Abstract
Colorectal cancer (CRC) is one of the leading causes of cancer mortality in Western civilization. Studies have shown that CRC arises as a consequence of the modification of genes that regulate important cellular functions. Deregulation of the WNT and RAS/MAPK/PI3K signaling pathways has been shown to be important in the early stages of CRC development and progression. Krüppel-like factor 5 (KLF5) is a transcription factor that is highly expressed in the proliferating intestinal crypt epithelial cells. Previously, we showed that KLF5 is a mediator of RAS/MAPK and WNT signaling pathways under homeostatic conditions and that it promotes their tumorigenic functions during the development and progression of intestinal adenomas. Recently, using an ultrahigh-throughput screening (uHTS) approach we identified a number of novel small molecules that have the potential to provide therapeutic benefits for colorectal cancer by targeting KLF5 expression. In the current study, we show that an improved analog of one of these screening hits, ML264, potently inhibits proliferation of CRC cells in vitro through modifications of the cell cycle profile. Moreover, in an established xenograft mouse model of colon cancer, we demonstrate that ML264 efficiently inhibits growth of the tumor within five days of treatment. We show that this effect is caused by a significant reduction in proliferation and that ML264 potently inhibits the expression of KLF5 and EGR1, a transcriptional activator of KLF5. These findings demonstrate that ML264, or an analog may hold a promise as a novel therapeutic agent to curb the development and progression of colorectal cancer.
Keywords: KLF5, ML264, colon cancer, xenografts
INTRODUCTION
Colorectal cancer (CRC) is the third most prevalent cancer in the United States with approximately 140,000 new cases and 50,000 deaths each year (1). The progression from normal intestinal epithelial tissue to metastatic neoplasm results from the disruption of multiple regulatory mechanisms involving critical signaling pathways that normally regulate proliferation, differentiation, migration, and apoptosis (2, 3). Previous research has coalesced around a model of CRC development whereby dysregulation of WNT and RAS/MAPK/PI3K signaling pathways is necessary for early development and progression (4). That the sequential impairment of each of these pathways is a requirement for developing intestinal neoplasia has been recently confirmed in an organoid system by Drost et al. (5). Despite this knowledge, few therapeutic strategies have been developed to specifically target components of these particular pathways and none have gained FDA approval. Our group has identified Krüppel-like factor 5 (KLF5), a WNT- and RAS/MAPK/PI3K-responsive molecule, as an important regulator of intestinal epithelial cell proliferation that is frequently overexpressed during intestinal tumorigenesis.
Krüppel-like factor 5 (KLF5) is a zinc-finger transcription factor highly expressed in the intestinal epithelium crypts (6). We previously demonstrated that transit-amplifying (TA) cells of the intestinal epithelium express high levels of KLF5 that co-localize with the proliferation marker, Ki-67 (6, 7). More recently, we demonstrated that KLF5 is also expressed in the crypt base columnar (CBC) cells of the intestinal crypts (8). CBC cells, which express the leucine-rich-repeat-containing G-protein-coupled receptor 5 (LGR5) gene, proved to be the dividing and long-lived intestinal stem cells (ISCs) that give rise to all lineages of the intestinal epithelium (9–12). Importantly we recently showed that KLF5 is essential for the proliferation and survival of LGR5-expressing ISCs (8). Mechanistically, we and others have shown that KLF5 expression is regulated by both the RAS/MAPK/PI3K and WNT signaling pathways, but via distinct mechanisms. While the RAS/MAPK/PI3K pathway exerts its action directly through early growth factor 1 (EGR1), a potent transcriptional activator of KLF5 (13, 14), the WNT pathway regulates the rate of the KLF5 protein degradation through GSK3β phosphorylation and a subsequent FBW7α-dependent degradation (15–17). Additionally, we have demonstrated that KLF5 is itself a positive regulator of both the RAS/MAPK and WNT signaling pathways. The RAS/MAPK pathway is affected by KLF5-mediated EGFR activation, while WNT signaling is stimulated by the ability of KLF5 to enhance β-catenin stability, nuclear localization and transcriptional activity (18, 19). Taken together, these observations illustrate the important role that KLF5 plays in the maintenance of homeostasis of intestinal epithelium by governing the activity of stem and TA cells.
Furthermore, we have shown that elevated levels of oncogenic KRAS, which is associated with hyperactive MAPK and PI3K signaling, result in the upregulation of KLF5 expression in both human primary colorectal cancers and transgenic mouse models (14, 20). KLF5 overexpression in turn potentiates the transforming activity of KRAS by increasing the proliferation rate of cells (20). Our group has also demonstrated that reduced Klf5 expression leads to reduced intestinal tumor formation in mice harboring a germline mutation in the tumor suppressor Apc, a crucial component of the WNT pathway, alone or in combination with activating KRAS mutations (18, 20, 21). Additionally, it has been recently demonstrated that KLF5 expressed in CBCs facilitates the oncogenic activity of mutated β-catenin promoting development of intestinal adenomas, while Klf5 deletion abrogates this process (22). Moreover, we have evidence that KLF5 expression levels are highest in cancer cells of colorectal cancer origin among the NCI60 panel of cancer cells (23). These lines of evidence suggest that small molecule compounds that decrease KLF5 expression could prove to be an effective therapeutic option for CRC.
We generated CRC cell lines stably expressing the luciferase reporter from the human KLF5 promoter and utilized these cells in an ultrahigh-throughput screening (uHTS) approach to identify compounds that modulated KLF5 expression (23, 24). Previously, we demonstrated that this screening method allows for specific identification of compounds that decrease KLF5 expression levels and that inhibit proliferation of CRC cell lines in in vitro systems (23, 24). Here, we show that ML264, a third-generation small molecule compound that arose from the first-generation of uHTS hits, potently inhibits KLF5 expression, decreases proliferation of CRC cell lines, and inhibits the growth of xenografts in a mouse model of primary tumor development.
MATERIALS AND METHODS
Cell lines and reagents
DLD-1 and HCT116 colorectal cancer cell lines were purchased from the American Type Culture Collection (ATCC). DLD-1 cells were maintained in RPMI1640 medium supplemented with 10% FBS and 1% penicillin/streptomycin, and HCT116 cells were maintained in McCoy's medium supplemented with 10% FBS and 1% penicillin/streptomycin. We routinely carry out morphology checks on all cell lines and we only passage the cell lines for three months. In addition, the cell lines were tested for Mycoplasma contamination. Furthermore, each experiment had appropriate controls to assure the behavior of tested cell lines. The compound ML264 was synthesized at The Scripps Research Institute in the laboratory of Dr. Thomas Bannister (25). The structure of ML264 compound and its synthesis pathway have been previously published (25). For in vitro experiments, ML264 was dissolved in dimethyl sulfoxide (DMSO, Fisher Scientific). For in vivo studies ML264 was dissolved in the vehicle solution: 80% dH2O, 10% DMSO and 10% Tween 80. The antibodies used for this study are listed in the Supplementary Table 1.
Cell proliferation, cell cycle and apoptosis assays
For cell proliferation experiments, DLD-1 and HCT116 cells were treated with 10µM ML264 or with vehicle (DMSO). Live cells were collected at 24, 48 and 72 hours post treatment and their numbers were determined by counting using a Coulter counter (Beckman Coulter). Each experiment was done in triplicate. In MTS assay, DLD-1 and HCT116 cells were treated with 10µM ML264 or with vehicle (DMSO). After 24, 48, and 72 hours of incubations, 20 µL of MTS solution (Promega, Cat. #G3582) was added to each well and an analysis was performed according to the manufacturer’s protocol. The measurement of the control (cells with medium and DMSO) was defined as 100% and the results from other measurements were calculated accordingly. Each experiment was done in sextuplicate. A cell cycle progression assay was performed as described previously (23). Each experiment was done in triplicate. The apoptosis rate was determined using the Alexa Fluor 488 Annexin V/Dead Cell Apoptosis Kit (Life Technologies, Cat. #V13241) according to the manufacturer’s instructions with analysis by flow cytometry. Each experiment was done in triplicate.
Western Blot analysis
Total protein was extracted from cells with Laemmli buffer and the analysis was performed as described previously (23).
