Abstract
Introduction
Trophoblast invasion establishes adequate blood flow between mother and fetus in early placental development. However, little is known about the cis-regulatory mechanisms underlying this important process. We aimed to identify enhancer elements that are active during trophoblast invasion, and build a trophoblast invasion gene-enhancer network.
Methods
We carried out ChIP-Seq for an enhancer-associated mark (H3k27Ac) at two time points during early placental development in mouse. One time point where invasion is at its peak (e7.5) and another time point shortly afterwards (e9.5). We use computational analysis to identify putative enhancers, as well as the transcription factor binding sites within them, that are specific to the time point of trophoblast invasion.
Results
We compared read profiles at e7.5 and e9.5 to identify 1,977 e7.5-specific enhancers. Within a subset of e7.5-specific enhancers, we discovered a cell migration associated regulatory code, consisting of three transcription factor motifs: AP1, Ets, and Tcfap2. To validate differential expression of the transcription factors that bind these motifs, we performed RNA-Seq in the same context. Finally, we integrated these data with publicly available protein-protein interaction data and constructed a trophoblast invasion gene-enhancer network.
Discussion
The data we generated and analysis we carried out improves our understanding of the regulatory mechanisms of trophoblast invasion, by suggesting a transcriptional code exists in the enhancers of cell migration genes. Furthermore, the network we constructed highlights novel candidate genes that may be critical for trophoblast invasion.
Keywords: trophoblast invasion, cis-regulation, gene-enhancer network
Introduction
In hemochorial placentation, trophoblast cells of the fetal placenta line the implantation site and invade the maternal decidua, enabling maternal spiral artery remodeling, and establishing adequate blood flow between mother and fetus [1–4]. Defects in this process of trophoblast invasion can lead to a number of disorders that have detrimental effects on both the mother and the child. Excessive trophoblast invasion may contribute to the pathogenesis of placenta creta, a severe pregnancy complication in which abnormally deep attachment of the placenta to the myometrium is observed [5–7]. Shallow invasion results in hypoxic trophoblast tissue, which can cause oxidative stress in the placenta and has been associated with pre-eclampsia and intrauterine growth restriction [8,9].
Yet the mechanisms underlying trophoblast invasion are poorly understood. Several genes and transcription factors (TFs) have been identified as important for regulating this process [2,10,11]. Nevertheless, little is known about how the genes are regulated, or about the enhancers to which the TFs bind to regulate gene expression. Enhancer regions (distal cis-regulatory regions that regulate spatiotemporal gene expression) are crucial to many processes, and often contribute to disease when disrupted [12,13]. Single nucleotide polymorphisms (SNPs) associated with placental disorders often reside in non-coding DNA, indicating the importance of non-coding enhancer elements in these disorders [14]. Next-generation sequencing of mouse [15,16] and human [17–19] placentas, as well as computational prediction [20], have been performed to identify putative placenta enhancer elements, but none of these has focused on the process of trophoblast invasion. Identifying enhancers genome-wide that contribute to the process of trophoblast invasion requires assaying placental tissue at an early time point, while invasion is taking place.
To identify enhancer elements that are important during trophoblast invasion from the fetal perspective, we carried out ChІP-Seq for H3k27Ac on mouse fetal placental tissue at e7.5 and e9.5. H3k27Ac is a histone modification that is associated with active enhancers [21]. E7.5 has been described as the time point for maximal trophoblast invasion in mice [22], while e9.5 is a late post-implantation stage that we used for comparison to isolate enhancers specific to e7.5 placentas and thus more likely to be crucial for the invasion process. While human and mouse placentation differ in certain respects, genetic manipulation of the mouse has been critical in providing insights into human placental development [4]. Furthermore, mouse and human placenta share homologous cell types and gene expression patterns [23]. For example, the outermost cell layer of the placenta is composed of trophoblast giant cells (TGCs) in mouse, and of extravillous cytotrophoblast cells in human, both of which display invasive behavior and express Matrix Metallopeptidase 9 (Mmp9) and Stimulated by Retinoic Acid 13 (Stral3) [23].
By comparing the enhancer landscape of the mouse placenta at e7.5 to that at e9.5, we were able to identify 1,977 enhancers with higher signal at e7.5. We then performed a motif scoring analysis and discovered three transcription factor (for AP1, Ets and Tcfap2) enriched within these enhancers. The subset of enhancers containing these three TF motifs is frequently associated with genes that are important for the invasion process in mouse and human. We performed transcriptome profiling to identify the specific TF family members likely binding to these enhancers. Finally, we incorporated protein-protein interaction data to construct a gene-enhancer network, which provides additional evidence for co-regulation of genes with enhancers containing the AP1, Ets, and Tcfap2 motifs, and highlights novel candidate genes that may be important for trophoblast invasion. By performing genomic analysis at two time points that are close in development, we were able to better understand regulatory mechanisms of a process that is particularly active during a small window of time.
Material and Methods
ChIP-Seq library construction and sequencing
Ectoplacental cone (EPC) tissue was dissected from embryos at e7.5, obtained from timed pregnant CD-1 mice (Charles Rivers Labs). The EPC was separated from the maternal decidua, embryo, and other extraembryonic tissue as described in [24]. At e9.5, the fetal portion of the placenta was dissected. ChIP was carried out on two biological replicates per time point. For each biological replicate at e7.5, EPCs from 25 pregnant mice (~10 embryos per mouse) were combined prior to chromatin isolation. For each biological replicate at e9.5, placentas from ~10 pregnant mice were combined prior to chromatin isolation. Chromatin isolation and immunoprecipitation were carried out as previously described [25], but with 5µg chromatin and 1.2µg of H3k27ac antibody (Abcam ab4729, lot: GR24371-1) for the immunoprecipitation assay.
ChIP-Seq libraries were prepared as described previously [26] using 2ng starting material for each biological replicate. For each biological replicate, 2ng of input DNA was also prepared as a control. Libraries were sequenced by the Stanford Center for Genomics and Personalized Medicine (SCGPM) using the Illumina GAIIx, and single-end 36 base pair reads.
ChIP-Seq data processing and peak-calling
GAIIx output was also analyzed by the SCGPM, using the Genome Analyzer Pipeline. Sequence tags that aligned uniquely to the mouse genome build mm9 with 0,1, or 2 mismatches, according to the ELAND alignment algorithm, were retained for further analysis. The number of sequence tags for each sample can be found in Supplementary Table SI.
Sequence tags from biological replicates were combined prior to peak calling. Peak calling was performed using GLITR [26] with default parameters, a 5% false discovery rate cutoff, and a maximum peak height >20. Data were further filtered to remove peaks that were also identified in input control data, and because of our interest in distal cis-regulatory elements, to remove peaks that overlapped exons (using the UCSC mm9.knownGene table) or that were within 2.5kb of a gene transcriptional start site.
Luciferase Assays
Mouse trophoblast giant cells (TGCs) were differentiated from trophoblast stem cells as described previously [20]. We did not select for a particular type of giant cell, so the TGCs likely represent a heterogeneous population as was found previously [27]. Putative enhancers were cloned into the pGL4.23 Ligation Independent Cloning vector, and transfection assays and luciferase assays were performed as described previously [20]. For cloning, peak coordinates were adjusted to respect conserved DNA boundaries based on Multiz alignments from Mouse to Rat and Human. Primers used to amplify genomic regions are listed in Supplementary Table S2.
Identification of enhancers with higher activity at e7.5 versus e9.5
In order to identify enhancers with the most differential activity between time points, we first combined the peak files of both time points, to obtain peaks that are active at e7.5 or e9.5. Overlapping peaks were merged. For each peak, a fold was calculated by normalizing the maximum number of overlapping tags at the peak by the number of tags that mapped to chromosome:
From the set of merged peaks, we then obtained: (1) those peaks that have higher signal at e7.5, as the peaks with the highest 25% peak-height fold difference at e7.5 compared to e9.5; and (2) those peaks that have higher signal at e9.5, as the peaks with the highest 25% peak-height fold difference compared to e7.5. Peaks that were not in set (1) or (2) were considered to have strong activity at both time points.
