Abstract
Platelets are critical for hemostasis, i.e. the body's ability to prevent blood loss at sites of vascular injury. They patrol the vasculature in a quiescent, non-adhesive state for approximately 10 days, after which they are removed from circulation by phagocytic cells of the reticulo-endothelial system. At sites of vascular injury, they promptly shift to an activated, adhesive state required for the formation of a hemostatic plug. The small GTPase RAP1 is a critical regulator of platelet adhesiveness. Our recent studies demonstrate that the antagonistic balance between the RAP1 regulators, CalDAG-GEFI and RASA3, is critical for the modulation of platelet adhesiveness, both in circulation and at sites of vascular injury. The RAP1 activator CalDAG-GEFI responds to small changes in the cytoplasmic calcium concentration and thus provides sensitivity and speed to the activation response, essential for efficient platelet adhesion under conditions of hemodynamic shear stress. The RAP1 inhibitor RASA3 ensures that circulating platelets remain quiescent by restraining CalDAG-GEFI-dependent RAP1 activation. Upon cellular stimulation, it is turned off by P2Y12 signaling to enable sustained RAP1 activation, required for the formation of a stable hemostatic plug. This review will summarize important studies that elucidated the signaling pathways that control RAP1 activation in platelets.
Introduction
Platelets are highly specialized blood cells evolved to secure the integrity of the cardiovascular system over a broad range of hemodynamic shear conditions[1]. Inhibitory signaling pathways ensure that platelets remain in a quiescent (non-adhesive) state as long as the endothelial lining is physically and biochemically intact. At sites of vascular injury, platelets employ Immunoreceptor Tyrosine-based Activation Motif (ITAM)-coupled receptors and G Protein-Coupled Receptors (GPCRs) to sense and respond to changes in their environment, such as the exposure of extracellular matrix (ECM) proteins and the activation of the coagulation system. Stimulation of these receptors triggers intracellular signaling cascades[2], including those dependent on elevated cytosolic calcium (Ca2+), which promote dramatic cytoskeletal changes, the secretion of granules and, most importantly, the conversion of integrins from a low- to a high-affinity state for their ligands (integrin inside-out activation) [3]. Integrins are the main platelet receptors that support platelet-matrix (platelet adhesion) and platelet-platelet interactions (platelet aggregation). αIIbβ3 integrin is by far the most abundant of the β1 integrins and β3 integrins expressed on the platelet surface. It facilitates the binding of various plasma proteins, including fibrinogen and von Willebrand factor (VWF), and it is crucial for platelet adhesion and aggregation. The formation of a stable hemostatic plug or a pathological thrombus requires sustained integrin inside-out activation, provided by co-stimulatory signaling via the autocrine/paracrine agonists thromboxane (Tx)A2 and ADP[4, 5]. ADP is released from platelet dense granules and supports sustained integrin activation by binding to the Gi-coupled receptor, P2Y12, the target of currently used anti-platelet drugs[6]. Studies by us and others identified a critical role for the small GTPase RAP1B in platelet activation and integrin-mediated cellular adhesion. This review will discuss how RAP1B and its known regulators, CalDAG-GEFI and RASA3, ensure that platelet integrin activation is rapid, sustained and tightly controlled.
RAP1 GTPases and platelet activation
Approximately 8% of the known proteins expressed in platelets are small GTPases and their regulators[7, 8]. The most abundant GTPases in platelets are two isoforms of the Ras-related protein (RAP) subfamily, RAP1B (~300,000 copies/platelet) and RAP1A (~125,000 copies/platelet). Like other small GTPases of the Ras superfamily, RAP proteins are molecular switches that cycle between an inactive GDP-bound state and an active GTP-bound state. Two classes of regulatory proteins control this switch. Guanine nucleotide exchange factors (GEFs) promote the activation by stimulating the exchange of GDP for GTP, and GTPase-activating proteins (GAPs) terminate the activation by catalyzing GTP hydrolysis[9].
In platelets, GTP-loading of RAP1 is stimulated by all known agonists[10, 11]. Upon engagement of agonist receptors, RAP1 translocates from the cytosolic leaflet of intracellular granules, where it is sequestered in resting platelets, to the plasma membrane[12, 13]. Activated RAP1 regulates multiple functional responses in platelets, most notably integrin activation [14, 15]. Genetic deletion in mice of the predominant RAP1 isoform, RAP1B, or inactivation of the main pathways leading to RAP1 activation markedly impaired integrin inside-out[15, 16] and outside-in[17, 18] signaling, granule secretion[18, 19], TxA2 generation[20], spreading[18, 19] and clot retraction[18, 19]. Consistent with the defective platelet activation response, these mice exhibited significantly prolonged bleeding times and a strong protection from experimental thrombosis[15, 21].
