Summary
The functional and molecular similarities and distinctions between human and murine astrocytes are poorly understood. Here we report the development of an immunopanning method to acutely purify astrocytes from fetal, juvenile, and adult human brains, and to maintain these cells in serum-free cultures. We found that human astrocytes have similar abilities to murine astrocytes in promoting neuronal survival, inducing functional synapse formation, and engulfing synaptosomes. In contrast to existing observations in mice, we found that mature human astrocytes respond robustly to glutamate. We next performed RNA-sequencing of healthy human astrocytes along with astrocytes from epileptic and tumor foci, and compared these to human neurons, oligodendrocytes, microglia, and endothelial cells. With these profiles, we identified novel human-specific astrocyte genes, and discovered a transcriptome-wide transformation between astrocyte precursor cells and mature post-mitotic astrocytes. These data represent some of the first cell type-specific molecular profiles of the healthy and diseased human brain.
Introduction
Constituting about 40% of all cells in the human brain, astrocytes have long been classified as mere passive support cells. Recent work, however, has demonstrated that murine astrocytes play many active roles and are critical for the development and function of the central nervous system (CNS) (Allen and Barres, 2009; Chung et al., 2015; Clarke and Barres, 2013; Hamilton and Attwell, 2010; Haydon and Nedergaard, 2014; Khakh and McCarthy, 2015; Molofsky et al., 2012). For example, purified neurons in culture have little ability to form functional synapses, which are powerfully promoted by astrocytes (Allen et al., 2012; Christopherson et al., 2005; Kucukdereli et al., 2011; Ullian et al., 2001). Murine astrocytes are not only important for the formation and function of synapses, but are also essential for the phagocytic elimination of synapses and the refinement of developing neural circuits (Chung et al., 2013). Since synapse formation, function, and elimination are key processes occurring during learning and memory, astrocytes are postulated to be an indispensible component in CNS plasticity. Additionally, murine astrocytes are required for neurotransmitter recycling (Rothstein et al., 1996), extracellular potassium homeostasis (Kuffler et al., 1966), regulation of blood flow (Attwell et al., 2010; Mulligan and MacVicar, 2004; Zonta et al., 2003), and providing energy substrates for neurons (Pellerin and Magistretti, 1994). Considering their central role in CNS physiology, it is not surprising that astrocyte dysfunction has been demonstrated or implicated in nearly all neurological disorders (Molofsky et al., 2012). Currently, the extent of our understanding of astrocyte physiology in health and disease is almost entirely restricted to observations in murine models, but how primary human and murine astrocytes compare at molecular and functional levels remains largely unknown.
Observational studies from postmortem human tissues have revealed that human astrocytes are much larger and more complex than their murine counterparts (Oberheim et al., 2006, 2009). More recently, transplantation of human glial progenitors into mouse brains has been shown to improve learning and memory (Han et al., 2013). These observations raise questions about how murine astrocyte physiology and function might extend to humans. Are there distinct properties of human astrocytes that contribute to human intelligence and cognition? Can we extend findings about astrocyte pathology in neurological disorders from mouse and rat studies to develop effective therapeutic approaches in humans?
A major hurdle in addressing these issues is the lack of a method to acutely purify human astrocytes and culture them in chemically-defined conditions. Current purification methods for human astrocytes require culturing dissociated nervous tissue in serum for days to weeks (McCarthy and de Vellis, 1980). In vivo, however, quiescent astrocytes do not contact serum except upon injury and blood-brain-barrier break down, and in vitro exposure to serum has been shown to induce irreversible reactive changes in astrocytes (Foo et al., 2011; Zamanian et al., 2012). Moreover, since serum-selection methods require a group of proliferating astrocyte progenitors, these protocols do not work efficiently to purify mature astrocytes from adult human brains. Because of these limitations, the transcriptome profile of mature resting human astrocytes has remained difficult to ascertain.
The method we present here is an immunopanning-based technique that utilizes an antibody targeted against HepaCAM (or GlialCAM), a surface protein expressed by human astrocytes, to generate purified (>95%) cultures of primary human astrocytes. With this method, we acutely purified astrocytes from over twenty fetal, juvenile, and adult human subjects, and then performed transcriptome profiling and functional studies to compare developmental and interspecies differences. We additionally purified each of the major CNS cell types and generated a user-friendly searchable online database to enable dissemination of human cell type specific gene expression data to the greater neuroscience community (http://www.BrainRNAseq.org).
Results
Acute purification and serum-free culture of fetal and mature human astrocytes
We obtained juvenile (8–18 years old) and adult (21–63 years old) human brain tissue samples from patients undergoing neurological surgeries with informed consent under a Stanford University Institutional Review Board approved protocol. The samples used in this study (except for the data shown in Figures 5 and 6 for epilepsy and glioblastoma) were small pieces of healthy temporal lobe cortices that were resected to gain access to deeper epileptic hippocampi (Figure 1A). In each of these cases, the temporal lobe cortex that was removed was considered normal based upon Magnetic Resonance Imaging (MRI), Electroencephalogram (EEG) studies, and pathological examination. Detailed information on patient diagnoses is listed in Table S1. We worked closely with the surgeons to minimize the time from tissue resection to the beginning of the purification protocol (within 1 hour). Furthermore, we generated all gene expression data by collecting RNA from cells immediately after purification to best represent their in vivo molecular profiles. In addition to surgical samples, we also obtained fetal brain tissue from elective pregnancy terminations at 17–20 gestational weeks under a Stanford University Institutional Review Board approved protocol. For fetal cases, we began purification within 5 hours after the procedures.
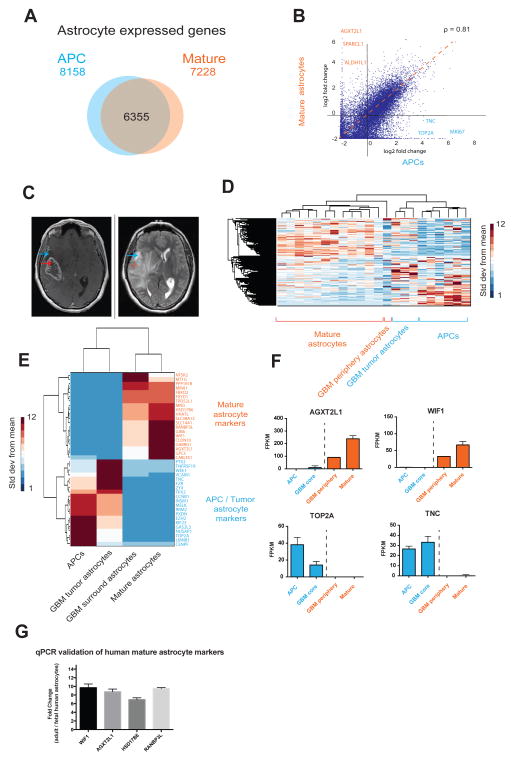
Figure 5.
The transcriptome of HepaCAM purified glioblastoma cells is more similar to APCs than mature astrocytes. A. Venn diagram of the number of genes expressed (FPKM>0.5) by APCs alone, mature astrocytes alone, and both APCs and mature astrocytes. Fetal, n=6 patients. Mature, n=12 patients. B. Scatter plot of gene expression by APCs and mature astrocytes. ρ represents the Spearman correlation coefficiency. C. MRI of a patient with a glioblastoma who underwent surgical resection shows the T1 weighted, heterogenously ring contrast enhancing core of the GBM (left) and the T2 hyperintensity of the surrounding brain (right), representing infiltrating tumor or edema. Blue arrows indicate a representative location of the peripheral tumor sample, and the red arrows indicate sample resected from the contrast-enhancing tumor core. D. Unsupervised hierarchical clustering of gene expression profiles of APCs, mature astrocytes as well as astrocytes from GBM core and peripheral regions using. Linkage; average, ColumnPdistance; Spearman, standardized across rows. Data represent standard deviation over means. E. Expression of genes that distinguish APCS and mature astrocytes in humans. Tumor core and surround astrocytes share expression patterns of APC and mature astrocyte markers, respectively. Tumor: n=3 patients. Tumor periphery: n=1 patient. F. Expression of some APC marker genes (blue) and mature astrocyte marker genes (red) in healthy and tumor samples. G. Differences in gene expression between adult and fetal human brain assessed with qPCR. Fold changes in gene expression were calculated to reflect enrichment in adult tissue (normalized to GAPDH levels).
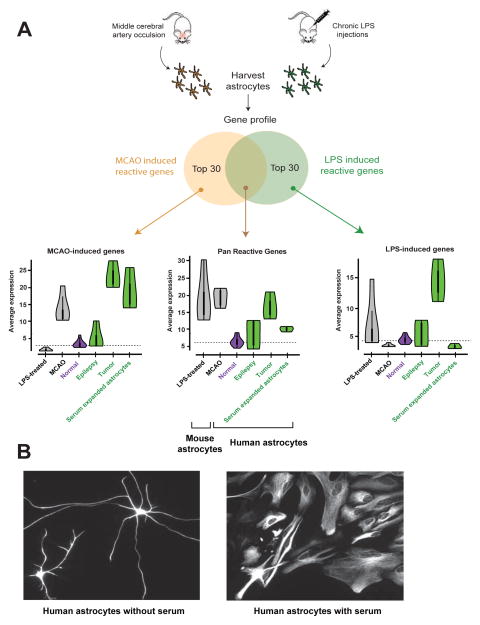
Figure 6.
Acutely purified human astrocytes display resting gene profiles. A. The Zamanian et al. dataset was used to identify the top 30 reactive astrocyte genes upregulated following MCAO injury, LPS infection, or both. Expression of this suite of ‘reactive’ genes was then probed in various astrocyte samples. The white dots in the Violin plots represent the median. Thick vertical black lines represent interquantile ranges. Horizontal dashed lines indicate median in normal human astrocytes for each condition. N=12, 4, 3, and 3 for normal, epilepsy, tumor, and serum-expanded astrocytes, respectively. B. Representative images of acutely purified human astrocytes grown in culture for 7 days in the absence (left) or presence (right) of serum. Scale bar: 20μm.
