Abstract
The significance of metal(oid)s as environmental pollutants has made them a priority in ecotoxicology, with the aim of minimizing exposure to animals or humans. Therefore, it is necessary to develop sensitive and inexpensive methods that can efficiently detect and monitor these pollutants in the environment. Conventional analytical techniques suffer from the disadvantages of high cost and complexity. Alternatively, prokaryotic or eukaryotic whole-cell biosensors (WCB) are one of the newest molecular tools employed in environmental monitoring that use the cell as an integrated reporter incorporating a reporter gene fused to a heavy metal responsive promoter. In the present paper, we report results from expressing, in the ciliate Tetrahymena thermophila, constructs consisting of the reporter gfp gene fused to the complete MTT1 or MTT5 protein coding regions under the transcriptional control of the MTT1 metallothionein promoter, which plays a critical role in heavy metal stress in this ciliate. When exposed to Cd2+, such cells overexpress both the GFP reporter transgene and the linked metallothionein gene. We report that, for the GFPMTT5 strain, this metallothionein overexpression results in marked resistance to cadmium toxicity (24h LC50 ~ 15 µM of Cd2+), compared to wild type cells (24h LC50 ~ 1.73 µM of Cd2+). These results provide the first experimental evidence that ciliate metallothioneins, like in other organisms, function to protect the cell against toxic metal ions. Because these strains may have novel advantages as WCBs, we have compared their properties to those of other previously reported Tetrahymena WCBs.
Keywords: Ciliate-metallothioneins, GFP reporter, heavy metals, Tetrahymena thermophila, whole-cell biosensor
Introduction
The importance of metals in biology is well known. Indeed, some of them are key components in many proteins necessary for cellular metabolism and growth (Lovley, 2000). However, certain metals are among the most abundant, persistent and toxic inorganic environmental pollutants. Due to their ecological, sanitary and even economic consequences, they have been considered priority environmental pollutants (Keith and Telliard, 1979). To monitor and minimize exposure of humans and other organisms to metal pollution, there is a pressing need to develop accurate metal detection assays for potentially polluted habitats. In environmental biomonitoring, global parameters such as bioavailability, toxicity and genotoxicity cannot be tested using molecular recognition or chemical analysis, but rather can only be assayed using whole cells.
Recently, the concept of "whole-cell biosensor" (WCB) has been introduced by several authors (Belkin, 2003; D´Souza, 2001; Park et al., 2013; Van der Meer and Belkin, 2010), as a very useful alternative to classical biosensors. A WCB uses the prokaryotic or eukaryotic cell as a single reporter incorporating both bioreceptor and transducer elements. Two types of bioassays using WCBs may be considered; turn off and turn on assays (Belkin, 2003). In turn on assays a quantifiable molecular reporter is fused to a specific gene promoter, known to be activated by the target chemical or environmental pollutant (such as metals). Both prokaryotic and eukaryotic microorganisms have been used to design WCBs for metals (Magrisso et al., 2008; Van der Meer and Belkin, 2010; Walmsley and Keenan, 2000). Among eukaryotic microorganisms, ciliates offer two specific advantages as environmental sensors: 1) they do not have a cell wall in their vegetative stage, which in other organisms can minimize the sensitivity to environmental pollutants as well as delay the cell response (Gutiérrez et al., 2008), and 2) as revealed by genome analysis in Tetrahymena thermophila and Paramecium tetraurelia (Aury et al., 2006; Eisen et al., 2006), ciliates and humans share more orthologs with each other than are shared between humans and the yeast Saccharomyces cerevisiae, or between T. thermophila and Plasmodium falciparum. For some applications, ciliates may therefore offer a good alternative to testing in animals (Gutierrez et al., 2003; 2008).
Metallothioneins (MTs) are metal-binding proteins involved in cellular protection from metal toxicity, and expression of the corresponding genes is accordingly induced by metals. The ciliate T. thermophila has five MT isoforms; three CdMTs (MTT1, MTT3 and MTT5) and two CuMTs (MTT2 and MTT4) (Díaz et al., 2007; Gutierrez et al., 2009; 2011). The CdMT genes are expressed at very different levels in response to metal stress (Díaz et al., 2007). In 2003, we proposed (Gutierrez et al., 2003) that the Tetrahymena MT promoters might be used to design metal biosensors, focusing on the MTT1 or MTT5 genes due to their rapid and strong induction by metals (Díaz et al., 2007; Shang et al., 2002).
Currently, the only ciliate-based WCBs rely on MT promoters from T. thermophila linked with the luciferase reporter gene, to detect heavy metals in aquatic or soil samples (Amaro et al., 2011). These T. thermophila WCBs (strains MTT1Luc and MTT5Luc), which have been validated using natural samples, exclusively detect bioavailable metals and demonstrate a high and differential sensitivity in both artificial and natural samples (Amaro et al., 2011).
In the present article, we introduce an alternative WCB using the green fluorescent protein (GFP) as a reporter gene and the CdMT promoter from the T. thermophila MTT1 gene (Amaro et al., 2011; Díaz et al., 2007). Another important difference between the novel WCBs and those previously reported is that metal exposure in the novel strains induces overexpression of metallothionein genes themselves. Thus, these gene fusions (PMTT1::GFP::MTT1/MTT5) have allowed us to characterize the effects of metallothionein overproduction in this ciliate model.
Materials and methods
Ciliate strains and cell culture
In this work the following Tetrahymena thermophila strains were used; CU248.1 (mpr1-1/mpr1-1; pm-S, mp-S, VII), B2086 (mpr1-1/mpr1-1; pm-S, mp-S, II) and SB1969 (chx1-1/chx1-1, mpr1-1/mpr1-1; pm-S, cy-S, II) (the last one was kindly supplied by E. Orias (University of California, Santa Barbara, USA). Micronuclear genotypes of these strains are homozygous for mpr1-1 (6-methyl-purine resistant) and chx1-1 (cycloheximide resistant), and macronuclear phenotypes are pm-S (paromomycin sensitive), mp-S (6-methyl-purine sensitive) and cy-S (cycloheximide sensitive). All strains were grown axenically at 30 ± 1 °C in SPPA medium (2 % proteose peptone (Difco), 0,1 % yeast extract (Difco), 0.2% glucose (Sigma), 0.003% Fe-EDTA (Sigma), supplemented with 250 µg/ml penicillin G and streptomycin sulfate (Sigma) and 0.25 µg/ml amphotericin B (Sigma) or PP210 medium (Diaz et al., 2007).