RNA analysis
Total RNA from DLD-1 and HCT116 cells was used for quantitative PCR. RNA was extracted using TRIzol Reagent (Life Technologies, Cat. #15596) according to the manufacturer’s instructions. Primers against human KLF5, EGR-1, CTNNB1, CCND1, CCNE1, CCNA2, CCNB1 and GAPDH were purchased from Qiagen. Their respective catalogue numbers are QT00074676, QT00218505, QT00077882, QT00495285, QT00041986, QT00014798, QT00006615 and QT00079247. Quantitative PCR was performed using the QuantiTect SYBR Green RT-PCR Kit (Qiagen, Cat. #204243) as per standard protocols. Observed CT values were then used to calculate fold change using the 2-ΔΔCt method of relative quantification (26). Human GAPDH was used as the housekeeping gene.
Immunofluorescence and immunohistochemistry
Tumors dissected from mice were first fixed in Bouin's fixative (50% ethanol +5% acetic acid in water) for 1 hour, then fixed overnight in 10% buffered formalin (Fisher Scientific, PA, USA). The tissues were then paraffin-embedded using an automated processor, sectioned at 5µm, collected onto charged slides and baked in a 65°C oven overnight, and were subsequently deparaffinized in xylene. Sections were incubated in a 2% hydrogen peroxide in methanol bath to block endogenous tissue peroxidases and were then rehydrated by incubation in a decreasing ethanol bath series (100%, 95%, 70%) followed by antigen retrieval in citrate buffer solution (10mM sodium citrate, 0.05% Tween-20, pH 6.0) at 120°C for 10 minutes using a decloaking chamber (Biocare Medical). Tissue sections were first incubated with blocking buffer (5% BSA in TBS-Tween) for 30 minutes at 37°C and then with primary antibody at 4°C overnight in a humidified chamber with gentle shaking. The list of primary antibodies used is shown in Supplementary Table 1. Sections were washed and incubated with secondary antibodies (HRP-conjugated or fluorescent-tagged) at the appropriate concentration for 30 minutes at 37°C. Betazoid DAB (Biocare Medical) was used to reveal IHC staining in tissues. For fluorescent sections, slides were washed after secondary antibody treatment and then stained with Hoechst (AnaSpec Inc.) and mounted with Prolong gold antifade (Life Technologies). Slides were analyzed under a Nikon Eclipse 90i microscope (Nikon) and representative photomicrographs were taken.
H&E staining
Histology of sections was observed upon staining 5µm sections that were fixed, paraffin-embedded, deparaffinized and rehydrated as mentioned before. Then they were stained with Hematoxylin Stain Solution, Gill 3 (Ricca Chemical Company) and Eosin Y (Sigma-Aldrich). Sections were dehydrated in an increasing series of ethanol bath (70%, 95%, 100%), cleared in xylene and mounted with Cytoseal XYL xylene-based mounting media (Thermo Scientific, Cat. #8312-4).
Mitotic figures quantification
In in vitro studies, DLD-1 cells were seeded onto slides with medium containing DMSO or 10µM ML264, fixed and stained with Hoechst (DNA labeling) after 24, 48 and 72 hours of treatment. To quantify the number of mitotic figures five fields, each with 100 cells were counted. For in vivo studies mitotic figures were counted in fifteen images of three vehicle- and ML264-treated animals using slides with H&E staining.
Ki-67 quantification
Ki-67 was quantified using images from three animals treated with vehicle and with ML264. We analyzed three images per animal per treatment using ImageJ software (27).
Xenografts
All mice studies were approved by the Stony Brook University Institutional Animal Care and Use Committee (IACUC). Nude mice were purchased from Jackson Laboratories (Bar Harbor). Animals were housed under specific pathogen-free conditions in ventilated and filtered cages under positive pressure. Xenograft tumors were generated by injecting subcutaneously 5×106 DLD-1 human colorectal cells into the right flank of 6–7 week old male nude mice. Tumor volume was determined by caliper measurement and calculated by established methods (28). When tumors reached a volume of about 100 mm3, mice were treated intraperitoneally (i.p.) with varying doses of ML264: 10 mg/kg daily, 10 mg/kg twice per day and 25 mg/kg twice per day, with each treatment regimen lasting for a duration of 10 days. The vehicle solution was used as the control treatment. Mice were monitored and weighed every two days. Experiments were terminated when the tumor’s greatest measurement reached 2 cm. Tumors were excised and retained for further analyses.
Statistical analysis
The analysis of in vitro experiments was performed with Student’s t-test. A value of p < 0.05 was considered significant. This analysis was performed using GraphPad Prism version 5.00 for Windows (GraphPad Software, Sand Diego, CA). A comparison of tumor progression between the different treatment regimens and the control group was performed at 5 and 10 days post-treatment by constructing linear mixed models for longitudinal data arising from every experiment. Autoregressive with order 1 structure was used to model the dependence among time points – the best structure compared with compound symmetric and unstructured. Pre-specified comparisons between treatment and vehicle group were made. A p-value less than 0.05 was considered statistically significant and analysis was performed using SAS 9.3 (SAS Institute Inc., Cary, NC).
RESULTS AND DISCUSSION
ML264 inhibits the growth of colorectal cancer cell lines in vitro
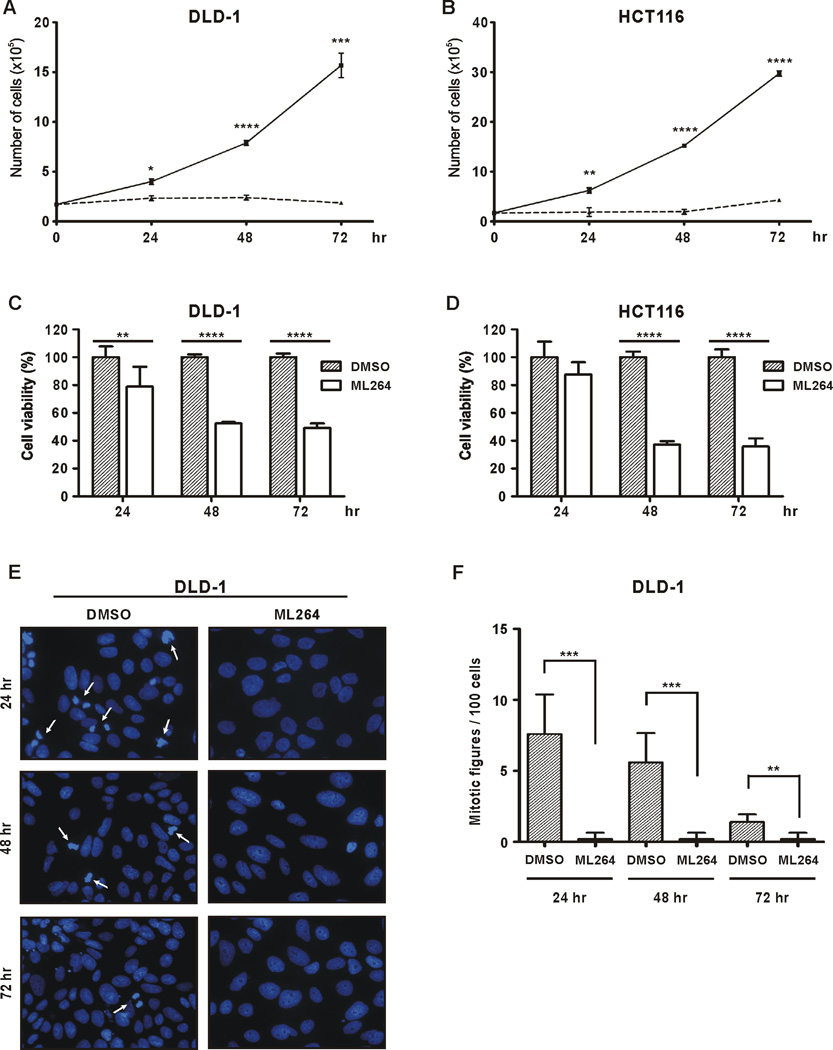
ML264 is a third-generation small molecule compound that has been developed with the goal of inhibiting the growth of colorectal cancer cells (CRCs) by reducing the activity of the oncogene KLF5 (25). We designed a series of experiments to test the efficacy of ML264 using DLD-1 and HCT116 CRC cell lines due to the relatively high levels of expression of KLF5 in these cells. We tested the effects of ML264 on the rate of cell proliferation of colon cancer cells lines DLD-1 (Fig. 1A) and HCT116 (Fig. 1B) over 72 hours. ML264 efficiently inhibited the rate of proliferation of both cell lines. A significant decrease in proliferation was evident within 24 hours of treatment (Figs. 1A and 1B) and by 72 hours the live cell numbers of ML264-treated and vehicle-treated cells differed by 15- to 30- fold. Additionally, we performed an MTS assay that allows the quantification of metabolically active cells. As shown in Figures 1C (DLD-1) and 1D (HCT116), both cell lines demonstrated a reduced level of viability that is in concordance with the results presented in Figures 1A and 1B.