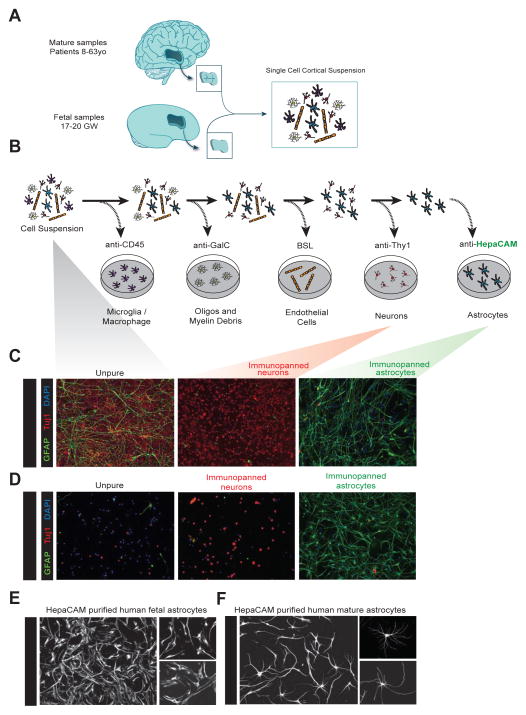
Figure 1.
Acute purification of fetal and mature human astrocytes. A. Juvenile and adult (8 to 63 years old) patient temporal lobe cortex tissue and fetal (17–20 gestational week) brain tissue is first dissociated into single cell suspensions. B. Schematics of immunopanning purification of cell types from human brain samples. C and D. Unpurified brain cells (left), Thy1-purified neurons (middle), and HepaCAM-purified astrocytes (right) from fetal (C) and mature (D) brains stained at 7div for neurons (TuJ1, red), astrocytes (GFAP, green), and nuclei of all cells (DAPI, blue). Scale bars: 100μm. E and F. Cultured human fetal (E) and mature (F) astrocytes grown in culture for 7 days and stained with GFAP. Scale bars: 100μm, insets 50 μm.
To develop a method for the purification of astrocytes, we first sought to identify an astrocyte-specific antibody that we could use to immunopan human cortical tissue. We mined our existing mouse astrocyte RNA-seq datasets (Zhang et al., 2014) for potential surface markers that were enriched in astrocytes, not expressed by radial glial cells (a major cellular constituent of fetal human brain), and where antibodies to the human antigen already existed. After screening numerous candidates, we settled on HepaCAM, a cell adhesion glycoprotein that is enriched in astrocytes. The immunopanning protocol involves passing a single cell suspension of dissociated tissue over a series of Petri dishes coated with antibodies directed against cell type specific antigens (Figure 1A, B). For juvenile and adult samples, our immunopanning protocol consisted of anti-CD45 antibodies to bind microglia (and macrophages), anti-GalC hybridoma to bind oligodendrocytes and myelin debris, anti-O4 hybridoma to bind oligodendrocyte precursor cells (OPCs), anti-Thy1 antibody to bind neurons, anti-HepaCAM antibody to bind astrocytes, and Banderiaea simplicifolia lectin 1 (BSL-1) to bind endothelial cells. We used a shortened antibody binding procedure for fetal samples because oligodendrocytes and myelin are not yet generated at 17–20 gestational weeks (Experimental Procedures and Supplemental Information). After cells were bound to an antibody-coated dish, we washed away loosely bound contaminating cells and then either detached cells via trypsin digestion for culture or scraped cells directly off the dish to extract RNA for RNA-seq.
We routinely obtained over 95% pure mature human astrocytes as determined by GFAP immunofluorescence (Figure 1D) and further validated these immunohistochemical purity estimates by RNA-seq (Figure 4 and see below). We refer to astrocytes purified from juvenile and adult brain tissue as “mature” astrocytes to distinguish from fetal astrocytes. Mature human astrocytes exhibited process-bearing morphology (Figure 1F, S1) and remained healthy for 3 weeks or longer in vitro when cultured in serum-free defined media containing the mitogen and survival factor HBEGF (Supplemental Information). Interestingly, while fetal human astrocytes proliferated robustly in vitro, mature astrocytes did not divide. We performed immunostaining of fetal and mature human astrocytes with an antibody against a mitotic cell marker, Ki67. While 14.9 ± 1.6% of fetal human astrocytes were actively proliferating, only 0.05 ± 0.1% of cells in mature human astrocyte culture were positive for Ki67. Similarly, we performed an EdU (5-ethynyl-2′-deoxyuridine) incorporation assay and detected EdU labeling in 15.0 ± 2.3% of fetal human astrocytes and only 0.4 ± 0.5% of cells in mature human astrocyte culture (Figure S2). This was further confirmed by mRNA expression of two mitotic markers, TOP2A and MKI67, which were exclusively present in fetal astrocytes (TOP2A: FPKM 38.2 ± 6.2 in fetal astrocytes 0.1 ± 0 in mature astrocytes; MKI67: FPKM 34.2 ± 3.9 in fetal astrocytes, 0.1 ± 0 in mature astrocytes. Data represent average ± SEM unless otherwise noted. Fetal, n= 6 patients. Mature, n=12 patients). Therefore, human astrocytes exist in two distinct developmental stages: a fetal, highly proliferative astrocyte precursor cell (APC) stage, and a mature, post-mitotic stage. Due to their proliferative nature, acutely purified human APCs can be frozen, stored, and defrosted at a later time. Frozen and defrosted APCs remain healthy and continue to proliferate in culture at similar rates (Figure S2), so experiments using immunopanned human APCs are not time-locked to tissue availability.
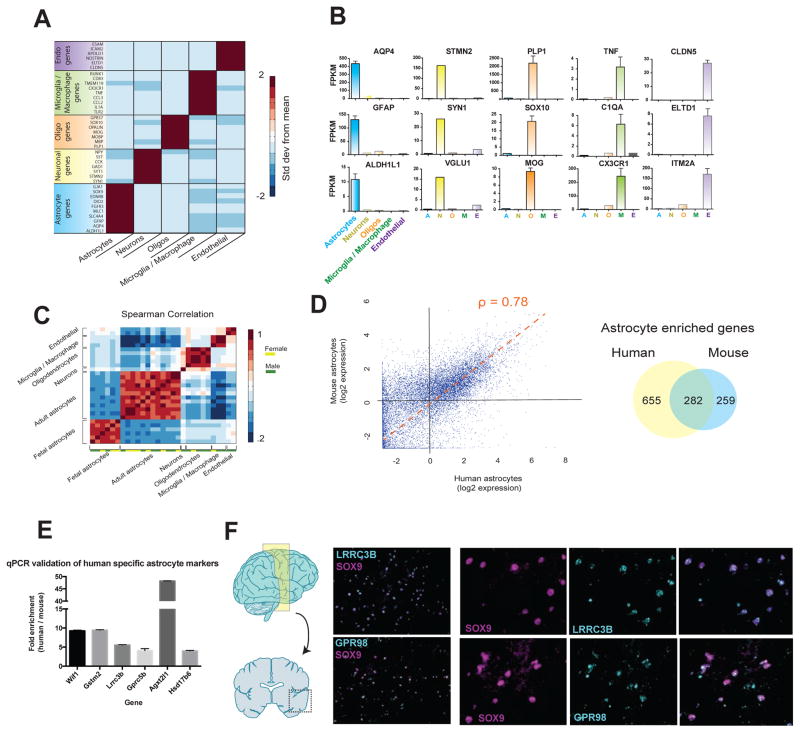
Figure 4.
RNA-seq transcriptome profiling of acutely purified human neurons, astrocytes, oligodendrocytes, microglia/macrophages, and endothelial cells. A. Expression of classic cell type-specific genes by acutely purified mature human neurons, astrocytes, oligodendrocytes, microglia/macrophages, and endothelial cells. Data represent standard deviation from mean across rows. N=12, 1, 5, 3, and 2 patients for astrocytes, neurons, oligodendrocytes, microglia, and endothelial cells, respectively. Age range: 8–63 years. B. Representative examples of the expression of cell type-specific genes by acutely purified cell samples. C. Spearman correlation between all fetal and mature samples. Fetal, n=6 patients. Age range: 18.0–18.5 gw. Mature, n=12, 1, 5, 3, and 2 patients for astrocytes, neurons, oligodendrocytes, microglia, and endothelial cells, respectively. Age range: 8–63 years. D. Scatterplot of human and mouse astrocyte gene expression. ρ represents the Spearman correlation coefficiency. Venn diagram indicates the number of astrocyte enriched genes (>4 fold, FPKM>5) for human and mouse astrocytes. Average expression data from all samples of mature human astrocytes were used for this analysis. Human, n=12 patients. Mouse, n=4 batches of astrocytes each comprising 3 mouse brains combined. Age range of human patients: 8–63 years. Age range of mice: 1–9 months. Fold enrichment in astrocytes is calculated as FPKM in astrocytes divided by the average FPKM in all the other cell types (neurons, oligodendrocytes, microglia, and endothelial cells). E. Differences in gene expression between purified human and mouse astrocytes measured using qPCR. Fold changes reflect enrichment of specific genes in human tissues. F. In situ hybridization performed on temporal lobe cortices from healthy patients (separate source from the RNAseq samples). Probes were designed against LRRC3B and GPR98 (cyan) and sections were counterstained against the astrocyte specific transcription factor, SOX9 (magenta). Scale bars: 100μm zoomed out, 50μm insets.
Human astrocytes promote neuron survival
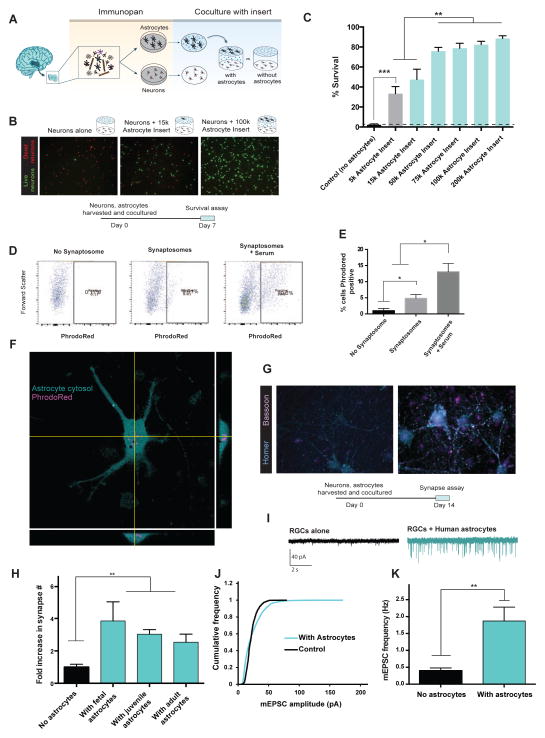
Our purification technique allowed us to investigate the functional capabilities of primary human astrocytes in vitro. We first tested whether human astrocytes could promote neuronal survival, a hallmark feature of mouse and rat astrocytes (Banker, 1980). We immunopanned human neurons and astrocytes (Figure 1A, B) and grew them together in the same cell culture wells on separate layers. The inserts separating the two cell types allowed secreted molecules to freely diffuse while preventing direct astrocyte to neuron contact (Figure 2A). After 7 days of co-culture, we stained live neurons with calcein-AM and dead neurons with ethidium homodimer-1 (Figure 2B). We found that human astrocytes strongly promoted neuronal survival in a dose-dependent manner (Neuronal survival: 2.2 ± 0.9% without astrocytes, 88.5 ± 2.4% with 200k astrocytes. n=8. Figure 2C.). Thus, human astrocytes share the ability of murine astrocytes to strongly promote neuronal survival in vitro.