Significance of overlap with mouse functional data sets
Mouse H3k27Ac data from 19 tissue types [15] were downloaded from the Ren Lab website: http://chromosome.sdsc.edu/mouse/download.html. The point coordinate in the H3k27Ac file was padded by ±500bp, and regions overlapping exons or within 2.5kb of a transcription start site in mm9 were removed.
To assess overlap significance of the 19 H3k27ac sets with the e7.5/e9.5 placenta peaks, we first created 10,000 shuffles of the peaks. For each shuffle, we counted the number of times each mouse tissue set was overlapped. We then calculated the fold enrichment, as the observed overlap divided by the mean shuffled overlap. The z-score was calculated as the (overlapobserved-overlapexpected)/(standard deviation).
Significance of overlap with human placenta data
DNAse I hypersensitive sites in human placenta [17,18] were provided by the Stamatoyannopoulos lab, and data from 6 samples were combined as described previously [20]. Peaks overlapping exons or within 2.5kb of a transcription start site in hg l9 were removed.
To assess overlap significance of e7.5 or e9.5 peaks with the DNAse I peaks, we first converted mouse peak coordinates to human (hgl9) coordinates using UCSCs liftover tool with default parameters. We then shuffled each e7.5 or e9.5 placenta peak set across the genome 10,000 times. For each shuffled set, we counted the number of times the DNAse I peaks were overlapped.
Transcription factor library curation
A transcription factor motif library was curated as described previously [28], with motifs added from additional studies and resources [29–34], resulting in a non-redundant set of 1,651 motifs.
Binding site prediction and enrichment analysis
To compare binding site occurrence across peaks at different time points, peak widths were all set to ±500bp from the peak center. Binding sites for TFs from the library were predicted using the PRISM excess conservation approach that has been described previously [28], with the following parameters: binding site prediction threshold of 0.9; binding sites were allowed to shift by 20bp relative to the reference; binding sites had to be present in human (hgl9); and the p-value of the observed motif score against motif shuffles in similarly conserved windows had to be ≤0.05. After obtaining binding site predictions for each TF, we counted the number of times each motif occurred in e7.5-specific enhancers, e9.5-specific enhancers (background set 1), and regions that were matched in GC content and length to the e7.5-specific enhancers (background set 2). We then calculated a p-value (using the hypergeometric test) and fold enrichment for each motif using occurrence counts in the e7.5 set versus each background set. A motif was considered significantly enriched at e7.5 if: it had a p-value<3e-6 (to account for 1,651 motifs tested) using each background; it had a fold enrichment ≥1.5 compared to each background; and it occurred in >10% of the e7.5-specific peaks. After discovering Ets and AP1 motifs within 100bp enriched in the peaks with higher signal at e7.5, we identified additional peaks with higher signal at e 7.5 with this pattern by reducing the binding site prediction threshold to 0.8, and extending peaks >1000bp back to their original length. For genome-wide predictions, a prediction threshold of 0.9 was used.
RNA-Seq library construction and sequencing
Placenta tissue was dissected at e7.5 and e9.5, as described above. RNA-Seq was carried out on 3 biological replicates per time point. For each biological replicate at e7.5, we pooled 5 EPCs, and for each biological replicate at e9.5, we pooled 2 placentas. RNA was isolated using the Qiagen RNeasy Micro Kit, following the manufacturer’s protocol. RNA quality was assessed using the Agilent Bioanalyzer, and all samples had a RIN score ≤ 8.7.
Samples were enriched for mRNA and prepared for sequencing using Illumina’s TruSeq Stranded mRNA Sample Prep Kit, following the manufacturer’s protocol, and starting with l|lg total RNA. All libraries were pooled and then sequenced by Elim Biopharmaceuticals, Inc. using the Illumina HiS eq 2500 with 100bp paired end reads. The pooled library sample was run over two sequencing lanes (technical replicates for each sample).
RNA-Seq data processing
Reads were aligned to mm9 using STAR v2.4.0 with default parameters [35]. Following alignment, reads from technical replicates were combined using the samtools merge function [36]. FPKMs were estimated, and genes with significant differential expression between e7.5 and e9.5 were determined using Cufflinks v2.2.1 and Cuffdiff v2.2.1 [37], with default parameters and a mask file specifying that rRNA, tRNA, and mitochondrial RNA from mm9 should be ignored. Genes with FPKM<1 were not considered to be expressed. Replicate clustering dendogram and FPKM distribution plots were generated using the cummeRbund package [38].
Genomic Region Enrichment Analysis (GREAT)
Enrichment analysis was performed using GREAT (http://great.stanford.edu/) v2.0.2 [39], with default parameters, unless otherwise noted in the text. We always required that at least ten genes in the term were hit. For the MGI Mouse Phenotype ontology, we used a version in which only single mutant gene to phenotype associations are included.
Cell migration gene network construction
Enhancers with the AP1/Ets/Tcfap2 pattern were associated with genes using regulatory domains defined through GREAT (http://great.stanford.edu/) v2.0.2 [39]. This gene set was used as input for string-db (http://string-db.org/) v9.1 [40]. We required medium-high confidence (.5) for predicted interactions, which are visualized in the confidence view. Interaction enrichment was calculated using the string-db function. For visualization only, genes with zero predicted/known interactions were removed. If a gene is annotated in the “regulation of cell migration” GO Biological Process term, or the “abnormal angiogenesis” MGI Mouse Phenotype term, the gene is considered a migration/angiogenesis gene.
Animal care and use
All animals were treated under protocol #18487 approved by Stanford University Institutional Animal Use and Care Committee.
