment NMR: a hawk-eyed view of biomolecular structure. Curr. Opin. Struct. Biol. 2005;15:563–570. doi: 10.1016/j.sbi.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 14.MacDonald D, Lu P. Residual dipolar couplings in nucleic acid structure determination. Curr. Opin. Struct. Biol. 2002;12:337–343. doi: 10.1016/s0959-440x(02)00328-7. [DOI] [PubMed] [Google Scholar]
- 15.Mollova ET, Hansen MR, Pardi A. Global structure of RNA determined with residual dipolar couplings. J. Am. Chem. Soc. 2000;122:11561–11562. [Google Scholar]
- 16.Tjandra N, Bax A. Measurement of dipolar contributions to (1)J(CH) splittings from magnetic-field dependence of J modulation in two-dimensional NMR spectra. J. Magn. Reson. 1997;124:512–515. doi: 10.1006/jmre.1996.1088. [DOI] [PubMed] [Google Scholar]
- 17.Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. Nuclear magnetic dipole interactions in field-oriented proteins—information for structure determination in solution. Proc. Natl. Acad. Sci. USA. 1995;92:9279–9283. doi: 10.1073/pnas.92.20.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williamson JR. Induced fit in RNA-protein recognition. Nat. Struct. Biol. 2000;7:834–837. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
- 19.Lilley DMJ. The Varkud satellite ribozyme. RNA. 2004;10:151–158. doi: 10.1261/rna.5217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dickerson RE, Goodsell DS, Kopka ML, Pjura PE. The effect of crystal packing on oligonucleotide double helix structure. J. Biomol. Struct. Dyn. 1987;5:557–579. doi: 10.1080/07391102.1987.10506413. [DOI] [PubMed] [Google Scholar]
- 21.Getz MM, Andrews AJ, Fierke CA, Al-Hashimi HM. Structural plasticity and Mg2+ binding properties of RNase P P4 from combined analysis of NMR residual dipolar couplings and motionally decoupled spin relaxation. RNA. 2006;13(2):251–266. doi: 10.1261/rna.264207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun X, Zhang Q, Al-Hashimi HM. Resolving fast and slow internal motions in the bulge containing stem-loop 1 of HIV-1 that are modulated by Mg2+ binding: role in the kissing-duplex structural transition. Nucleic Acids Res. 2007;35(5):1698–1713. doi: 10.1093/nar/gkm020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Musselman C, et al. Impact of static and dynamic A-form heterogeneity on the determination of RNA global structural dynamics using NMR residual dipolar couplings. J. Biomol. NMR. 2006;36:235–249. doi: 10.1007/s10858-006-9087-9. [DOI] [PubMed] [Google Scholar]
- 24.Al-Hashimi HM, et al. Concerted motions in HIV-1 TAR RNA may allow access to bound state conformations: RNA dynamics from NMR residual dipolar couplings. J. Mol. Biol. 2002;315:95–102. doi: 10.1006/jmbi.2001.5235. [DOI] [PubMed] [Google Scholar]
- 25.Zacharias M, Hagerman PJ. Bulge-induced bends in RNA—quantification by transient electric birefringence. J. Mol. Biol. 1995;247:486–500. doi: 10.1006/jmbi.1995.0155. [DOI] [PubMed] [Google Scholar]
- 26.Zacharias M, Hagerman PJ. The influence of symmetric internal loops on the flexibility of RNA. J. Mol. Biol. 1996;257:276–289. doi: 10.1006/jmbi.1996.0162. [DOI] [PubMed] [Google Scholar]
- 27.Varani G, Aboulela F, Allain FHT. NMR investigation of RNA structure. Prog. Nucl. Magn. Reson. Spectrosc. 1996;29:51–127. [Google Scholar]
- 28.Wijmenga SS, van Buuren BNM. The use of NMR methods for conformational studies of nucleic acids. Prog. Nucl. Magn. Reson. Spectrosc. 1998;32:287–387. [Google Scholar]