Figure 2.
Functional characterization of human astrocytes. A. Schematics of co-culture experiments. 17–20 gestation week fetal human astrocytes and neurons were purified by immunopanning, grown in the same wells separated by porous inserts. B. Calcein stain of live neurons (green) and ethidium homodimer stain of dead neurons (red) in the presence and absence of astrocytes. Scale bars: 100μm C. Quantification of survival rate. N=7–8 images per condition. Data represent mean ± SEM in all the figures unless otherwise noted. **, p<0.01. ***, p<0.001. Two-tail unpaired T-test was used for data in C, H, and K. D. Human astrocytes engulf synaptosomes in vitro. FACS plot of human fetal astrocytes incubated without synaptosomes, with synaptosomes, or with synaptosomes and 5% serum. E. Percentages of synaptosome-positive astrocytes. *, p=0.05, one-tailed Wilcoxon rank sum test. n=3 patients. F. Confocal image of a human fetal astrocyte stained with Vibrant CFDA (cyan) engulfing PhrodoRed labeled synaptosomes (magenta) Scale bar: 20μm. G. Retinal ganglion cells form more synapses in the presence of human astrocytes. Cyan: immunofluorescence of post-synaptic marker, Homer. Magenta: immunofluorescence of pre-synaptic marker, Bassoon. Scale bar: 10μm. H. The number of synapses (Homer/Bassoon double positive puncta) in the presence and absence of human astrocytes. N=10–20 images per patient. Each image contains on average two cells. Fetal: 1 patient, age: 18 gw. Juvenile: 3 patients, age: 8, 13, 18 yo. Adult: 3 patients, age: 26, 41, 47 yo. **, p<0.01. I. Representative traces of whole-cell patch clamp recordings from RGCs cultured with or without feeder layers of human astrocytes in the presence of TTX. Few mEPSCs were observed without feeder layers of human astrocytes. J. Human astrocytes significantly increased the amplitude of mEPSCs (p=.0001 Kolmogorov-Smirnov test, n= 488 and 2837 mEPSCs from 16 and 18 cells for no astrocyte and with astrocytes respectively). K. The frequency of mEPSCs was significantly increased in the presence of human astrocytes (Two-tailed Wilcoxon rank sum test. p=0.004. n = 16–18 neurons per condition).
Human astrocytes promote synapse formation
To assess whether human astrocytes, like rat astrocytes, promote synapse formation, we co-cultured human astrocytes with rat retinal ganglion cells (RGCs) using inserts to separate the two cell types. We chose rat RGCs because our group has previously developed a culture medium that supports RGC survival in the absence of astrocytes (Figure S3C, D) (Meyer-Franke et al., 1995; Winzeler and Wang, 2013). After co-culturing human astrocytes with RGCs for 14–17 days, we performed double immunostaining for the presynaptic marker Bassoon and the postsynaptic marker Homer to quantify colocalized synaptic puncta. At the immunohistological level we found that human astrocytes increased the number of synapses three to four fold as we have previously reported for rat astrocytes. This was true for astrocytes of all developmental ages including fetal, juvenile, and adult time points (Figure 2H. Normalized synapse number: 1.0 ± 0.2 without astrocytes; 3.8 ± 1.2 with fetal astrocytes, 3.1 ± 0.3 with juvenile astrocytes, and 2.5 ± 0.5 with adult astrocytes. N=10–20 images per patient. Each image contains on average two cells. Fetal: 1 patient, age: 18 gw. Juvenile: 3 patients, age: 8, 13, 18 yo. Adult: 3 patients, age: 26, 41, 47 yo.). In addition to increasing the number of synapses, we also found that the area of colocalization of the Homer/Bassoon positive puncta were significantly increased when human astrocytes were present (Average colocalization pixel overlap = 16.2 ± 0.6 without astrocytes and 20.8 ± 0.5 with astrocytes. N=1065 and 3212 synapses, in the absence and presence of astrocytes, respectively. Data were obtained using astrocytes from 7 patients. Age: 18gw, 8, 13, 18, 26, 41, and 47 yo).
To next examine whether the synapses induced by human astrocytes were functional and to further validate the synaptogenic effects of human astrocytes, we performed patch-clamp recordings of neurons cultured with or without astrocyte feeder layers. Consistent with our immunohistochemical observations, we found that human astrocytes significantly increased the amplitude (rightward shift in the cumulative probability plot of miniature excitatory postsynaptic currents (mEPSC) amplitude, p=0.0001, Kolmogorov-Smirnov test; mean amplitude of RGCs alone: 21.7 ± 0.4 pA, RGCs with human astrocytes: 23.7 ± 0.3 pA, n= 488 and 2837 mEPSCs from n=16 and 18 cells for no astrocyte and with astrocytes respectively, Figure 2J) and frequency of mEPSCs (RGCs alone 0.4 ± 0.1 Hz; RGCs with human astrocytes: 1.9 ± 0.4 Hz, p= 0.004. n=15 and 23 cells for no astrocyte and with astrocytes respectively. Figure 2I,K). Taken together, these results demonstrate that human astrocytes strongly induce the formation and function of excitatory synapses.
Human astrocytes engulf synaptosomes
Recent studies have found that mouse astrocytes are involved in eliminating extra synapses to help refine neural circuits (Chung et al., 2013). To address whether human astrocytes share similar capabilities, we performed an in vitro synaptosome engulfment assay. We purified synaptosomes and labeled them with a fluorescent dye, PhrodoRed. PhrodoRed is non-fluorescent at neutral pH and fluoresces brightly in acidic environments. It therefore allowed us to detect engulfed synaptosomes that had been trafficked to lysosomes with minimal background fluorescence from sticky synaptosomes attached to the astrocyte surface. To assess the amount of phagocytosis in human astrocytes, we fed PhrodoRed labeled synaptosomes to purified astrocytes either in the presence of astrocyte-conditioned medium (ACM) or serum. Although serum contains high concentrations of bridging molecules that are required for phagocytosis, it is also known to induce reactive astrogliosis. Therefore, we wondered whether astrocyte-conditioned media alone, in the absence of serum, would be sufficient to induce phagocytosis (although serum is only added to cells for the 3 hour duration of the engulfment assay). After adding the labeled synaptosomes, we performed confocal imaging (Figure 2F) and fluorescence assisted cell sorting (FACS) to analyze the percentage of cells with PhrodoRed fluorescence. While synaptosome phagocytosis was greatest with serum present, human astrocytes incubated with ACM alone also significantly engulfed synaptosomes. (Figure 2D–F, percentage of PhrodoRed positive cells: no synaptosome 1.0 ± 0.4%; with synaptosomes 4.8 ± 1.1%; with synaptosomes and serum 13.0±3.3%. n=3 patients.).
Human astrocytes have larger territories and more complex morphologies than rat astrocytes in vitro
Previous studies found that human astrocytes are larger than mouse and rat astrocytes in vivo (Oberheim et al., 2006, 2009). We wondered whether the size of human astrocytes is determined by intrinsic qualities of the astrocytes or via non-cell autonomous mechanisms. To distinguish between these possibilities, we purified human and rat astrocytes by immunopanning, cultured them in identical serum-free culture media at comparable densities, and quantified the length of processes at 5–6 days in vitro (div). We found that the total arborization length of human astrocytes were significantly greater than rat astrocytes in vitro (Figure S1. Average total human astrocyte process length: 566 ± 56 μm. Average total rat astrocyte process length: 290 ± 53μm. Human: n= 20 cells from 5 patients. Rat: n=10 cells from 6 animals.). Additionally, human astrocytes had on average almost twice the number of branches as rat astrocytes (Human: 8.5 ± 1.1, rat: 4.5 ± 0.5) and territories 8.6 times the size of rat astrocytes (Human: 51 ± 13 ×103 μm2. Rat: 5.9 ± 2.9×103 μm2.). These data are consistent with in vivo measurements (Oberheim et al., 2009). Therefore, intrinsic differences contribute, at least in part, to the larger size of human astrocytes compared with rat astrocytes.
Human astrocytes exhibit distinct calcium response properties compared with murine astrocytes
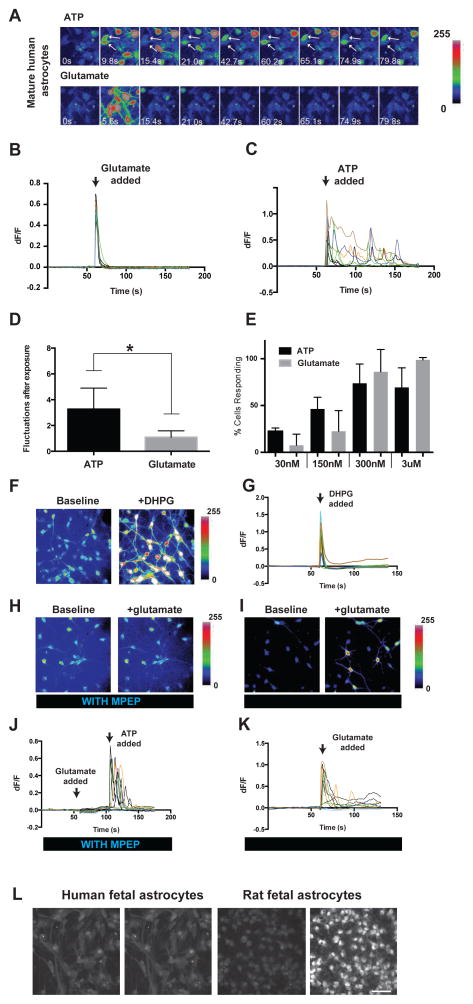
Mouse and rat astrocytes respond to sensory input, synaptic release of glutamate, and extracellular ATP via elevations of intracellular calcium concentrations (Ding et al., 2013; Nimmerjahn et al., 2009; Paukert et al., 2014; Schummers et al., 2008). These calcium transients have been proposed to be important for synaptic transmission, plasticity, and regulation of blood flow (Agulhon et al., 2010; Hamilton and Attwell, 2010; Khakh and McCarthy, 2015; Nedergaard, 1994; Parpura et al., 1994). To investigate whether human astrocytes also respond to these extracellular cues, we performed calcium imaging studies using the calcium sensitive dye Fluo-4 AM on acutely purified human astrocytes. We found that at both fetal and adult stages, human astrocytes responded robustly to ATP (Figure 3A, C, E).