Results
E7.5 and e9.5 H3k27Ac peaks are near placental development genes
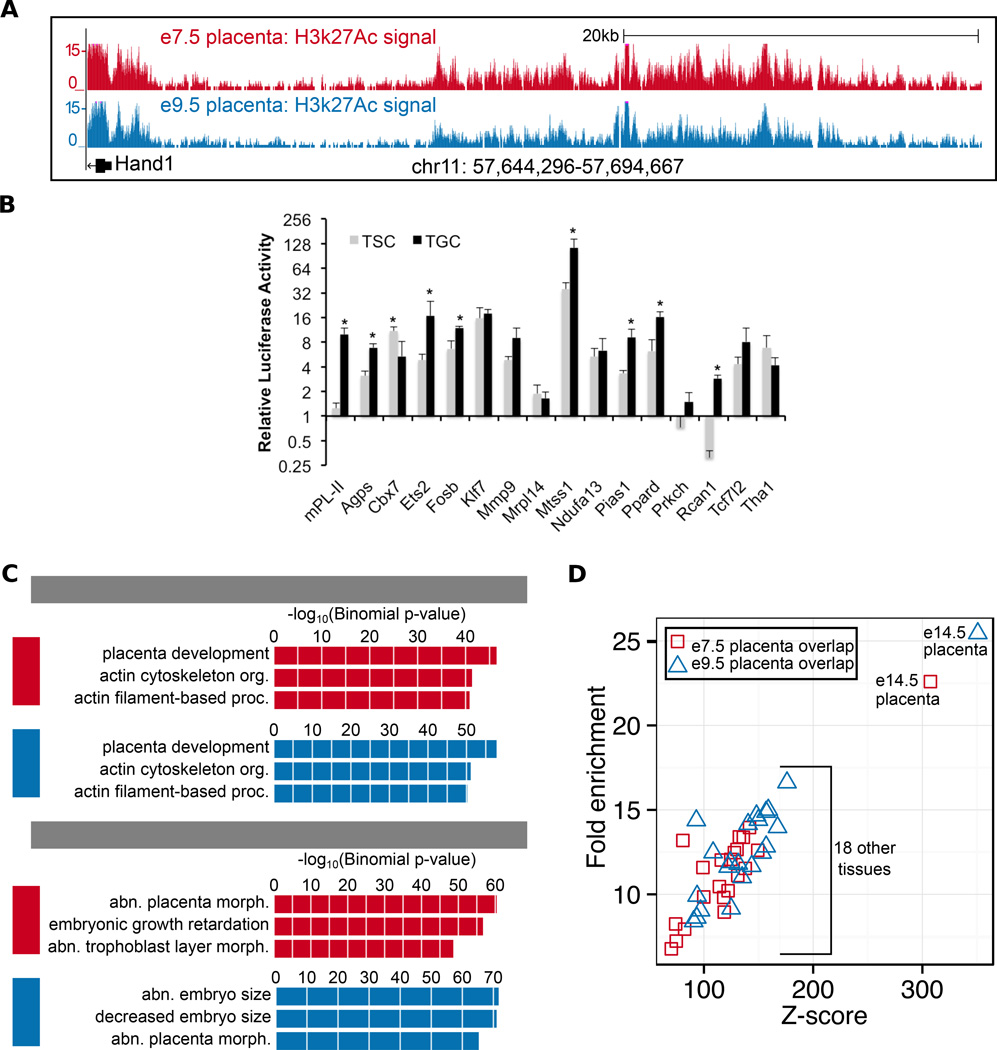
In order to identify enhancer elements active during early placental development, we carried out ChlP-Seq for an enhancer-associated mark, H3k27Ac, on e7.5 and e9.5 fetal placentas. Following data processing and peak-calling (see Methods), we identified 7,224 peaks (putative enhancers) at e7.5 and 7,620 peaks at e9.5. Enhancers were assayed in placental tissue, and therefore enhancer peaks are expected to be near placental genes. Indeed, we observe high H3k27Ac signal upstream of genes known to be important for placental development, such as Heart and Neural Crest Derivatives Expressed 1 (Handl) [41,42] (Figure 1A). We also confirmed that putative enhancers are able to drive gene activity in a relevant placenta cell type by performing luciferase assays in mouse trophoblast giant cells (TGCs). We tested 15 putative enhancers from the union of the e7.5 and e9.5 peaks, 14 of which are associated with genes that have previously been shown to be expressed in TGCs (Supplementary Table S3) [43]. We also tested the mouse placental lactogen II (mPL-II) enhancer as a positive control [20,44]. Of these 15 putative enhancers, 13 (87%) are able to drive luciferase activity more than 2-fold compared to an empty vector control (Figure 1B). To investigate specificity of the enhancers, we also measured activity in trophoblast stem cells (TSCs). Of the 13 enhancers that are able to drive luciferase activity in TGCs, 11 (85%) have higher activity on average in TGCs, compared to 2 (15%) that have higher activity on average in TSCs; and 7 (54%) have significantly higher activity in TGCs, compared to 1 (8%) that has significantly higher activity in TSCs (p-value≤05) (Figure 1B).
Figure 1. Characterization of H3k27Ac peaks in e7.5 and e9.5 placentas.
(A) Genome browser showing high H3k27Ac signal at both time points upstream of a known placental development gene, Handl. (B) 13/15 (87%) H3k27Ac marked putative enhancers drive luciferase activity in Trophoblast Giant Cells (TGCs). 11/13 (85%) active TGC enhancers have higher activity in TGCs on average compared to Trophoblast Stem Cells (TSCs). * Significantly different activity between TSCs and TGCs (p-value.05). mPL-II serves as a positive control. Error bars represent the standard deviation of triplicates. (C) Functional enrichment analysis showing that peaks at both time points are generally associated with placental genes. Top three terms in GREAT for GO Biological Process and MGI Mouse Phenotype ontologies are shown. (D) E7.5 and e9.5 peaks show most significant overlap with mouse ENCODE H3k27Ac data in el 4.5 placenta compared to 18 other tissues.
To determine whether enhancer peaks are generally enriched near placental genes, we used GREAT, a genomic regions functional enrichment analysis tool. For both time points, the top term for the GO Biological Process ontology is “placenta development”, and for the MGI: Mouse Phenotype ontology, placenta and embryonic growth terms make up the top 3 (Figure 1C). Other top terms enriched in the GO Biological Process ontology are related to cytoskeletal integrity, which has been shown to be important for differentiation of invasive trophoblast giant cells [45,46]. The functional enrichment analysis test not only serves as a global quality control, demonstrating that the H3k27Ac peaks are generally associated with placental genes, but also provides many avenues for future research, as most of these enhancers, and the pathways they are working together to regulate, have not yet been characterized in placental development.
E7.5 and e9.5 H3k27Ac peaks have significant overlap with other placenta datasets
To further assess the quality of each data set, and their association with placental tissue, we calculated the peak overlaps with H3k27Ac peaks from 19 tissue types, including el 4.5 placenta, generated through the mouse ENCODE project [15]. As expected, the e7.5 and e9.5 placenta H3k27Ac peaks show the most significant overlap with the ENCODE el4.5 placenta peaks (Figure 1D). While the e7.5 and e9.5 placenta peaks overlap significantly with the 32,467 el 4.5 peaks, they are not subsumed, and each set has numerous unique peaks relative to the el 4.5 peaks (2,772 at e7.5 and 2,401 at e9.5).
To confirm the relevance of our mouse data to human, we first determined that, at each time point, almost 80% of the peaks have sequence conservation in human (77% at e7.5 and 78% at e9.5). Moreover, after mapping mouse peaks to orthologous human coordinates, the peaks are still enriched near placental genes (Supplementary Figure S1 A), and the peaks overlap with human placenta (85–113 days) DNAse-Seq data more than would be expected by chance (p-value<10−4 , z-score>78) (Supplementary Figure S1B). Again, while the overlap is significant, many peaks do not overlap with the DNAse-Seq data (4,000 at e7.5 and 4,096 at e9.5).
E7.5-specific enhancers are near trophoblast invasion genes
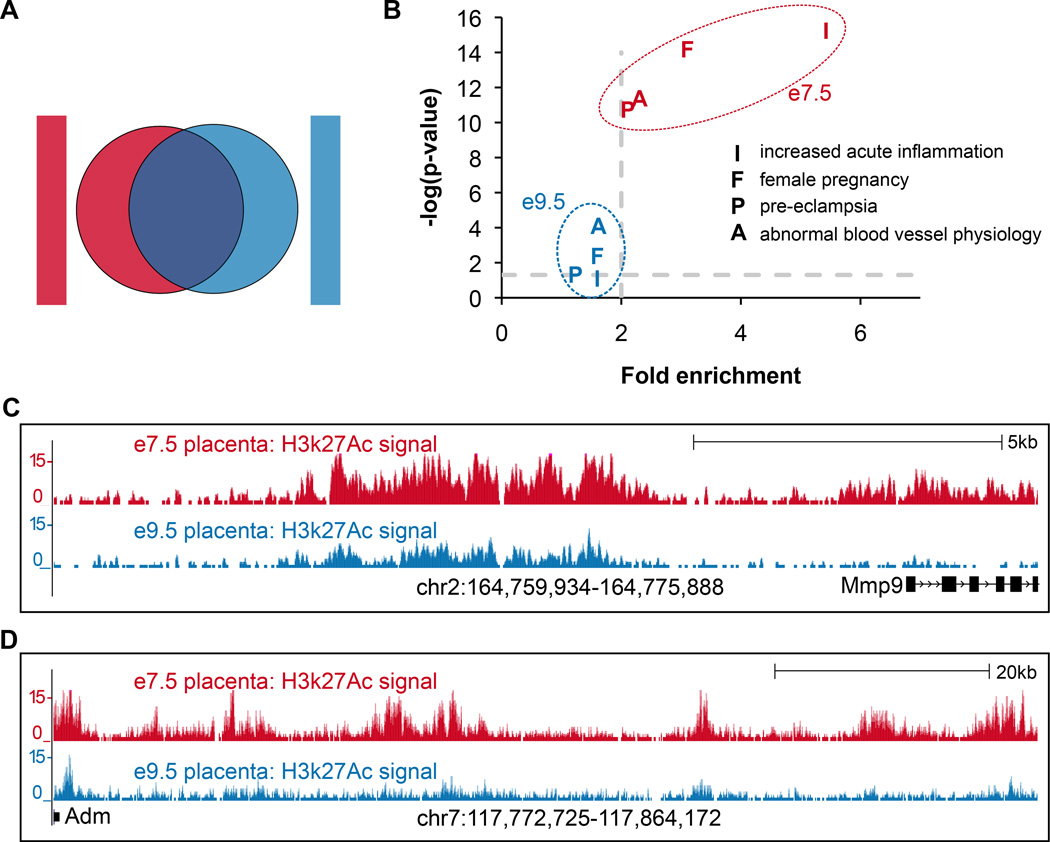
We were interested in enhancers more likely to be involved in trophoblast invasion, so we identified peaks that have the highest relative H3k27Ac signal at e7.5 versus e9.5 (see Methods). This procedure identified 1,977 peaks that have higher signal at e7.5 (hereafter, e7.5-specific enhancers) (Figure 2A). While the top GREAT terms for the full set of e7.5 enhancer peaks were related to general placental development, we see more specific terms for the e7.5-specific enhancers: increased acute inflammation (p-value= l.11×10−13); abnormal blood vessel physiology (p-value=5.11 ×10−10); and pre-eclampsia (p-value=4.48×10−10) (Figure 2B).