- 29.Furtig B, Richter C, Wohnert J, Schwalbe H. NMR spectroscopy of RNA. ChemBioChem. 2003;4:936–962. doi: 10.1002/cbic.200300700. [DOI] [PubMed] [Google Scholar]
- 30.Al-Hashimi HM, Patel DJ. Residual dipolar couplings: synergy between NMR and structural genomics. J. Biomol. NMR. 2002;22:1–8. doi: 10.1023/a:1013801714041. [DOI] [PubMed] [Google Scholar]
- 31.Al-Hashimi HM, Gorin A, Majumdar A, Gosser Y, Patel DJ. Towards structural genomics of RNA: rapid NMR resonance assignment and simultaneous RNA tertiary structure determination using residual dipolar couplings. J. Mol. Biol. 2002;318:637–649. doi: 10.1016/S0022-2836(02)00160-2. [DOI] [PubMed] [Google Scholar]
- 32.Olson WK, et al. A standard reference frame for the description of nucleic acid base-pair geometry. J. Mol. Biol. 2001;313:229–237. doi: 10.1006/jmbi.2001.4987. [DOI] [PubMed] [Google Scholar]
- 33.Neidle S. Oxford Handbook of Nucleic Acid Structure. New York: Oxford University Press; 1999. [Google Scholar]
- 34.McCallum SA, Pardi A. Refined solution structure of the iron-responsive element RNA using residual dipolar couplings. J. Mol. Biol. 2003;326:1037–1050. doi: 10.1016/s0022-2836(02)01431-6. [DOI] [PubMed] [Google Scholar]
- 35.Bondensgaard K, Mollova ET, Pardi A. The global conformation of the hammerhead ribozyme determined using residual dipolar couplings. Biochemistry. 2002;41:11532–11542. doi: 10.1021/bi012167q. [DOI] [PubMed] [Google Scholar]
- 36.Sibille N, Pardi A, Simorre JP, Blackledge M. Refinement of local and long-range structural order in theophylline-binding RNA using C-13-H-1 residual dipolar couplings and restrained molecular dynamics. J. Am. Chem. Soc. 2001;123:12135–12146. doi: 10.1021/ja011646+. [DOI] [PubMed] [Google Scholar]
- 37.Leeper TC, Athanassiou Z, Dias RLA, Robinson JA, Varani G. TAR RNA recognition by a cyclic peptidomimetic of Tat protein. Biochemistry. 2005;44:12362–12372. doi: 10.1021/bi0510532. [DOI] [PubMed] [Google Scholar]
- 38.Richards RJ, et al. Structural study of elements of Tetrahymena telomerase RNA stem-loop IV domain important for function. RNA. 2006;12:1475–1485. doi: 10.1261/rna.112306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bothner-By AA. In: Encyclopedia of Nuclear Magnetic Resonance. Grant DM, Harris RK, editors. Chichester: Wiley; 1995. pp. 2932–2938. [Google Scholar]
- 40.Prestegard JH, Al-Hashimi HM, Tolman JR. NMR structures of biomolecules using field oriented media and residual dipolar couplings. Q. Rev. Biophys. 2000;33:371–424. doi: 10.1017/s0033583500003656. [DOI] [PubMed] [Google Scholar]
- 41.Tolman JR, Ruan K. NMR residual dipolar couplings as probes of biomolecular dynamics. Chem. Rev. 2006;106:1720–1736. doi: 10.1021/cr040429z. [DOI] [PubMed] [Google Scholar]
- 42.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dingley AJ, Grzesiek S. Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide (2)J(NN) couplings. J. Am. Chem. Soc. 1998;120:8293–8297. [Google Scholar]
- 44.Pervushin K, et al. NMR scaler couplings across Watson-Crick base pair hydrogen bonds in DNA observed by transverse relaxation optimized spectroscopy. Proc. Natl. Acad. Sci. USA. 1998;95:14147–14151. doi: 10.1073/pnas.95.24.14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saupe A. Recent results in field of liquid crystals. Angew. Chem. Int. Edn. 1968;7:97–112. [Google Scholar]
