Abstract
Obesity is a global epidemic that results in significant morbidity and mortality. Mutations in the melanocortin 4 receptor (MC4R) gene, which codes for a G‐protein‐coupled receptor responsible for postprandial satiety signaling, have been associated with monogenic obesity. The prevalence of obesity is on the increase in South Africa, and it is hypothesized that mutations in MC4R are a contributing factor. The aim of this study was to perform a retrospective assessment of the relationship between allelic variants of MC4R and BMI in a South African study cohort. DNA was isolated from a demographically representative cohort of 297 individuals and the entire MC4R gene sequenced by Sanger sequencing. Eight previously reported MC4R variants were identified in 42 of the 297 (14.1%) study participants. The most frequently observed MC4R alleles were V103I (4.0%), I170V (1.5%), and I198I (1.2%), while the remaining five variants together constituted 1.18%. Five compound heterozygotes were also detected. Although MC4R variants were rare, the majority of variation was observed in individuals of Black African ancestry. No statistically significant associations with BMI were reported. Given that lifestyle interventions have limited success in decreasing obesity, there is an urgent need to perform large‐scale population studies to further elucidate the molecular underpinnings of this disease.
Keywords: Genotype–phenotype correlation, melanocortin 4 receptor, obesity, South Africa
Introduction
Obesity (body mass index [BMI] >30 kg/m2) is recognized as a chronic disease by the World Health Organization (WHO). It has reached pandemic proportions (Swinburn et al. 2011) and the prevalence continues to increase in the majority of African countries, particularly in individuals living in urban areas. In several parts of South Africa, the prevalence of combined overweight and obesity (BMI > 25) in economically active (18–65 years) Black African women has reached the alarming figure of 75% (Van der Merwe and Pepper 2006).
Genetic susceptibility to the development of obesity has been well documented. In this regard, it is estimated that the BMI of an individual is heritable in 40–70% of cases (Cheung and Mao 2012). Although obesity is most commonly polygenic, rare monogenic forms do exist, and the affected genes described thus far include leptin, the leptin receptor, pro‐opiomelanocortin (POMC), single‐minded 1 (SIM1), neurotrophic tyrosine kinase receptor type‐2 (NTRK2), and the melanocortin 4 receptor (Dubern and Clement 2012).
The melanocortin 4 Receptor (MC4R) is a G‐protein‐coupled receptor with the primary function of regulating food intake following the binding of the agonist alpha melanocyte stimulating hormone (α‐MSH). Once bound, the production of a satiety signal through the activation of the cyclic AMP (cAMP) second messenger system occurs (List and Habener 2003). Agouti‐related protein (AGRP) is an MC4R antagonist and inverse agonist and when bound produces an orexigenic signal (Govaerts et al. 2005; Chen et al. 2006).
MC4R deficiency is the most common monogenic form of obesity (Farooqi et al. 2003). Variations in the MC4R gene (OMIM *155541) have a population prevalence of at least one in 2000 (0.05%), are found in 0.5–1% of obese adults and are accountable for 6% of all severe cases of the disease starting in childhood (Dubern et al. 2001; Farooqi et al. 2003). These low percentages indicate that the variants have a low epidemiological significance. However, with regard to the symptomatic individual, these mutations should be considered highly significant. Individuals harboring MC4R mutations will preferentially transmit their mutations to their offspring with an 82% frequency, and individuals that carry pathogenic mutations have a 4.5‐fold increased risk of developing obesity when compared to noncarriers (Hinney et al. 2003; MacKenzie 2006). The frequency of MC4R variants is dependent on ethnicity, and differing ethnic backgrounds contribute to variability in the penetrance of MC4R mutations (Tao 2005; MacKenzie 2006). Recently, the role of gender has also been implicated in the penetrance of obesity‐related variants in black South African adolescents (Lombard et al. 2012).
MC4R mutations result in an increase in fat mass, lean mass, linear growth, extensive hyperinsulinaemia, an increase in bone mineral density, hyperphagia in early childhood and possibly binge‐eating disorder (Dubern et al. 2001; Cone 2005). The role of MC4R variants in the latter is however controversial (Branson et al. 2003; Lubrano‐Berthelier et al. 2006). Individuals with MC4R mutations also present with an elevated prevalence of the metabolic syndrome, which includes an increase in peripheral fat mass ratio, type 2 diabetes, dyslipidaemia, and hypertension (Potoczna et al. 2004). Adult subjects harboring MC4R mutations have an increased risk of becoming obese, and with respect to gender, it is estimated that these variants may account for up to four and 9.5 kg/m2 increases in BMI in males and females, respectively (Dempfle et al. 2004).