Figure 2. Identification of e7.5-specific enhancers.
(A) Venn diagram showing grouping of peaks into those with higher signal at e7.5 (e 7.5-specific enhancers), and those with higher signal at e9.5 (e9.5-specific enhancers). (B) GREAT terms showing that enrichment for e7.5-specific enhancers are related to pregnancy disorders and inflammation. Gray dashed lines indicate default GREAT cutoffs. (C) Genome browser showing a putative enhancer upstream of Mmp9, a gene that is important for cell invasion. (D) Genome browser showing dense e7.5-specific activity upstream of Adm, a gene that is important for trophoblast invasion.
Among the e7.5-specific enhancers are those that are likely important for trophoblast invasion. For example, Mmp9 is a gene that is known to be relevant for trophoblast invasion in mouse and human [47–49]. In addition to its contribution to trophoblast invasion, MMP9 protein expression is weak or absent in pre-eclamptic placentas compared to placentas from normal pregnancies [50]. However, distal cis-regulatory elements that may be regulating this gene in the placenta have never before been identified. Our analysis identified one such element, with strong enhancer activity at e7.5 compared to e9.5, in an unstudied region upstream of Mmp9 (Figure 2C). This region was confirmed to drive luciferase activity in TGCs (Figure 1B), which have previously been shown to express Mmp9 itself (Supplementary Table S3) [43]. Identifying genomic regions such as these, with e7.5-specific enhancers that are associated with known invasion genes, is important for understanding the mechanisms underlying trophoblast invasion.
Dense e7.5-specific enhancer activity near trophoblast invasion genes
Genes flanked by a high density of enhancer activity are thought to be important for cell identity, and are thought to be the most relevant in the context being assayed [51–55]. In an analysis guided by the same concept, we searched for genes with dense e7.5-specific enhancer activity, and identified genes that have more e7.5-specific enhancers than expected by chance. For each gene, we used GREAT to calculate a p-value for the observed number of e7.5-specific enhancers per gene, normalized to the length of the gene’s regulatory domain. Using this analysis, we were able to identify genes that have a surprisingly high density of e7.5-specific enhancers in their regulatory domains. The top five genes (by GREAT p-value), as well as the functions for which they have been studied, are shown in Table 1. For example, this analysis identified Adrenomedullin (Adm), which is known to be important for the invasion process [56,57] (Figure 2D). However, distal enhancers that are important for regulating this gene in the context of trophoblast invasion have never before been identified. We highlight additional genes and their novel enhancers, such as Prostaglandin 12 (Prostacyclin) Synthase (Ptgis), as likewise potentially important to trophoblast invasion. Ptgis knockout mice have vascular disorders, mildly increased blood pressure, and increased blood urea nitrogen and creatinine levels [58], however the role of this gene specifically in trophoblast invasion, to our knowledge, has not been investigated.
Table 1. Genes with multiple e7.5-specific enhancers.
Genes with the most significant number of e7.5-specific enhancer peaks, sorted by GREAT p-value, which adjusts for predicted gene regulatory domain length.
| Gene Symbol |
p-value | Number of peaks |
Functions (PMID) |
|---|---|---|---|
| Plac8 | 1.48×10−18 | 11 |
|
| Adm | 9.10×10−17 | 10 |
|
| Ptgis | 1.19×10−14 | 9 |
|
| Tcfap2c | 2.14×10−13 | 11 |
|
| Sept9 | 9.71×10−13 | 11 |
|
Motif analysis reveals transcriptional code in e7.5-specific peaks
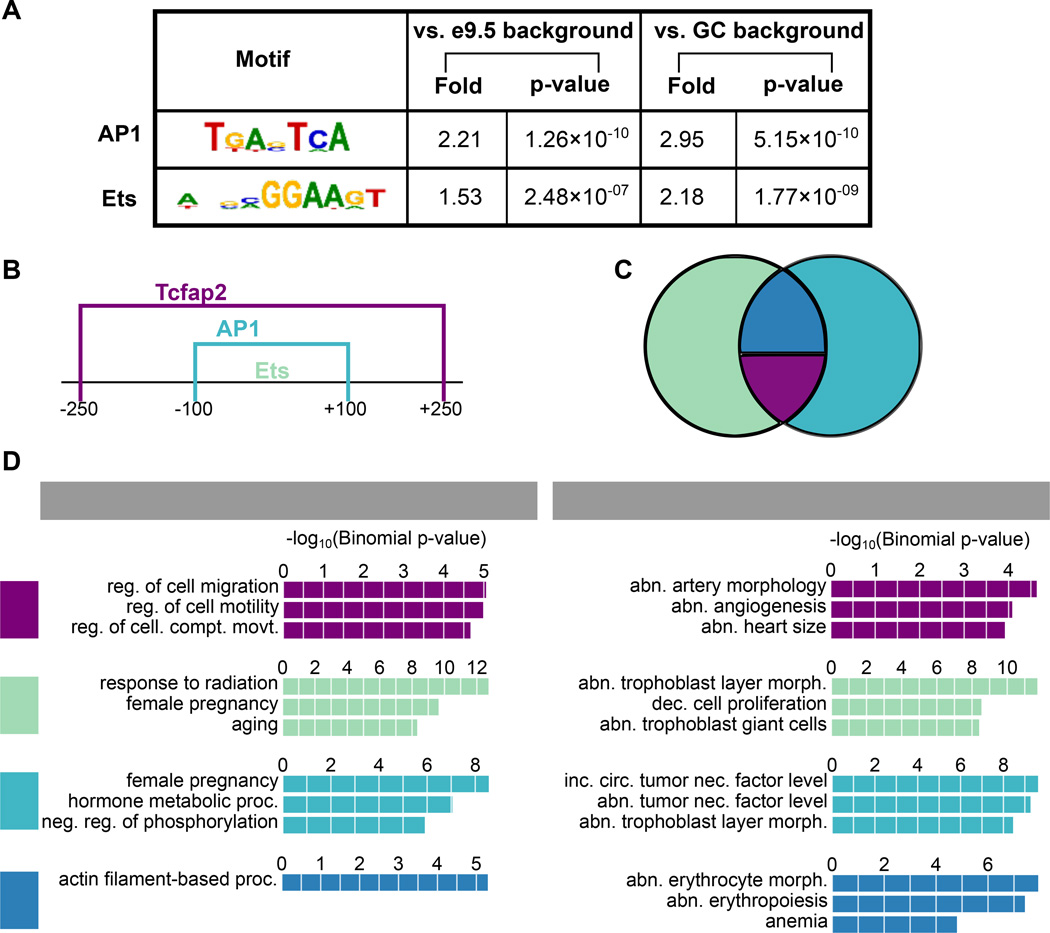
In order to determine whether a subset of e7.5-specific enhancers are co-regulated by a common set of TFs, we searched for TF motifs that occur significantly more frequently in e7.5-specific enhancers than in either of two background sets. The first background set consisted of genomic regions matched in GC content and length to e7.5-specific enhancers. The second background was the set of peaks with higher signal at e9.5 (e9.5-specific enhancers),. To search for motif occurrence in all three sets, we scanned a library of 1,651 non-redundant motifs and used a scoring metric described previously [28], requiring that predictions be conserved in mouse and human (see Methods). We then calculated a p-value for the significance of each motif occurring within e7.5-specific enhancers, relative to each of the two background sets (see Methods).