CalDAG-GEFI: a critical RAP-GEF and accelerator of platelet activation
Pharmacological and genetic studies at the turn of the century demonstrated that two kinetically distinct pathways regulate RAP1 activation in platelets. Rapid RAP1 activation is triggered by an increase in intracellular Ca2+ concentrations[10], while sustained RAP1 activation requires signaling by protein kinase C (PKC)[22], the Gi-coupled receptor for ADP, P2Y12, and phosphatidylinositol 3-kinases (PI3K)[23–25]. The molecular nature of the GEFs and GAPs regulating RAP1 activity in platelets, however, remained elusive.
The work by Shattil and colleagues was the first to suggest an important role for the calcium-sensing GEF, CalDAG-GEFI (Rasgrp2), in RAP1 activity regulation in platelets. Using ES cell-derived megakaryocytes they demonstrated a good correlation between the expression level of this candidate protein and RAP1 and integrin activation in these cells[26]. Shortly after these pioneering studies, we used Caldaggef1 knockout mice to establish a fundamental role for CalDAG-GEFI in Ca2+-dependent RAP1 activation in platelets[27]. Platelets lacking CalDAG-GEFI exhibited a marked aggregation defect to various agonists, including ADP and collagen, while a more robust aggregation response was observed in response to stimulation with thrombin. A very similar aggregation profile was recently described for human platelets isolated from patients with severe bleeding due to a point mutation in the catalytic GEF domain of CalDAG-GEFI[28].
CalDAG-GEFI is the predominant platelet RAP-GEF and the only member of the RASGRP family to be expressed in the platelet/megakaryocytic lineage (Fig.1)[7, 29]. RASGRPs typically consist of a N-terminal catalytic GEF domain and a C-terminal regulatory region comprising a pair of EF hand domains and a C1 domain (Fig.2). However the catalytic specificity and the activity regulation of CalDAG-GEFI are the most divergent within this family, as CalDAG-GEFI has RAP, but not RAS, exchange activity in vivo[30], and structural rearrangements induced by the binding of Ca2+ to the EF domains are critical for its activation[31]. The EF hand domains have very high affinity for Ca2+ (KD~80nM)[31] and thus provide remarkable sensitivity towards minor changes in the cytoplasmic Ca2+ concentration, which in resting platelets was measured at ~20–50nM. The C1 domain, however, is insensitive to the second messenger diacylglycerol (DAG)[32, 33], but it contributes to optimal CalDAG-GEFI-dependent RAP1 activation by a hitherto unknown mechanism[21]. Platelets from Caldaggef1−/− mice[27] or from patients expressing an inactive CalDAG-GEFI mutant[28] a) failed to aggregate in response to calcium ionophore, while they reacted to the DAG mimetic PMA (phorbol 12-myristate 13-acetate), b) did not respond to threshold doses of physiological agonists (lack of sensitivity) and c) exhibited a delayed activation response to high doses of strong agonists (lack of speed). Rapid agonist-induced elevation in cytosolic Ca2+ concentrations is what drives near-immediate platelet activation, which is essential for efficient platelet adhesion in the presence of hemodynamic shear forces. Thus platelets lacking the sensitivity and speed provided by CalDAG-GEFI failed to form three-dimensional thrombi particularly at arterial shear rates, and Caldaggef1−/− mice were strongly protected from FeCl3-induced carotid artery thrombosis[21] and collagen-[27] or immune complex-induced[34] pulmonary embolism. However a defective CalDAG-GEFI-dependent pathway does not prevent platelet activation completely. Both mouse and human platelets lacking functional CalDAG-GEFI could undergo a slow but sustained activation that supported the formation of small three-dimensional thrombi at conditions of low shear flow conditions, such as those found in the venous system[21]. This residual RAP1 activation was inhibited by antagonists of PKC or P2Y12. An important element of this CalDAG-GEFI-independent pathway was discovered very recently (see below).