Functional defects resulting from mutations in the MC4R gene that are responsible for obesity include decreased or absent ligand binding; decreased cell surface receptor expression (because of intracellular retention of mutant receptors); incorrect protein folding (which results in the receptor not being released from the endoplasmic reticulum); and reduced signal transduction (Tao and Segaloff 2005; Chen et al. 2006). It has been suggested that the most common functional defect is a reduction in the constitutive activity of the receptor (Chen et al. 2006).
MC4R mutations have a co‐dominant pattern of inheritance, with modulation of penetrance and expressivity (Farooqi et al. 2003). Thus, homozygotes are known to be more obese than heterozygotes. However, certain MC4R variants appear to have a recessive pattern of inheritance, while others lead to the production of a receptor that is indistinguishable from the wild‐type (Farooqi et al. 2000).
Over 160 variants have been described for the MC4R gene to date (Hinney et al. 2013). Limited data regarding obesity risk alleles and their association with BMI exists in South Africa. It was therefore the aim of this study to assess the relationship between MC4R variants and BMI in a South African study cohort.
Materials and Methods
Study population
A sample cohort of the South African population was established from three groups of unrelated individuals. The first group included a random collection of individuals from the general population (n = 99); the second group comprised individuals that were being treated for diabetes at a tertiary hospital (n = 144); and the third group was established from individuals that had been assessed for obesity and its co‐morbidities by an endocrinologist (n = 54). The study (including informed consent forms and the proposed participant recruitment strategy) was approved by the Research Ethics Committee of the Faculty of Health Sciences of the University of Pretoria, approval numbers 102/2005, 135/2006, S103/2006 and S142/2007. The study was conducted in accordance with the Declaration of Helsinki. Age, gender, racial group, height and body weights were recorded on the day of sample collection.
Sequence analysis of the MC4R gene
DNA was extracted from peripheral blood buffy coats, using a Genomic DNA Purification kit (Fermentas Life Sciences, Inc., Hanover, MD). PCR amplification of the MC4R gene was performed using custom designed primers MC4 EF (5′‐GCT CTG GAC TTG TGA CAT TTA C‐3′) and MC4 ER (5′‐CCA GTA CCC TAC ACG GAA GAG‐3′). The cycling conditions were as follows: 95°C/5 min; 30 cycles of 94°C/30 s, 58°C/30 s, 72°C/2 min; and a final elongation step of 72°C for 10 min. Reactions were carried out in a total volume of 50 μL, which included 2 μL (~100 ng/μL) genomic DNA, 1.25 U GoTaq® Flexi DNA Polymerase (Promega, Madison, WI), 10 μL 5× buffer, 25 pmol of each primer, 200 μmol/L of each dNTP, and 1.5 mmol/L MgCl2. Amplicons were visualized on 1% agarose gels and purified, using the DNA Clean and Concentrator ‐5 kit (Zymo Research Corporation©, Irvine, CA, 2005–2006).
Sequencing reactions were performed using the ABI Big Dye Terminator Cycle Sequencing kit version 3.1 (Applied Biosystems Inc., Foster City, CA) and electrophoresed by Inqaba Biotechnical Industries Pty (Ltd), South Africa. Cycling conditions were as follows: 94°C/2 min; 30 cycles of 94°C/10 s, 50°C/10 s, 60°C/4 min. Primers used included the PCR primers as externals and two newly designed internal primers: MC4 IF 5′–GCA GTG GAC AGG TAC TTT ACT ATC–3′ and MC4 IR 5′–GTC ATA ATG TTA TGG TAC TG–3′. Reactions were carried out in a total volume of 10 μL, which included 0.75 μL purified PCR product, 1.5 pmol primer, 2.25 μL 5× dilution buffer, and 1 μL ABI Prism Big Dye Terminator mix, v3.1 (Applied Biosystems). The sequencing products were cleaned using the ZR‐96 DNA Sequencing Clean‐up kit™ (Zymo Research Corporation© Irvine, CA, 2005–2006). Products were analyzed on the Applied Biosystems/Hitachi 3130 × 1 Genetic Analyser. The MC4R gene was sequenced bi‐directionally in every patient in order to confirm the observed variants. These variants were confirmed by a second round of sequencing.
Electropherograms were edited using FinchTV Version 1.4.0 (Copyright© 2004–2006, Geospiza Inc.) and alignments made with the MC4R sequence available from Genbank (Accession number NG_016441.1). Alignments were carried out using CLC Free Workbench Version 4.0.1 (CLC bio A/S 2005) with gap settings as follows: gap open cost: 10; gap extension cost: 1; end gap cost: as any other. The alignment was set at the “Slow (very accurate)” setting.
Statistical analysis
Statistical analysis was performed using The Statistical Package for the Social Sciences version 22.0 (SPSS Inc., Chicago, IL). Sample distributions were tested for normality according to the Shapiro–Wilk Test. The Mann–Whitney U‐test was used when comparing the distribution of BMI of two groups.