The only two TF motif families that were significantly enriched in the e7.5-specific enhancers, relative to both backgrounds, represent the DNA binding domains of the AP1 and Ets transcription factor families (Figure 3 A). Interestingly, peaks that contain both of these TF motifs are specifically enriched near cell migration genes, and the highest enrichment is seen when the motifs are within 100bp of each other (Supplementary Figure S2).
Figure 3. Motif scoring in e7.5-specific enhancers.
(A) AP1 and Ets TF family motifs are the only motifs enriched in e7.5-specific enhancers compared to e9.5-specific enhancers and a GC/length matched background. (B) Pattern identified in e7.5-specific enhancers: AP1 motif within 100bp of Ets motif, and Tcfap2 motif within 250bp of Ets motif. (C) Overlap between Ets and AP1 group represents Ets and AP1 motifs within lOObp of one another. 108 e7.5-specific enhancers contain the AP1/Ets/Tcfap2 pattern from (B). (D) Functional enrichment analysis from each of the groups in (C) shows that e7.5-specific enhancers with the AP1/Ets/Tcfap2 pattern are enriched specifically for cell migration/angiogenesis like terms. Top three terms with fold ≥3 from GREAT are shown.
We next broadened our search for the AP1/Ets motif pattern in the e7.5-specific enhancers by lowering our motif match threshold, and extending the e7.5-specific enhancer region we were searching in (see Methods). This increased our set size from 48 e7.5-specific enhancers to 234 e7.5-specific enhancers with AP1 and Ets within 100 bp (AP1/Ets enhancers). These 234 enhancers preserve the significant association with cell migration genes (term: regulation of cell migration, p-value=4.78×l0−6).
To identify any additional TFs working with API and Ets, we searched for other motifs enriched near API or Ets in the AP1/Ets enhancers, compared to genomic regions matched in GC content and length to AP1/Ets enhancers. The only motif that was enriched in this analysis represents the DNA binding domain of the Tcfap2 family (p-value=1.4 ×l0−6 ), which was within 250bp of Ets (Figure 3B). In total, 108 of the AP1/Ets enhancers also have Tcfap2 within 250bp of Ets (AP1/Ets/Tcfap2 enhancers) (Figure 3C). Interestingly, these e7.5-specific enhancers, containing all three TF motifs, are enriched specifically for terms relating to processes linked with trophoblast invasion (regulation of cell migration, abnormal angiogenesis), unlike the other subsets of enhancers (Figure 3D).
Transcriptional code generalizes in a genome-wide scan
We wanted to determine, using statistical methods, whether the TF motifs occurred in a particular arrangement AP1/Ets/Tcfap2 enhancers, as some enhancers contain rigid arrangements of TF motifs [12]. To this end, for each pair of motifs in these enhancers, we counted the number of instances each motif occurred at a particular distance from the other (taking strand into account) [34], and found that the distance and orientation between each pair of motifs is variable, with counts at each distance never exceeding 5 (Supplementary Figure S3). This statistical analysis demonstrates that when considered as a group, the TF motifs in the AP1/Ets/Tcfap2 enhancers are not in stringent positions relative to one another, and therefore the transcriptional code cannot be specified further with these data.
We next determined whether the general pattern we identified is specific to trophoblast migration/invasion, or whether it could be used to identify enhancers associated with cell migration genes in other contexts. To do so, we looked genome-wide for the motif pattern of AP1 and Ets within 100bp of one another, and Tcfap2 within 250bp of Ets (Figure 3B). We identified 922 instances of this pattern across the genome. Strikingly, these putative enhancers are highly enriched near cell migration genes (Table 2). We used functional data to identify a code for trophoblast migration/invasion, and this code persists at the genome-wide level, indicating that we identified a transcriptional code that could be generally important for the migration process.
Table 2. Regions with AP1/Ets/Tcfap2 pattern are generally associated with cell migration genes.
Top 10 terms in GO Biological Process ontology for genome-wide predictions of API /Kts/Tcfar2 nattern from Figure 3B
| Term | p-value | Fold enrichment |
Number of regions |
|---|---|---|---|
| Positive regulation of cell differentiation | 4.49×10−12 | 2.00 | 112 |
| Negative regulation of cell proliferation | 7.16×10−11 | 2.06 | 94 |
| Regulation of cell motility | 8.36×10−11 | 2.17 | 83 |
| Regulation of cell migration | 2.38×10−10 | 2.15 | 81 |
| Regulation of cellular component movement | 6.14×10−10 | 2.07 | 84 |
| Regulation of locomotion | 7.64×10−10 | 2.03 | 87 |
| Positive regulation of locomotion | 5.29×10−9 | 2.30 | 60 |
| Cell fate commitment | 9.88×10−9 | 2.19 | 64 |
| Positive regulation of cellular component movement | 2.16×10−8 | 2.26 | 57 |
| Positive regulation of cell motility | 2.69×10−8 | 2.27 | 56 |
RNA-Seq validates differential expression of predicted TFs
We identified three TF motifs to be overrepresented in e7.5-specific enhancers associated with trophoblast invasion genes. However, there are multiple protein family members that can all recognize similar DNA binding domains, and therefore have very similar TF motifs. To determine which TFs likely bind to the e7.5-specific enhancers, we sought to identify the TF family members that are the most highly expressed in e7.5 placentas, and the most up-regulated compared to e9.5 placentas.
To this end, we performed RNA-Seq on e7.5 and e9.5 fetal placentas for three biological replicates at each of the two time points. As expected, the replicates from each time point cluster together (Supplementary Figure S4A), and the e7.5 and e9.5 FPKM (fragments per kilobase of exon per million fragments mapped) distributions are similar (Supplementary Figure S4B). Our H3k27Ac data and RNA-Seq data are well correlated, since the e7.5-specific enhancers are significantly associated with genes that are up-regulated at e7.5, but not with genes up-regulated at e9.5, in the RNA-Seq data (Supplementary Table S4).
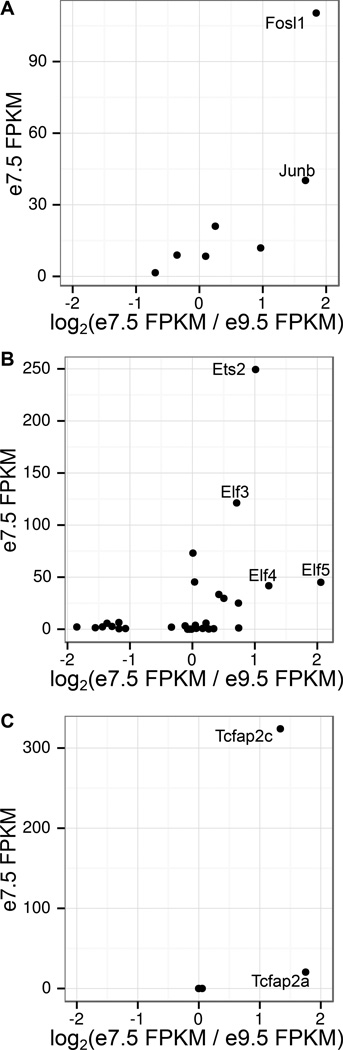
Finally, in order to determine the TF most likely binding to the AP1/Ets/Tcfap2 enhancers, we focused on the expression levels for each member of these TF families. Based on the e7.5 FPKM and the fold difference (e7.5 FPKM/e9.5 FPKM), the TFs most likely binding to migration enhancers at e7.5 are Fosl1, Ets2, and Tcfap2c (Figure 4). These expression-based predictions are promising, as each of the three TFs has been implicated in placental development [59–63], and Fosll has already been further implicated in trophoblast invasion [62]. Our analysis suggests that all three factors are working together to regulate trophoblast invasion, and identifies the specific enhancers to which they may bind.