Figure 3.
Calcium responses of human astrocytes in vitro. A. Representative images of calcium responses to 3μM ATP (top) and glutamate (bottom) of mature human astrocytes (21 yo). Time is labeled in each image. Arrows point to two cells showing calcium fluctuations in response to ATP. B, C. ΔF/F of all cells in (A). Each ΔF/F trace represents a single cell. D. Average number of calcium fluctuations in human astrocytes after exposure to ATP or glutamate. N=11 movies. Concentration of ATP and Glutamate: 30nM–3μM. Two-tailed Wilconxon rank-sum test. *, p<0.05. E. Percentage of cells responding to ATP or glutamate at different concentrations. Cells with ΔF/F >5% are counted as responsive. N=3–5 movies per condition. Each movie has 5–20 cells. Data were collected from 2 patients 21 and 26 years of age, respectively. F. Fluo4-AM fluorescence of adult human astrocytes before and after addition of DHPG (200μM). G. ΔF/F of adult human astrocytes before and after addition of DHPG. H. Fluo4-AM fluorescence of adult human astrocytes before and after addition of 3 μM glutamate in the presence of 25 μM MPEP. I. Fluo4-AM fluorescence of adult human astrocytes before and after addition of 3 μM glutamate after washing out MPEP for 30 minutes. J. ΔF/F of adult human astrocytes before and after addition of 3 μM glutamate and 3 μM ATP in the presence of 25 μM MPEP. Arrowheads represent addition of glutamate or ATP K. ΔF/F of adult human astrocytes before and after addition of 3 μM glutamate after washing out MPEP. L. Representative images of calcium responses to 3μM glutamate in fetal human (17–20 gestation week, left) and rat (E18.5, right) astrocytes. Scale bar: 50μm.
We next wondered whether human astrocytes would respond to glutamate. Despite previous observations that adult mouse astrocytes are unresponsive to glutamate receptor agonists (Sun et al., 2013), we found that adult human astrocytes readily responded to glutamate at sub-micromolar concentrations (Figure 3A–E). To better understand the glutamate sensitivity of human astrocytes, we sought to identify which receptor might mediate glutamate responsiveness. We found low, but nonnegligent levels of mGluR5 mRNA in human astrocytes (FPKM=0.34 ± 0.06, n=9, age range =21–63 years), which is consistent with previous reports in human and mouse (Sun et al., 2013). We first tested the effect of the group I mGluR agonist, dihydroxyphenylglycine (DHPG) on adult human astrocytes, and found that DHPG administration reliably induced robust calcium responses (Figure 3F,G). To further dissect which group I mGluR mediates the glutamate response in adult human astrocytes, we applied the mGluR5-specific antagonist 2-methyl-6-phenylethynyl-pyridine (MPEP) to adult human astrocytes prior to the addition of glutamate. The administration of MPEP completely blocked any glutamate response (Figure 3H,J). It is noteworthy that human astrocytes still responded robustly to ATP stimulation in the presence of MPEP, indicating that MPEP does not interfere with calcium storage or release machinery in astrocytes (Figure 3J). After washout of MPEP for 30 minutes, the cells again robustly responded to glutamate (Figure 3IK).
To assess the development of glutamate responsiveness in human astrocytes, we compared the glutamate responses of fetal human and rat astrocytes. Astrogenesis commences shortly before 17–20 gestation week in humans and embryonic day 18.5 (E18.5) in rats. We therefore considered these stages developmentally comparable in these two species. In contrast to rat fetal (E18.5) astrocytes, which readily responded to glutamate at sub-micromolar concentrations, fetal human astrocytes did not respond to glutamate at micromolar or even milimolar concentrations (Percentage of cells responding to 30nM, 300nM, 800nM, 1.7μM, and 3μM glutamate: 5%, 70%, 70%, 100%, and 100%, respectively for rat fetal astrocytes; 0% at all concentrations tested for human fetal astrocytes. n=20 cells per condition. Figure 3L). Therefore, human astrocytes likely acquire the ability to respond to glutamate at a stage later than mid-gestation.
Although adult human astrocytes responded to both ATP and glutamate, closer examination of the calcium transients indicated that ATP and glutamate elicited responses with distinct temporal patterns. Independent of concentration, glutamate stimulation consistently produced a synchronous ‘all or none’ rise in intracellular calcium levels across most astrocytes that then quickly decreased (within 10 seconds) to baseline levels (Figure 3B, C). In contrast, the onset of calcium response to ATP stimulation was asynchronous among the astrocytes in the imaging chamber. After the initial calcium elevation, some cells exhibited fluctuations in calcium concentrations that lasted for minutes after initial ATP stimulation (Figure 3A, C). It remains unclear whether ATP and glutamate elicit calcium responses with similar temporal dynamics in murine astrocytes.
Transcriptomes of acutely purified human neurons, astrocytes, oligodendrocytes, microglia, and endothelial cells
The human brain is composed of neurons, glia (astrocytes, oligodendrocytes, microglia) and vascular cells. Each of these cell types has a distinct role to play in the nervous system, and each can be characterized by a unique molecular repertoire necessary for cell-specific functions. To construct a transcriptome database of purified human neurons, astrocytes, oligodendrocytes, microglia, and endothelial cells, we acutely-isolated each cell type by immunopanning (Figure 1), extracted RNA, and performed RNA-seq (31 ± 4 million pair-end 150bp reads per sample). RNA was collected immediately at the conclusion of the purification protocol to best reflect the true in vivo molecular profiles. Although we took great effort to minimize the duration of time between tissue harvest and RNA collection, gene expression may change during dissociation and purification. Therefore, we collected RNA from cortical tissue immediately after resection and compared this data to RNA collected from the same cortical sample after dissociating the tissue and letting it sit for the duration of a typical purification timeline (within about 5 hours from tissue resection). We found a strong correlation between the two samples, with only 1.49% of the genes changing greater than 4-fold in either direction during the tissue dissociation protocol (294/19713 genes). Those genes on this small list largely constitute immediate early genes from the acute macrophage response to injury (Table S2 and S5). We cross-referenced this list with our downstream analyses to ensure these genes did not confound the interpretation of our data.
To first assess the purity of our immunopanning-isolated cell samples, we probed the transcriptome data for expression of well-known cell-specific genes for astrocytes (e.g. GFAP, ALDH1L1, SOX9, AQP4), neurons (e.g. VGLUT1, STMN2, SYT1, SYN1), oligodendrocytes (e.g. PLP1, MOG, SOX10, MBP), microglia/macrophages (e.g. C1QA, CX3CR1, CCL3, TNF), and endothelial cells (e.g. CLDN5, ELTD1, ITM2A, ESAM) (Figure 4A, B, and Table S4). The expression of these classical cell-specific markers each demonstrated definitive cell type selectivity in their corresponding cell types with undetectable or low level of expression by other cell populations not known to express these markers (Figure 4A, B and Table S3. N=12, 1, 5, 3, and 2 patients for astrocytes, neurons, oligodendrocytes, microglia, and endothelial cells, respectively). To assess the extent of individual differences between human patients, we examined the correlation between replicates of identical cell types across different patients (n=23, age range: 8–63 years) and found high correlations (0.83 ± 0.02, Spearman correlation) between cell type replicates and low correlations (0.34 ± .013) among samples of differing cell types (Figure 4C). We next specifically examined the variation between replicates of human astrocyte samples (0.89 ± 0.05). To determine the degree to which variation among these samples was due to variability in the panning procedure as opposed to inherent inter-patient differences, we performed RNA-seq on four whole cortex samples (specimens that had not been immunopanned) from separate individuals. We found almost identical correlations between whole cortex samples (0.89 ± 0.08) and what we observed in the immunopanned astrocyte samples, suggesting that the variance across astrocyte samples can largely be attributed to small, inherent levels of variation across individuals. As expected, mouse astrocyte samples from the inbred strain (C57BL6) demonstrated a slightly higher correlation (0.94 ± 0.04) than samples from different human patients.
Additionally, we performed unsupervised hierarchical clustering and found that samples of identical cell types clustered together, rather than clustering by patient (Figure S4). Together these data suggest that transcriptome differences between cell types are not obscured by individual genetic and/or environmental variations between subjects. To disseminate this human cell type specific gene expression data to the greater neuroscience community, we generated a user-friendly searchable online database of the gene expression profiles of all acutely purified human cell types (http://www.BrainRnaSeq.org).
Transcriptome differences between human APCs and mature astrocytes
The molecular and cellular changes that occur during astrocyte maturation are poorly understood. To examine gene expression changes associated with astrocyte maturation, we compared transcriptomes of APCs and mature human astrocytes. Whereas a large number of genes were expressed both by APCs and mature astrocytes (Figure 5A, B, fetal, n=6 patients. Mature, n=12 patients), a substantial contingent (>2000) of genes were differentially expressed between the two stages (Table 1). For example, APCs expressed high levels of proliferative genes such as MKI67 and TOP2A, confirming their immature properties (Figure 5B, F). In contrast, a number of genes were only expressed by mature astrocytes, such as gap junction genes GJA1 and GJB6, the human neuropsychiatric disorder associated gene AGXT2L1, and a Wnt pathway inhibitory factor WIF1. These genes provide new markers that could be used for the discrimination of developing and mature human astrocytes. To take an unbiased approach to find pathways unique to APCs and mature astrocytes, we identified all significantly differentially expressed (DE) genes between the two developmental stages (using volcano plots with cutoffs at p=.01 and 4-fold ratios, Figure S5C) and used these DE genes to run a Gene Ontology (GO) term analysis with DAVID bioinformatics resources (Huang et al., 2009). As astrocytes mature, we found significant down regulation of cell cycle genes, and a significant upregulation in genes involved with nerve impulse transmission, cell-cell signaling, fatty acid metabolism, cell adhesion, and ion homeostasis (full list of GO terms and DE genes in Tables S5, S6). We performed identical volcano plot and DAVID analyses for subsequent comparisons between cell types and species (Figure S5 B–E and Tables S5, S6).