Results
Study population
Two hundred and ninety‐seven (n = 297) individuals were recruited for the study and evaluated for variants in the MC4R gene. The final study cohort was largely representative of the South African population, and comprised Black Africans (63.0%), White Caucasians (22.2%), Indians (7.7%), and individuals of mixed ancestry (7.1%). The study population was predominantly female (60.6%), with a mean age of 47.5 years. The mean BMI of the study cohort was 30.80 kg/m2 (standard deviation = 9.32). The distribution according to BMI classification (as defined by the WHO) and population group is illustrated in Figure 1. Given that the cohort was established from three different defined groups ‐ control, diabetic and obese – it was possible to recruit participants with BMIs in each of the WHO defined BMI classification groups (Fig. 1A), with nearly 75% of participants being either of healthy weight, preobese, or obese class I. This recruitment strategy did however introduce a certain level of bias, as evidenced by a non‐normal distribution of BMI (Shapiro–Wilk P < 0.001). With respect to racial groups (Fig. 1B), and with the exception of the Indian cohort, more than 70% of each of the recruited population groups was at least pre‐obese (BMI ≥ 25.00), while at least 45% were obese (BMI ≥ 30.00).
Figure 1.
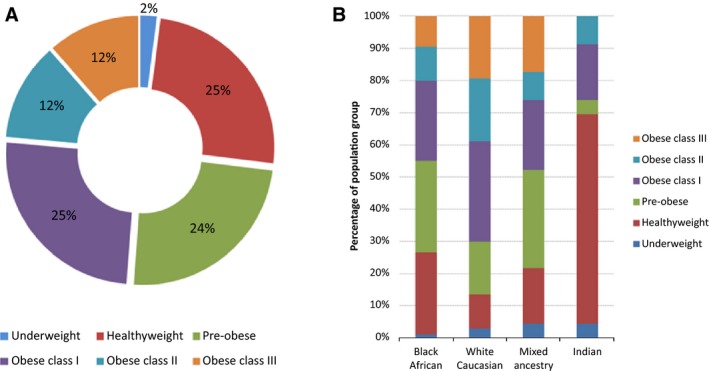
Distribution of the study cohort according to body mass index (BMI) classification. (A) Distribution according to BMI classification of the entire cohort. (B) Percentage distribution of BMI classification according to racial group. BMI classifications assigned according to the World Health Organisation (WHO): Underweight, BMI < 18.50; Healthy weight, BMI = 18.50–24.99; Pre‐obese, BMI = 25.00–29.99; Obese class I, BMI = 30.00–34.99; Obese class II, BMI = 35.00–39.99; and Obese class III, BMI ≥ 40.00.
Sequence analysis of the MC4R gene
DNA sequence analysis of the entire MC4R gene revealed the presence of eight previously reported single nucleotide variants (SNVs, Fig. 2). The most commonly identified MC4R variant, V103I, was present at an allelic frequency of 4.04%. The next most frequent alleles were I170V (1.52%) and I198I variant (1.18%). The remaining five alleles were rare, and constituted an overall frequency of 1.18% (R7H, S36T, R165Q, F202L, and I251L). Five compound heterozygote individuals were identified in this study, three of whom harbored the F202L and I198I variants, while the fourth presented with the I170V and V103I alleles, and the fifth with V103I and I251L.
Figure 2.
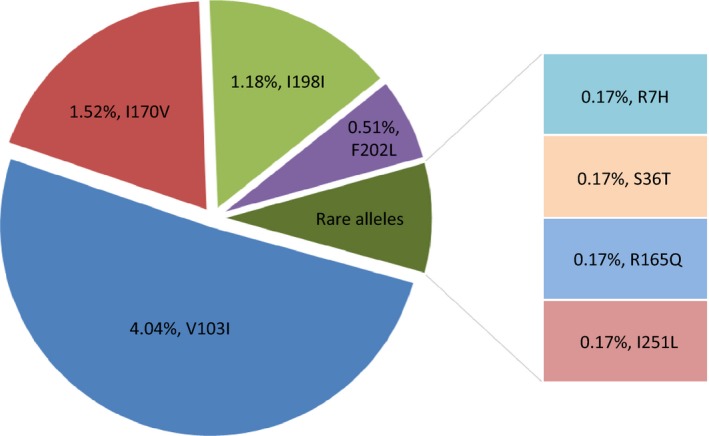
Distribution of MC4R allele frequencies in the total study cohort.
Genotype‐phenotype correlation
When stratifying the study population according to racial groups and aligning this to the presence of MC4R variants (Table 1), it was apparent that the Black African ancestry individuals harbored the greatest degree of allelic variation when considering both presence and overall frequency of SNVs (9.89%). Only wild‐type MC4R variants were reported in the cohort of Indian ancestry, while the overall frequency of alleles in the White Caucasian and Mixed Ancestry cohorts was 1.07% and 1.60%, respectively. Although the Mixed Ancestry study population was limited to 23 individuals, it appeared that a greater degree of MC4R variation was observed in this population when compared to the White Caucasian cohort (Table 1).