Figure 1. ML264 inhibits proliferation of colorectal cancer cell lines.
(A) DLD-1 and (B) HCT116 cells were seeded in 6 well plate format with medium containing DMSO or 10µM ML264. Twenty-four, forty-eight and seventy two hours post-treatment cells were counted using a cell counter. The solid lines represent control and the dotted lines treatment with ML264. Data represent mean ± S.D. (n=3). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. (C) DLD-1 and (D) HCT116 cells were seeded in 96 well plate format with medium containing DMSO or 10µM ML264. Twenty-four, forty-eight and seventy two hours post-treatment cells were analyzed using MTS assay. The measurement of the control (cells with medium and DMSO) was defined as 100% and the results from other measurements were calculated accordingly. Data represent mean ± S.D. (n=6). **p < 0.01, ****p < 0.0001. (E) DLD-1 cells were seeded onto slides with medium containing DMSO or 10µM ML264, fixed and stained with Hoechst (DNA labeling) after 24, 48 and 72 hours of treatment. White arrows marked cells undergoing mitosis. (F) Quantitative representation of (C). Five fields, each with 100 cells were counted. Data represent mean ± S.D. (n=5). **p < 0.01, ***p < 0.001.
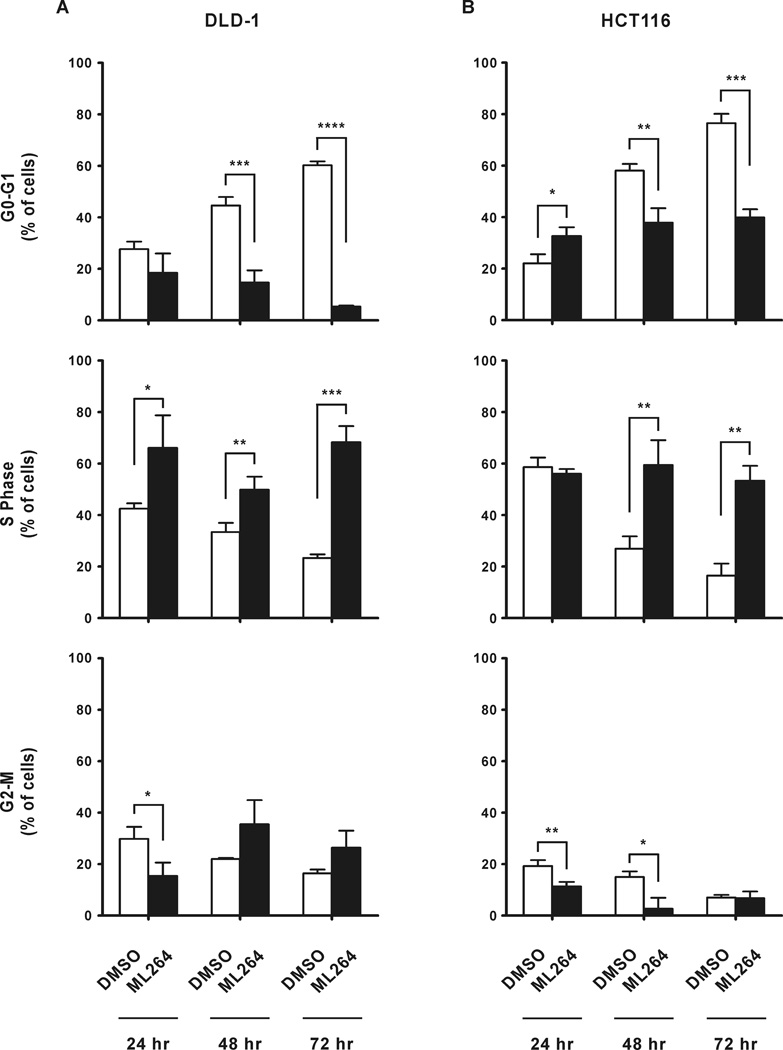
To determine the mechanism by which ML264 inhibits cancer cell proliferation, we first investigated the effect of ML264 on the number of mitotic figures in DLD-1 cells, cultured in the presence of ML264 or vehicle, over a three-day period. As seen in Figures 1E and 1F, ML264 treatment significantly reduced the number of cells undergoing mitosis in DLD-1 cells at 24, 48 and 72 hours. This result suggests that ML264 either prevents cells from entering into or progressing through mitosis. Next, we treated DLD-1 and HCT116 cells with ML264 or with vehicle for up to 72 hours and assessed the cell cycle profile or rate of apoptosis by staining with propidium iodide (PI) alone (Figs. 2A and 2B) or in combination with Annexin V (Suppl. Figs. 1A and 1B), respectively. ML264 treatment led to a significant decrease in the population of cells in G0/G1 phase at 48 and 72 hours for both cell lines (Figs. 2A and 2B, top panels). In ML264-treated DLD-1 cells, there was a significant increase in the S-phase population at 24 hours, which persisted until 72 hours (Fig. 2A, middle panel). In ML264-treated HCT116 cells an increase in S-phase cells was seen at 48 hours that persisted through 72 hours (Fig. 2B, middle panel). Additionally, we noted changes in the number of ML264-treated cells in the G2/M phase. In DLD-1 cells, an initial decrease in G2/M cells was seen at 24 hours and this was followed by an increase in this population at 48 and 72 hours (Fig. 2A, bottom panel). In contrast, ML264-treated HCT116 cells showed a decrease in G2/M population at 24 and 48 hours (Fig. 2B, bottom panel). Furthermore, Annexin V/PI staining of these cells demonstrated only a modest, though significant, increase in apoptotic cells following ML264 treatment (Suppl. Figs. 1A and 1B; Annexin V and PI double-positive fraction). Early apoptotic cells (Annexin V single-positive) increased in ML264 treated DLD-1 cells at 48 hours and in HCT116 cells at 48 and 72 hours. In both DLD-1 and HCT116 cells, late apoptotic cells (Annexin V and PI double-positive fraction) increased. In DLD-1 cells the increase was noted at 48 hours and in HCT116 at 48–72 hours post-treatment with ML264. It is unlikely that these modest changes in apoptotic cells explain the dramatic difference in cell number that was initially observed. Our data indicate that inability of DLD-1 and HCT116 cells to enter mitosis could be a consequence of S-phase arrest upon ML264 treatment. Thus, these results suggest that ML264 inhibits cell proliferation mainly by modifying cell cycle progression, though the induction of apoptosis may also contribute to this phenotype.
Figure 2. ML264 modifies cell cycle progression in colorectal cancer cell lines.