Reporter constructs in pVGFMTT1 and pVGFMTT5 plasmids
The fusion proteins GFP-MTT1 and GFP-MTT5 were generated by directional cloning of the PCR-amplified MTT1 or MTT5 genes into the ApaI/XhoI site of the pVGF.MTT vector (Cowan et al., 2005) which includes the ORF of GFP (Green Fluorescent Protein) under the control of the MTT1 promoter. The MTT1 and MTT5 genes were amplified from T. thermophila genomic DNA using the primer pairs MTT1A (5’CATCTCGAGATGGATAAAGTTAATAGCTGTTGC3’) / MTT1B (5’CTGGGGCCCTCATTTACAACATTAACAAGTCTA3’) and MTT5A (5’CATCTCGAGATGGATAAAATTTCTGGTGAAAGC3’) / MTT5B (5’CTGGGGCCCTCAGCAACTACCTCCAGG3’), respectively. These primers provided the ApaI/XhoI restriction site to the 5’ and 3’ ends of the amplified sequence for ligation into pVGF.MTT vector. Correct ligation was verified by direct sequencing (3730 DNA Analyzer Applied Biosystems, with the Big-dye™ kit (Applied Biosystems). Therefore, the obtained recombinant plasmids pVGFMTT1 or pVGFMTT5 carried out the reporter constructs PMTT1 ::GFP::MTT1 and PMTT1::GFP::MTT5, respectively.
Tetrahymena transformation and expression of GFP-MT reporter proteins
Each recombinant plasmid (pVGFMTT1 or pVGFMTT5) was introduced by electroporation into conjugating T. thermophila cells (mating pairs of CU428.1 and B2086) using the ECM 600 electroporator (BTX Inc., San Diego, CA) as previously described (Gaertig and Gorovsky, 1992). The transformant cells (named GFPMTT1 or GFPMTT5) were selected after testing paramomycin (Sigma) resistance at 120 µg/ml. After 3 days at 30°C the transformants were screened for GFP expression by fluorescent microscopy after Cd2+ exposure with 9 ×10−7 M CdCl2 (Sigma) for 2 h at 30°C in PP210 medium (Diaz et al., 2007) or 0.01 M TrisHCl buffer pH 6.8.
Turn on bioassays and flow cytometry quantification
Log-phase cultures (1–3 × 105 cells/ml) of T. thermophila transformants strains to be used in turn on bioassays were washed and resuspended in 0.01 M TrisHCl buffer pH 6.8. Two ml aliquots of cells were transferred to a sterile 24-well plate and exposed to different Cd2+ concentrations (80 nM – 15 µM range) for 2 or 24 hours at 30 °C. After Cd2+ exposure, cells were harvested and resuspended in 500 µl aliquots in 0.01 M TrisHCl buffer, and GFP-MT fusion protein expression was detected by fluorescence microscopy. To quantify the GFP-fluorescence signal, flow cytometry was performed on a FACScaliburg flow cytometer (Becton Dickinson) equipped with Cell Quest software. Red fluorescence due to Iodine propidium (IP) fluorophore was used to estimate dead cells, and was collected in the FL3 channel (670 nm LP), while the green fluorescence from GFP was collected in the FL1 channel (530/30 nm bandpass filter). Four types of controls were used: first, samples with untreated transformant cells without IP (used to calibrate the cytometer); 2nd, untreated non-transformant cells + IP (to establish basal mortality not due to metal exposure); 3rd, untreated transformant cells + IP (to determine basal GFP gene expression), and; 4th, cells fixed with 100% ethanol +IP (to calibrate the signal for 100% mortality).
Fluorescence microscopy and transmission electron microscopy (TEM)
For fluorescence microscopy, cells were fixed with 1 ml of 4% paraformaldehyde (Merck) for 30 min in the dark. Cells were then washed 2× with 50 mM HEPES buffer pH 7 and observed under the epifluorescent microscope Axioplan 2 (Carl-Zeiss), using green fluorescent filters (480/520-30 nm). Living cells were also observed by fluorescent microscopy, after depositing cells directly on poly-lysine treated slides (Sigma). For TEM analysis the protocol from Dentler (2000) was applied. Both Cd2+ treated and untreated transformant and non-transformant cells were fixed with 2.5% glutaraldehyde in 100 mM sodium cacodylate buffer pH 7.2 for 1 h. Then they were washed three times in cacodylate buffer (TAAB) and postfixed with 1% OsO4 (TAAB) in cacodylate buffer for 45 min on ice. Fixed cells were washed three times (2–3 min each time) with deionized water, and stained with 1% uranyl acetate (TAAB) for 1h. Dehydration of cells was carried out by an acetone series of increasing concentration, and then the samples were embedded in Embed 812 (TAAB) resin, according to the commercial supplied protocol. Thin sections were stained with 1% toluidine blue to check cell structure preservation. Ultrathin sections were obtained by microtome Ultracut (Reichert-Jung), stained with 2% uranyl acetate and lead citrate, and observed in a JEM1010 (Jeol) transmission electron microscope at 80 kV.
DNA isolation and standard PCR reactions
Total Tetrahymena DNA was isolated from 50 ml log-phase cultures as described in Hamilton and Orias (2000). Bacterial plasmids were isolated with QIAprep Spin Miniprep kit (Qiagen). All standard PCR reactions were carried out in the thermocycler Mastercycler gradient (Eppendorf). PCR mixtures were carried out as described in Díaz et al., (2007). PCR products were analyzed by standard 1.5 % agarose gel electrophoresis.