- 46.Losonczi JA, Andrec M, Fischer MWF, Prestegard JH. Order matrix analysis of residual dipolar couplings using singular value decomposition. J. Magn. Reson. 1999;138:334–342. doi: 10.1006/jmre.1999.1754. [DOI] [PubMed] [Google Scholar]
- 47.Tolman JR, Al-Hashimi HM, Kay LE, Prestegard JH. Structural and dynamic analysis of residual dipolar coupling data for proteins. J. Am. Chem. Soc. 2001;123:1416–1424. doi: 10.1021/ja002500y. [DOI] [PubMed] [Google Scholar]
- 48.Zweckstetter M, Bax A. Evaluation of uncertainty in alignment tensors obtained from dipolar couplings. J. Biomol. NMR. 2002;23:127–137. doi: 10.1023/a:1016316415261. [DOI] [PubMed] [Google Scholar]
- 49.Pervushin K, Riek R, Wider G, Wuthrich K. Transverse relaxation-optimized spectroscopy (TROSY) for NMR studies of aromatic spin systems in C-13-labeled proteins. J. Am. Chem. Soc. 1998;120:6394–6400. [Google Scholar]
- 50.Ying JF, Grishaev A, Bryce DL, Bax A. Chemical shift tensors of protonated base carbons in helical RNA and DNA from NMR relaxation and liquid crystal measurements. J. Am. Chem. Soc. 2006;128:11443–11454. doi: 10.1021/ja061984g. [DOI] [PubMed] [Google Scholar]
- 51.Hansen AL, Al-Hashimi HM. Insight into the CSA tensors of nucleobase carbons in RNA polynucleotides from solution measurements of residual CSA: towards new long-range orientational constraints. J. Magn. Reson. 2006;179:299–307. doi: 10.1016/j.jmr.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 52.Grishaev A, Ying JF, Bax A. Pseudo-CSA restraints for NMR refinement of nucleic acid structure. J. Am. Chem. Soc. 2006;128:10010–10011. doi: 10.1021/ja0633058. [DOI] [PubMed] [Google Scholar]
- 53.Briggman KB, Tolman JR. De novo determination of bond orientations and order parameters from residual dipolar couplings with high accuracy. J. Am. Chem. Soc. 2003;125:10164–10165. doi: 10.1021/ja035904+. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Q, Throolin R, Pitt SW, Serganov A, Al-Hashimi HM. Probing motions between equivalent RNA domains using magnetic field induced residual dipolar couplings: accounting for correlations between motions and alignment. J. Am. Chem. Soc. 2003;125:10530–10531. doi: 10.1021/ja0363056. [DOI] [PubMed] [Google Scholar]
- 55.Chen Y, et al. Structure of stem-loop IV of Tetrahymena telomerase RNA. EMBO J. 2006;25:3156–3166. doi: 10.1038/sj.emboj.7601195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Q, Sun XY, Watt ED, Al-Hashimi HM. Resolving the motional modes that code for RNA adaptation. Science. 2006;311:653–656. doi: 10.1126/science.1119488. [DOI] [PubMed] [Google Scholar]
- 57.Hansen MR, Mueller L, Pardi A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat. Struct. Biol. 1998;5:1065–1074. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- 58.Kontaxis G, Clore GM, Bax A. Evaluation of cross-correlation effects and measurement ofone-bond couplings in proteins with short transverse relaxation times. J. Magn. Reson. 2000;143:184–196. doi: 10.1006/jmre.1999.1979. [DOI] [PubMed] [Google Scholar]
- 59.Redfield AG. The theory of relaxation processes. Adv. Mag. Reson. 1965;1:1–32. [Google Scholar]
- 60.de Alba E, Tjandra N. On the accurate measurement of amide one-bond N-15-H-1 couplings in proteins: effects of cross-correlated relaxation, selective pulses and dynamic frequency shifts. J. Magn. Reson. 2006;183:160–165. doi: 10.1016/j.jmr.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 61.Boisbouvier J, Bryce DL, O’Neil-Cabello E, Nikonowicz EP, Bax A. Resolution-optimized NMR measurement of D-1(CH), D-1(CH) and D-2(CH) residual dipolar couplings in nucleic acid bases. J. Biomol. NMR. 2004;30:287–301. doi: 10.1007/s10858-005-1846-5. [DOI] [PubMed] [Google Scholar]