Table 1.
Top gene expression differences between APC and mature astrocytes. Genes are ranked by fold change in expression between APC and mature stages. The top and bottom 20 genes are listed. Genes with FPKM<10 in both APC and mature stages are excluded to highlight highly expressed genes that are more likely to be useful markers for APC or mature astrocytes. All genes have >2 fold enrichment in astrocytes over other cell types.
| Top mature astrocyte genes | Top APC genes | ||||||
|---|---|---|---|---|---|---|---|
| Gene | Expression in fetal astrocytes | Expression in mature astrocytes | Fold change | Gene | Expression in fetal astrocytes | Expression in mature astrocytes | Fold change |
| AGXT2L1 | 0.1 | 238.6 | 1988.6 | HIST1H2AI | 57.9 | 0.1 | 578.9 |
| S100A1 | 0.1 | 84.9 | 849.4 | HIST1H3E | 51.0 | 0.1 | 509.6 |
| SLC14A1 | 0.1 | 29.3 | 334.6 | HIST1H3B | 176.6 | 0.6 | 292.1 |
| TMEM176A | 0.1 | 27.1 | 270.6 | HIST1H1B | 23.4 | 0.1 | 234.0 |
| TMX2 | 0.1 | 25.9 | 259.1 | PPDPF | 26.4 | 0.2 | 119.3 |
| HHATL | 0.1 | 24.9 | 237.6 | TPX2 | 22.0 | 0.2 | 93.9 |
| PADI2 | 0.1 | 26.8 | 225.3 | NUSAP1 | 27.4 | 0.3 | 90.4 |
| TLR4 | 0.1 | 18.7 | 212.9 | HIST1H2AC | 39.0 | 0.5 | 76.8 |
| HSD17B6 | 0.1 | 20.5 | 204.6 | HIST2H2AC | 68.9 | 1.3 | 53.2 |
| CHI3L1 | 0.1 | 15.2 | 172.2 | TNC | 26.6 | 0.6 | 45.5 |
| NUDT3 | 0.1 | 13.5 | 135.2 | KIF15 | 17.2 | 0.5 | 32.7 |
| FBXO2 | 0.1 | 10.8 | 115.1 | HIST1H1A | 45.9 | 1.5 | 30.7 |
| NTSR2 | 0.1 | 10.8 | 107.5 | HIST1H2BC | 215.2 | 13.2 | 16.3 |
| ALDH1L1 | 0.1 | 10.8 | 82.5 | FABP5 | 23.9 | 1.5 | 15.5 |
| ALDOC | 4.8 | 371.0 | 77.6 | DTYMK | 10.3 | 0.7 | 14.8 |
| SLC1A2 | 34.5 | 1987.9 | 57.6 | RAB11B | 10.9 | 0.8 | 13.5 |
| RYR3 | 0.3 | 16.0 | 55.0 | HES6 | 21.6 | 2.1 | 10.3 |
| GABRA2 | 0.9 | 46.1 | 51.2 | LRIG3 | 13.3 | 1.4 | 9.6 |
| CPE | 16.0 | 657.2 | 41.0 | E2F5 | 10.1 | 1.4 | 7.2 |
| GLUL | 21.0 | 826.9 | 39.4 | MPPED2 | 16.0 | 2.3 | 7.0 |
Similarities between APCs and glioblastoma astrocytes
The molecular profiles of APCs appear overwhelmingly immature and proliferative, much like what might be expected for astrocytes that comprise late stage glioblastomas in adults. Therefore, we wondered how astrocytes originating from glioblastoma tumor samples might compare to our fetal and mature resting astrocyte profiles. We used HepaCAM to purify astrocytes from resected glioblastoma multiforme (GBM) tumors and subsequently sequenced these samples to obtain their molecular profiles. GBMs are aggressive and infiltrative tumors that do not have clear margins, so we chose to purify two populations of cells from the GBM; those that enhance with gadolinium contrast on an MRI scan (referred to as tumor core), and those that are non contrast enhancing (tumor periphery) (Figure 5C). Although not involved with the core of the tumor itself, the tumor periphery cells are still considered abnormal because as GBM cells infiltrate the normal brain, the brain tissue surrounding the contrast-enhancing ring is edematous as a reaction to tumor involvement. As expected, the HepaCAM purified cells from the tumor periphery and core both clustered with APCs and mature astrocytes, but interestingly, the tumor core regions of the GBM clustered more closely to APCs as compared to the tumor periphery samples, which clustered alongside the mature astrocytes (Figure 5D – F, S4C, fetal, n=6. Tumor core, n=3. Tumor periphery, n=1).
Comparing human and mouse astrocyte transcriptomes
We next investigated the extent to which human and mouse astrocytes share similar gene expression profiles. Since existing RNA-seq data from mouse astrocytes was previously collected by FACS (Fluorescence Assisted Cell Sorting) sorting from transgenic lines (Zhang et al., 2014), we collected new mouse astrocyte samples using an identical procedure to our human astrocytes, including the use of the monoclonal HepaCAM antibody for immunopanning. To ensure that HepaCAM panned mouse astrocytes were comparable to our previous method of purifying mouse astrocytes via FACS sorting ALDH1L1-EGFP mice, we collected and compared aged-matched FACS sorted ALDH1L1-EGFP astrocytes with HepaCAM panned astrocytes. Again using volcano plots with DE cutoffs of p=.01 and 4-fold ratio changes, we could detect only 55 total DE genes (0.3%) between the two different methods of mouse astrocyte purification (Figure S5A and Table S6). We then compared the RNA-seq based transcriptomes of human and mouse HepaCAM purified astrocytes and found significant overlap including all of the classic astrocyte-specific genes (e.g. GFAP, ALDH1L1, GLUL, AQP4, SLC1A2, and SLC1A3). Using a threshold of 4-fold enrichment (calculated as expression in astrocytes divided by the average expression level in all other cell types—neurons, oligodendrocytes, microglia, and endothelial cells), 52% of mouse astrocyte-enriched genes were similarly enriched in human astrocytes. Conversely, only 30% of human astrocyte-enriched genes were identified as astrocyte-enriched in mice (Figure 4D and Table 2. Human, n=12 patients. Mouse, n=4 batches of astrocytes each made from 3 mouse brains combined). These trends were consistent across a range of thresholds from 2-fold to 50-fold (data not shown). We next examined whether human-mouse differences in gene expression were more prominent in specific cell types. We found almost identical transcriptome-level differences between human and mouse in each of the brain cell types that we examined, and these values were also comparable to non-neural tissues such as liver and muscle cells (data not shown).
Table 2.
Top astrocyte-enriched genes shared by humans and mice, and those only present in human or mouse astrocytes. Shared astrocyte genes are expressed in both human and mouse astrocytes (FPKM > 0.5) and enriched by at least 4 fold over the average expression in neurons, oligodendrocytes, microglia/macrophages, and endothelial cells. Species-specific astrocyte genes have FPKM values < 1 in astrocytes from the non-enriched species, and those that are also astrocyte enriched are indicated with asterisks. Top 20 genes are listed for all categories (ranked by fold enrichment).
| Human and Mouse Astrocyte Specific Markers | Genes in human astrocytes not found in mouse astrocytes | Genes in mouse astrocytes not found in human astrocytes | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene symbol |
Human expression |
Mouse expression |
Gene symbol |
Human astrocyte expression |
Mouse astrocyte expression |
Fold enrichment in human astrocytes / mouse astrocytes |
Gene symbol | Human astrocyte expression |
Mouse astrocyte expression |
Fold enrichment in mouse astrocytes / human astrocytes |
| GJA1 | 560.3 | 351.5 | WIF1* | 66.9 | 0.1 | 668.7 | PLA2G7* | 0.2 | 138.0 | 866.5 |
| SLC39A12 | 79.3 | 88.6 | PMP2 | 80.1 | 0.1 | 663.8 | H3F3A | 0.1 | 56.2 | 561.9 |
| GJB6 | 106.0 | 88.8 | GSTM2* | 43.6 | 0.1 | 331.1 | THRSP | 0.1 | 36.1 | 360.6 |
| BMPR1B | 27.0 | 24.7 | LRRC3B* | 29.7 | 0.1 | 245.0 | ITIH3* | 0.1 | 28.5 | 285.2 |
| SLC14A1 | 29.3 | 37.8 | HSD17B6* | 20.5 | 0.1 | 204.6 | LXN* | 0.2 | 61.2 | 265.0 |
| PAMR1 | 23.1 | 23.8 | FAM198B* | 20.0 | 0.1 | 198.9 | VCAM1* | 0.2 | 45.6 | 226.6 |
| RGS20 | 17.9 | 52.6 | PNMA1 | 26.1 | 0.1 | 187.7 | NNAT | 0.1 | 21.8 | 218.3 |
| FGFR3 | 38.4 | 182.4 | MRVI1* | 18.3 | 0.1 | 183.2 | TNFRSF19 | 0.2 | 37.9 | 217.6 |
| MGST1 | 72.4 | 18.1 | RYR3* | 16.0 | 0.1 | 160.0 | NGEF | 0.1 | 22.1 | 211.9 |
| SLC4A4 | 97.6 | 199.4 | GSTM3 | 19.3 | 0.1 | 144.1 | SNAPC5 | 0.1 | 19.6 | 195.7 |
| AQP4 | 450.0 | 198.4 | APOC2 | 15.3 | 0.1 | 137.4 | CHST2 | 0.3 | 57.3 | 195.0 |
| F3 | 126.1 | 112.6 | RGN | 18.1 | 0.1 | 133.5 | MAGOH | 0.1 | 18.0 | 180.0 |
| MLC1 | 15.3 | 242.3 | ACBD7* | 21.6 | 0.2 | 132.3 | ROMO1 | 0.1 | 16.9 | 169.3 |
| ELOVL2 | 41.2 | 34.2 | RGR* | 12.5 | 0.1 | 115.6 | AKR1B10 | 0.1 | 16.2 | 161.9 |
| SLC1A2 | 1987.9 | 1087.4 | AMY2B | 11.5 | 0.1 | 114.5 | FAM176A | 0.1 | 16.4 | 160.5 |
| CLU | 384.2 | 769.3 | NDUSF5 | 27.3 | 0.2 | 112.3 | IL18* | 0.7 | 107.8 | 154.8 |
| ACSBG1 | 19.6 | 140.8 | STOX1* | 18.1 | 0.2 | 107.3 | POU3F3 | 0.1 | 15.1 | 150.6 |
| TTYH1 | 16.4 | 360.5 | GPR98* | 50.3 | 0.5 | 97.1 | FAM173A | 0.1 | 16.2 | 145.9 |
| ATP1B2 | 468.7 | 550.7 | AADAT* | 10.7 | 0.1 | 79.2 | RNF5 | 0.2 | 24.6 | 142.9 |
| SOX9 | 50.5 | 57.7 | CALCOCO2 | 10.5 | 0.1 | 75.4 | TSPAN2 | 0.1 | 13.5 | 134.4 |
Denotes a gene that is also enriched >4 fold in astrocytes over other CNS cell types
One further concern about interpreting interspecies differences is that the human data is exclusively derived from temporal lobe, in comparison to whole cortex data from mouse. To address this potential confounder, we utilized the Allen Brain Atlas dataset (Hawrylycz et al., 2012) to determine all genes that are enriched in temporal lobe vs other cortical areas and found no overlap with our candidate human vs mouse enriched genes (Table 2 and S7). Despite the homology between human and mouse genomes, there are a number of human genes and long non-coding RNAs without mouse orthologs. We found that some of these human-specific genomic elements are expressed in cell type-specific patterns (Table S8). Further investigation of these candidates may provide novel insights into new properties that have evolved from mouse to human in various CNS cell types.