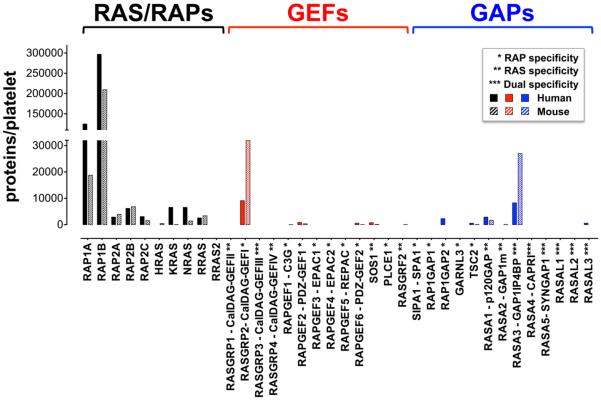
Fig.1. RAS/RAPs, GEFs and GAPs protein expression in platelets.
Estimated protein levels based on the most comprehensive quantitative proteomic analysis of human[7] (full bars) and mouse[29] (striped bars) platelets. CalDAG-GEFI and RASA3 are the most abundant RAP regulators expressed in both human and mouse platelets.
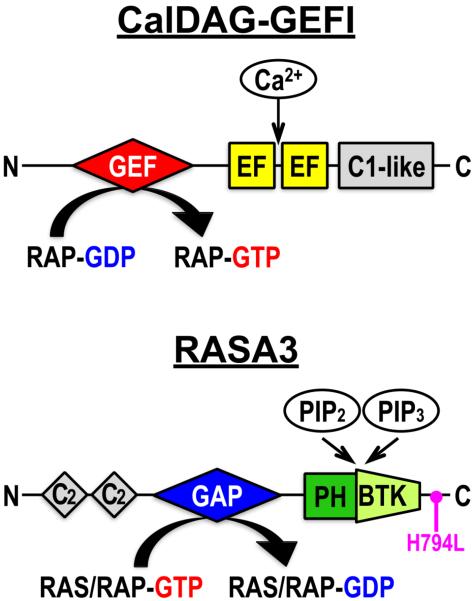
Fig.2. Domain structure of the RAP1 regulators, CalDAG-GEFI and RASA3.
CalDAG-GEFI consists of a RAP-specific catalytic guanine nucleotide exchange factor (GEF) domain, a pair of Ca2+ binding EF hand domains (EF) and a C1-like domain of hitherto unknown function that is necessary for optimal RAP-GEF activity. RASA3 consists of a catalytic GTPase-activating protein (GAP) domain specific for RAS and RAP, flanked by a pair of C2 domains and by a PH/Btk domain that binds phosphatidylinositol 4,5-diphosphate (PIP2) and phosphatidylinositol 3,4,5-triphosphate (PIP3). The Rasa3hlb mutant mouse strain is characterized by a single point mutation (H794L) in the C-terminal region of RASA3. GTP (guanosine triphosphate), GDP (guanosine diphosphate).
The RAP-GAP RASA3: a critical inhibitor of platelet activation and the missing link in the P2Y12-RAP1 signaling pathway
Important information on the molecular nature of this missing RAP1 regulator came from unbiased studies of the platelet proteome and transcriptome[7, 29, 35, 36]. These studies demonstrated that the most abundant RAP-GAP in platelets (Fig.1) is a protein called RASA3 (also known as GAP1IP4BP, GAPIII, R-Ras GAP). RASA3 was originally purified from the plasma membrane of human platelets in 1994[37] but its role in platelet biology was elucidated only recently with studies in transgenic mouse models[38].
RASA3 belongs to a subfamily [39] of four GAP1 proteins (GAP1m/RASA2, RASA3, RASAL1 and CAPRI/RASA4) structurally characterized by a GAP domain flanked N-terminally by two tandem C2 domains and C-terminally by a pleckstrin homology domain linked to a 26 amino acid Btk motif (PH/Btk) (Fig.2). The catalytic domain is a conventional RAS-GAP domain but it is capable of stimulating the GTPase activity of both RAS- and RAP-GTPases depending on the cellular context [40]. This catalytic plasticity is mediated by the N- and C-terminal flanking regions, which orient the RAP-GTP catalytic residues in a way that allows RAP to hijack the RAS-GAP catalytic machinery[41, 42]. The C2 domains of RASA3 are non-canonical and thus do not bind Ca2+ or phospholipids[43]. They have currently no other known function besides their essential role in support of the RAP-GAP activity. On the contrary, the PH/Btk domain, in addition to its steric role in the catalytic reaction, is crucial for RASA3 plasma membrane localization and is presumed to dynamically regulate the GAP activity in response to cellular stimulation[44].