Table 1.
MC4R allelic frequencies according to this study population and from the 1000 Genomes Project
| MC4R variant | This study (%) | 1000 Genomes (%)a | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Black African (n = 187) | White Caucasian (n = 66) | Mixed ancestry (n = 21) | Indian (n = 23) | Global | Total African (n = 661) | African continentb (n = 504) | European (n = 503) | American (n = 347) | East Asian (n = 504) | South Asian (n = 489) | |
| R7H (rs142837166) | 0.27 | 0 | 0 | 0 | – | – | – | – | – | – | – |
| S36T | 0.27 | 0 | 0 | 0 | – | – | – | – | – | – | – |
| V103I (rs2229616) | 5.08 | 0.80 | 0.54 | 0 | 1.62 | 1.97 | 1.98 | 0.70 | 0.87 | 1.79 | 2.45 |
| R165Q | 0.27 | 0 | 0 | 0 | – | – | – | – | – | – | – |
| I170V (rs121913560) | 2.14 | 0 | 0.27 | 0 | 0.08 | 0.30 | 0.40 | 0 | 0 | 0 | 0 |
| I198I (rs61741819) | 1.34 | 0 | 0.54 | 0 | 1.28 | 4.46 | 4.27 | 0 | 0.29 | 0 | 0.31 |
| F202L (rs138281308) | 0.54 | 0 | 0.27 | 0 | 0.28 | 1.06 | 1.09 | 0 | 0 | 0 | 0 |
| I251L (rs52820871) | 0 | 0.27 | 0 | 0 | 0.26 | 0 | 0 | 0.90 | 0.43 | 0 | 0.10 |
| Total | 9.89 | 1.07 | 1.60 | 0 | 3.52 | 7.80 | 7.74 | 1.59 | 1.59 | 1.79 | 2.86 |
1000 Genomes frequency data determined via ENSEMBL Variant Effect Predictor (VEP).
Includes only populations residing on the African continent, namely the Esan from Nigeria; Gambian in Western Division, The Gambia; Luhya in Webuye, Kenya; Menda in Sierra Leone; Yoruba in Ibadan, Nigeria.
When aligning the presence of MC4R alleles to BMI (Fig. 3), we observed that study participants heterozygous for I170V (n = 8) and I198I (n = 4) had BMIs that were mostly over 30.0 kg/m2, and hence were collectively classified as obese class I according to the WHO. The five individuals that were compound heterozygotes for MC4R variants presented with a median BMI of 34.80 kg/m2, but with a noticeably wide distribution ranging from 27.20 to 49.45 kg/m2. With particular interest in these individuals, the BMI distribution was compared to the wild‐type group by means of a Mann–Whitney U test, which reported no statistically significant difference (P = 0.069). No further groups were compared.
Figure 3.
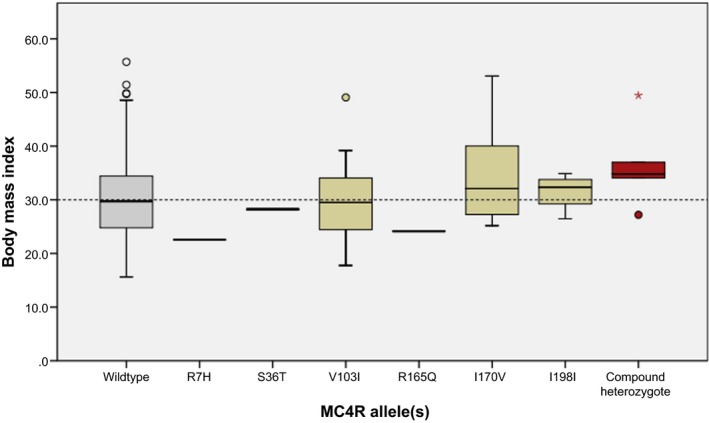
Body mass index (BMI ) distribution of study participants harboring MC4R variants. Wildtype refers to individuals with no SNVs in MC4R; R7H through to I198I represents BMI of individuals heterozygous for these variants; compound heterozygotes refers to individuals harboring I198I and F202L (n = 3), V103I and I170V (n = 1), and V103I and I251L (n = 1). Since BMI data points for compound heterozygotes are represented independently and excluded from the heterozygous data sets, no box‐whiskers for individuals harboring F202L and I251L are shown. One individual, who was morbidly obese (BMI = 125.99 kg/m2), was excluded from the V103I data set to avoid skewing of the central tendency. Box‐whisker parameters: central tendency = median; upper and lower line of box = interquartile range; whiskers = 1.5 times the inter‐quartile range; outliers and extreme outliers are represented by open circles and asterisks, respectively.