Total RNA isolation and quantitative real time RT-PCR
Total RNA was isolated according to the protocol supplied by the RNeasy Mini kit (Qiagen). To remove possible DNA contamination, all samples were treated with DNase I (RNase free) (Ambion). Total RNA samples were analyzed using denaturing 1.2 % agarose gels according to (Sambrook and Russell, 2001). The cDNA synthesis was carried out using 5 µg RNA, according to the protocol supplied by 1st Strand cDNA Synthesis kit (AMV, Roche). cDNA samples were amplified in duplicate 96 microtiter plates (Applied Biosystems). Each PCR reaction (20 µl total volume) contained: 10 µl of 1× SYBR Green PCR Master Mix (Applied Biosystems), 1 µl of each primer (300 nM final concentration), 4 µl of H2O and 5 µl of a 1/10 cDNA dilution (about 125 ng). The primers used were ATUBQ1 (5´TGTCGTCCCCAAGGAT3´) and ATUBQ2 (5´GTTCTCTTGGTCTTGATGGT3´) for the ATU1 gene (α-tubulin) which was used as an endogenous control gene, MTT5QA (5’TGTGTCGGTTCAGGAGAAGGA3’) and MTT5QB (5’AGGTCCGCATTTACATTCAGCTT3’) for the MTT5 gene, and GFPQA (5’CTACCCGAGCACCCAGTC3’) and GFPQB (5’ACGAACTCCAGCAGGAC3’) for gfp gene. Primers were designed using the software Primer Express v.2 (Applied Biosystems). All PCR products were confirmed by direct sequencing (3730 DNA Analyzer Applied Biosystems, with the Big-dye™ kit (Applied Biosystems). Real time PCR reactions were carried out in an ABI PRIMS 7900 HT Real Time PCR System (Applied Biosystems), and the thermal cycling protocol was as described in Díaz et al., (2007). The specificity of each primer was tested by quantitative PCR (a unique PCR product was obtained for all primers as determined by melting curve analyses). For each gene studied and the endogenous control (ATU1), standard curves were obtained using 10-fold cDNA dilutions and determining the Ct (cycle threshold) values. The standard line parameters (amplification efficiency (E), the slope of trend line (S) and the correlation coefficient (R2) were 2.04, −3.22 and 0.99 for ATU1; 2.19, −2.94 and 0.99 for gfp; 1.99, − 3,34 and 1 for MTT5, respectively. These data represent the mean obtained from four independent experiments (each performed in duplicates). Quantification of relative gene expression was carried out according to the Standard-curve quantification method (Larionov et al. 2005).
Results and Discussion
GFPMTT1 and GFPMTT5 reporter strains
Several promoters from T. thermophila metallothionein genes, namely the CdMT MTT1 or MTT5 promoters and the CuMT MTT2 promoter, have been shown to be excellent biotechnological tools to drive heterologous protein expression in T. thermophila, due to their fast and strong induction by low concentrations of metals (Amaro et al. 2011; Boldrin et al. 2008; Shang et al. 2002). To further exploit the favorable features of these promoters, we incorporated them into constructs to create new potential WCBs that use GFP as a reporter fused to an T. thermophila MTT gene.
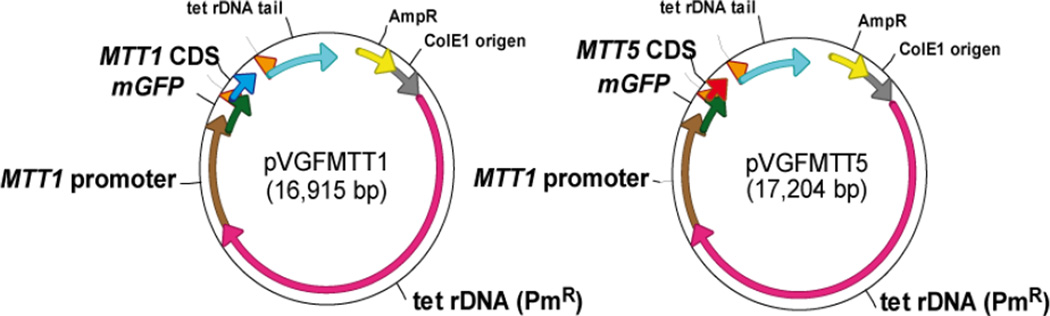
To create the reporter constructs pVGFMTT1 and pVGFMTT5, the GFP::MTT1 or GFP::MTT5 reporter gene fusions were placed under the transcriptional control of the T. thermophila MTT1 promoter in the pVGF.MTT expression vector. The resulting plasmids, named pVGFMTT1 and pVGFMTT5 (Fig. 1), were introduced into T. thermophila cells by electroporation as described in Methods. And the two generated strains were named GFPMTT1 (containing the pVGFMTT1 reporter plasmid) and GFPMTT5 (with the pVGFMTT5 plasmid).
Fig. 1.
Schematic representation of plasmids with reporter constructs. pVGFMTT1 (16.9 Kb) and pVGFMTT5 (17.2 Kb).
Turn on bioassay using the GFPMTT5 strain
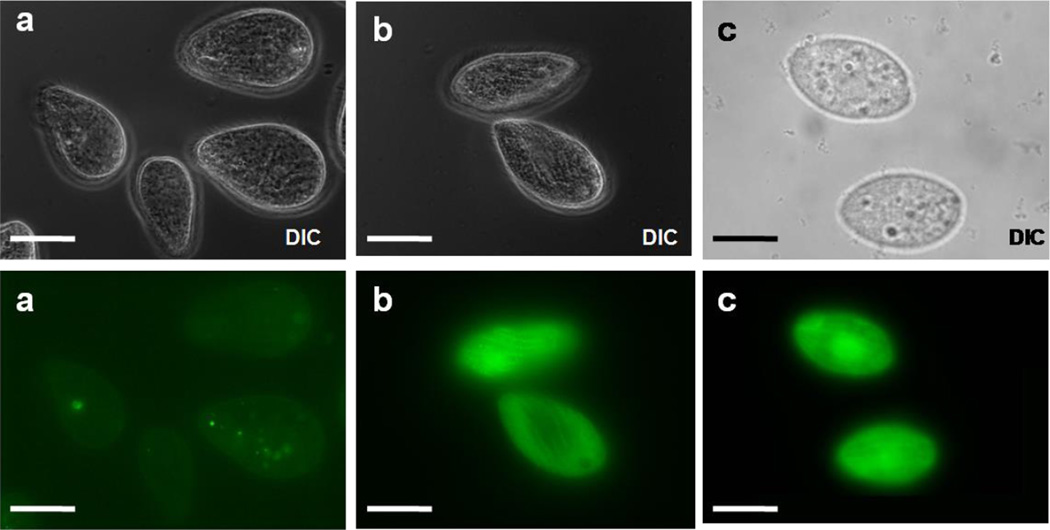
Strains GFPMTT1 and GFPMTT5 were exposed to 0.9 µM Cd2+ in 0.01 M Tris-HCl buffer pH 6.8 for 0.5, 1 or 2 hours and examined by fluorescence microscopy to determine the minimal exposure time that results in a strong and fast GFP signal. We selected 2h as optimal incubation time for bioassays because it produced the strongest GFP fluorescence signal for both strains. Since MTT1 promoter responds within 30 minutes to Cd2+ exposure (Shang et al., 2000), the 2h incubation time may be due to the fact that GFP folding and maturation require 95 minutes (Tsien, 1998). As shown in Fig. 2, incubation with Cd2+ (2h) dramatically induced the production of green fluorescence (509 nm) in both experimental strains when compared to controls that were not treated with Cd2+. Very low levels of fluorescence emission were detected in absence of Cd2+ probably due to basal expression from the MTT1 promoter (Díaz et al. 2007).