The PH/Btk domain of RASA3 is unique as it binds with similarly high affinity to phosphatidylinositol 4,5-diphosphate (PIP2) (KD=0.8±0.5 μM) and phosphatidylinositol 3,4,5-triphosphate (PIP3) (KD=0.5±0.2 μM). PIP2 is abundant in the plasma membrane of both resting and activated cells and mediates RASA3 constitutive localization to this membrane compartment[44]. PIP3 is the catalytic product of PI3K-mediated phosphorylation of PIP2. PIP3 can drive plasma membrane localization of an engineered RASA3 unable to bind PIP2[45]. It is not yet clear if and how PIP3 regulates RASA3 GAP activity. Since RASA3 binds with high affinity also the water-soluble cognate head group of PIP3, inositol 1,3,4,5-tetrakisphosphate (IP4), it has been argued that its GAP activity may be regulated by a competition between the binding of IP4 and membrane phosphoinositides[46]. However RASA3 has been shown to localize exclusively to the plasma membrane, even in conditions where IP4 concentrations are high[44]. Thus the most likely regulatory mechanism of RASA3 involves PIP3 and does not require membrane detachment. Recent studies, employing two-color super-resolution imaging, have shown that PIP2 and PIP3 cluster into distinct membrane microdomains[47]. Future studies need to address if PIP2/PIP3-mediated regulation of RASA3 involves compartmentalization into different sub-regions of the plasma membrane and/or conformational changes of the protein.
To investigate its role in platelet biology, we generated mice with deletion of Rasa3 in the platelet/megakaryocyte lineage[38]. Unexpectedly, these mice exhibited a high embryonic/neonatal lethality, similar to that observed in germline knockout mice[38] or mice expressing a catalytically inactive mutant of the protein (Rasa3ΔGAP)[48]. Thus, while studies in the Rasa3ΔGAP mice suggested that the high lethality might be caused by a defect in vascular cells, our studies favour a critical role for RASA3 in platelets/megakaryocytes during embryonic development. Consistent with this conclusion, we found that Rasa3 knockout mice are severely thrombocytopenic and that knockout embryos exhibit a marked vascular mixing phenotype, similar to that observed in other mouse models of severe thrombocytopenia or impaired platelet function[49]. To understand the cause of the thrombocytopenia and the role of RASA3 in platelets, our group employed a mutant mouse strain (Rasa3hlb) derived from a forward genetic screen in C57BL/6 mice, which is characterized by a single point mutation (H794L) in the C-terminal region of RASA3[38]. Platelets from Rasa3hlb homozygous mice exhibit a markedly reduced RASA3 protein level and a hypomorphic phenotype, characterized by Mendelian birth rates and a severe, but incomplete, thrombocytopenia (29±18 × 103 platelets/μl). Compared to controls, the few Rasa3hlb mutant platelets present in circulation were younger and bigger in size, had a short half-life and, consistently with the role of RASA3 as a RAP-GAP, had elevated RAP1 and integrin activation both at baseline and in stimulated conditions. The latter observation prompted us to cross Rasa3hlb mice with mice lacking CalDAG-GEFI, a major regulator of RAP1 activation in platelets. Deletion of CalDAG-GEFI reverted the platelet phenotype in Rasa3hlb mice, including the pre-activated state, the high turnover, and the severe thrombocytopenia. In contrast to studies in Rasa3ΔGAP mice[50], we did not find any gross abnormalities in the morphology of MKs and in their ability to form pro-platelets.
The studies outlined above suggest that RASA3 is critical to maintain circulating platelets in a quiescent, non-adhesive state. At sites of vascular injury, however, this negative feedback would be a liability for platelets during hemostatic plug formation. Thus, we speculated that RASA3 activity is downregulated during platelet activation, and that the required signal is provided by the CalDAG-GEFI-independent, PKC/P2Y12-dependent signaling pathway (see above). Consistent with this hypothesis, we observed that Rasa3 mutant platelets do not require feedback activation via the P2Y12 pathway, and that their ability to aggregate is not affected by inhibitors of P2Y12[38]. Interestingly, RAP1-dependent integrin activation in Rasa3 mutant platelets was also insensitive to inhibitors of PI3K[38], suggesting that conversion from PIP2 to PIP3, mediated by PI3K, is critical in the activity regulation of this important RAP-GAP.