Figure 4. RNA-Seq data confirms expression of AP1, Ets, and Tcfap2 TF family members.
Plots showing e7.5 FPKM vs. log2(e7.5 FPKM / e9.5 FPKM) for (A) Ap1, (B) Ets, and (C) Tcfap2 family of transcription factors. TFs with the highest e7.5 FPKM and highest log2(e7.5 FPKM / e9.5 FPKM) are strong candidates for binding AP1/Ets/Tcfap2 enhancers during trophoblast invasion.
Incorporating protein-protein interaction data to construct a trophoblast invasion gene-enhancer network
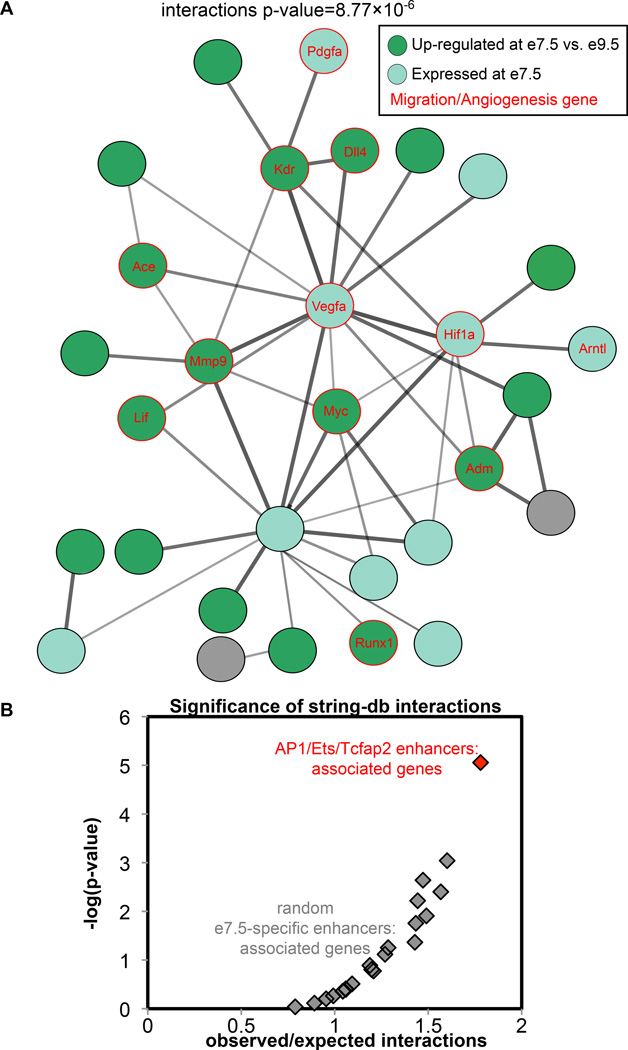
If the AP1/Ets/Tcfap2 enhancers that we identified do in fact regulate genes in a common pathway, then the observed interactions between the regulated genes, including in contexts other than trophoblast invasion where they may also work together, should be higher than expected by chance. To assess this, we first assigned each of the 108 AP1/Ets/Tcfap2 enhancers to genes, based on GREAT gene regulatory domains [39]. We then used this gene set as input to string-db [40], a database for known and predicted protein-protein interactions. Indeed, this set of genes is highly enriched for interactions (p-value=8.77×10−6 ), determined using the string-db functionality for calculating significance of interactions between the input set of genes (Supplementary Figure S5).
The string-db network also highlights a core set of interactions (the largest component in Figure S5), which consists of multiple genes known to be important in cell migration/angiogenesis in various contexts (Figure 5A). More than half of these genes (18/30, 60%) are significantly up-regulated in our RNA-Seq data at e7.5 compared to e9.5, providing further evidence for co-regulation of genes with AP1/Ets/Tcfap2 enhancers. This network of genes and their cognate AP1/Ets/Tcfap2 enhancers not only further suggests the genes are co-regulated, but also highlights additional candidate genes that may be involved in the process of trophoblast invasion.
Figure 5. Genes with AP1/Ets/Tcfap2 enhancers are enriched for known/predicted interactions.
(A) Known and predicted interactions between genes with AP1/Ets/Tcfap2 enhancers were generated using string-db. Our gene set is highly enriched for interactions, with migration/angiogenesis genes at the core of the network. Thicker lines indicate higher confidence in interactions. (B) Genes with associated AP1/Ets/Tcfap2 enhancers are more enriched for interactions than random subsets of genes with associated e7.5-specific enhancers (p-value<05).
To ensure that the interaction enrichment was specific to the AP1/Ets/Tcfap2 enhancers and not simply all e7.5-specific enhancers, we also performed this test on random subsets of genes (matched in size to the number of genes with an associated AP1/Ets/Tcfap2 enhancer) from the list of genes with associated e7.5-specific enhancers. The genes with AP1/Ets/Tcfap2 enhancers are also enriched for interactions when compared to random sets of genes with e7.5-specific enhancers (p-value<.05) (Figure 5B).
Discussion
We performed ChIP-Seq with H3k27Ac at two time points during early mouse placental development, in order to identify enhancers important for trophoblast invasion. By comparing the enhancer landscape at e7.5 to that at e9.5, we were able to identify 1,977 e7.5-specific enhancers. Many of these enhancers are upstream of genes known to be important for trophoblast invasion, revealing the cis-regulatory mechanisms by which these genes are likely regulated during trophoblast invasion.
We identified a TF motif code consisting of motifs for the API, Ets, and Tcfap2 families of transcription factors within a subset of e7.5-specific enhancers. Interestingly, this set of enhancers is most significantly associated with genes known to be important in processes linked to trophoblast invasion. In order to identify the most likely TF family member binding to this set of enhancers, we performed RNA-Seq, and determined that Fosl1, Ets2, and Tcfap2c are strong candidates for co-regulating the trophoblast invasion genes. While these three TFs are known to be important in the placenta, this enhancer transcriptional code and the corresponding network of migration genes it is regulating are novel.
It is possible that other members of the AP1, Ets, and Tcfap2 TF families can bind enhancers of trophoblast invasion genes. For example, AP1 is a dimer, and we predict one component of the dimer is Fosll. Our data suggest that Fosll may be dimerizing with Junb, however Fosl1 could also be dimerizing with TFs that are not differentially expressed between the two time points. For the Ets family of transcription factors, we see that Elf3, Elf4, and Elf5 are also differentially expressed between e7.5 and e9.5 placentas. This could indicate that these TFs have overlapping roles with Ets2; recently it has been shown that Ets2 can partially rescue Elf5−/− mutants, which have defects in extraembryonic tissue [64]. In order to fully understand the interplay among these TF family members, and their specific contributions to trophoblast invasion, additional genetic studies will be needed.
When searching genome-wide for the same regulatory code that we identified in e7.5-specific enhancers, by using predictions across the entire genome, we find hundreds of additional putative enhancer elements that are also enriched near known migration genes. This indicates that the AP1/Ets/Tcfap2 regulatory code is likely generally important for the process of cell migration. Cell migration is important in many contexts, such as during gastrulation, in the intestine, and in the immune system [65]. The process also contributes to vascular disease, cancer, and other pathological processes [65]. Therefore, it would be interesting to follow up on the AP1/Ets/Tcfap2 regulatory code not only in the context of trophoblast invasion, but also in cell migration in other contexts.