Fig. 2.
Fluorescence micrographs. (a) Control. (b) GFPMTT1 strain. (c) GFPMTT5 strain. (b, c) after 0.9 µM Cd2+ (2h treatment). DIC: differential interference contrast. Bars: 25 µm.
Since both strains showed similar values of GFP signal after Cd2+ induction, we therefore pursued further analysis using just one strain, GFPMTT5. Fluorescence microscopy examination showed that GFP fluorescence appeared uniformly distributed throughout the cytoplasm after 2h of induction with Cd2+ (Fig. 2). Similar results were observed after 24h of metal exposure (data not shown), except that the fluorescence at 24h was stronger, probably due to accumulation of the GFP reporter.
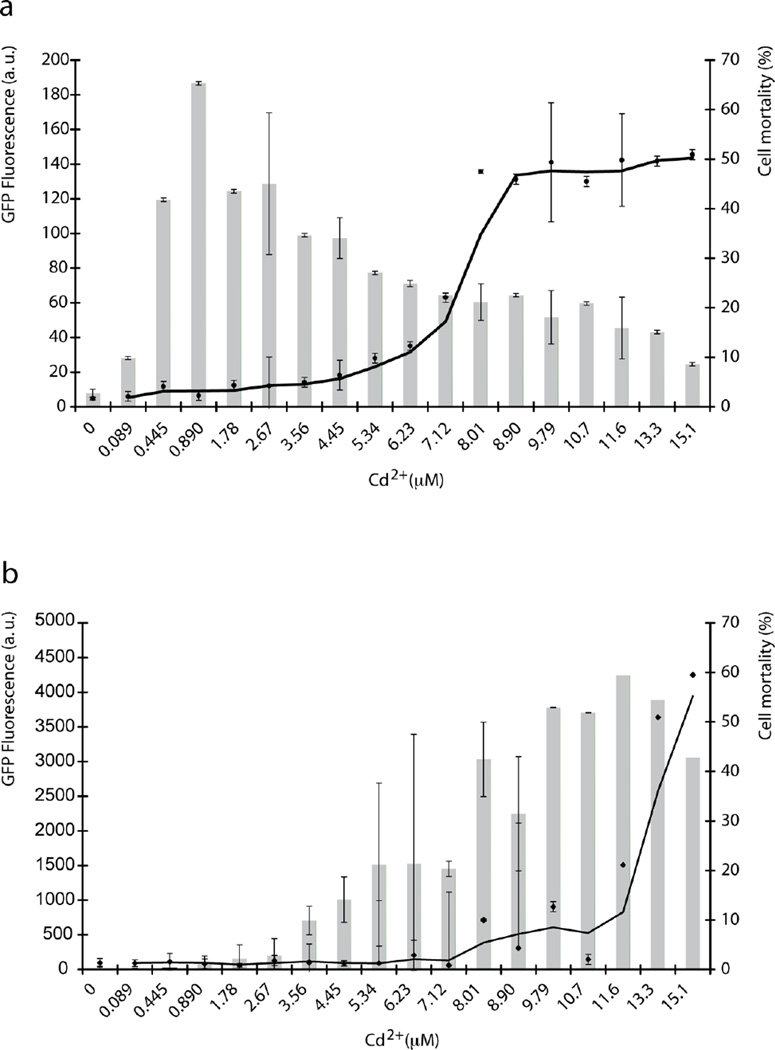
Fluorescence signal of GFPMTT5 strain was quantified by flow cytometry after incubation with different Cd2+ concentrations (89 nM – 15 µM) for 2 h (Fig. 3a). In the absence of metal, a low background level of GFP fluorescence (7.4 arbitrary units) was detected. The lowest Cd2+ concentration causing a noticeable GFP signal was 0.44 µM. Emission of fluorescence increased with increasing concentrations of Cd2+ to reach the maximum levels for 0.5 – 2.7 µM Cd2+. The highest fluorescence within this range was obtained after 2 h exposure to about 0.9 µM Cd2+ (Fig. 3a). At higher Cd2+ concentrations the GFP signal started to decrease, probably because of the toxicity of this metal. Cell mortality remained less than 10% up to 5.3 µM Cd2+, but protein biosynthesis may be inhibited at lower Cd2+ levels, thus affecting GFP accumulation (Fig. 3a). Beyond 8 µM Cd2+ about 50% mortality was observed and the fluorescence fell to a minimum at 14–15 µM Cd2+. Notwithstanding the toxicity, at all Cd2+ concentrations the treated cells showed a fluorescent signal considerable higher than background levels (untreated controls). Cadmium could be detected at concentrations as low as 0.44 µM using the GFPMTT5 strain as a WCB, according to the dose-response curve (Fig 3a).
Fig. 3.
Flow cytometry analysis (histograms) of the GFPMTT5 strain exposed to different Cd2+ concentrations (89 nM – 15 µM). (a) 2h or (b) 24h treatment. GFP fluorescence was quantified in arbitrary units (au). Continuous line represents the % of cell mortality. Data are the average of three different experiments. Error bars represent the standard deviation.
To evaluate whether long-term exposure to Cd2+ affects the viability of the GFPMTT5 cells, we also measured GFP fluorescence levels after 24 h of incubation with the same range of Cd2+ concentrations (Fig. 3b). Under these conditions, the GFP fluorescence levels were considerably higher than those detected for 2h of metal exposure. The lowest metal concentration inducing a significant fluorescence was 0.9 µM Cd2+. The fluorescence increased with increasing metal concentrations, and no mortality was detected up to 8 µM Cd2+. The highest fluorescence was reached at 11.56 µM Cd2+, whereas the production of GFP decreased at higher Cd2+ concentrations likely due to metal toxicity (Fig. 3b).