- 62.Prestegard JH, Kishore AI. Partial alignment of biomolecules: an aid to NMR characterization. Curr. Opin. Chem. Biol. 2001;5:584–590. doi: 10.1016/s1367-5931(00)00247-7. [DOI] [PubMed] [Google Scholar]
- 63.Clore GM, Starich MR, Gronenborn AM. Measurement of residual dipolar couplings of macromolecules aligned in the nematic phase of a colloidal suspension of rod-shaped viruses. J. Am. Chem. Soc. 1998;120:10571–10572. [Google Scholar]
- 64.Hansen MR, Hanson P, Pardi A. Filamentous bacteriophage for aligning RNA, DNA, and proteins for measurement of nuclear magnetic resonance dipolar coupling interactions. Methods Enzymol. 2000;317:220–240. doi: 10.1016/s0076-6879(00)17017-x. [DOI] [PubMed] [Google Scholar]
- 65.Ottiger M, Tjandra N, Bax A. Magnetic field dependent amide N-15 chemical shifts in a protein-DNA complex resulting from magnetic ordering in solution. J. Am. Chem. Soc. 1997;119:9825–9830. [Google Scholar]
- 66.Kung HC, Wang KY, Goljer I, Bolton PH. Magnetic alignment of duplex and quadruplex DNAs. J. Mag. Reson. Series B. 1995;109:323–325. doi: 10.1006/jmrb.1995.9987. [DOI] [PubMed] [Google Scholar]
- 67.Tjandra N, Omichinski JG, Gronenborn AM, Clore GM, Bax A. Use of dipolar H-1-N-15 and H-1-C-13 couplings in the structure determination of magnetically oriented macromolecules in solution. Nat. Struct. Biol. 1997;4:732–738. doi: 10.1038/nsb0997-732. [DOI] [PubMed] [Google Scholar]
- 68.Al-Hashimi HM, et al. Field- and phage-induced dipolar couplings in a homodimeric DNA quadruplex, relative orientation of G center dot(C-A) triad and G-tetrad motifs and direct determination of C2 symmetry axis orientation. J. Am. Chem. Soc. 2001;123:633–640. doi: 10.1021/ja003379y. [DOI] [PubMed] [Google Scholar]
- 69.Zweckstetter M, Bax A. Prediction of sterically induced alignment in a dilute liquid crystalline phase: aid to protein structure determination by NMR. J. Am. Chem. Soc. 2000;122:3791–3792. [Google Scholar]
- 70.Ravishanker G, Swaminathan S, Beveridge DL, Lavery R, Sklenar H. Conformational and helicoidal analysis of 30 Ps of molecular-dynamics on the D(Cgcgaattcgcg) double helix—curves, dials and windows. J. Biomol. Struct. Dyn. 1989;6:669–699. doi: 10.1080/07391102.1989.10507729. [DOI] [PubMed] [Google Scholar]
- 71.Dickerson RE. DNA bending: the prevalence of kinkiness and the virtues of normality. Nucleic Acids Res. 1998;26:1906–1926. doi: 10.1093/nar/26.8.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lu XJ, El Hassan MA, Hunter CA. Structure and conformation of helical nucleic acids: analysis program (SCHNAaP) J. Mol. Biol. 1997;273:668–680. doi: 10.1006/jmbi.1997.1346. [DOI] [PubMed] [Google Scholar]
- 74.Bansal M, Bhattacharyya D, Ravi B. NUPARM and NUCGEN: software for analysis and generation of sequence dependent nucleic acid structures. Comput. Appl. Biosci. 1995;11:281–287. doi: 10.1093/bioinformatics/11.3.281. [DOI] [PubMed] [Google Scholar]
- 75.Valafar H, Prestegard JH. REDCAT: a residual dipolar coupling analysis tool. J. Magn. Reson. 2004;167:228–241. doi: 10.1016/j.jmr.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 76.Wei YF, Werner MH. iDC: a comprehensive toolkit for the analysis of residual dipolar couplings for macromolecular structure determination. J. Biomol. NMR. 2006;35:17–25. doi: 10.1007/s10858-006-9009-x. [DOI] [PubMed] [Google Scholar]