(A) DLD-1 and (B) HCT116 cells were seeded in 60 mm plate format with medium containing DMSO or 10µM ML264. Twenty-four, forty-eight and seventy two hours post-treatment cells were collected for cell cycle analysis with propidium iodide. Each experiment was performed in triplicate and data is shown as mean ± S.D. (n=3). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
The impact of ML264 on signaling of the RAS/MAPK/PI3K and WNT pathways
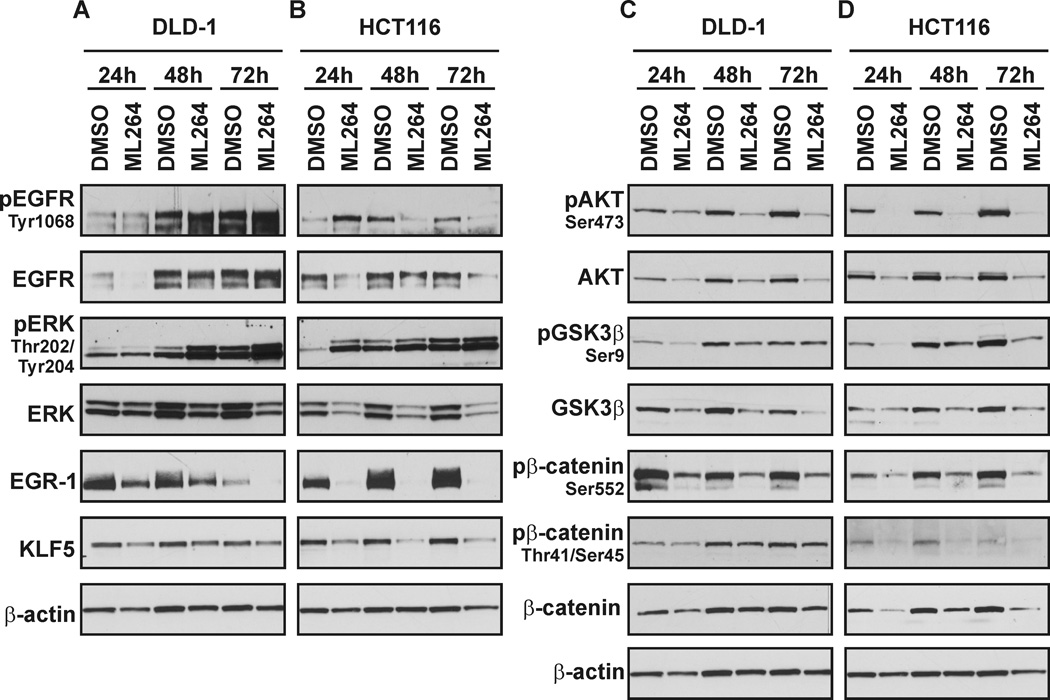
ML264 and related analogs emerged from a screen that was designed to identify small molecule that inhibit the expression of oncogenic KLF5 and that therefore would inhibit the growth of CRCs (23). We investigated how ML264 affects the expression levels of proteins involved in several of the signaling pathways (MAPK, WNT and PI3K) that regulate either KLF5 expression and activity or the progression through the cell cycle (14, 24, 29, 30). We collected protein and RNA samples over a period of three days from DLD-1 and HCT116 CRCs that had been treated with vehicle or with ML264 and analyzed them by Western blot and by qRT-PCR. As seen in Figures 3A and 3B, the protein levels of p-EGFR and EGFR are differentially modulated by ML264 in a cell line-dependent manner. In DLD-1 cells, the basal levels of EGFR protein are not significantly changed, yet there is an increase in the phosphorylation status of EGFR at 48 and 72 hours post-treatment with ML264 as compared to control (Fig. 3A). On the other hand, we observed a slight decrease in the basal levels of EGFR in HCT116 cells treated with ML264 over the three-day period and a decrease of its phosphorylation status at 48 and 72 hours (Fig. 3B). The basal levels of ERK decreased and this decline was accompanied by an up-regulation of its phosphorylated form, in both DLD-1 and HCT116 cells. However, both ML264-treated cell lines showed a decrease in the protein levels of KLF5 that parallels a reduction in the levels of the transcription factor early growth response 1 (EGR1), a direct activator of KLF5 expression (Figs. 3A and 3B) (13).
Figure 3. Inhibitory effects of ML264 on protein levels of selected components of the MAPK, WNT and PI3K signaling pathways.
DLD-1 and HCT116 cells were seeded with medium containing DMSO or 10µM ML264. Twenty-four, forty-eight and seventy two hours post-treatment cells were collected for protein analysis. (A) DLD-1 and (B) HCT116 – Representative Western blots of selected components of MAPK signaling pathway, (C) DLD-1 and (D) HCT116 – Representative Western blots of selected components of PI3K and WNT signaling pathways.
Previously, we and others have shown that KLF5 plays an important role in regulating the activity of β-catenin, while KLF5 in turn is regulated by the WNT and PI3K signaling pathways (18, 22, 24). Our Western blot analyses show a down-regulation of the basal levels of AKT and GSK3β in both cell lines following ML264 treatment, a decrease that was accompanied by a similar decline in pAKT and pGSK3β levels (Figs. 3C and 3D). We also observed a decrease in the levels of the active, nuclear form of β-catenin, which is phosphorylated at serine 552. This reduction in β-catenin phosphorylated at serine 552 is likely the result of the decrease in AKT levels, as this kinase is known to regulate the phosphorylation status of serine 552 (31, 32). Additionally, we have noticed that in DLD-1 cells treated with DMSO β-catenin phosphorylated at serine 552 predominantly localizes to the cells that are undergoing mitosis and that it is absent in the DLD-1 cells treated with ML264 for the three day period (data not shown). Furthermore, we have analyzed the levels of β-catenin that is phosphorylated at threonine 41/serine 45, which is a pool of β-catenin destine for degradation as well as the levels of total β-catenin (33). No significant difference in total β-catenin and β-catenin phosphorylated at threonine 41/serine 45 were observed in ML264-treated DLD-1 cells. That observation, combined with the fact that levels of GSK3β were reduced, suggests that the degradation pathway of β-catenin is not up regulated upon ML264 treatment in DLD-1 cells. In contrast to DLD-1, HCT116 cells show reduced levels of both total β-catenin and the levels of β-catenin phosphorylated at threonine 41/serine 45 following ML264 treatment.
Based on our results, ML264 affects the RAS/MAPK signaling pathway primarily by decreasing the levels of EGR1 and KLF5. A preliminary qPCR analysis showed that there is a decrease in the mRNA levels of both genes already after eight hours of treatment with ML264 relative to control (data not shown). This implies that ML264 may directly impact the transcription of EGR1 and KLF5 by targeting a common regulator or alternatively that ML264 may inhibit the expression of EGR1, which in turns diminishes the expression level of KLF5. As shown, ML264 modifies cycle cell progression by inhibiting the progression through mitosis and by S-phase block (Fig. 2). Consequently, the changes in the PI3K/AKT pathway activity can have a dual origin. It has been previously shown that PI3K/AKT pathway can regulate the progression of cells through G1/S and G2/M phases. On the other hand, the modification of cell cycle progression may in turn influence expression levels of AKT. Our data demonstrates that ML264 in DLD-1 and HCT116 cells decreased the levels of β-catenin phosphorylated at serine 552 and additionally in HCT116 ML264 treatment reduced the levels of total β-catenin (Figs. 3C and 3D). The reduced levels of β-catenin phosphorylated at serine 552 could be a result of down regulation of the expression level of AKT, and its reduced activation. The exact mechanism by which ML264 regulates the RAS/MAPK/PI3K and WNT signaling pathways is still unknown. Ongoing studies that pertain to the analysis of the protein and RNA levels of the components of the RAS/MAPK/PI3K and WNT pathways at much shorter intervals (e.g. hourly) are currently under way.
Altogether, these results suggest that ML264 inhibits the MAPK pathway by reducing EGR1 and KLF5 levels and further that this causes reduced levels of active β-catenin (β-catenin Ser552), which is associated with the down regulation of AKT.