Interestingly, the GFPMTT5 strain was significantly more resistant to cadmium toxicity than previously characterized wild type strains of T. thermophila. Indeed, at 24h of Cd2+ exposure flow cytometry analysis revealed that the LC50 value for the GFPMTT5 strain (LC50 ~ 15 µM Cd2+) was about 9 fold higher than the LC50 for the previously reported SB1969 wild type strain (~ 1.73 µM) (Gallego et al. 2007). The increased cadmium resistance of GFPMTT5 might be explained by the higher MTT5 gene copy number, since this strain contains both the endogenous MTT5 copies (~ 45 macronuclear copies per cell) and the plasmid-encoded copies which are GFP-MTT5 fusions, which can number in the thousands (Cowan et al., 2005; Yao and Yao, 1991). As a result, exposure to Cd2+ induces the overproduction of the MTT5 protein in GFPMTT5 cells. In addition flow cytometry data suggest that the GFP does not affect the Cd2+-binding function of the MTT5, since the GFP-MTT5 fusion protein is able to chelate Cd2+ ions. These data constitute the first experimental evidence that Tetrahymena Cd-metallothionein MTT5 (Díaz et al. 2007) functions like metallothioneins in other living systems, sequestering toxic metal ions and so protecting the cell against these important environmental pollutants.
Expression analysis of GFP-MTT5 fusion gene by quantitative RT-PCR
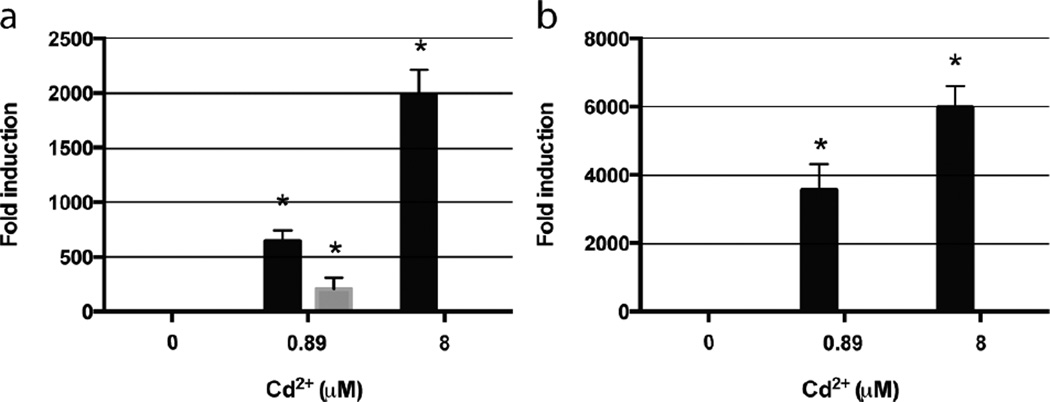
To test if increased Cd2+ resistance of the GFPMTT5 strain correlates with higher GFP-MTT5 gene expression levels, we analyzed by quantitative RT-PCR the expression level of both genes (gfp and MTT5) after 2h of Cd2+ exposure. As shown in Fig. 4a, cadmium exposure (0.9 µM) induced MTT5 gene expression in both strains (SB1969 and GFPMTT5). However, MTT5 mRNA levels were significantly more abundant in the GFPMTT5 strain, as expected due to increased gene copy number as discussed previously. The increase in MTT5 transcript level in GFPMTT5 compared to SB1969 was roughly 400-fold (Fig. 4a). While 8 µM Cd2+ was lethal for SB1969 cells, that concentration was tolerated by GFPMTT5. Under those conditions the levels of MTT5 transcript were roughly 2,000-fold higher than mRNA levels of α-tubulin gene (housekeeping gene control) (Fig. 4a). As a positive control, the expression of the gfp gene (included in the GFP::MTT5 reporter construct) was similarly quantified by RT-PCR. As expected, the gfp expression was dramatically increased under Cd2+ exposure (Fig. 4b). The RT-PCR values from the gfp transcripts were considerably higher (about 3–5 fold) than those obtained from the MTT5 gene (Fig. 4). This difference is probably due to the different lengths of the mRNAs transcribed from pVGFPMTT5. The gfp ORF is closer to the promoter (PMTT1) than the MTT5 ORF, so the former may be included in a subset of incomplete transcripts that lack a full length MTT5 sequence.
Fig. 4.
Relative expression levels of MTT5 (a) and gfp (b) genes from SB1969 (control strain) (grey bars) and GFPMTT5 strain (black bars), after Cd2+ exposure for 2h. Gene expression levels are shown relative to an untreated control (which is set at 1 ± 0.0 for every gene). Expression was normalized relative to the endogenous ATU1 (α-tubulin) gene. Each bar of the histogram corresponds to an average value of two independent experiments. Error bars represent the standard deviation. (*) : Significantly different from the control at p< 0.05.