Validation of genes differentially expressed by human and mouse
We next examined the validity of our RNA-seq transcriptome datasets by orthologous methods. We first examined a subset of genes identified by our RNA-seq data to be differentially expressed by human and mouse cells using PCR on cDNA generated from whole mouse and human brain samples. PCR amplification of all of the genes we examined (WIF1, MRVI1, APOC2, GLDN, GPR98, PMP2, CNDP1, and MAG) showed species selective expression patterns as predicted by RNA-seq (Figure S6). Next, we used qPCR to validate a subset of the top gene candidates differentially expressed between fetal vs adult human and mouse vs human astrocytes and found consistent agreement with our RNA-seq findings in each gene that we investigated (Figure 4E and 5G).
Additionally, we used in situ hybridization to examine the spatial expression of two human astrocyte-enriched genes on human brain sections. We combined fluorescence in situ hybridization with probes designed against two of the top human-specific astrocyte candidates, GPR98 and LRRC3B, and immunofluorescence with an antibody against SOX9, an astrocyte-specific nuclear protein. We found strong colocalization between our RNA probes and SOX9 staining, providing evidence that GPR98 and LRRC3B are both highly enriched in human astrocytes (Figure 4F) as predicted by the RNA-Seq data.
Reactive changes of human astrocytes in neurological disorders
Previous methods of enriching human astrocytes (the McCarthy de Vellis or MD method) in serum-based cultures involve long-term exposure to a non-physiological environment, and serum exposure in culture has been shown to induce irreversible reactive changes in astrocytes (Foo et al., 2011; Zamanian et al., 2012). Furthermore, astrocytes grown in vitro via serum-selection methods demonstrate unusual polygonal fibroblast-like morphologies that are not present in vivo, whereas our acutely purified astrocytes grown in serum-free medium exhibit extensive process bearing morphologies (Figures 1, S3). To test whether exposure to serum could elicit morphological changes in acutely purified human astrocytes, we added serum to our cultures of immunopanned human astrocytes and quickly noted a transition from a process-bearing morphology to the polygonal fibroblast-like morphology typically found in serum-cultured cells (Figure 6B).
To examine whether the transcriptome profiles of acutely purified human astrocytes more closely resembled resting or reactive astrocytes, we probed the RNA-seq dataset of human astrocytes for reactive astrocyte genes. Previous reports of mouse reactive astrocytes have described two distinct types of reactive phenotypes, a bacterial lipopolysaccharide (LPS) infection induced “A1” phenotype, and an ischemia induced “A2” phenotype (Zamanian et al., 2012). We cross-referenced the top 30 genes that have been shown in astrocytes to be induced by ischemia alone, LPS alone, or both injury models with our acutely purified human astrocytes. We found only low expression of these reactive astrocyte markers in our preparations of acutely-purified human astrocytes (Figure 6A, n=12 patients). In contrast, we found that human astrocytes obtained using serum selection methods from two commercial sources, Lonza and Sciencell (Lonza astrocytes, n=1, and Sciencell astrocytes, n=2) (Malik et al., 2014), expressed high levels of these reactive astrocyte markers.
What are the transcriptome-wide changes that affect human astrocytes in neurological diseases? Numerous postmortem immunofluorescence studies of patients with a variety of neurological conditions have demonstrated up-regulation of a single reactive astrocyte marker, GFAP (Beach and McGeer, 1988), but a transcriptome level examination of reactive changes in human astrocytes has not yet been performed. To examine disease-induced gene expression changes in human astrocytes, we acutely purified astrocytes from brain samples taken from regions involved in epilepsy and glioblastoma. We then performed RNA-seq to compare transcriptomes of disease-affected astrocytes with healthy human astrocytes. Much like in serum-cultured astrocytes, we found significant increases in the expression of reactive astrocyte genes in disease-affected astrocytes (Figure 6A and Table S6). These results support numerous morphological observations that human astrocytes undergo robust reactive changes in epilepsy and glioblastoma.
Discussion
We have developed a method to acutely purify human astrocytes and to grow these cells in serum-free conditions in vitro. This method allowed us for the first time to directly investigate the functions of human astrocytes. Like mouse astrocytes, we found that human astrocytes strongly promote neuronal survival, synapse formation, and engulf synapses. We also found that human astrocytes retain their larger size in vitro and that adult human astrocytes demonstrate robust mGluR5-mediated calcium responses to glutamate stimulation. We next performed RNA-seq transcriptome profiling of purified human neurons, astrocytes, oligodendrocytes, microglia, and endothelial cells and established a database that serves as a road map for understanding cell type specific gene function in the human brain. By comparing human and mouse astrocyte transcriptome profiles, we found large numbers of genes shared by astrocytes of both species as well as a gene expression signature specific to human astrocytes. We further demonstrated that acutely purified human astrocytes from normal brain samples exhibit resting astrocyte gene expression profiles and that epilepsy and tumor robustly induce reactive changes in human astrocytes.
Application of new immunopanning purification protocol for studies of human brain cell types
The ability to purify and culture human astrocytes opens many new avenues of investigation. The vast majority of drug candidates for neurological disorders succeed in mouse model studies but fail in human clinical trials (Kola and Landis, 2004). Thus, there is a strong need for human cell culture models for disease mechanism studies and drug testing. Acutely purified human astrocytes can be cultured for several weeks in vitro and maintain functional properties at these later time points (Figure S1, S3). It should be noted, however, that the transcriptome profiles of ‘resting’ human astrocytes are derived from acutely purified astrocytes and not those in culture.
Since astrocytes have been found to be key players in many neurological disorders, improvement of human astrocyte culture methods may lead to new discoveries in the treatment of neurological disorders. Previously developed serum-based culture methods generate human astrocytes that express reactive astrocyte genes and are already in a disease-like state. Therefore, it is difficult to identify additional disease-induced changes above the background reactive changes with these cells. The data presented here of acutely purified human astrocytes provides a baseline by which to compare changes in gene expression in diseased vs. healthy brains.
Calcium responses in human astrocytes
The glutamate response of adult human astrocytes is surprising in light of a recent study on mouse astrocytes that reported that only young mouse astrocytes respond to a glutamate receptor agonist (Sun et al., 2013). Like Sun et al., we found low mRNA levels of mGluR5 in adult human astrocytes, but pursued pharmacological studies to examine whether this low expression might mediate the robust glutamate response of adult human astrocytes. Our pharmacological experiments suggest that human astrocyte glutamate signaling occurs via mGluR5. The difference in the ability of adult human and mouse astrocytes to respond to extracellular glutamate suggests that adult human astrocytes may have evolved an improved capability to detect and potentially respond to synaptic activity. Whether such a difference contributes to unique cognitive abilities of humans remains an intriguing question.
Two distinct stages of human astrocyte development
The assembly of neural circuits and maturation of the brain involves precisely orchestrated changes of neurons and glia. The distinct characteristics of neural progenitors and mature neurons, oligodendrocytes precursor cells and mature oligodendrocytes have been well defined. Astrocyte maturation, however, is much less understood. Mouse astrocytes are generated from radial glial cells during late embryogenesis and early postnatal development, and continue to divide and populate the brain during the first two postnatal weeks (Ge et al., 2012; Tien et al., 2012). After proceeding through a poorly understood maturation process, astrocytes lose their abilities to divide, develop elaborate processes that ensheath synapses, and surround blood vessels to form the blood-brain-barrier. Investigating how molecular profiles of human astrocytes mature throughout development is critical not only for understanding brain development but for gaining insights into pathologies where astrocytes acquire fetal phenotypes (i.e. glioblastoma) (Figure 5).
In this study, we found that human astrocytes exist in at least two distinct states: a fetal highly proliferative astrocyte precursor cell (APC) state and a postnatal (childhood and adult) non-proliferative mature state, bearing much similarity with the oligodendrocyte precursor cells (OPCs) and mature oligodendrocyte stage. As has been previously observed in mouse astrocytes (Ge et al., 2012; Tien et al., 2012), astrocytes isolated from fetal human brains had a vastly higher proliferative ability than mature astrocytes. We did not observe proliferation (or the presence of mitotic genes) in astrocytes isolated from older human brains, even in the presence of high levels of the mitogen HBEGF. Our transcriptome profiling of fetal, juvenile, and adult human astrocytes provides a molecular description of the numerous changes that occur during astrocyte maturation. As human astrocytes mature, they turn on genes encoding synapse inducing proteins (SPARCL1, Glypicans), gap junction proteins (GJA1, GJB6), proteins for neural transmitter recycling (SLC1A2, SLC1A3, GLUL), secreted proteins (CLU, SPON1), a thyroid hormone metabolism enzyme (DIO2), and a variety of transmembrane receptors (FGFR3, and GABRA2). We cross-referenced a published dataset of human brain gene expression across developmental stages (Kang et al., 2011) and found that mature astrocytes are hardly present prior to birth in most of the regions in the human brain. The vast majority of mature astrocyte markers rapidly increase in expression soon after birth, peaking around 6 months to 1 year of age. Interestingly, changes of synapse density in the developing human cortex follows an identical time course (Huttenlocher, 1979; Huttenlocher et al., 1982). Furthermore, a phagocytic receptor required for astrocyte-mediated synapse elimination, MERTK (Chung et al., 2013), is not expressed by fetal astrocytes but later turns on and reaches peak levels around 6 months to 1 year of age. Concurrently, synapse density begins to decline after 1 year of age in humans (Huttenlocher, 1979; Huttenlocher et al., 1982). The precise temporal correspondence of human astrocyte maturation and synapse density, as well as previously reported roles of astrocytes in synapse formation and elimination suggests that astrocytes may trigger maturation of neural circuitry. Consistent with this notion, a number of genes associated with neurodevelopmental disorders including autism and schizophrenia are enriched in astrocytes (data not shown). An important question for future study is to understand the intrinsic or extrinsic signaling mechanisms that trigger astrocyte maturation. Alterations in the timing of this maturation could potentially underlie neurodevelopmental disorders.