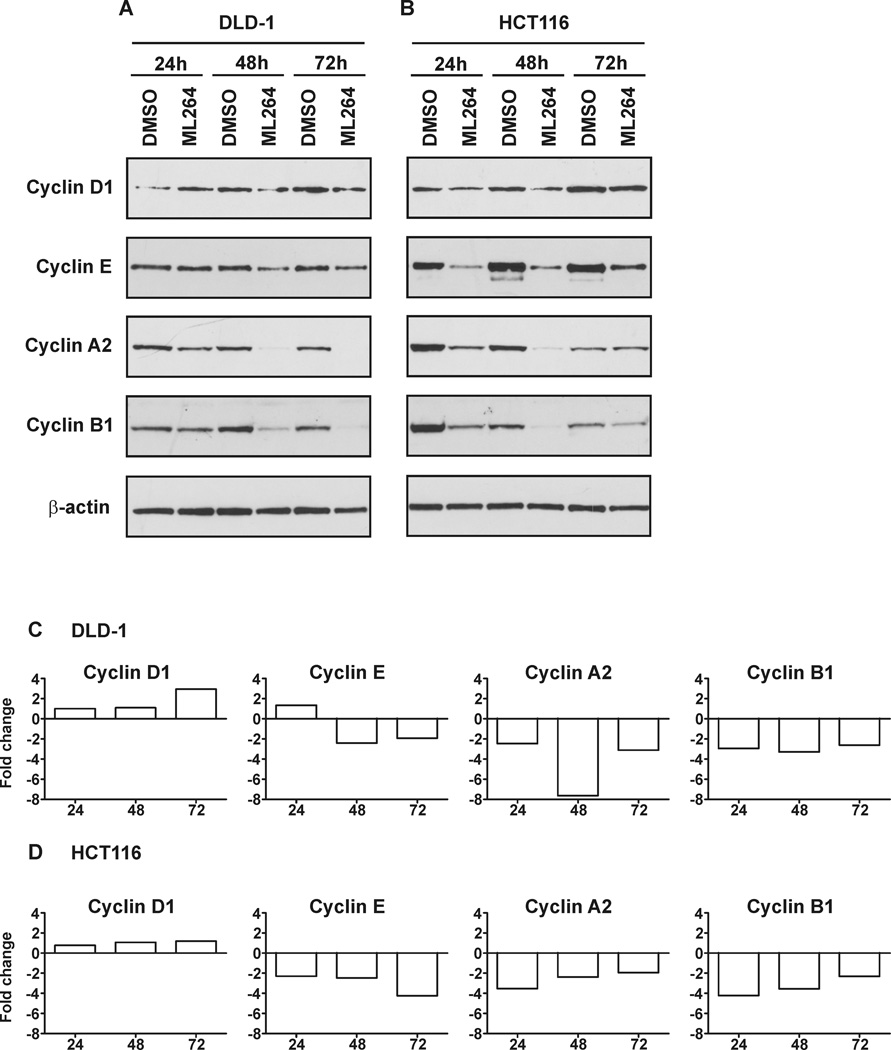
ML264 negatively regulates the expression of cyclins
As shown earlier, treatment with ML264 altered the cell cycle progression of DLD-1 and HCT116 cells as compared to control (Fig. 2). It has been previously shown that KLF5 is a pro-proliferative factor in CRCs and that it regulates the expression levels of cyclins D1 and B (7, 14, 34–38). We therefore analyzed the expression levels of cyclins D1, E, A2, and B1 in DLD-1 and HCT116 cells that were treated with ML264 or with vehicle in a manner corresponding to that shown in Fig. 2. As shown in Figures 4A and 4B, cyclins E, A2 and B1 are down regulated in both cell lines with changes evident even in the first day of treatment with ML264. However, cyclin D1 levels are differentially regulated in these cell lines. In HCT116 cells, we observed a decrease in cyclin D1 levels at 24 and 48 hours after ML264 treatment (Fig. 4B). However, this trend was not apparent in ML264-treated DLD-1 cells (Fig. 4A). We next investigated whether the changes in protein levels were the result of decreased transcription of the corresponding cyclin genes. As shown in Figures 4C and 4D, we failed to see a significant change in the cyclin D1 mRNA levels for either cell line that was treated with ML264 (with exception of DLD-1 cells after 72 hours treatment). However, mRNA levels for cyclins E, A2 and B1 in both cell lines were down regulated during the three-day treatment with ML264. Here, we showed that ML264 affects cell cycle progression by increasing the population of the cells in the S-phase and by inhibiting cells from entering mitosis or from transitioning through mitosis (Fig. 2). The rate of cell proliferation can be affected in many ways, for example by perturbing the cell cycle, by arrest, and/or by promoting apoptosis (39, 40), and is associated with cell cycle progression and is regulated by cyclins (e.g. A2, B1 and E1). Our results (Figs. 4A–4D) show that ML264 affects the expression levels of RNA and protein of cyclin B1 (which is responsible for the transition through mitosis), cyclin A2 (which regulates G1/S and G2/M transition) and cyclin E1 (which plays a role in the G1/S transition) (41). Previously we demonstrated that KLF5 is a positive transcriptional regulator of cyclin D1 and cyclin B1 (37). Current data suggests that upon treatment with ML264, cyclin B1 expression levels are decreased, which may be a direct effect of KLF5 down regulation. Additionally, it has been shown that KLF5 regulates the promoter activity of cyclin E while promoting proliferation of vascular smooth muscle (42). The relationship between KLF5 and cyclin A2 has not been studied until now. Taken together, these results show that ML264 decreases the levels of EGR1 and KLF5 and modulates the cell cycle progression by perturbing the levels of cyclins.
Figure 4. ML264 modulates expression levels of cyclins.
DLD-1 and HCT116 cells were seeded with medium containing DMSO or 10µM ML264. Twenty-four, forty-eight and seventy two hours post-treatment cells were collected for protein and RNA analysis. (A) DLD-1 and (B) HCT116 - Representative image of Western blots. (C) DLD-1 and (D) HCT116 – RNA analysis. Fold change is calculated in comparison to vehicle (DMSO) treated cells as described in Materials and Methods section.
ML264 inhibits the growth of DLD-1 tumor xenografts in nude mice
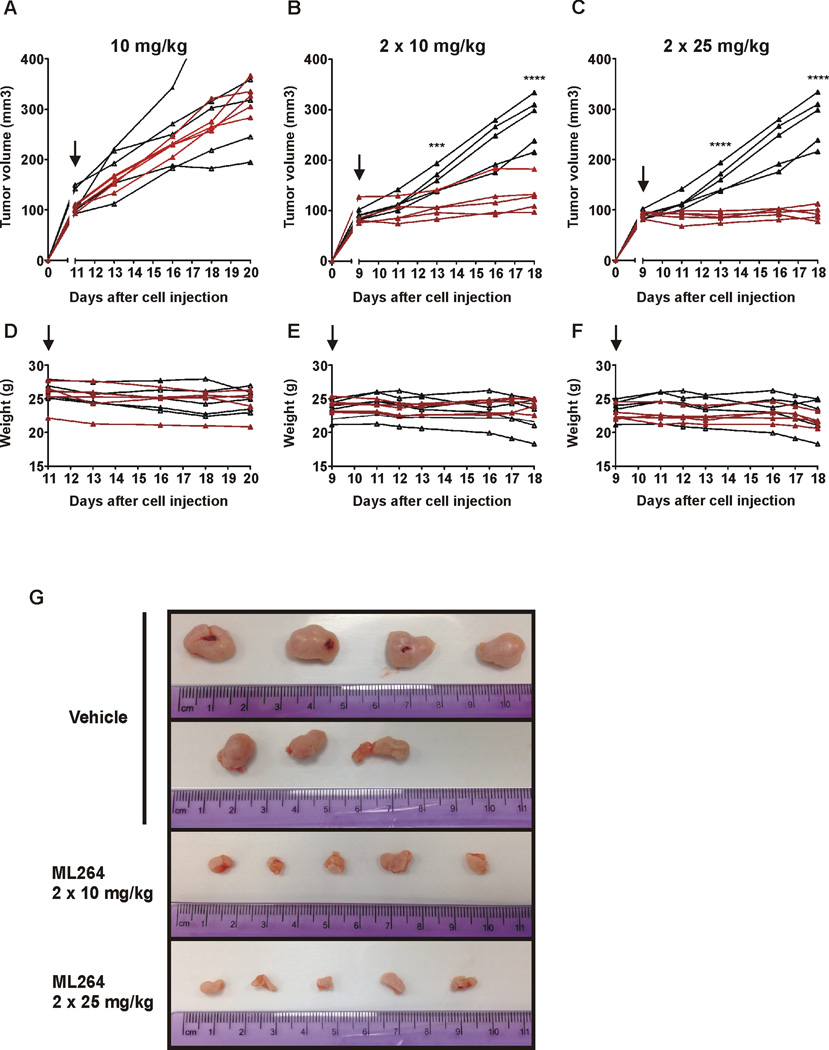
Subsequent to find that ML264 inhibits proliferation of CRC cell lines in vitro, we evaluated its effectiveness in inhibiting growth of tumor xenografts in nude mice. In these experiments, DLD-1 cells were subcutaneously injected into nude mice until a tumor volume of approximately 100 mm3 was achieved. The mice were then injected intraperitoneally (IP) for ten days with ML264 according to the following regimens: 10 mg/kg (once per day), 10 mg/kg (twice per day) and 25 mg/kg (twice per day). In all cases controls were similarly maintained, DLD-1 cells were subcutaneously injected into nude mice until a tumor volume of approximately 100 mm3 was achieved and mice were then injected IP for ten days with vehicle, as described in the Materials and Methods section. As shown in Figure 5A, single daily injections of ML264 at 10 mg/kg did not significantly affect tumor growth. However, twice daily injections of ML264 at 10 mg/kg or 25 mg/kg resulted in significant reductions in tumor growth (Figs. 5B and 5C), and this effect could be detected as early as two days after the first injection. The data also show that there is a concentration-dependent effect of ML264 on the tumor volume. Statistical analysis of tumor growth revealed significant tumor size reduction in mice treated twice daily with ML264 compared to those receiving only vehicle at day 5 and 10. It is noteworthy that none of the treatment regimens affected the weight of the mice (Figs. 5D–5F). At the end of ten days of treatment we harvested tumors from the treated and control mice, photographed the tumors, and sectioned them for further analysis. The photographs shown in Figure 5G clearly demonstrate the dose-dependent differences in tumor size resulting from ML264 treatment as compared to controls.
Figure 5. ML264 inhibits the growth of DLD-1-derived tumor xenografts in nude mice model.