Model of RAP1-dependent platelet regulation
In summary, these studies demonstrate that a tight regulation of the antagonistic balance between the RAP1 regulators, CalDAG-GEFI and RASA3, is critical for both the prevention of premature platelet activation in circulation as well as for hemostatic plug formation at sites of vascular injury. In platelets that patrol healthy vessels, active RASA3 in the plasma membrane is required to antagonize the highly sensitive CalDAG-GEFI/RAP1 signaling module and to maintain the cells in a quiescent state (Fig.3, left panel). Upon vascular injury (Fig.3, central panel), platelet stimulation via GPCRs and ITAM-coupled receptors results in increased levels of cytosolic Ca2+ and the release of ADP from storage granules. CalDAG-GEFI is activated by small changes in the cytosolic Ca2+ concentration, leading to the rapid activation of RAP1 and αIIbβ3 integrin. CalDAG-GEFI signaling, however, is reversible and returns to baseline when cytosolic Ca2+ levels normalize. Thus, to sustain integrin activation and to facilitate hemostatic plug formation, RASA3 activity must be reduced as part of the activation response. The required signal is provided upon ADP engagement of the P2Y12 receptor and signaling by PI3K. Finally, our studies suggest that inhibitors of P2Y12, such as clopidogrel bisulfate, protect from thrombosis and impair hemostasis primarily by preventing the inactivation of RASA3 and thus prohibiting sustained RAP1 signaling (Fig.3, right panel).
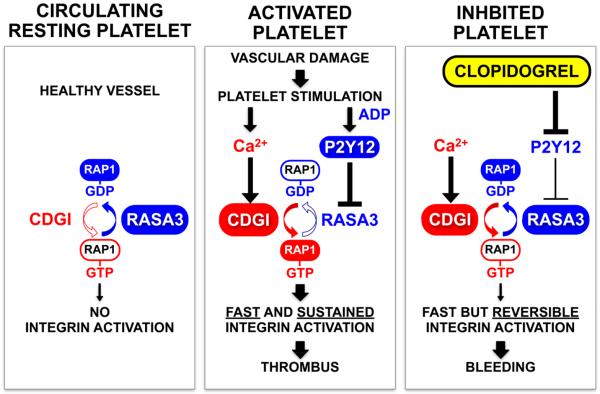
Fig.3. Schematic model of RAP1-dependent platelet regulation.
Left panel. In platelets circulating healthy vessels RASA3 is active to restrain uncontrolled RAP1 activation and maintain a quiescent, non-adhesive state (no integrin activation). Central panel. At sites of vascular injury, platelets stimulation results in an increase in the cytosolic Ca2+ levels and the release of ADP from dense granules. Ca2+ activates CalDAG-GEFI, which mediates rapid GTP-loading of RAP1. ADP stimulates the P2Y12 receptor, which leads to decreased RASA3 function and sustained RAP1 signaling. Fast and sustained RAP1/integrin activation results in the formation of stable three-dimensional thrombi over a broad range of hemodynamic shear conditions. Right panel. Inhibitors to P2Y12, such as Clopidogrel, prevent the inactivation of RASA3, prohibit sustained RAP1 signaling and destabilize the growing thrombus.
There are many unanswered questions concerning RAP1 signaling in platelets. For example, it will be interesting to see if and how platelet function is affected by RAP GEFs and GAPs other than CalDAG-GEFI and RASA3, which are expressed at low levels in these cells. Important information is also missing with regard to the activity regulation of CalDAG-GEFI and RASA3 by engagement of their regulatory domains or posttranslational modifications within these proteins. As for RAP itself, it will be interesting to determine if there are functional redundancies as well as unique contributions of the various RAP isoforms to platelet activation, and which protein effectors link RAP to its downstream platelet responses, especially talin-dependent integrin activation.
In addition to these more fundamental questions, we need more studies on the clinical relevance of alterations in platelet RAP1 signaling. For example, some inherited disorders of platelet function or number may, at least in part, be explained by gene variations in key players of the RAP pathway. Platelet hypo- or hyper-reactivity observed among individuals or as a result of certain disease states may be the result of inter-individual or acquired variability in the antagonistic balance between CalDAG-GEFI and RASA3 (expression and/or activity). If successful these studies could pave the way to novel and more personalized approaches to anti-platelet therapy.