By comparing placental tissue at e7.5 and e9.5, our differential analysis could be identifying patterns in cell types that comprise a larger portion of the e7.5 tissue, such as invasive trophoblast cells. While the e7.5 tissue we dissected contains invasive trophoblast cells, and cells on their way to becoming invasive trophoblast cells, the tissue is still heterogeneous and the proportion of each cell population differs at e7.5 and e9.5. However, because invasive cells do comprise a larger proportion of the tissue we dissected at e7.5, the analysis methods we used likely isolate processes that are specific to the invasive cells. Nevertheless, it would be useful to perform similar studies on a single cell basis, or on purified cell populations, in order to better characterize the invasive cells in vivo.
We have demonstrated how the wealth of data obtained from next-generation sequencing can be used to study a specific process, trophoblast invasion, by performing analyses to bring thousands of peaks down to tens of putative enhancers driven by three transcription factors, which may be some of the most important enhancers for that process. The rich genomic data sets we produced could also be used to study other processes, such as metabolic processes that may be more active in e9.5 placenta. More generally, functional genomic studies of two or more time points can help isolate a process in a given tissue that is highly active at a particular time point. Computational analysis can then be performed to help understand the regulatory mechanisms underlying that process, and drive the most exciting hypotheses for further experimental follow-up.
We used this approach to focus on the process of trophoblast invasion, which is most active in e7.5 placenta. We found enhancers sharing a specific TF motif pattern associated with cell migration genes allowing us to construct a trophoblast invasion gene-enhancer network. Beyond the TFs that we identified in the trophoblast invasion gene enhancers, many other components of the gene-enhancer network are worth future investigation. The network we constructed opens up multiple avenues of research that will help us understand in more detail the mechanisms regulating trophoblast invasion.
Supplementary Material
Highlights.
A genome-wide in vivo enhancer map from e7.5 and e9.5 placenta is obtained
E7.5-specific enhancers are associated with trophoblast invasion genes
A transcriptional code is identified in enhancers associated with migration genes
A trophoblast invasion gene-enhancer network is constructed
Acknowledgements
We thank Xiujun Fan and Nihar Nayak for technical assistance; Emin Maltepe for providing trophoblast stem cells; Julie Baker for her advice and guidance; Harendra Guturu, James Notwell, Bruce Schaar, and Aaron Wenger for manuscript advice; Bejerano lab members for feedback and technical assistance throughout this project; Ziming Weng and the Stanford Center for Genomics and Personalized Medicine; Bob Thurman (John Stamatoyannopoulos Lab) for human placenta DNase-Seq data, and Michael Hiller for the MGI Phenotype Single KO ontology.
Funding
This work was supported by the Eunice Kennedy Shriver National Institute Of Child Health & Human Development of the National Institutes of Health under Award Number K99HD079545 to G.T., by the A. P. Giannini Foundation Postdoctoral Research Fellowship to G T, and by a Packard Fellowship and Microsoft Research Fellowship to G B. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funders/funding agencies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ain R, Canham LN, Soares JM. Gestation stage-dependent intrauterine trophoblast cell invasion in the rat and mouse: novel endocrine phenotype and regulation. Dev. Biol. 2003;260:176–190. doi: 10.1016/s0012-1606(03)00210-0. [DOI] [PubMed] [Google Scholar]
- 2.Lunghi L, Ferretti ME, Medici S, Biondi C, Vesce F. Control of human trophoblast function. Reprod. Biol. Endocrinol. 2007;5:6. doi: 10.1186/1477-7827-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maltepe E, Bakardjiev AI, Fisher SJ. The placenta: transcriptional, epigenetic, and physiological integration during development. J. Clin. Invest. 2010;120:1016–1025. doi: 10.1172/JCI41211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.John R, Hemberger M. A placenta for life. Reprod. Biomed. Online. 2012;25:5–11. doi: 10.1016/j.rbmo.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 5.Kim K-R, Jun S-Y, Kim J-Y, Ro JY. Implantation site intermediate trophoblasts in placenta cretas. Mod. Pathol. Off. J. U. S. Can. Acad. Pathol. Inc. 2004;17:1483–1490. doi: 10.1038/modpathol.3800210. [DOI] [PubMed] [Google Scholar]
- 6.Stanek J, Drummond Z. Occult placenta accreta: the missing link in the diagnosis of abnormal placentation. Pediatr. Dev. Pathol. Off. J. Soc. Pediatr. Pathol. Paediatr. Pathol. Soc. 2007;10:266–273. doi: 10.2350/06-10-0174.1. [DOI] [PubMed] [Google Scholar]
- 7.Tantbirojn P, Crum CP, Parast MM. Pathophysiology of placenta creta: the role of decidua and extravillous trophoblast. Placenta. 2008;29:639–645. doi: 10.1016/j.placenta.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Kaufmann P, Black S, Huppertz B. Endovascular Trophoblast Invasion: Implications for the Pathogenesis of Intrauterine Growth Retardation and Preeclampsia. Biol. Reprod. 2003;69:1–7. doi: 10.1095/biolreprod.102.014977. [DOI] [PubMed] [Google Scholar]
- 9.Huppertz B. Placental origins of preeclampsia: challenging the current hypothesis. Hypertension. 2008;51:970–975. doi: 10.1161/HYPERTENSIONAHA.107.107607. [DOI] [PubMed] [Google Scholar]
- 10.Knofler M. Critical growth factors and signalling pathways controlling human trophoblast invasion. Int. J. Dev. Biol. 2010;54:269–280. doi: 10.1387/ijdb.082769mk. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knöfler M, Pollheimer J. IFPA Award in Placentology lecture: molecular regulation of human trophoblast invasion. Placenta. 2012;33(Suppl):S55–S62. doi: 10.1016/j.placenta.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spitz F, Furlong EEM. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 13.Sakabe NJ, Savic D, Nobrega MA. Transcriptional enhancers in development and disease. Genome Biol. 2012;13:238. doi: 10.1186/gb-2012-13-1-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tuteja G, Cheng E, Papadakis H, Bejerano G. PESNPdb: a comprehensive database of SNPs studied in association with pre-eclampsia. Placenta. 2012;33:1055–1057. doi: 10.1016/j.placenta.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 15.Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chuong EB, Rumi MAK, Soares MJ, Baker JC. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat. Genet. 2013;45:325–329. doi: 10.1038/ng.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kellis M, Wold B, Snyder MP, Bernstein BE, Kundaje A, Mannov GK, et al. Defining functional DNA elements in the human genome. Proc. Natl. Acad. Sci. U. S. A. 2014;1ll:6131–6138. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tuteja G, Moreira KB, Chung T, Chen J, Wenger AM, Bejerano G. Automated discovery of tissue-targeting enhancers and transcription factors from binding motif and gene function data. PLoS Comput. Biol. 2014;10:e1003449. doi: 10.1371/journal.pcbi.1003449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U. S. A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cross JC, Werb Z, Fisher SJ. Implantation and the placenta: key pieces of the development puzzle. Science. 1994;266:1508–1518. doi: 10.1126/science.7985020. [DOI] [PubMed] [Google Scholar]
- 23.Hemberger M, Cross JC. Genes governing placental development. Trends Endocrinol. Metab TEM. 2001;12:162–168. doi: 10.1016/s1043-2760(01)00375-7. [DOI] [PubMed] [Google Scholar]
- 24.Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manip. Mouse Embryo. 3rd ed. Cold Spring Harbor Laboratory Press; 2003. Isolation, Culture, and Manipulation of Postimpantation Embryos; pp. 220–224. [Google Scholar]