DLD-1 cells were subcutaneously injected into nude mice for the development of xenograft tumors. Mice were treated with vehicle only or with ML264 as follows: daily with 10mg/kg (A, D), twice per day with 10mg/kg (B, E) or twice per day with 25mg/kg (C, F) as detailed in Materials and Methods. Black lines label vehicle-treated mice, and red lines label ML264-treated mice. Black arrows point to the start of injections of vehicle or ML264. Tumor volume is depicted in A–C and mice weight in D–F. The asterisks (***) and (****) indicate a significant difference (p < 0.001) and (p < 0.0001), respectively, between the ML264-treated and vehicle-treated groups. (G) Photographic images of the tumor collected at the end of 10 days treatment as shown in (B) and (C).
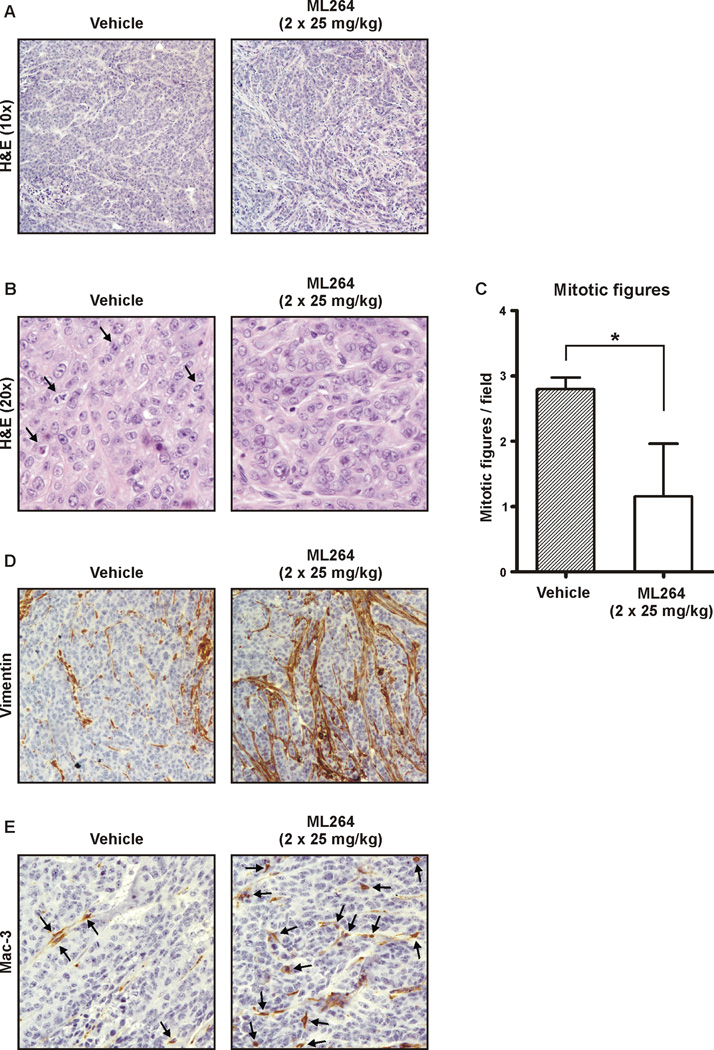
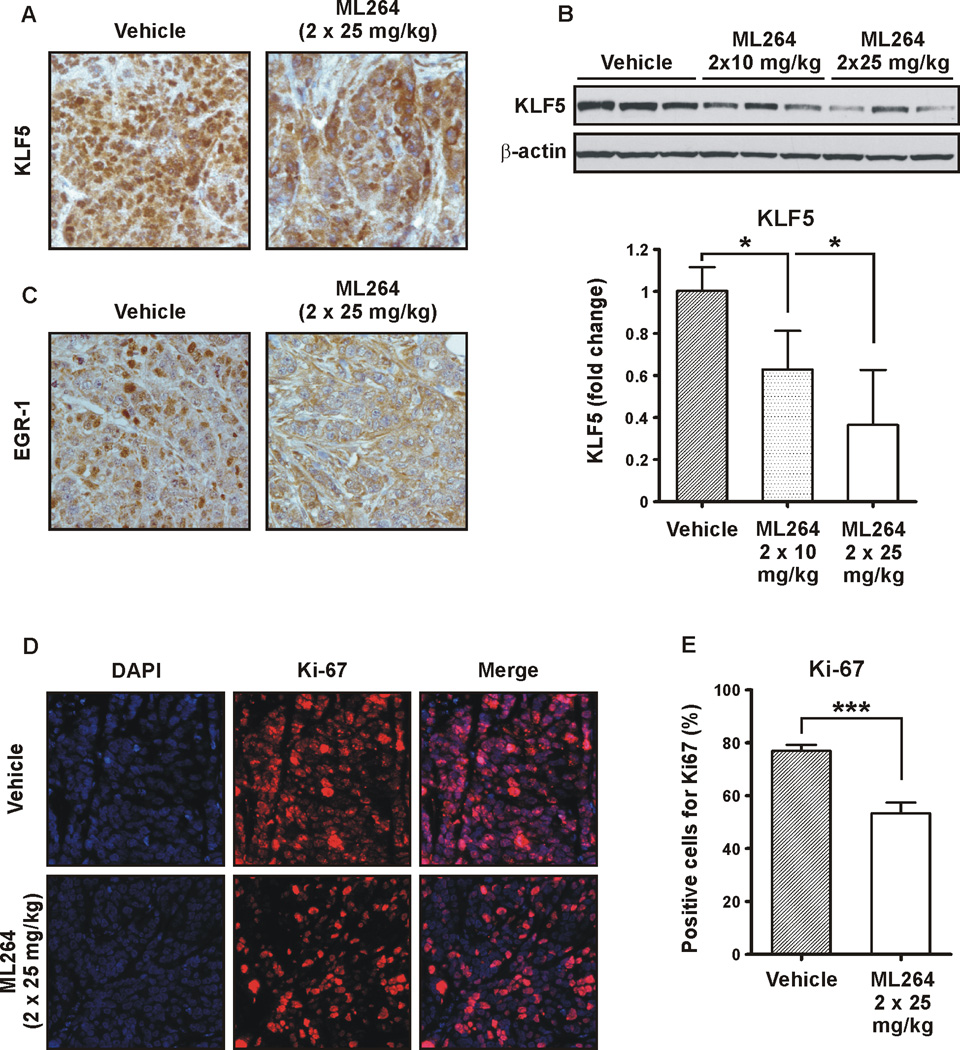
Subsequently, we performed H & E staining of the tumors (Fig. 6A). As shown in Figures 6B and 6C, there is a significant reduction in the number of mitotic figures in xenografts from mice treated twice daily with ML264 at 25 mg/kg. This explains the lack of xenograft growth in ML264-treated mice (Fig. 5C). The initial histological analysis indicated that there is an increase in the level of fibrosis and inflammation in the xenografts treated with ML264 in comparison to vehicle-treated. Thus, we stained the tumors for vimentin to detect fibroblasts associated with fibrosis. Here, we observed a significantly increased density of fibroblasts in the ML264-treated xenografts (Fig. 6D). Consequently, we stained the tissues for the presence of inflammatory cells. As shown in Figure 6E, xenografts treated with ML264 had increased levels of mononuclear phagocytes staining for Mac-3. As KLF5 is pro-proliferative marker and a potential target for ML264’s activity, we next investigated the impact of ML264 on KLF5 levels in these xenografts. As seen using immunohistochemistry (Fig. 7A and Supplementary Fig. 2), there is a reduction in KLF5 levels and an absence of KLF5 from the nuclei in the xenografts from mice treated with ML264. This result was corroborated by a Western blot analysis for KLF5, shown in Figure 7B. We also examined these xenografts for EGR1, a major transcriptional regulator of KLF5, and saw a significant reduction of EGR1 in xenografts from mice that had been treated with ML264 (Fig. 7C and Supplementary Fig. 3). Furthermore, we performed staining for Ki-67, a well-characterized marker of cell proliferation (43), to assess the proliferation status of the xenografts with or without treatment. As shown in Figures 7D and 7E, the levels of Ki-67 staining are significantly reduced in xenografts from mice that were treated with ML264 as compared to those from control mice.
Figure 6. Histology and immunohistochemistry of DLD-1-derived tumor xenografts treated with ML264.