- 77.Skrynnikov NR, et al. Orienting domains in proteins using dipolar couplings measured by liquid-state NMR: differences in solution and crystal forms of maltodextrin binding protein loaded with beta-cyclodextrin. J. Mol. Biol. 2000;295:1265–1273. doi: 10.1006/jmbi.1999.3430. [DOI] [PubMed] [Google Scholar]
- 78.Al-Hashimi HM, et al. Variation of molecular alignment as a means of resolving orientational ambiguities in protein structures from dipolar couplings. J. Magn. Reson. 2000;143:402–406. doi: 10.1006/jmre.2000.2049. [DOI] [PubMed] [Google Scholar]
- 79.Tang RS, Draper DE. Bulge loops used to measure the helical twist of RNA in solution. Biochemistry. 1990;29:5232–5237. doi: 10.1021/bi00474a003. [DOI] [PubMed] [Google Scholar]
- 80.Bhattacharyya A, Murchie AIH, Lilley DMJ. RNA bulges and the helical periodicity of double-stranded-RNA. Nature. 1990;343:484–487. doi: 10.1038/343484a0. [DOI] [PubMed] [Google Scholar]
- 81.Bhattacharyya A, Lilley DMJ. Single base mismatches in DNA—long-range and short-range structure probed by analysis of axis trajectory and local chemical-reactivity. J. Mol. Biol. 1989;209:583–597. doi: 10.1016/0022-2836(89)90596-2. [DOI] [PubMed] [Google Scholar]
- 82.Riordan FA, Bhattacharyya A, McAteer S, Lilley DMJ. Kinking of RNA helices by bulged bases, and the structure of the human-immunodeficiency-virus transactivator response element. J. Mol. Biol. 1992;226:305–310. doi: 10.1016/0022-2836(92)90947-i. [DOI] [PubMed] [Google Scholar]
- 83.Tang RS, Draper DE. On the use of phasing experiments to measure helical repeat and bulge loop-associated twist in RNA. Nucleic Acids Res. 1994;22:835–841. doi: 10.1093/nar/22.5.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tang RS, Draper DE. Bend and helical twist associated with a symmetrical internal loop from 5S ribosomal-RNA. Biochemistry. 1994;33:10089–10093. doi: 10.1021/bi00199a036. [DOI] [PubMed] [Google Scholar]
- 85.Kim HD, et al. Mg2+-dependent conformational change of RNA studied by fluorescence correlation and FRET on immobilized single molecules. Proc. Natl. Acad. Sci. USA. 2002;99:4284–4289. doi: 10.1073/pnas.032077799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rueda D, et al. Diffusely bound Mg2+ ions slightly reorient stems I and II of the hammerhead ribozyme to increase the probability of formation of the catalytic core. Biochemistry. 2003;42:9924–9936. doi: 10.1021/bi0347757. [DOI] [PubMed] [Google Scholar]
- 87.Al-Hashimi HM, Pitt SW, Majumdar A, Xu WJ, Patel DJ. Mg2+-induced variations in the conformation and dynamics of HIV-1 TAR RNA probed using NMR residual dipolar couplings. J. Mol. Biol. 2003;329:867–873. doi: 10.1016/s0022-2836(03)00517-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Woodson SA. Metal ions and RNA folding: a highly charged topic with a dynamic future. Curr. Opin. Chem. Biol. 2005;9:104–109. doi: 10.1016/j.cbpa.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 89.Ippolito JA, Steitz TA. A 1.3-angstrom resolution crystal structure of the HIV-1 trans-activation response region RNA stem reveals a metal ion-dependent bulge conformation. Proc. Natl. Acad. Sci. USA. 1998;95:9819–9824. doi: 10.1073/pnas.95.17.9819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Du Z, Lind KE, James TL. Structure of TAR RNA complexed with a Tat-TAR interaction nanomolar inhibitor that was identifiedby computational screening. Chem. Biol. 2002;9:707–712. doi: 10.1016/s1074-5521(02)00151-5. [DOI] [PubMed] [Google Scholar]