Ultrastructural analysis
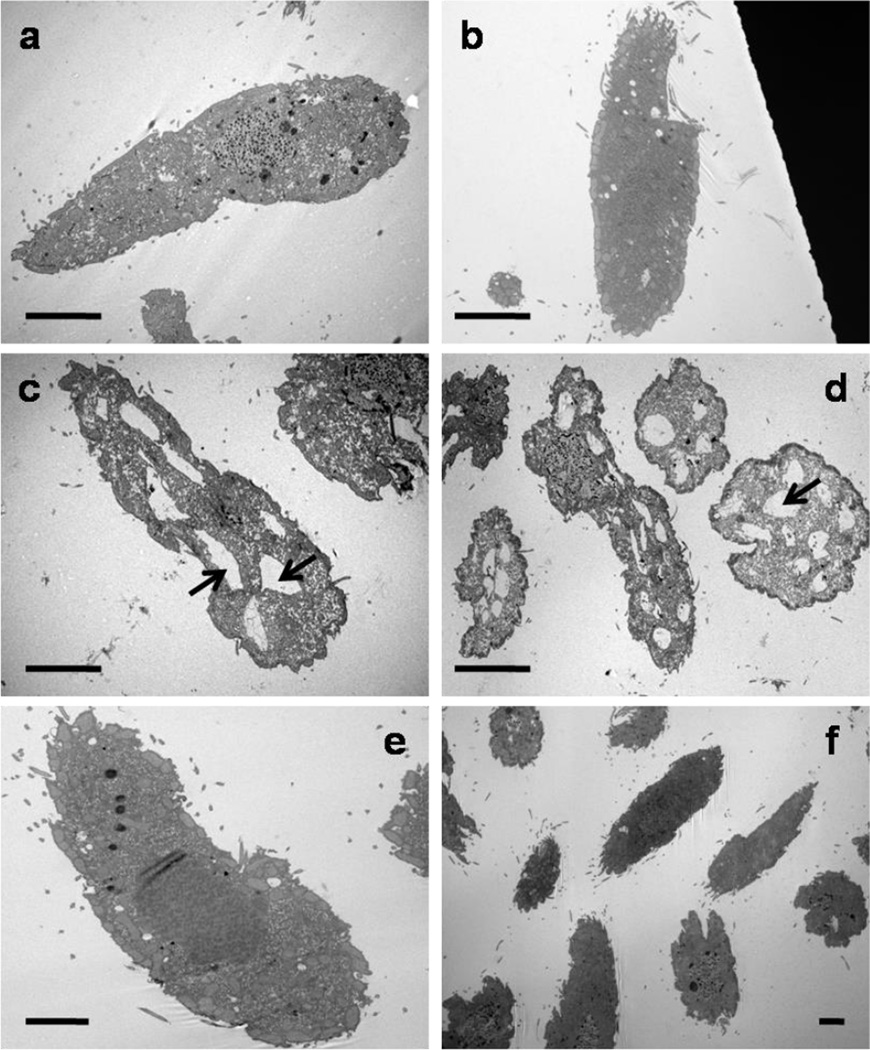
The results described above indicate that wild type T. thermophila (SB1969) experiences metal toxicity at much lower levels of cadmium than the GFPMTT5 strain. To validate this difference at the morphological level, we performed electron microscopic analysis of both strains under a variety of metal stress conditions. Cells were exposed to Cd2+ at 0.9 or 8 µM for either 2 or 24h. As shown in Fig. 5, untreated SB1969 and GFPMTT5 cells were morphologically similar (Fig. 5a, 5b). However, after treatment for 2h with 0.9 µM Cd2+, SB1969 cells showed intense vacuolization (7–10 vacuoles/cell in a longitudinal section) in the majority of cells (Fig. 5c, 5d), while the GFPMTT5 strain treated with 8 µM Cd2+ (2h) or 0.9 µM Cd2+ (24h) retained the appearance of untreated cells (Fig. 5e, 5f). We detected no changes in the structure of mitochondria, micro- or macronuclei, or cilia, in either strain under any condition tested.
Fig. 5.
Transmission electron microscopy micrographs of SB1969 (control strain) and GFPMTT5 strain, after Cd2+ treatments (0.9 or 8 µM during 2 or 24h) or without metal treatment. (a, b) SB1969 and GFPMTT5 strains without Cd2+ treatment. (c, d) SB1969 strain treated with 0.9 µM Cd2+ for 2h. Arrows indicate an intense vacuolization. (e, f) GFPMTT5 strain after 0.9 µM (24h) or 8 µM (2h) Cd2+ treatments, respectively. Bars: 10 µm.
Cytoplasmic vacuolization seems to be a common response to heavy metal exposure. It has been documented in Cd2+ treated microalgae (Nassiri et al. 1997; Visviki and Rachlin 1994) and in diverse ciliate species including the genus Tetrahymena (Coppellotti 1994; Dunlop and Chapman 1981; Martin-Gonzalez et al. 2006; Piccinni and Albergoni 1996; Pyne et al. 1983). Metals can change membrane permeability and therefore perturb the cellular osmotic equilibrium (Valko et al. 2005), which may explain the intensive cell vacuolization. The lack of vacuolization in the GFPMTT5 strain is consistent with the hypothesis that increased levels of MTT5 product successfully chelate the otherwise toxic Cd2+ and protect the cell from downstream effects.
GFPMTT1 / GFPMTT5 strains as potential whole cell biosensors for heavy metal monitoring
In 2011, we introduced the first WCB based on a ciliate metallothionein promoter (MTT1 or MTT5 gene) and the eukaryotic luciferase as a reporter gene (Amaro et al. 2011). These WCBs could rapidly detect bioavailable heavy metals in environmental aquatic or soil samples. Based on a comparison with other microbial metal cell biosensors, Tetrahymena WCBs (MTT1Luc or MTT5Luc) offer the most sensitive metal biosensors currently available among eukaryotic microorganisms, which for some applications are also more sensitive than available bacterial WCBs (Amaro et al. 2011). The GFPMTT1 and GFPMTT5 strains have three main differences compared with MTT1Luc or MTT5Luc: 1- a different reporter gene (gfp), while MTT1 or MTT5Luc strains have the luciferase gene; 2- the complete MTT1 or MTT5 ORF fused to GFP, and; 3- the GFP reporter constructs are expressed from the plasmid pVGF.MTT, while in Luc strains the reporter constructs are integrated in the macronuclear genome (Amaro et al. 2011).
A comparison (Table 1) between both types of Tetrahymena WCBs based on CdMT promoters suggests that each presents certain advantages and disadvantages.
Table 1.
Advantages and disadvantages of Tetrahymena WCBs based on metallothionein promoters with different reporter genes.
| Biosensor | Advantages | Disadvantages |
|---|---|---|
| GFPMTT5 / GFPMTT1 | No exogenous substrates required. | Minimal exposure time : 2h |
| Cells viable up to 15 µM Cd2+ | Minimum detectable Cd2+ concentration: 445 nM | |
| MTT1Luc / MTT5Luc (Amaro et al. 2011) | Minimal exposure time : 1h | Exogenous substrate required (D-luciferin) |
| Minimum detectable Cd2+ concentration: 5–25 nM | Bioluminiscence until 5 µM Cd2+ | |
The use of gfp as a reporter gene has the advantage that the reporter signal (fluorescence) can be detected in vivo directly (without adding exogenous substrates or cofactors). It can be easily detected by fluorescence microscopy and quantified by flow cytometry. Furthermore, the gfp ORF was fused with the complete MTT1 or MTT5 ORF in a multicopy plasmid, resulting in a higher metal tolerance in these strains. As a result, these WCBs might be effective at a higher level of Cd2+ concentrations. While Cd2+ concentrations ≥ 5 µM (2h treatment) are lethal for both SB1969 (wild type strain) and MTT1Luc (recombinant strain), the LC50 for GFPMTT5 is about 15 µM, so these GFP based biosensors might be very useful to test polluted samples with high metal concentrations.