DLD-1 cells were subcutaneously injected into nude mice for development of xenograft tumors. Mice were treated with vehicle only or with ML264 at 25mg/kg as detailed in Materials and Methods. (A) and (B) Hematoxylin and eosin (H&E)-stained histological images of DLD-1-derived xenografts treated with vehicle (left panel) and treated with ML264 twice per day at 25mg/kg (right panel) for ten days at magnification 10× and 20×, respectively. Black arrows label mitotic figures in panel B. (C) Quantitative representation of (B). Three fields were counted for each condition. Data represent mean ± S.D. (n=3). *p < 0.05. (D) Representative images of immunohistochemistry for vimentin in DLD-1-derived xenografts treated with vehicle (left panel) and treated with ML264 twice per day at 25mg/kg (right panel) for ten days. (E) Representative images of immunohistochemistry for Mac-3 in DLD-1-derived xenografts treated with vehicle (left panel) and treated with ML264 twice per day at 25mg/kg (right panel) for ten days. Black arrows label Mac-3 positive staining.
Figure 7. ML264 treatment reduced the expression levels of KLF5 and EGR1 in DLD-1-derived tumor xenografts.
(A) Representative immunohistochemistry staining of KLF5 in DLD-1-derived xenografts treated with vehicle (left panel) and treated with ML264 twice per day at 25mg/kg (right panel) for ten days. (B) Western blot analysis (top panel) and the quantitative analysis (bottom panel) of KLF5 levels in DLD-1-derived xenografts treated with vehicle and treated with ML264 twice per day at 10mg/kg and 25mg/kg for ten days. Results shown from three independent experiments. Data represent mean ± S.D. (n=3). *p < 0.05. (C) Representative immunohistochemistry staining of EGR1 in DLD-1-derived xenografts treated with vehicle (left panel) and treated with ML264 twice per day at 25mg/kg (right panel) for ten days. (D) Immunofluorescence staining of Ki-67 (proliferative marker). Top panel – vehicle treated mice, Bottom panel – ML264-treated mice. (E) Quantitative representation of (D). Three fields were counted for each condition. Data represent mean ± S.D. (n=3). ***p < 0.001.
In the present study, we have shown that ML264 efficiently inhibits the growth of CRCs in vitro (Fig. 1) and in vivo (Figs. 5, 6 and 7). We were able to show that in an in vitro system ML264 regulates the cell cycle progression while inducing apoptosis minimally (Fig. 2 and Supplementary Fig. 1). Interestingly, we have noticed that the arrest of cells in S-phase and the induction of apoptosis (Fig. 2 and Supplementary Fig. 1) are accompanied by increased levels of phosphorylated ERK and by decreased levels of phosphorylated AKT (Fig. 3). This phenomenon has been reported previously and has an important implications for the approach, particularly in the development of combinatorial drug treatments that target the MAPK signaling pathway (44–46). Our results demonstrate that ML264 significantly inhibits cellular proliferation, and mitosis in particular, in both in vitro and in vivo systems, as shown by analyses of a) mitotic figures in DLD-1 cells treated in culture (Figs. 1E and 1F) and b) mitotic figures and Ki-67 staining of DLD-1-derived xenografts (Figs. 6B, 6C, 7D and 7E). Our studies showed that ML264 efficiently modulates cell cycle progression and affects the expression pattern of cyclin B1, A2 and E (Figs. 1, 2 and 4). It has been previously shown that EGR1 and KLF5 regulate progression through the cell cycle by modifying the levels of cyclins D1 and B1, also that KLF5 affects cyclin E (37, 42). It is feasible that ML264 acts by decreasing the levels of EGR1 and KLF5, inhibiting the expression of the mentioned cyclins, and interrupting the cell cycle progression.
ML264 originated from an ultra-high throughput screen to identify compounds that inhibit the proliferation of CRCs by targeting the expression of KLF5 (25). In these experiments, we have demonstrated that ML264 is capable of reducing the expression levels of KLF5 in cells treated in culture (Figs. 3A and 3B) and in a tumor xenograft model of tumorigenesis (Figs. 7A and 7B and Supplementary Fig. 2). Moreover, we have found that this compound has a profound effect on the expression levels of EGR1, a direct transcriptional regulator of KLF5 (Figs. 3A, 3B, 7C, and Supplementary Fig. 3). At present, the exact mechanism by which ML264 inhibits KLF5 is unknown. Studies are currently under way to assess the mechanism of action and define if it has either a direct or indirect effect on KLF5 expression.
Our in vitro Drug Metabolism and Pharmacokinetics (DMPK) studies suggested that ML264 would have high stability to first pass metabolism due to its high stability upon exposure to hepatic microsomes. ML264 also does not inhibit the activity of cytochrome P450 isoenzymes, indicative of its potential for safety with respect to drug-drug interactions. Importantly, it was inactive against a selected panel of 47 kinases and 67 protein targets of therapeutic and /or toxicological interest (25). Our in vivo DMPK studies demonstrated that ML264 has 2-hour half-life in mice. Interestingly, ML264 also displays 47% oral bioavailability in mice, a very promising feature for compound development with potential therapeutic advantages. Ongoing studies in our laboratory aim to identify the direct target of ML264 and to test its role in modulating the early steps of development of intestinal adenomas using ApcMin/+/KRASV12G mice models.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Dr. Jie Yang from the Department of Preventive Medicine, Stony Brook University, for assistance regarding biostatistical analysis of the tumor growth. We would like to thank Research Flow Cytometry Core in the Department of Pathology, Stony Brook University for assistance with data analysis. This work was supported by grants from the National Institutes of Health (DK052230, CA084197 and CA172113 to V.W. Yang, CA172113 to A.B. Bialkowska, CA172517 to M. García-Barros, CA172113 and MH084512 to T.D. Bannister).
Footnotes
There are no conflicts of interest.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Arnold CN, Goel A, Blum HE, Boland CR. Molecular pathogenesis of colorectal cancer: implications for molecular diagnosis. Cancer. 2005;104:2035–2047. doi: 10.1002/cncr.21462. [DOI] [PubMed] [Google Scholar]
- 3.Mariani F, Sena P, Roncucci L. Inflammatory pathways in the early steps of colorectal cancer development. World journal of gastroenterology : WJG. 2014;20:9716–9731. doi: 10.3748/wjg.v20.i29.9716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fearon ER. Molecular genetics of colorectal cancer. Annual review of pathology. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 5.Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43–47. doi: 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
- 6.McConnell BB, Ghaleb AM, Nandan MO, Yang VW. The diverse functions of Kruppel-like factors 4 and 5 in epithelial biology and pathobiology. BioEssays : news and reviews in molecular, cellular and developmental biology. 2007;29:549–557. doi: 10.1002/bies.20581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nandan MO, Ghaleb AM, Liu Y, Bialkowska AB, McConnell BB, Shroyer KR, et al. Inducible intestine-specific deletion of Kruppel-like factor 5 is characterized by a regenerative response in adult mouse colon. Developmental biology. 2014;387:191–202. doi: 10.1016/j.ydbio.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nandan MO, Ghaleb AM, Bialkowska AB, Yang VW. Kruppel-like factor 5 is essential for proliferation and survival of mouse intestinal epithelial stem cells. Stem cell research. 2015;14:10–19. doi: 10.1016/j.scr.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barker N, van Es JH, Jaks V, Kasper M, Snippert H, Toftgard R, et al. Very long-term self-renewal of small intestine, colon, and hair follicles from cycling Lgr5+ve stem cells. Cold Spring Harbor symposia on quantitative biology. 2008;73:351–356. doi: 10.1101/sqb.2008.72.003. [DOI] [PubMed] [Google Scholar]
- 10.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 11.Barker N, Clevers H. Lineage tracing in the intestinal epithelium. Current protocols in stem cell biology. 2010;Chapter 5(Unit5A 4) doi: 10.1002/9780470151808.sc05a04s13. [DOI] [PubMed] [Google Scholar]