- 91.Faber C, Sticht H, Schweimer K, Rosch P. Structural rearrangements of HIV-1 Tat-responsive RNA upon binding of neomycin. B. J. Biol. Chem. 2000;275:20660–20666. doi: 10.1074/jbc.M000920200. [DOI] [PubMed] [Google Scholar]
- 92.Aboul-ela F, Karn J, Varani G. The structure of the human-immunodeficiency-virus type-1 Tar RNA reveals principles of RNA recognition by Tat protein. J. Mol. Biol. 1995;253:313–332. doi: 10.1006/jmbi.1995.0555. [DOI] [PubMed] [Google Scholar]
- 93.Aboul-ela F, Karn J, Varani G. Structure of HIV-1 TAR RNA in the absence of ligands reveals a novel conformation of the trinucleotide bulge. Nucleic Acids Res. 1996;24:3974–3981. doi: 10.1093/nar/24.20.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Puglisi JD, Tan RY, Calnan BJ, Frankel AD, Williamson JR. Conformation of the Tar RNA-arginine complex by Nmr-spectroscopy. Science. 1992;257:76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- 95.Murchie AI, et al. Structure-based drug design targeting an inactive RNA conformation: exploiting the flexibility of HIV-1 TAR RNA. J. Mol. Biol. 2004;336:625–638. doi: 10.1016/j.jmb.2003.12.028. [DOI] [PubMed] [Google Scholar]
- 96.Pitt SW, Majumdar A, Serganov A, Patel DJ, Al-Hashimi HM. Argininamide binding arrests global motionsin HIV-1 TAR RNA: comparison with Mg2+-induced conformational stabilization. J. Mol. Biol. 2004;338:7–16. doi: 10.1016/j.jmb.2004.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pitt SW, Zhang Q, Patel DJ, Al-Hashimi HM. Evidence that electrostatic interactions dictate the ligand-induced arrest of RNA global flexibility. Angew. Chem. Int. Edn. 2005;44:3412–3415. doi: 10.1002/anie.200500075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Casiano-Negroni A, Sun X, Al-Hashimi HM. Probing Na(+)-induced changes in the HIV-1 TAR conformational dynamics using NMR residual dipolar couplings: New insights into the role of counterions and electrostatic interactions in adaptive recognition. Biochemistry. in the press doi: 10.1021/bi700335n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Saupe A, Englert G. High-resolution nuclear magnetic resonance spectra of orientated molecules. Phys. Rev. Lett. 1963;11:462–464. [Google Scholar]
- 100.Cornell WD, et al. A 2nd generation force-field for the simulation of proteins, nucleic-acids, and organic-molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 101.Clowney L, et al. Geometric parameters in nucleic acids: nitrogenous bases. J. Am. Chem. Soc. 1996;118:509–518. [Google Scholar]
- 102.Getz MM, Sun X, Casiano-Negroni A, Zhang Q, Al-Hashimi HM. NMR studies of RNA dynamics and structural plasticity using NMR residual dipolar couplings. Biopolymers. in the press doi: 10.1002/bip.20765. [DOI] [PubMed] [Google Scholar]
- 103.Davis B, et al. Rational design of inhibitors of HIV-1 TAR RNA through the stabilisation of electrostatic “hot spots”. J. Mol. Biol. 2004;336:343–356. doi: 10.1016/j.jmb.2003.12.046. [DOI] [PubMed] [Google Scholar]
- 104.Miclet E, O’Neil-Cabello E, Nikonowicz EP, Live D, Bax A. H-1-H-1 dipolar couplings provide a unique probe of RNA backbone structure. J. Am. Chem. Soc. 2003;125:15740–15741. doi: 10.1021/ja0388212. [DOI] [PubMed] [Google Scholar]