Acknowledgements
This work was supported by the American Heart Association (14EIA18910004) and NIH grants R01 HL121650 and P01 HL120846.
Footnotes
Conflict of interest The authors have declared that no conflict of interest exists.
References
- 1.Ruggeri ZM. Platelet adhesion under flow. Microcirculation. 2009;16:58–83. doi: 10.1080/10739680802651477. doi: 10.1080/10739680802651477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stegner D, Nieswandt B. Platelet receptor signaling in thrombus formation. J Mol Med. 2010;89:109–121. doi: 10.1007/s00109-010-0691-5. doi: 10.1007/s00109-010-0691-5. [DOI] [PubMed] [Google Scholar]
- 3.Kim C, Ye F, Ginsberg MH. Regulation of integrin activation. Annu Rev Cell Dev Biol. 2011;27:321–345. doi: 10.1146/annurev-cellbio-100109-104104. doi: 10.1146/annurev-cellbio-100109-104104. [DOI] [PubMed] [Google Scholar]
- 4.Wei AH, Schoenwaelder SM, Andrews RK, Jackson SP. New insights into the haemostatic function of platelets. Br J Haematol. 2009;147:415–430. doi: 10.1111/j.1365-2141.2009.07819.x. doi: 10.1111/j.1365-2141.2009.07819.x. [DOI] [PubMed] [Google Scholar]
- 5.Jackson SP. Arterial thrombosis--insidious, unpredictable and deadly. Nat Med. 2011;17:1423–1436. doi: 10.1038/nm.2515. doi: 10.1038/nm.2515. [DOI] [PubMed] [Google Scholar]
- 6.Cattaneo M. P2Y12 receptors: structure and function. J Thromb Haemost. 2015;13(Suppl 1):S10–6. doi: 10.1111/jth.12952. doi: 10.1111/jth.12952. [DOI] [PubMed] [Google Scholar]
- 7.Burkhart JM, Vaudel M, Gambaryan S, Radau S, Walter U, Martens L, Geiger J, Sickmann A, Zahedi RP. The first comprehensive and quantitative analysis of human platelet protein composition allows the comparative analysis of structural and functional pathways. Blood. 2012;120:e73–82. doi: 10.1182/blood-2012-04-416594. doi: 10.1182/blood-2012-04-416594. [DOI] [PubMed] [Google Scholar]
- 8.Burkhart JM, Gambaryan S, Watson SP, Jurk K, Walter U, Sickmann A, Heemskerk JWM, Zahedi RP. What can proteomics tell us about platelets? Circ Res. 2014;114:1204–1219. doi: 10.1161/CIRCRESAHA.114.301598. doi: 10.1161/CIRCRESAHA.114.301598. [DOI] [PubMed] [Google Scholar]
- 9.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: Critical Elements in the Control of Small G Proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 10.Franke B, Akkerman JW, Bos JL. Rapid Ca2+-mediated activation of Rap1 in human platelets. The EMBO Journal. 1997;16:252–259. doi: 10.1093/emboj/16.2.252. doi: 10.1093/emboj/16.2.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guidetti GF, Torti M. The Small GTPase Rap1b: A Bidirectional Regulator of Platelet Adhesion Receptors. J Signal Transduct. 2012;2012:412089. doi: 10.1155/2012/412089. doi: 10.1155/2012/412089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagata K, Nozawa Y. A low M(r) GTP-binding protein, Rap1, in human platelets: localization, translocation and phosphorylation by cyclic AMP-dependent protein kinase. Br J Haematol. 1995;90:180–186. doi: 10.1111/j.1365-2141.1995.tb03398.x. [DOI] [PubMed] [Google Scholar]
- 13.Berger G, Quarck R, Tenza D, Levy-Toledano S, de Gunzburg J, Cramer EM. Ultrastructural localization of the small GTP-binding protein Rap1 in human platelets and megakaryocytes. Br J Haematol. 1994;88:372–382. doi: 10.1111/j.1365-2141.1994.tb05033.x. [DOI] [PubMed] [Google Scholar]
- 14.Bertoni A, Tadokoro S, Eto K, Pampori N, Parise LV, White GC, Shattil SJ. Relationships between Rap1b, affinity modulation of integrin alpha IIbbeta 3, and the actin cytoskeleton. J Biol Chem. 2002;277:25715–25721. doi: 10.1074/jbc.M202791200. doi: 10.1074/jbc.M202791200. [DOI] [PubMed] [Google Scholar]