Discussion
Allelic variation of MC4R
Forty two of the 297 individuals (14.1%) analyzed presented with MC4R variants. No homozygous genotypes were observed in this study population, while five compound heterozygotes were identified, three of whom harbored the I198I and F202L alleles in combination. The fourth and fifth compound heterozygotes were both genotyped with the common V103I allele in combination with either I170V or I251L.
At 9.89%, the overall frequency of MC4R variants was highest in individuals of African ancestry. When compared to data derived from the 1000 Genomes Project (Table 1), this frequency appeared to be higher than the overall frequency in African populations (7.80%) and those residing on the African continent (7.74%). Moreover, the frequencies of each MC4R variant were also observed to differ considerably. For example, V103I was present in this study in Black Africans (5.08%) at more than twice the frequency reported in Africans globally (1.97%) and Africans living in Africa (1.98%). I170V, which was present at 2.14% in Black Africans in this study, is only present at 0.30% and 0.40% in total Africans and African residing elsewhere on the continent, respectively. The most frequent MC4R allele reported in Africans from the 1000 Genomes study was I198I, which was present at 4.46% in all Africans and 4.27% in Africans residing on the African continent. However, this is at least three times greater than the 1.34% reported in the Black Africans recruited in this study.
The frequencies of MC4R variants in White Caucasian study participants was similar to that reported in European and American populations of the 1000 Genomes Project. Interestingly, the South Asian population of the 1000 Genomes Project reported an overall frequency of 2.86% of MC4R variants, which is higher than that seen in the European (1.59%) and American (1.59%) populations. This is surprising, considering that no variants were reported in the Indian individuals participating in this study, albeit that a small sample size of n = 23 was recruited. Given the extensive admixture of the South African Mixed Ancestry population, it would not be reasonable to compare allelic frequencies to any populations from the 1000 Genomes Project.
Genotype‐phenotype correlation and allelic penetrance
A great deal of controversy exists regarding the phenotypic expression of MC4R variants. Major contributors to this controversy are (1) the varying phenotypic penetrance of putative pathogenic variants among individuals; (2) the role of other genes in the pathogenesis of obesity; and (3) influence of the environment. An additional complicating factor is the differing approaches used by researchers when performing functional studies.
By using in silico prediction tools as a starting point (Table 2), two MC4R variants that are most likely to be associated with an obesity phenotype are I170V and R165Q. With respect to I170V, Tao and Segaloff originally reported that this variant had no impact on receptor function when it came to cell surface expression, agonist binding and cAMP production (Tao and Segaloff 2005). However, in 2006, Lubrano‐Berthelier and colleagues reported contradictory data, which implicated this SNV in a loss‐of‐function phenotype due to decreased cell surface expression (Lubrano‐Berthelier et al. 2006). In silico prediction of the impact of I170V via SIFT (Sorting Intolerant From Tolerant) and POLYPHEN (Polymorphism Phenotyping) reports “Tolerated (0.06)” and “Benign (0.084)” effects on the receptor. However, a “Deleterious” 0.64 score was generated from CONDEL (CONsensus DELeteriousness score). In the present study cohort, the I170V variant was identified in nine individuals, all of whom presented with BMIs of greater than 25.00 kg/m2 and a median BMI of 32.08 kg/m2 (Fig. 3). It appears therefore that I170V is indeed a contributing factor to the pathogenesis of obesity in this South Africa cohort.
Table 2.
In silico prediction of MC4R variants reported in this study
| MC4R allele | In silico prediction | ||
|---|---|---|---|
| SIFT | POLYPHEN | CONDEL | |
| R7H (rs142837166) | Tolerated ‐ low confidence (0.9) | Benign (0) | Neutral (0.47) |
| S36T | Tolerated (0.47) | Benign (0.001) | Neutral (0.44) |
| V103I (rs2229616) | Tolerated (0.5) | Benign (0.279) | Neutral (0.40) |
| R165Q | Deleterious (0) | Possibly damaging (0.598) | Deleterious (0.63) |
| I170V (rs121913560) | Tolerated (0.06) | Benign (0.084) | Deleterious (0.64) |
| I198I (rs61741819) | NA | NA | – |
| F202L (rs138281308) | Tolerated (1) | Benign (0.005) | Neutral (0.43) |
| I251L (rs52820871) | Tolerated (1) | Benign (0.022) | Neutral (0.26) |
SIFT, sorting intolerant from tolerant, data accessed via ENSEMBL Variant Effect Predictor; POLYPHEN, polymorphism phenotyping, data accessed via ENSEMBL Variant Effect Predictor; CONDEL, CONsensus DELeteriousness score (González‐Pérez and López‐Bigas 2011).