- 105.Miclet E, Boisbouvier J, Bax A. Measurement of eight scalar and dipolar couplings for methine-methylene pairs in proteins and nucleic acids. J. Biomol. NMR. 2005;31:201–216. doi: 10.1007/s10858-005-0175-z. [DOI] [PubMed] [Google Scholar]
- 106.O’Neil-Cabello E, Bryce DL, Nikonowicz EP, Bax A. Measurement of five dipolar couplings from a single 3D NMR multiplet applied to the study of RNA dynamics. J. Am. Chem. Soc. 2004;126:66–67. doi: 10.1021/ja038314k. [DOI] [PubMed] [Google Scholar]
- 107.Ottiger M, Delaglio F, Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J. Magn. Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 108.Vallurupalli P, Moore PB. Measurement of H2′-C2′and H3′-C3′ dipolar couplings in RNA molecules. J. Biomol. NMR. 2002;24:63–66. doi: 10.1023/a:1020620016891. [DOI] [PubMed] [Google Scholar]
- 109.Yan JL, Corpora T, Pradhan P, Bushweller JH. MQ-HCN-based pulse sequences for the measurement of (13)C1′-(1)H1′, (13)C1′-N-15, (1)H1′-N-15, (13)C1′-(13)C2′, (1)H1′-(13)C2′, (13)C6/8-(1)H6/8, (13)C6/8-N-15, (1)H6/8- N-15, (13)C6-(13)C5, (1)H6-(13)C5 dipolar couplings in C-13, N-15-labeled DNA (and RNA) J. Biomol. NMR. 2002;22:9–20. doi: 10.1023/a:1013876105589. [DOI] [PubMed] [Google Scholar]
- 110.Zidek L, Wu HH, Feigon J, Sklenar V. Measurement of small scalar and dipolar couplings in purine and pyrimidine bases. J. Biomol. NMR. 2001;21:153–160. doi: 10.1023/a:1012435106858. [DOI] [PubMed] [Google Scholar]
- 111.Brutscher B, Boisbouvier J, Pardi A, Marion D, Simorre JP. Improved sensitivity and resolution in H-1-C-13 NMR experiments of RNA. J. Am. Chem. Soc. 1998;120:11845–11851. [Google Scholar]
- 112.Jaroniec CP, Boisbouvier J, Tworowska I, Nikonowicz EP, Bax A. Accurate measurement of 15N-13C residual dipolar couplings in nucleic acids. J Biomol NMR. 2005;31:231–241. doi: 10.1007/s10858-005-0646-2. [DOI] [PubMed] [Google Scholar]
- 113.Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 114.Ottiger M, Bax A. Characterization of magnetically oriented phospholipid micelles for measurement of dipolar couplings in macromolecules. J. Biomol. NMR. 1998;12:361–372. doi: 10.1023/a:1008366116644. [DOI] [PubMed] [Google Scholar]
- 115.Sass J, et al. Purple membrane induced alignment of biological macromolecules in the magnetic field. J. Am. Chem. Soc. 1999;121:2047–2055. [Google Scholar]
- 116.Koenig BW, et al. NMR measurement of dipolar couplings in proteins aligned by transient binding to purple membrane fragments. J. Am. Chem. Soc. 1999;121:1385–1386. [Google Scholar]
- 117.Tycko R, Blanco FJ, Ishii Y. Alignment of biopolymers in strained gels: a new way to create detectable dipole-dipole couplings in high-resolution biomolecular NMR. J. Am. Chem. Soc. 2000;122:9340–9341. [Google Scholar]
- 118.Sass HJ, Musco G, Stahl SJ, Wingfield PT, Grzesiek S. Solution NMR of proteins within polyacrylamide gels: diffusional properties and residual alignment by mechanical stress or embedding of oriented purple membranes. J. Biomol. NMR. 2000;18:303–309. doi: 10.1023/a:1026703605147. [DOI] [PubMed] [Google Scholar]
- 119.Ruckert M, Otting G. Alignment of biological macromolecules in novel nonionic liquid crystalline media for NMR experiments. J. Am. Chem. Soc. 2000;122:7793–7797. [Google Scholar]
- 120.Alvarez-Salgado F, Desvaux H, Boulard Y. NMR assessment of the global shape ofa non-labelled DNA dodecamer containing a tandemof G-T mismatches. Magn. Reson. Chem. 2006;44:1081–1089. doi: 10.1002/mrc.1902. [DOI] [PubMed] [Google Scholar]