- 25.Tuteja G, Jensen ST, White P, Kaestner KH. Cis-regulatory modules in the mammalian liver: composition depends on strength of Foxa2 consensus site. Nucleic Acids Res. 2008;36:4149–4157. doi: 10.1093/nar/gkn366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tuteja G, White P, Schug J, Kaestner KH. Extracting transcription factor targets from ChIP-Seq data. Nucleic Acids Res. 2009;37:ell3. doi: 10.1093/nar/gkp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simmons DG, Fortier AL, Cross JC. Diverse subtypes and developmental origins of trophoblast giant cells in the mouse placenta. Dev. Biol. 2007;304:567–578. doi: 10.1016/j.ydbio.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 28.Wenger AM, Clarke SL, Guturu H, Chen J, Schaar BT, McLean CY, et al. PRISM offers a comprehensive genomic approach to transcription factor function prediction. Genome Res. 2013;23:889–904. doi: 10.1101/gr.139071.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pachkov M, Balwierz PJ, Arnold P, Ozonov E, van Nimwegen E. SwissRegulon, a database of genome-wide annotations of regulatory sites: recent updates. Nucleic Acids Res. 2013;41:D214–D220. doi: 10.1093/nar/gks1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kulakovskiy IV, Medvedeva YA, Schaefer U, Kasianov AS, Vorontsov IE, Bajic VB, et al. HOCOMOCO: a comprehensive collection of human transcription factor binding sites models. Nucleic Acids Res. 2013;41:D195–D202. doi: 10.1093/nar/gks1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, et al. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 33.Jankowski A, Szczurek E, Jauch R, Tiuryn J, Prabhakar S. Comprehensive prediction in 78 human cell lines reveals rigidity and compactness of transcription factor dimers. Genome Res. 2013;23:1307–1318. doi: 10.1101/gr.154922.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guturu H, C Doxey A, Wenger AM, Bejerano G. Structure-aided prediction of mammalian transcription factor complexes in conserved non-coding elements. Philos. Trans. R Soc. Lond. B. iol. Sci. 2013;368:20130029. doi: 10.1098/rstb.2013.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinforma. Oxf Engl. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinforma. Oxf. Engl. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goff L, Trapnell C, Kelley D. cummeRbund: Analysis, exploration, manipulation, and visualization of Cufflinks high-throughput sequencing data. R Package Version. 2.8.2. 2013 [Google Scholar]
- 39.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riley P, Anson-Cartwright L, Cross JC. The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nat. Genet. 1998;18:271–275. doi: 10.1038/ng0398-271. [DOI] [PubMed] [Google Scholar]
- 42.C Scott I, Anson-Cartwright L, Riley P, Reda D, Cross JC. The HAND1 basic helix-loop-helix transcription factor regulates trophoblast differentiation via multiple mechanisms. Mol. Cell. Biol. 2000;20:530–541. doi: 10.1128/mcb.20.2.530-541.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hannibal RL, Chuong EB, Rivera-Mulia JC, Gilbert DM, Valouev A, Baker JC. Copy Number Variation Is a Fundamental Aspect of the Placental Genome. PLoS Genet. 2014;10:1004290. doi: 10.1371/journal.pgen.1004290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun Y, Duckworth ML. Identification of a Placental-Specific Enhancer in the Rat Placental Lactogen II Gene That Contains Binding Sites for Members of the Ets and AP-1 (Activator Protein 1) Families of Transcription Factors. Mol. Endocrinol. 1999;13:385–399. doi: 10.1210/mend.13.3.0243. [DOI] [PubMed] [Google Scholar]
- 45.Parast MM, Aeder S, Sutherland AE. Trophoblast giant-cell differentiation involves changes in cytoskeleton and cell motility. Dev. Biol. 2001;230:43–60. doi: 10.1006/dbio.2000.0102. [DOI] [PubMed] [Google Scholar]
- 46.Choi HJ, Sanders TA, Tormos KV, Ameri K, Tsai JD, Park AM, et al. ECM-dependent FIJF induction directs trophoblast stem cell fate via LIMK1-mediated cytoskeletal rearrangement. PloS One. 2013;8:e56949. doi: 10.1371/journal.pone.0056949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Librach CL, Werb Z, Fitzgerald ML, Chiu K, Corwin NM, Esteves RA, et al. 92-kD type IV collagenase mediates invasion of human cytotrophoblasts. J. Cell Biol. 1991;113:437–449. doi: 10.1083/jcb.113.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alexander CM, Hansell EJ, Behrendtsen O, Flannery ML, Kishnani NS, Hawkes SP, et al. Expression and function of matrix metalloproteinases and their inhibitors at the maternal-embryonic boundary during mouse embryo implantation. Dev. Camb. Engl. 1996;122:1723–1736. doi: 10.1242/dev.122.6.1723. [DOI] [PubMed] [Google Scholar]
- 49.Whiteside EJ, Jackson MM, Herington AC, Edwards DR, Harvey MB. Matrix metalloproteinase-9 and tissue inhibitor of metalloproteinase-3 are key regulators of extracellular matrix degradation by mouse embryos. Biol. Reprod. 2001;64:1331–1337. doi: 10.1095/biolreprod64.5.1331. [DOI] [PubMed] [Google Scholar]
- 50.Shokry M, Omran OM, Hassan HI, Elsedfy GO, Hussein MRA. Expression of matrix metalloproteinases 2 and 9 in human trophoblasts of normal and preeclamptic placentas: preliminary findings. Exp. Mol. Pathol. 2009;87:219–225. doi: 10.1016/j.yexmp.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 51.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pott S, Lieb JD. What are super-enhancers? Nat. Genet. 2015;47:8–12. doi: 10.1038/ng.3167. [DOI] [PubMed] [Google Scholar]
- 54.Wenger AM, Clarke SL, Notwell JH, Chung T, Tuteja G, Guturu H, et al. The Enhancer Landscape during Early Neocortical Development Reveals Patterns of Dense Regulation and Co-option. PLoS Genet. 2013;9:e1003728. doi: 10.1371/journal.pgen.1003728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Galhardo M, Berninger P, Nguyen T-P, Sauter T, Sinkkonen L. Cell type-selective disease-association of genes under high regulatory load. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang X, Green KE, Yallampalli C, Dong YL. Adrenomedullin enhances invasion by trophoblast cell lines. Biol. Reprod. 2005;73:619–626. doi: 10.1095/biolreprod.105.040436. [DOI] [PubMed] [Google Scholar]
- 57.Li M, Schwerbrock NMJ, Lenhart PM, Fritz-Six KL, Kadmiel M, Christine KS, et al. Fetal-derived adrenomedullin mediates the innate immune milieu of the placenta. J. Clin. Invest. 2013;123:2408–2420. doi: 10.1172/JCI67039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yokoyama C, Yabuki T, Shimonishi M, Wada M, Hatae T, Ohkawara S, et al. Prostacyclin-deficient mice develop ischemic renal disorders, including nephrosclerosis and renal infarction. Circulation. 2002;106:2397–2403. doi: 10.1161/01.cir.0000034733.93020.bc. [DOI] [PubMed] [Google Scholar]
- 59.Kuckenberg P, Kubaczka CH, Schorle H. The role of transcription factor Tcfap2c/TFAP2C in trophectoderm development. Reprod. Biomed. Online. 2012;25:12–20. doi: 10.1016/j.rbmo.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 60.Werling U, Schorle H. Transcription factor gene AP-2 gamma essential for early murine development. Mol. Cell. Biol. 2002;22:3149–3156. doi: 10.1128/MCB.22.9.3149-3156.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamamoto H, Flannery ML, Kupriyanov S, Pearce J, McKercher SR, Henkel GW, et al. Defective trophoblast function in mice with a targeted mutation of Ets2. Genes Dev. 1998;12:1315–1326. doi: 10.1101/gad.12.9.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Renaud SJ, Kubota K, Rumi MAK, Soares JM. The FOS transcription factor family differentially controls trophoblast migration and invasion. J. Biol. Chem. 2014;289:5025–5039. doi: 10.1074/jbc.M113.523746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kent LN, Rumi MAK, Kubota K, Lee D-S, Soares JM. FOSL1 is integral to establishing the maternal-fetal interface. Mol. Cell. Biol. 2011;31:4801–4813. doi: 10.1128/MCB.05780-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Donnison M, Broadhurst R, Pfeffer PL. Elf5 and Ets2 maintain the mouse extraembryonic ectoderm in a dosage dependent synergistic manner. Dev. Biol. 2015;397:77–88. doi: 10.1016/j.ydbio.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 65.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, et al. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.