R165Q has been reported to result in a 15 to 90‐fold reduction in receptor activity when in the presence of endogenous agonists, although activity with synthetic ligands was reduced by 2 to 9‐fold (Ma et al. 2004; Xiang et al. 2006). The variant was identified in 10 out of 300 obese Pima Indians and showed a strong correlation with the development of early onset obesity (Ma et al. 2004). It also affects cell surface expression of the receptor and has generally been described as a rare allele. SIFT, POLYPHEN, and CONDEL scores of 0, 0.598, and 0.63, respectively, all point toward a likely deleterious effect of this variant. In contrast to this evidence, the single individual found to be heterozygous for R165Q in this study was of a healthy weight (BMI = 24.14 kg/m2).
The most frequently reported allele in this study cohort, V103I (4.04%), has been reported at similar frequencies in both obese and nonobese subjects (Dubern et al. 2001; Geller et al. 2004; Rong et al. 2006). When functionally characterized in vitro, a V103I receptor shows similarity to the wild‐type receptor and has been implicated in a gain‐of‐function phenotype that results in protection of the carrier against obesity (Geller et al. 2004). This was later supported in a large population‐based study and meta‐analysis of 29,563 individuals, where the authors report statistically significant evidence for an 18% lower risk of obesity in the presence of V103I (Young et al. 2007). Within the investigated cohort, V103I was present in individuals from all BMI classification groups (Fig. 3), confirming the lack of consensus regarding its role in obesity.
Several rare MC4R alleles were identified in this South African study. I198I was detected in seven individuals, of which three were present in compound heterozygosity with F202L. Although I198I is a synonymous variant of MC4R, it has been reported to be present in both obese and non‐obese individuals (Rettenbacher et al. 2007; Deliard et al. 2013). The F202L variant was identified in three individuals, and was exclusively present in combination with I198I. BMIs for these three individuals were 27.20 kg/m2 (preobese), 34.08 kg/m2 (obese class I), and 49.45 kg/m2 (obese class III). F202L is located in the fifth transmembrane domain of MC4R, and when functionally characterized in vitro, decreased basal receptor activity was observed (Tao and Segaloff 2005). In contrast, F202L was subsequently reported to have no influence on MC4R activity (Xiang et al. 2010), being present in both obese and nonobese individuals (Jacobson et al. 2002; Farooqi et al. 2003).
The R7H, S36T, and I251L SNVs were each reported in single cases with BMIs of 22.55, 28.23 and 34.80 kg/m2, respectively. R7H has been implicated in reduced MC4R function in vitro, due to a weakened receptor response to α–MSH (Lubrano‐Berthelier et al. 2003; Hinney et al. 2013). S36T was originally reported in a single obese individual and has been demonstrated in two independent studies to have no influence on MC4R activity (Larsen et al., 2005; Hughes et al., 2009). The I251L variant has been shown to have no functional implication on MC4R activity (Xiang et al. 2006) and has been reported to be present in both healthy and obese populations (Vaisse and Clement 2000; Dubern et al. 2001; Hinney et al. 2003).
Five study participants harboring two different MC4R variants were identified in this study. The median BMI for these compound heterozygote individuals was 34.80 kg/m2, which borders on an obese class II classification, and which appeared to be higher than the majority of individuals with wild‐type MC4R genotypes. This difference was not found to be statistically significant (P = 0.069). However, as evidenced in this study and in the literature, genetic variation of MC4R is generally observed at a very low frequency, and hence a considerably larger study cohort would be required to identify genotype–phenotype associations and differences with statistical significance.
Concluding Remarks
Data from this investigation confirms the presence of previously reported MC4R variants in a South African cohort. It is widely accepted that MC4R has a part to play in the in the pathogenesis of obesity; however, the incomplete penetrance of pathogenic MC4R variants, the polygenic nature of obesity and the influence of environmental factors (including lifestyle) all contribute to our inability to establish a definitive molecular diagnosis for this complex disease. Since lifestyle interventions have limited success in decreasing obesity, there is an urgent need to elucidate the molecular underpinnings of this disease – on a scale similar to that described by Speliotes and colleagues, in which nearly 250,000 individuals were investigated (Speliotes et al. 2011).
Conflict of Interest
None declared.
Acknowledgments
This study was supported by funds from the South African Medical Research Council, the National Research Foundation of South Africa, the National Health Laboratory Services, and the Institute for Cellular and Molecular Medicine, University of Pretoria.