- 12.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 13.Chen C, Zhou Y, Zhou Z, Sun X, Otto KB, Uht RM, et al. Regulation of KLF5 involves the Sp1 transcription factor in human epithelial cells. Gene. 2004;330:133–142. doi: 10.1016/j.gene.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 14.Nandan MO, Yoon HS, Zhao W, Ouko LA, Chanchevalap S, Yang VW. Kruppel-like factor 5 mediates the transforming activity of oncogenic H-Ras. Oncogene. 2004;23:3404–3413. doi: 10.1038/sj.onc.1207397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu N, Li H, Li S, Shen M, Xiao N, Chen Y, et al. The Fbw7/human CDC4 tumor suppressor targets proproliferative factor KLF5 for ubiquitination and degradation through multiple phosphodegron motifs. The Journal of biological chemistry. 2010;285:18858–18867. doi: 10.1074/jbc.M109.099440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bialkowska AB, Liu Y, Nandan MO, Yang VW. A colon cancer-derived mutant of Kruppel-like factor 5 (KLF5) is resistant to degradation by glycogen synthase kinase 3beta (GSK3beta) and the E3 ubiquitin ligase F-box and WD repeat domain-containing 7alpha (FBW7alpha) The Journal of biological chemistry. 2014;289:5997–6005. doi: 10.1074/jbc.M113.508549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao D, Zheng HQ, Zhou Z, Chen C. The Fbw7 tumor suppressor targets KLF5 for ubiquitin-mediated degradation and suppresses breast cell proliferation. Cancer Res. 2010;70:4728–4738. doi: 10.1158/0008-5472.CAN-10-0040. [DOI] [PubMed] [Google Scholar]
- 18.McConnell BB, Bialkowska AB, Nandan MO, Ghaleb AM, Gordon FJ, Yang VW. Haploinsufficiency of Kruppel-like factor 5 rescues the tumor-initiating effect of the Apc(Min) mutation in the intestine. Cancer Res. 2009;69:4125–4133. doi: 10.1158/0008-5472.CAN-08-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McConnell BB, Kim SS, Bialkowska AB, Yu K, Sitaraman SV, Yang VW. Kruppel-like factor 5 protects against dextran sulfate sodium-induced colonic injury in mice by promoting epithelial repair. Gastroenterology. 2011;140:540 e2–549 e2. doi: 10.1053/j.gastro.2010.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nandan MO, McConnell BB, Ghaleb AM, Bialkowska AB, Sheng H, Shao J, et al. Kruppel-like factor 5 mediates cellular transformation during oncogenic KRAS-induced intestinal tumorigenesis. Gastroenterology. 2008;134:120–130. doi: 10.1053/j.gastro.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nandan MO, Ghaleb AM, McConnell BB, Patel NV, Robine S, Yang VW. Kruppel-like factor 5 is a crucial mediator of intestinal tumorigenesis in mice harboring combined ApcMin and KRASV12 mutations. Molecular cancer. 2010;9:63. doi: 10.1186/1476-4598-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakaya T, Ogawa S, Manabe I, Tanaka M, Sanada M, Sato T, et al. KLF5 regulates the integrity and oncogenicity of intestinal stem cells. Cancer Res. 2014;74:2882–2891. doi: 10.1158/0008-5472.CAN-13-2574. [DOI] [PubMed] [Google Scholar]
- 23.Bialkowska AB, Crisp M, Bannister T, He Y, Chowdhury S, Schurer S, et al. Identification of small-molecule inhibitors of the colorectal cancer oncogene Kruppel-like factor 5 expression by ultrahigh-throughput screening. Molecular cancer therapeutics. 2011;10:2043–2051. doi: 10.1158/1535-7163.MCT-11-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bialkowska AB, Du Y, Fu H, Yang VW. Identification of novel small-molecule compounds that inhibit the proproliferative Kruppel-like factor 5 in colorectal cancer cells by high-throughput screening. Molecular cancer therapeutics. 2009;8:563–570. doi: 10.1158/1535-7163.MCT-08-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bialkowska A, Crisp M, Madoux F, Spicer T, Knapinska A, Mercer B, et al. Probe Reports from the NIH Molecular Libraries Program. Bethesda (MD): 2010. ML264: An Antitumor Agent that Potently and Selectively Inhibits Kruppel-like Factor Five (KLF5) Expression: A Probe for Studying Colon Cancer Development and Progression. [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim JH, Alfieri AA, Kim SH, Young CW. Potentiation of radiation effects on two murine tumors by lonidamine. Cancer Res. 1986;46:1120–1123. [PubMed] [Google Scholar]
- 29.Usui S, Sugimoto N, Takuwa N, Sakagami S, Takata S, Kaneko S, et al. Blood lipid mediator sphingosine 1-phosphate potently stimulates platelet-derived growth factor-A and -B chain expression through S1P1-Gi-Ras-MAPK-dependent induction of Kruppel-like factor 5. The Journal of biological chemistry. 2004;279:12300–12311. doi: 10.1074/jbc.M305025200. [DOI] [PubMed] [Google Scholar]
- 30.Yang Y, Goldstein BG, Nakagawa H, Katz JP. Kruppel-like factor 5 activates MEK/ERK signaling via EGFR in primary squamous epithelial cells. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2007;21:543–550. doi: 10.1096/fj.06-6694com. [DOI] [PubMed] [Google Scholar]
- 31.Fang D, Hawke D, Zheng Y, Xia Y, Meisenhelder J, Nika H, et al. Phosphorylation of beta-catenin by AKT promotes beta-catenin transcriptional activity. The Journal of biological chemistry. 2007;282:11221–11229. doi: 10.1074/jbc.M611871200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He XC, Yin T, Grindley JC, Tian Q, Sato T, Tao WA, et al. PTEN-deficient intestinal stem cells initiate intestinal polyposis. Nature genetics. 2007;39:189–198. doi: 10.1038/ng1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, et al. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- 34.Chen C, Benjamin MS, Sun X, Otto KB, Guo P, Dong XY, et al. KLF5 promotes cell proliferation and tumorigenesis through gene regulation and the TSU-Pr1 human bladder cancer cell line. International journal of cancer Journal international du cancer. 2006;118:1346–1355. doi: 10.1002/ijc.21533. [DOI] [PubMed] [Google Scholar]
- 35.Du JX, Yun CC, Bialkowska A, Yang VW. Protein inhibitor of activated STAT1 interacts with and up-regulates activities of the pro-proliferative transcription factor Kruppel-like factor 5. The Journal of biological chemistry. 2007;282:4782–4793. doi: 10.1074/jbc.M603413200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y, Wen JK, Dong LH, Zheng B, Han M. Kruppel-like factor (KLF) 5 mediates cyclin D1 expression and cell proliferation via interaction with c-Jun in Ang II-induced VSMCs. Acta pharmacologica Sinica. 2010;31:10–18. doi: 10.1038/aps.2009.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nandan MO, Chanchevalap S, Dalton WB, Yang VW. Kruppel-like factor 5 promotes mitosis by activating the cyclin B1/Cdc2 complex during oncogenic Ras-mediated transformation. FEBS letters. 2005;579:4757–4762. doi: 10.1016/j.febslet.2005.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McConnell BB, Kim SS, Yu K, Ghaleb AM, Takeda N, Manabe I, et al. Kruppel-like factor 5 is important for maintenance of crypt architecture and barrier function in mouse intestine. Gastroenterology. 2011;141:1302–1313. 13 e1–16 e1. doi: 10.1053/j.gastro.2011.06.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elmore S. Apoptosis: a review of programmed cell death. Toxicologic pathology. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vermeulen K, Van Bockstaele DR, Berneman ZN. The cell cycle: a review of regulation, deregulation and therapeutic targets in cancer. Cell proliferation. 2003;36:131–149. doi: 10.1046/j.1365-2184.2003.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson DG, Walker CL. Cyclins and cell cycle checkpoints. Annual review of pharmacology and toxicology. 1999;39:295–312. doi: 10.1146/annurev.pharmtox.39.1.295. [DOI] [PubMed] [Google Scholar]
- 42.Shi HJ, Wen JK, Miao SB, Liu Y, Zheng B. KLF5 and hhLIM cooperatively promote proliferation of vascular smooth muscle cells. Molecular and cellular biochemistry. 2012;367:185–194. doi: 10.1007/s11010-012-1332-9. [DOI] [PubMed] [Google Scholar]
- 43.Gerdes J, Lemke H, Baisch H, Wacker HH, Schwab U, Stein H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. Journal of immunology. 1984;133:1710–1715. [PubMed] [Google Scholar]
- 44.Guan TJ, Qin FJ, Du JH, Geng L, Zhang YY, Li M. AICAR inhibits proliferation and induced S-phase arrest, and promotes apoptosis in CaSki cells. Acta pharmacologica Sinica. 2007;28:1984–1990. doi: 10.1111/j.1745-7254.2007.00675.x. [DOI] [PubMed] [Google Scholar]
- 45.Yadav V, Varshney P, Sultana S, Yadav J, Saini N. Moxifloxacin and ciprofloxacin induces S-phase arrest and augments apoptotic effects of cisplatin in human pancreatic cancer cells via ERK activation. BMC Cancer. 2015;15:581. doi: 10.1186/s12885-015-1560-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan Z, Guo W, Yang J, Li L, Wang M, Lei Y, et al. PNAS-4, an Early DNA Damage Response Gene, Induces S Phase Arrest and Apoptosis by Activating Checkpoint Kinases in Lung Cancer Cells. The Journal of biological chemistry. 2015;290:14927–14944. doi: 10.1074/jbc.M115.658419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.