However, GFPMTT5 / GFPMTT1 biosensors also have certain disadvantages compared with the previously reported MTT1Luc / MTT5Luc biosensors (Amaro et al. 2011). GFP must mature to become fully fluorescent, a process requiring several autocatalytic reactions (Tsien 1998). Thus, the reporter output following exposure to metals cannot be measured before 2 h. On the other hand, the bioluminescence signal from MTT1Luc / MTT5Luc strains is high enough to be readily detected after 1h of incubation with the sample. Another important difference is the sensitivity of detection or the minimal amount of metal that a biosensor is able to detect. For MTT1Luc the minimum detectable Cd2+ concentration was 25–50 nM (Amaro et al. 2011), while for the GFPMTT5 strain the level is about 445 nM. Several studies confirm that WCBs based on bioluminescence are more sensitive than those based on GFP (fluorescence) (Hakkila et al. 2002; van der Meer et al. 2004). However, Kohlmeier et al. (2007) have reported that when using a LIF-CS (Laser- induced fluorescence confocal spectroscopy) system to quantify fluorescence, the sensitivity of the GFP as reporter was similar to that of biosensors based on bioluminescence (LucFF or LuxFF genes).
Importantly, both types of biosensors based on Tetrahymena CdMT promoters have a Cd2+ sensitivity lower than the maximum permitted concentration of this toxic metal in soil (8.9 – 27 µM Cd2+) as established by the European Directive (86/278/CEE). However, like other prokaryotic or eukaryotic WCBs, Tetrahymena WCBs do not distinguish unambiguously between different heavy metals. The Tetrahymena CdMT promoters, from both the MTT1 or MTT5 genes, preferentially respond to Cd2+, but they are also inducible by other metals (Amaro et al. 2011; Díaz et al. 2007).
Acknowledgments
This work was supported by the Grant CGL2008-00317/BOS (MINECO) to JCG and by NIH GM077607 to APT. A PhD scholarship from Complutense University (UCM) was awarded to FA.
Abbreviations
- GFP
Green Fluorescent Protein
- IP
Iodine Propidium
- MT
metallothionein
- RT-PCR
Retro-Transcription-Polymerase chain reaction
- TEM
Transmission Electron Microscopy
- WCB
Whole-Cell Biosensor
References
- Amaro F, Turkewitz AP, Martin-Gonzalez A, Gutierrez JC. Whole-cell biosensors for detection of heavy metal ions in environmental samples based on metallothionein promoters from Tetrahymena thermophila. Microb. Biotechnol. 2011;4:513–522. doi: 10.1111/j.1751-7915.2011.00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aury JM, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Ségurens B, Daubin V, Anthouard V, Aiach N, Arnaiz O, Billaut A, Beisson J, Blanc I, Bouhouche K, Câmara F, Duharcourt S, Guigo R, Gogendeau D, Katinka M, Keller AM, Kissmehl R, Klotz C, Koll F, Le Mouël A, Lepère G, Malinsky S, Nowacki M, Nowak JK, Plattner H, Poulain J, Ruiz F, Serrano V, Zagulski M, Dessen P, Bétermier M, Weissenbach J, Scarpelli C, Schächter V, Sperling L, Meyer E, Cohen J, Wincker P. Global trends of the whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- Belkin S. Microbial whole-cell sensing systems of environmental pollutants. Curr. Opin. Microbiol. 2003;6:206–212. doi: 10.1016/s1369-5274(03)00059-6. [DOI] [PubMed] [Google Scholar]
- Boldrin F, Santovito G, Formigari A, Bisharyan Y, Cassidy-Hanley D, Clark TG, Piccinni E. MTT2, a copper-inducible metallothionein gene from Tetrahymena thermophila. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2008;147:232–240. doi: 10.1016/j.cbpc.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Coppellotti O. Effects of cadmium on Uronema marinum (Ciliophora, Scuticociliatida) from Antartica. Acta Protozool. 1994;33:159–167. [Google Scholar]
- Cowan AT, Bowman GR, Edwards KF, Emerson JJ, Turkewitz AP. Genetic, genomic and functional analysis of the granule lattice proteins in Tetrahymena secretory granules. Mol. Biol. Cell. 2005;16:4046–4060. doi: 10.1091/mbc.E05-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentler W. Fixation of Tetrahymena cells for electron microscopy. In: Asai DJ, Forney JD, editors. Tetrahymena thermophila. Methods in Cell Biology. Vol. 62. London: Academic Press; 2000. pp. 323–331. [DOI] [PubMed] [Google Scholar]
- Díaz S, Amaro F, Rico D, Campos V, Benitez L, Martin-Gonzalez A, Hamilton EP, Orias E, Gutierrez JC. Tetrahymena metallothioneins fall into two discrete subfamilies. PloS ONE. 2007;2:e291. doi: 10.1371/journal.pone.0000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D´Souza SF. Microbial biosensors. Biosens. Bioelectron. 2001;49:337–353. doi: 10.1016/s0956-5663(01)00125-7. [DOI] [PubMed] [Google Scholar]
- Dunlop S, Chapman G. Detoxification of zinc and cadmium by the freshwater protozoan Tetrahymena pyriformis. II Growth experiments and ultrastructural studies on sequestration of heavy metals. Environ. Research. 1981;24:264–274. doi: 10.1016/0013-9351(81)90185-7. [DOI] [PubMed] [Google Scholar]
- Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, Tallon LJ, Delcher AL, Salzberg SL, Silva JC, Haas BJ, Majoros WH, Farzad M, Carlton JM, Smith RK, Jr, Garg J, Pearlman RE, Karrer KM, Sun L, Manning G, Elde NC, Turkewitz AP, Asai DJ, Wilkes DE, Wang Y, Cai H, Collins K, Stewart BA, Lee SR, Wilamowska K, Weinberg Z, Ruzzo WL, Wloga D, Gaertig J, Frankel J, Tsao CC, Gorovsky MA, Keeling PJ, Waller RF, Patron NJ, Cherry JM, Stover NA, Krieger CJ, del Toro C, Ryder HF, Williamson SC, Barbeau RA, Hamilton EP, Orias E. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PloS Biol. 2006;4:c286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertig J, Gorovsky MA. Efficient mass transformation of Tetrahymena thermophila by electroporation of conjugants. Proc. Natl. Acad. Sci. USA. 1992;89:9196–9200. doi: 10.1073/pnas.89.19.9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego A, Martin-Gonzalez A, Ortega R, Gutierrez JC. Flow cytometry assessment of cytotoxicity and reactive oxygen species generation by single and binary mixtures of cadmium, zinc and copper on populations of the ciliated protozoan Tetrahymena thermophila. Chemosphere. 2007;68:647–661. doi: 10.1016/j.chemosphere.2007.02.031. [DOI] [PubMed] [Google Scholar]
- Gutierrez JC, Amaro F, Martin-Gonzalez A. From heavy metal-binder to biosensors: ciliate metallothioneins discussed. BioEssays. 2009;31:805–816. doi: 10.1002/bies.200900011. [DOI] [PubMed] [Google Scholar]
- Gutierrez JC, Amaro F, Diaz S, de Francisco P, Cubas LL, Martin-Gonzalez A. Ciliate metallothioneins: unique microbial eukaryotic heavy-metal-binder molecules. J. Biol. Inorg. Chem. 2011;16:1025–1034. doi: 10.1007/s00775-011-0820-9. [DOI] [PubMed] [Google Scholar]
- Gutierrez JC, Martin-Gonzalez A, Diaz S, Ortega R. Ciliates as potential source of cellular and molecular biomarker/biosensors for heavy metal pollution. Eur. J. Protistol. 2003;39:461–467. [Google Scholar]
- Gutierrez JC, Martin-Gonzalez A, Diaz S, Amaro F, Ortega R, Gallego A, Lucas MP. Ciliates as cellular tools to study the eukaryotic cell – heavy metal interactions. In: Brown SE, Welton WC, editors. Heavy Metal Pollution. New York: Nova Science Publishers; 2008. pp. 1–44. [Google Scholar]
- Hamilton EP, Orias E. Genetically mapping new mutants and cloned genes. In: Asai DJ, Forney JD, editors. Tetrahymena thermophila. Methods in Cell Biology. Vol. 62. London: Academic Press; 2000. pp. 265–280. [DOI] [PubMed] [Google Scholar]
- Keith LH, Telliard WA. Priority pollutants: a perspective view. Env. Sci. Tech. 1979;13:416–423. [Google Scholar]
- Kohlmeier S, Mancuso M, Tecon R, Harms H, van der Meer JR, Wells M. Bioreporters: gfp versus lux revisited and single-cell response. Biosens. Bioelectron. 2007;22:1578–1585. doi: 10.1016/j.bios.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Larionov A, Krause A, Miller W. A standard curve based method for relative real time PCR data processing. BMC Bioinformatics. 2005;6:62. doi: 10.1186/1471-2105-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley DR. Environmental Microbe-Metal Interactions. Washington DC: ASM Press; 2000. [Google Scholar]
- Magrisso S, Erel Y, Belkin S. Microbial reporters of metal bioavailability. Microbial. Biotech. 2008;1:320–330. doi: 10.1111/j.1751-7915.2008.00022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Gonzalez A, Diaz S, Borniquel S, Gallego A, Gutierrez JC. Cytotoxicity and bioaccumulation of heavy metals by ciliated protozoa isolated from urban wastewater treatment plants. Res. Microbiol. 2006;157:108–118. doi: 10.1016/j.resmic.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Nassiri Y, Mansot JL, Wery J, Ginsburger-Vogel T, Amiard JC. Ultrastructural and electron energy loss spectroscopy studies of sequestration mechanisms of Cd and Cu in the marine diatom Skeletonema costatum. Arch. Environ. Contam. Toxicol. 1997;33:147–155. doi: 10.1007/s002449900236. [DOI] [PubMed] [Google Scholar]
- Park M, Tsai S-L, Chen W. Microbial biosensors: Engineered microorganisms as the sensing machinery. Sensors. 2013;13:5777–5795. doi: 10.3390/s130505777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccinni E, Albergoni V. Cadmium detoxification in Protists. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 1996;113C:141–147. [Google Scholar]
- Pyne CK, Iftode F, Curgy J. The effects of cadmium on the growth pattern and ultrastructure of the ciliate Tetrahymena thermophila. Biol. Cel. 1983;48:121–132. [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Shang Y, Song X, Bowen J, Corstanje R, Gao Y, Gaertig J, Gorovsky MA. A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA. 2002;97:3734–3739. doi: 10.1073/pnas.052016199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien RY. The green fluorescent protein. Annu. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- Valko M, Morris H, Cronin MT. Metals, toxicity and oxidative stress. Curr. Med. Chem. 2005;12:1161–1208. doi: 10.2174/0929867053764635. [DOI] [PubMed] [Google Scholar]
- Van der Meer JR, Belkin S. Where microbiology meets microengineering: design and applications of reporter bacteria. Nat. Rev. Microbiol. 2010;8:511–522. doi: 10.1038/nrmicro2392. [DOI] [PubMed] [Google Scholar]
- Visviki I, Rachlin JW. Acute and chronic exposure of Dunaliella salina and Chlamydomonas bullosa to copper and cadmium: effects on growth. Arch. Environ. Contam. Toxicol. 1994;26:149–153. doi: 10.1007/BF00224798. [DOI] [PubMed] [Google Scholar]
- Walmsley RM, Keenan P. The eukaryote alternative: advantages of using yeasts in place of bacteria in microbial biosensor development. Biotechnol. Bioprocess Eng. 2000;5:387–394. [Google Scholar]
- Yao M-C, Yao C-H. Transformation of Tetrahymena to cycloheximide resistance with a ribosomal protein gene through sequence replacement. Proc. Natl. Acad. Sci. USA. 1991;88:9493–9497. doi: 10.1073/pnas.88.21.9493. [DOI] [PMC free article] [PubMed] [Google Scholar]