- 15.Chrzanowska-Wodnicka M, Smyth SS, Schoenwaelder SM, Fischer TH, White GC. Rap1b is required for normal platelet function and hemostasis in mice. J Clin Invest. 2005;115:680–687. doi: 10.1172/JCI22973. doi: 10.1172/JCI22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cifuni SM, Wagner DD, Bergmeier W. CalDAG-GEFI and protein kinase C represent alternative pathways leading to activation of integrin alphaIIbbeta3 in platelets. Blood. 2008;112:1696–1703. doi: 10.1182/blood-2008-02-139733. doi: 10.1182/blood-2008-02-139733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernardi B, Guidetti GF, Campus F, Crittenden JR, Graybiel AM, Balduini C, Torti M. The small GTPase Rap1b regulates the cross talk between platelet integrin alpha2beta1 and integrin alphaIIbbeta3. Blood. 2006;107:2728–2735. doi: 10.1182/blood-2005-07-3023. doi: 10.1182/blood-2005-07-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang G, Xiang B, Ye S, Chrzanowska-Wodnicka M, Morris AJ, Gartner TK, Whiteheart SW, White GC, Smyth SS, Li Z. Distinct roles for Rap1b protein in platelet secretion and integrin αIIbβ3 outside-in signaling. J Biol Chem. 2011;286:39466–39477. doi: 10.1074/jbc.M111.239608. doi: 10.1074/jbc.M111.239608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stefanini L, Boulaftali Y, Ouellette TD, Holinstat M, Désiré L, Leblond B, Andre P, Conley PB, Bergmeier W. Rap1-Rac1 circuits potentiate platelet activation. Arterioscler Thromb Vasc Biol. 2012;32:434–441. doi: 10.1161/ATVBAHA.111.239194. doi: 10.1161/ATVBAHA.111.239194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stefanini L, Roden RC, Bergmeier W. CalDAG-GEFI is at the nexus of calcium-dependent platelet activation. Blood. 2009;114:2506–2514. doi: 10.1182/blood-2009-04-218768. doi: 10.1182/blood-2009-04-218768. [DOI] [PubMed] [Google Scholar]
- 21.Stolla M, Stefanini L, Roden RC, Chavez M, Hirsch J, Greene T, Ouellette TD, Maloney SF, Diamond SL, Mortimer Poncz, et al. The kinetics of αIIbβ3 activation determines the size and stability of thrombi in mice: implications for antiplatelet therapy. Blood. 2011;117:1005–1013. doi: 10.1182/blood-2010-07-297713. doi: 10.1182/blood-2010-07-297713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franke B, van Triest M, de Bruijn KM, van Willigen G, Nieuwenhuis HK, Negrier C, Akkerman JW, Bos JL. Sequential regulation of the small GTPase Rap1 in human platelets. Molecular and Cellular Biology. 2000;20:779–785. doi: 10.1128/mcb.20.3.779-785.2000. doi: 10.1128/MCB.20.3.779-785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woulfe D, Jiang H, Mortensen R, Yang J, Brass LF. Activation of Rap1B by G(i) family members in platelets. J Biol Chem. 2002;277:23382–23390. doi: 10.1074/jbc.M202212200. doi: 10.1074/jbc.M202212200. [DOI] [PubMed] [Google Scholar]
- 24.Lova P, Paganini S, Sinigaglia F, Balduini C, Torti M. A Gi-dependent pathway is required for activation of the small GTPase Rap1B in human platelets. J Biol Chem. 2002;277:12009–12015. doi: 10.1074/jbc.M111803200. doi: 10.1074/jbc.M111803200. [DOI] [PubMed] [Google Scholar]
- 25.Lova P, Paganini S, Hirsch E, Barberis L, Wymann M, Sinigaglia F, Balduini C, Torti M. A selective role for phosphatidylinositol 3,4,5-trisphosphate in the Gi-dependent activation of platelet Rap1B. J Biol Chem. 2003;278:131–138. doi: 10.1074/jbc.M204821200. doi: 10.1074/jbc.M204821200. [DOI] [PubMed] [Google Scholar]