Astrocyte transcriptome changes in epilepsy and glioblastoma
Our immunopanning technique allowed us to purify and profile astrocytes from diseased environments. At the transcriptomic level, our data corroborates numerous morphological observational studies of astrocytes in these conditions. We found over four hundred differentially expressed genes between healthy astrocytes and those from sclerotic hippocampus, and similar numbers from those in glioblastoma. A careful examination of the gene expression changes in these disease states may potentially generate new insights into the role of astrocytes in these diseases.
Species differences in astrocyte properties
To compare the transcriptomic landscape of human and mouse astrocytes, we began by performing an unbiased genome-wide comparison of the two species. We found over 600 human astrocyte-enriched genes that were not similarly enriched in mouse astrocytes. Some of these genes may provide insight into the special physiological and functional properties of human astrocytes. For example we found that the ryanodine receptor 3 (Ryr3) is highly expressed by human astrocytes but not by mouse astrocytes. Ryr3 is a calcium permeable ion channel located on the membrane of the endoplasmic reticulum (ER), where calcium is stored inside cells. It is activated by elevation of cytoplasmic calcium concentration and releases calcium from the internal store in a process called “calcium activated calcium release” in muscle cells (Gordienko and Bolton, 2002). The presence of Ryr3 in human astrocytes might amplify elevation of intracellular calcium concentration and allow fast propagation of calcium transients along astrocyte processes by releasing calcium from spatially discrete ER compartments. We also found human specific expression of MRVI1, a protein that binds the IP3 receptor and regulates intracellular calcium stores (Figure S6). These findings are consistent with a study reporting differences in the propagation of calcium transients between human and mouse astrocyte networks (Oberheim et al., 2009).
As the evidence accumulates for the importance of glia, and particularly astrocytes, in health and disease, the purification method and transcriptome datasets presented here will be an invaluable resource for investigating the biology of human astrocytes and for investigating new treatment approaches for neurological disorders.
Experimental Procedures
Human tissue
Human brain tissue was obtained with informed consent under a Stanford University Institutional Review Board approved protocol. Juvenile and adult human brain tissue was obtained from surgeries for treating epilepsy and tumors. All experiments described in this study, except the reactive astrocyte gene induction studies (Figure 5 and 6, epilepsy, tumor, and tumor peripheral data), were performed with by-and-large normal temporal lobe cortex resected in order to access deeper hippocampus areas involved in epilepsy. The cortical tissue was determined as normal by EEG and MRI. For assessment of reactive astrocyte gene expression we used sclerotic hippocampus specimens involved in epileptic foci and cortical specimens from glioblastoma core and peripheral regions, defined as contrast enhancing and non-contrast enhancing regions, respectively. Tissue was immersed in 4°C Neurobasal medium and transferred to the lab for tissue dissociation within 1 hour after resection. Fetal human brain tissue was obtained with informed consent following elective pregnancy termination under a Stanford University Institutional Review Board approved protocol. Tissue was immersed in 4°C RPMI medium and transferred to the lab for tissue dissociation within 5 hours after the procedure.
Vertebrate animals
All procedures involving animals were conducted in conformity with Stanford University guidelines that are in compliance with national and state laws and policies. We used mice for RNA-seq transcriptome profiling, in situ hybridization and PCR validation experiments of the RNA-seq data. We used rats for in vitro experiments including morphology measurement and calcium imaging because of higher yield and consistency of rat astrocyte culture compared to mouse astrocyte culture.
Purification of mature human astrocytes, neurons, oligodendrocytes, microglia/macrophages, and endothelial cells
For a detailed purification protocol, see Supplemental Information. Briefly, we dissected out grey matter from mature human brain specimens and incubated the tissue in 20 unit/ml papain at 34°C for 100 minutes and washed with a protease inhibitor stock solution. We then gently triturated the tissue and then added the single cell suspension to a series of plastic petri dish pre-coated with cell type specific antibodies and incubated for 10 – 30 minutes each at room temperature. Unbound cells were transferred to the subsequent petri dish while the dish with bound cells was rinsed 8 times with PBS to wash away loosely bound contaminating cell types. The antibodies used include anti-CD45 to capture microglia/macrophages, anti-GalC hybridoma supernatant to harvest oligodendrocytes, anti-O4 hybridoma to harvest oligodendrocytes precursor cells, anti-Thy1 (CD90) to harvest neurons, anti-HepaCAM to harvest astrocytes, and finally Banderiaea simplicifolia lectin 1 (BSL-1) to harvest endothelial cells. For RNA-seq, cell samples were scraped off the panning dish directly with Qiazol reagent (Qiagen). For cell culture and in vitro experiments, astrocytes bound to the anti-HepaCAM antibody coated dishes were incubated in a trypsin solution and incubated at 37°C for 5–10 minutes and gently squirted off the plate. We then spun down the astrocytes and plated them on poly-D-lysine coated plastic coverslips in a Neurobasal-DMEM based serum-free medium (the composition is detailed in Supplemental Information). We replaced half of the volume with fresh medium every 3–4 days to maintain the cultures. Fetal human astrocytes and neurons were purified in a similar protocol to the above-mentioned procedure with the modifications detailed in the Supplemental Information.
RNA-seq library construction and sequencing
Total RNA was extracted using the miRNeasy kit (Qiagen) under the protocols of the manufacturer. The quality was assessed by Bioanalyzer. Samples with RNA integrity number higher than 8 were used for library construction. We used the Ovation® RNA-seq system V2 (Nugen 7102) to perform first and second-strand cDNA synthesis. We then used the Next Ultra RNA-seq library prep kit for Illumina (NEB E7530) and NEBNext® multiplex oligos for Illumina® (NEB E7335 E7500) to perform end repair, adaptor ligation, and 5–6 cycles of PCR enrichment according to manufacturer’s instructions. The quality of the libraries was assessed by bioanalyzer and qPCR and high quality libraries were sequenced by the Illumina NextSeq sequencer to obtain 150bp pair-end reads. See detailed protocol in Supplemental Information.
RNA-seq read mapping, transcript assembly, and expression level estimation
We analyzed RNA-seq reads with the Galaxy web-platform (http://usegalaxy.org). The FASTQ files were first groomed using the FASTQ groomer and then mapped using TopHat2, which invokes Bowtie as an internal read mapper. The paired end option was selected and human genome version 19 (hg19) and mouse genome version 9 (mm 9) was used as the reference genome for the human and mouse RNA-seq data, respectively. We then ran Cufflinks to assemble transcripts and estimate expression level as fragments per kilobase of transcript sequence per million mapped fragments (FPKM). We deposited the RNA-seq data in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO, accession number GSE73721).
Calcium imaging
Cells were incubated for 15 minutes with 2μM Fluo 4 AM (Invitrogen, F-14201) and then washed with PBS and replaced with growth medium. Images were taken at 0.7s intervals and analyzed with ImageJ.
Immunocytochemistry
Cultured cells were fixed with 4% PFA for 10 minutes at room temperature, permeablized and blocked with 10% goat serum with 0.2% Triton-X100. See Supplemental Information for antibody details.
Electrophysiology
Whole-cell patch-clamp recordings from cultured RGC neurons were performed at room temperature in an isotonic saline solution. Miniature excitatory postsynaptic currents (mEPSCs) were recorded in TTX (1μM, Alomone) from a holding potential of −70 mV. Series resistance was monitored throughout the recording and was <20 MΩ. Data were sampled at 50 kHz and filtered at 1 kHz. See detailed protocol in Supplemental Information.
Synapse formation assay
We purified rat RGCs by sequential immunopanning to greater than 99% purity and cultured them in serum-free medium as previously described (Winzeler and Wang, 2013). Human astrocytes were plated on inserts and co-cultured with RGCs for 14–17 days. For quantification of structural synapses, RGCs were fixed and stained with antibodies against the presynaptic marker Bassoon and postsynaptic marker Homer. Synapse number and size were quantified by a custom-written Matlab program (available upon request). See detailed protocol in Supplemental Information.
In situ hybridization
Full-length human cDNA expression plasmids were obtained from Dharmocon. Digoxigenin (DIG) labeled single-stranded antisense riboprobes were prepared and fresh frozen 12 μm thick brain sections were processed as previously described (Zhang et al., 2014) (Supplemental Information).