References
- Branson, R. , Potoczna N., Kral J. G., Lentes K.‐U., Hoehe M. R., and Horber F. F.. 2003. Binge eating as a major phenotype of melanocortin 4 receptor gene mutations. N. Engl. J. Med. 348:1096–1103. [DOI] [PubMed] [Google Scholar]
- Chen, M. , Celik A., Georgeson K. E., Harmon C. M., and Yang Y.. 2006. Molecular basis of melanocortin‐4 receptor for AGRP inverse agonism. Regul. Pept. 136:40–49. [DOI] [PubMed] [Google Scholar]
- Cheung, W. W. , and Mao P.. 2012. Recent advances in obesity: genetics and beyond. ISRN Endocrinol. 2012:536905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone, R. D. 2005. Anatomy and regulation of the central melanocortin system. Nat. Neurosci. 8:571–578. [DOI] [PubMed] [Google Scholar]
- Deliard, S. , Panossian S., Mentch F. D., Kim C. E., Hou C., Frackelton E. C., , et al. 2013. The missense variation landscape of FTO, MC4R, and TMEM18 in obese children of African Ancestry. Obesity (Silver Spring). 21:159–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempfle, A. , Hinney A., Heinzel‐Gutenbrunner M., Raab M., Geller F., Gudermann T., et al. 2004. Large quantitative effect of melanocortin‐4 receptor gene mutations on body mass index. J. Med. Genet. 41:795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubern, B. , and Clement K.. 2012. Leptin and leptin receptor‐related monogenic obesity. Biochimie 94:2111–2115. [DOI] [PubMed] [Google Scholar]
- Dubern, B. , Clément K., Pelloux V., Froguel P., Girardet J. P., Guy‐Grand B., et al. 2001. Mutational analysis of melanocortin‐4 receptor, agouti‐related protein, and alpha‐melanocyte‐stimulating hormone genes in severely obese children. J. Pediatr. 139:204–209. [DOI] [PubMed] [Google Scholar]
- Farooqi, I. S. , Yeo G. S. H., Keogh J. M., Aminian S., Jebb S. A., Butler G., et al. 2000. Dominant and recessive inheritance of morbid obesity associated with melanocortin 4 receptor deficiency. J. Clin. Invest. 106:271–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooqi, I. S. , Keogh J. M., Yeo G. S. H., Lank E. J., Cheetham T., and O'Rahilly S.. 2003. Clinical spectrum of obesity mutations in the melanocortin 4 receptor gene. N. Engl. J. Med. 34:1085–1095. [DOI] [PubMed] [Google Scholar]
- Geller, F. , Reichwald K., and Dempfle A.. 2004. Melanocortin‐4 receptor gene variant I103 is negatively associated with obesity. Am. J. Genet. 74:572–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González‐Pérez, A. , and López‐Bigas N.. 2011. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score. Condel. Am. J. Hum. Genet. 88:440–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govaerts, C. , Srinivasan S., Shapiro A., Zhang S., Picard F., Clement K., et al. 2005. Obesity‐associated mutations in the melanocortin 4 receptor provide novel insights into its function. Peptides 26:1909–1919. [DOI] [PubMed] [Google Scholar]
- Hinney, A. , Hohmann S., Geller F., Vogel C., Hess C., Wermter A.‐K., et al. 2003. Melanocortin‐4 receptor gene: case‐control study and transmission disequilibrium test confirm that functionally relevant mutations are compatible with a major gene effect for extreme obesity. J. Clin. Endocrinol. Metab. 88:4258–4267. [DOI] [PubMed] [Google Scholar]
- Hinney, A. , Volckmar A.‐L., and Knoll N.. 2013. Melanocortin‐4 receptor in energy homeostasis and obesity pathogenesis. Elsevier Inc, Amsterdam, The Netherlands. [DOI] [PubMed] [Google Scholar]
- Hughes, D. A. , Hinney A., Brumm H., Wermter A.‐K., Biebermann H., Hebebrand J., et al. 2009. Increased constraints on MC4R during primate and human evolution. Hum. Genet. 124:633–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson, P. , Ukkola O., Rankinen T., Snyder E. E., Leon A. S., Cowan G. S., et al. 2002. Melanocortin 4 receptor sequence variations are seldom a cause of human obesity: the Swedish obese subjects, the HERITAGE Family Study, and a Memphis Cohort. J. Clin. Endocrinol. Metab. 87:4442–4446. [DOI] [PubMed] [Google Scholar]
- Larsen, L. H. , Echwald S. M., Sørensen T. I. A., Andersen T., and Wulff B. S.. 2005. Prevalence of mutations and functional analyses of melanocortin 4 receptor variants identified among 750 men with juvenile‐onset obesity. J. Endocrinol. Metab. 90:219–224. [DOI] [PubMed] [Google Scholar]
- List, J. F. , Ph D., and Habener J. F.. 2003. Editorials defective melanocortin 4 receptors in hyperphagia. N. Engl. J. Med. 20:1160–1163. [DOI] [PubMed] [Google Scholar]
- Lombard, Z. , Crowther N. J., van der Merwe L., Pitamber P., Norris S., and Ramsay M.. 2012. Appetite regulation genes are associated with body mass index in black South African adolescents: a genetic association study. BMJ Open 2:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubrano‐Berthelier, C. , Cavazos M., Dubern B., Shapiro A., Stunff C. L., Zhang S., et al. 2003. Molecular genetics of human obesity‐associated MC4R mutations. Ann. N. Y. Acad. Sci. 994:49–57. [DOI] [PubMed] [Google Scholar]
- Lubrano‐Berthelier, C. , Dubern B., Lacorte J.‐M., Picard F., Shapiro A., Zhang S., et al. 2006. Melanocortin 4 receptor mutations in a large cohort of severely obese adults: prevalence, functional classification, genotype‐phenotype relationship, and lack of association with binge eating. J. Clin. Endocrinol. Metab. 91:1811–1818. [DOI] [PubMed] [Google Scholar]
- Ma, L. , Tataranni P., Bogardus C., and Baier L.. 2004. Melanocortin 4 receptor gene variation is associated with severe obesity in Pima Indians. Diabetes 53:2–5. [DOI] [PubMed] [Google Scholar]
- MacKenzie, R. G. 2006. Obesity‐associated mutations in the human melanocortin‐4 receptor gene. Peptides 27:395–403. [DOI] [PubMed] [Google Scholar]
- van der Merwe, M.‐T. , and Pepper M. S.. 2006. Obesity in South Africa. Obes. Rev. 7:315–322. [DOI] [PubMed] [Google Scholar]
- Potoczna, N. , Branson R., Kral J. G., Piec G., Steffen R., Ricklin T., et al. 2004. Gene variants and binge eating as predictors of comorbidity and outcome of treatment in severe obesity. J. Gastrointest. Surg. 8:971–981; discussion 981–2. [DOI] [PubMed] [Google Scholar]
- Rettenbacher, E. , Tarnow P., Brumm H., Prayer D., Wermter A.‐K., Hebebrand J., et al. 2007. A novel non‐synonymous mutation in the melanocortin‐4 receptor gene (MC4R) in a 2‐year‐old Austrian girl with extreme obesity. Exp. Clin. Endocrinol. Diabetes 115:7–12. [DOI] [PubMed] [Google Scholar]
- Rong, R. , Tao Y.‐X., Cheung B. M. Y., Xu A., Cheung G. C. N., and Lam K. S. L.. 2006. Identification and functional characterization of three novel human melanocortin‐4 receptor gene variants in an obese Chinese population. Clin. Endocrinol. (Oxf) 65:198–205. [DOI] [PubMed] [Google Scholar]
- Speliotes, E. K. , Willer C. J., Berndt S. I., Monda K. L., Thorleifsson G., Jackson A. U., et al. 2011. Association analyses of 249,796 individuals reveal eighteen new loci associated with body mass index. Nat. Genet. 42:937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinburn, B. A. , Sacks G., Hall K. D., McPherson K., Finegood D. T., Moodie M. L., et al. 2011. The global obesity pandemic: shaped by global drivers and local environments. Lancet 378:804–814. [DOI] [PubMed] [Google Scholar]
- Tao, Y.‐X. 2005. Molecular mechanisms of the neural melanocortin receptor dysfunction in severe early onset obesity. Mol. Cell. Endocrinol. 239:1–14. [DOI] [PubMed] [Google Scholar]
- Tao, Y. , and Segaloff D. L.. 2005. Functional analyses of melanocortin‐4 receptor mutations identified from patients with binge eating disorder and nonobese or obese subjects. J. Clin. Endocrinol. Metab. 90:5632–5638. [DOI] [PubMed] [Google Scholar]
- Vaisse, C. , and Clement K.. 2000. Melanocortin‐4 receptor mutations are a frequent and heterogeneous cause of morbid obesity. J. Clin. Invest. 106:253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, Z. , Litherland S. A., Sorensen N. B., Proneth B., Wood M. S., Shaw A. M., et al. 2006. Pharmacological characterization of 40 human melanocortin‐4 receptor polymorphisms with the endogenous proopiomelanocortin‐derived agonists and the agouti‐related protein (AGRP) antagonist. Biochemistry 45:7277–7288. [DOI] [PubMed] [Google Scholar]
- Xiang, Z. , Proneth B., Dirain M. L., Litherland S. A., and Haskell‐Luevano C.. 2010. Pharmacological characterization of 30 human melanocortin‐4 receptor polymorphisms with the endogenous proopiomelanocortin‐derived agonists, synthetic agonists, and the endogenous agouti‐related protein antagonist. Biochemistry 49:4583–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, E. H. , Wareham N. J., Farooqi S., Hinney A., Hebebrand J., Scherag A., et al. 2007. The V103I polymorphism of the MC4R gene and obesity: population based studies and meta‐analysis of 29 563 individuals. Int. J. Obes. (Lond) 31:1437–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]