Supplementary Material
Acknowledgments
We would like to thank Maria Fabian, Yingmei Li, Annie Abay, Tom Babcock, Andrew Olson, Stanford University Neurosurgery Brain Bank, and Stanford Neuroscience Microscopy Service for technical assistance, Won-suk Chung for help with synaptosome engulfment assay, Mariko Bennett and Chris Bennett for help with in situ hybridization, the Barres laboratory for helpful discussions, and Nenad Sestan for brain tissue. This study was supported by National Institute of Health grants: R01NS081703 and R01MH099555, and a grant from the JPB foundation to B.A.B., Walter V. and Idun Berry Fellowship and K99NS089780 to Y.Z., and T32GM007365 with additional support from Stanford School of Medicine and its Medical Scientist Training Program and F30MH106261 to S. S. We thank Vincent and Stella Coates and Dr. Miriam and Sheldon G. Adelson Medical Research Foundation for their generous support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agulhon C, Fiacco TA, McCarthy KD. Hippocampal short- and long-term plasticity are not modulated by astrocyte Ca2+ signaling. Sci (New York, NY) 2010;327:1250–1254. doi: 10.1126/science.1184821. [DOI] [PubMed] [Google Scholar]
- Allen NJ, Barres BA. Neuroscience: Glia - more than just brain glue. Nature. 2009;457:675–677. doi: 10.1038/457675a. [DOI] [PubMed] [Google Scholar]
- Allen NJ, Bennett ML, Foo LC, Wang GX, Chakraborty C, Smith SJ, Barres BA. Astrocyte glypicans 4 and 6 promote formation of excitatory synapses via GluA1 AMPA receptors. Nature. 2012;486:410–414. doi: 10.1038/nature11059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attwell D, Buchan AM, Charpak S, Lauritzen M, MacVicar BA, Newman EA. Glial and neuronal control of brain blood flow. Nature. 2010;468:232–243. doi: 10.1038/nature09613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banker GA. Trophic interactions between astroglial cells and hippocampal neurons in culture. Science. 1980;209:809–810. doi: 10.1126/science.7403847. [DOI] [PubMed] [Google Scholar]
- Beach TG, McGeer EG. Lamina-specific arrangement of astrocytic gliosis and senile plaques in Alzheimer’s disease visual cortex. Brain Res. 1988;463:357–361. doi: 10.1016/0006-8993(88)90410-6. [DOI] [PubMed] [Google Scholar]
- Christopherson KS, Ullian EM, Stokes CCA, Mullowney CE, Hell JW, Agah A, Lawler J, Mosher DF, Bornstein P, Barres BA. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell. 2005;120:421–433. doi: 10.1016/j.cell.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Chung WS, Clarke LE, Wang GX, Stafford BK, Sher A, Chakraborty C, Joung J, Foo LC, Thompson A, Chen C, et al. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature. 2013;504:394–400. doi: 10.1038/nature12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W-S, Allen NJ, Eroglu C. Astrocytes Control Synapse Formation, Function, and Elimination. Cold Spring Harb Perspect Biol. 2015:a020370. doi: 10.1101/cshperspect.a020370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke LE, Barres BA. Emerging roles of astrocytes in neural circuit development. Nat Rev Neurosci. 2013;14:311–321. doi: 10.1038/nrn3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, O’Donnell J, Thrane AS, Zeppenfeld D, Kang H, Xie L, Wang F, Nedergaard M. α1-Adrenergic receptors mediate coordinated Ca2+ signaling of cortical astrocytes in awake, behaving mice. Cell Calcium. 2013:1–9. doi: 10.1016/j.ceca.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo LC, Allen NJ, Bushong EA, Ventura PB, Chung WS, Zhou L, Cahoy JD, Daneman R, Zong H, Ellisman MH, et al. Development of a method for the purification and culture of rodent astrocytes. Neuron. 2011;71:799–811. doi: 10.1016/j.neuron.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge WP, Miyawaki A, Gage FH, Jan YN, Jan LY. Local generation of glia is a major astrocyte source in postnatal cortex. Nature. 2012;484:376–380. doi: 10.1038/nature10959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordienko DV, Bolton TB. Crosstalk between ryanodine receptors and IP 3 receptors as a factor shaping spontaneous Ca 2+-release events in rabbit portal vein myocytes. J Physiol. 2002;542:743–762. doi: 10.1113/jphysiol.2001.015966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton NB, Attwell D. Do astrocytes really exocytose neurotransmitters? Nat. Rev Neurosci. 2010;11:227–238. doi: 10.1038/nrn2803. [DOI] [PubMed] [Google Scholar]
- Han X, Chen M, Wang F, Windrem M, Wang S, Shanz S, Xu Q, Oberheim NA, Bekar L, Betstadt S, et al. Forebrain Engraftment by Human Glial Progenitor Cells Enhances Synaptic Plasticity and Learning in Adult Mice. Cell Stem Cell. 2013;12:342–353. doi: 10.1016/j.stem.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon PG, Nedergaard M. How Do Astrocytes Participate in Neural Plasticity? Cold Spring Harb. Perspect Biol. 2014;7 doi: 10.1101/cshperspect.a020438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huttenlocher PR. Synaptic density in human frontal cortex — Developmental changes and effects of aging. Brain Res. 1979;163:195–205. doi: 10.1016/0006-8993(79)90349-4. [DOI] [PubMed] [Google Scholar]
- Huttenlocher PR, de Courten C, Garey LJ, Van der Loos H. Synaptogenesis in human visual cortex — evidence for synapse elimination during normal development. Neurosci Lett. 1982;33:247–252. doi: 10.1016/0304-3940(82)90379-2. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AMM, Pletikos M, Meyer KA, Sedmak G, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh BS, McCarthy KD. Astrocyte calcium signaling: from observations to functions and the challenges therein. Cold Spring Harb Perspect Biol. 2015;7:a020404. doi: 10.1101/cshperspect.a020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nat. Rev Drug Discov. 2004;3:711–715. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- Kucukdereli H, Allen NJ, Lee AT, Feng A, Ozlu MI, Conatser LM, Chakraborty C, Workman G, Weaver M, Sage EH, et al. Control of excitatory CNS synaptogenesis by astrocyte-secreted proteins Hevin and SPARC. Proc Natl Acad Sci. 2011;108:E440–E449. doi: 10.1073/pnas.1104977108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuffler SW, Nicholls JG, Orkand RK. Physiological properties of glial cells in the central nervous system of amphibia. J Neurophysiol. 1966;29:768–787. doi: 10.1152/jn.1966.29.4.768. [DOI] [PubMed] [Google Scholar]
- Malik N, Wang X, Shah S, Efthymiou AG, Yan B, Heman-Ackah S, Zhan M, Rao M. Comparison of the gene expression profiles of human fetal cortical astrocytes with pluripotent stem cell derived neural stem cells identifies human astrocyte markers and signaling pathways and transcription factors active in human astrocytes. PLoS One. 2014;9:e96139. doi: 10.1371/journal.pone.0096139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy KD, de Vellis J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J Cell Biol. 1980;85:890–902. doi: 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Franke A, Kaplan MR, Pfrieger FW, Barres BA. Characterization of the signaling interactions that promote the survival and growth of developing retinal ganglion cells in culture. Neuron. 1995;15:805–819. doi: 10.1016/0896-6273(95)90172-8. [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Krenick R, Ullian E, Tsai H-H, Deneen B, Richardson WD, Barres BA, Rowitch DH. Astrocytes and disease: a neurodevelopmental perspective. Genes Dev. 2012;26:891–907. doi: 10.1101/gad.188326.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan SJ, MacVicar BA. Calcium transients in astrocyte endfeet cause cerebrovascular constrictions. Nature. 2004;431:195–199. doi: 10.1038/nature02827. [DOI] [PubMed] [Google Scholar]
- Nedergaard M. Direct signaling from astrocytes to neurons in cultures of mammalian brain cells. Sci (New York, NY) 1994;263:1768–1771. doi: 10.1126/science.8134839. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn A, Mukamel EA, Schnitzer MJ. Motor behavior activates Bergmann glial networks. Neuron. 2009;62:400–412. doi: 10.1016/j.neuron.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberheim NA, Wang X, Goldman S, Nedergaard M. Astrocytic complexity distinguishes the human brain. Trends Neurosci. 2006;29:547–553. doi: 10.1016/j.tins.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Oberheim NA, Takano T, Han X, He W, Lin JHC, Wang F, Xu Q, Wyatt JD, Pilcher W, Ojemann JG, et al. Uniquely hominid features of adult human astrocytes. J Neurosci. 2009;29:3276–3287. doi: 10.1523/JNEUROSCI.4707-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parpura V, Basarsky TA, Liu F, Jeftinija K, Jeftinija S, Haydon PG. Glutamate-mediated astrocyte-neuron signalling. Nature. 1994;369:744–747. doi: 10.1038/369744a0. [DOI] [PubMed] [Google Scholar]
- Paukert M, Agarwal A, Cha J, Doze VA, Kang JU, Bergles DE. Norepinephrine controls astroglial responsiveness to local circuit activity. Neuron. 2014;82:1263–1270. doi: 10.1016/j.neuron.2014.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellerin L, Magistretti PJ. Glutamate uptake into astrocytes stimulates aerobic glycolysis: a mechanism coupling neuronal activity to glucose utilization. Proc Natl Acad Sci U S A. 1994;91:10625–10629. doi: 10.1073/pnas.91.22.10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein JD, Dykes-Hoberg M, Pardo CA, Bristol LA, Jin L, Kuncl RW, Kanai Y, Hediger MA, Wang Y, Schielke JP, et al. Knockout of glutamate transporters reveals a major role for astroglial transport in excitotoxicity and clearance of glutamate. Neuron. 1996;16:675–686. doi: 10.1016/s0896-6273(00)80086-0. [DOI] [PubMed] [Google Scholar]
- Schummers J, Yu H, Sur M. Tuned responses of astrocytes and their influence on hemodynamic signals in the visual cortex. Sci (New York, NY) 2008;320:1638–1643. doi: 10.1126/science.1156120. [DOI] [PubMed] [Google Scholar]
- Sun W, McConnell E, Pare JF, Xu Q, Chen M, Peng W, Lovatt D, Han X, Smith Y, Nedergaard M. Glutamate-Dependent Neuroglial Calcium Signaling Differs Between Young and Adult Brain. Sci (New York, NY) 2013;339:197–200. doi: 10.1126/science.1226740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien AC, Tsai HH, Molofsky AV, McMahon M, Foo LC, Kaul A, Dougherty JD, Heintz N, Gutmann DH, Barres BA, et al. Regulated temporal-spatial astrocyte precursor cell proliferation involves BRAF signalling in mammalian spinal cord. Development. 2012;139:2477–2487. doi: 10.1242/dev.077214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullian EM, Sapperstein SK, Christopherson KS, Barres BA. Control of synapse number by glia. Sci (New York, NY) 2001;291:657–661. doi: 10.1126/science.291.5504.657. [DOI] [PubMed] [Google Scholar]
- Winzeler A, Wang JT. Purification and Culture of Retinal Ganglion Cells from Rodents. Cold Spring Harb Protoc. 2013;2013 doi: 10.1101/pdb.prot074906. pdb.prot074906 – pdb.prot074906. [DOI] [PubMed] [Google Scholar]
- Zamanian JL, Xu L, Foo LC, Nouri N, Zhou L, Giffard RG, Barres BA. Genomic analysis of reactive astrogliosis. J Neurosci. 2012;32:6391–6410. doi: 10.1523/JNEUROSCI.6221-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. An RNA-Sequencing Transcriptome and Splicing Database of Glia, Neurons, and Vascular Cells of the Cerebral Cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonta M, Angulo MC, Gobbo S, Rosengarten B, Hossmann K-A, Pozzan T, Carmignoto G. Neuron-to-astrocyte signaling is central to the dynamic control of brain microcirculation. Nat Neurosci. 2003;6:43–50. doi: 10.1038/nn980. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.