- 26.Eto K, Murphy R, Kerrigan SW, Bertoni A, Stuhlmann H, Nakano T, Leavitt AD, Shattil SJ. Megakaryocytes derived from embryonic stem cells implicate CalDAG-GEFI in integrin signaling. Proc Natl Acad Sci USA. 2002;99:12819–12824. doi: 10.1073/pnas.202380099. doi: 10.1073/pnas.202380099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crittenden JR, Bergmeier W, Zhang Y, Piffath CL, Liang Y, Wagner DD, Housman DE, Graybiel AM. CalDAG-GEFI integrates signaling for platelet aggregation and thrombus formation. Nat Med. 2004;10:982–986. doi: 10.1038/nm1098. doi: 10.1038/nm1098. [DOI] [PubMed] [Google Scholar]
- 28.Canault M, Ghalloussi D, Grosdidier C, Guinier M, Perret C, Chelghoum N, Germain M, Raslova H, Peiretti F, Morange PE, et al. Human CalDAG-GEFI gene (RASGRP2) mutation affects platelet function and causes severe bleeding. J Exp Med. 2014;211:1349–1362. doi: 10.1084/jem.20130477. doi: 10.1084/jem.20130477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeiler M, Moser M, Mann M. Copy Number Analysis of the Murine Platelet Proteome Spanning the Complete Abundance Range. Molecular & Cellular Proteomics. 2014;13:3435–3445. doi: 10.1074/mcp.M114.038513. doi: 10.1074/mcp.M114.038513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawasaki H, Springett GM, Toki S, Canales JJ, Harlan P, Blumenstiel JP, Chen EJ, Bany IA, Mochizuki N, Ashbacher A, et al. A Rap guanine nucleotide exchange factor enriched highly in the basal ganglia. Proc Natl Acad Sci USA. 1998;95:13278–13283. doi: 10.1073/pnas.95.22.13278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwig JS, Vercoulen Y, Das R, Barros T, Limnander A, Che Y, Pelton JG, Wemmer DE, Roose JP, Kuriyan J. Structural analysis of autoinhibition in the Ras-specific exchange factor RasGRP1. eLife. 2013;2:e00813. doi: 10.7554/eLife.00813. doi: 10.7554/eLife.00813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Irie K, Masuda A, Shindo M, Nakagawa Y, Ohigashi H. Tumor promoter binding of the protein kinase C C1 homology domain peptides of RasGRPs, chimaerins, and Unc13s. Bioorganic & Medicinal Chemistry. 2004;12:4575–4583. doi: 10.1016/j.bmc.2004.07.008. doi: 10.1016/j.bmc.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 33.Johnson JE, Goulding RE, Ding Z, Partovi A, Anthony KV, Beaulieu N, Tazmini G, Cornell RB, Kay RJ. Differential membrane binding and diacylglycerol recognition by C1 domains of RasGRPs. Biochem J. 2007;406:223. doi: 10.1042/BJ20070294. doi: 10.1042/BJ20070294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amirkhosravi A, Boulaftali Y, Robles-Carrillo L, Meyer T, McKenzie SE, Francis JL, Bergmeier W. CalDAG-GEFI deficiency protects mice from FcγRIIa-mediated thrombotic thrombocytopenia induced by CD40L and β2GPI immune complexes. J Thromb Haemost. 2014;12:2113–2119. doi: 10.1111/jth.12748. doi: 10.1111/jth.12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rowley JW, Oler AJ, Tolley ND, Hunter BN, Low EN, Nix DA, Yost CC, Zimmerman GA, Weyrich AS. Genome-wide RNA-seq analysis of human and mouse platelet transcriptomes. Blood. 2011;118:e101–11. doi: 10.1182/blood-2011-03-339705. doi: 10.1182/blood-2011-03-339705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simon LM, Edelstein LC, Nagalla S, Woodley AB, Chen ES, Kong X, Ma L, Fortina P, Kunapuli S, Holinstat M, et al. Human platelet microRNA-mRNA networks associated with age and gender revealed by integrated plateletomics. Blood. 2014;123:e37–45. doi: 10.1182/blood-2013-12-544692. doi: 10.1182/blood-2013-12-544692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cullen PJ, Patel Y, Kakkar VV, Irvine RF, Authi KS. Specific binding sites for inositol 1,3,4,5-tetrakisphosphate are located predominantly in the plasma membranes of human platelets. Biochem J. 1994;298(Pt 3):739–742. doi: 10.1042/bj2980739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stefanini L, Paul DS, Robledo RF, Chan ER, Getz TM, Campbell RA, Kechele DO, Casari C, Piatt R, Caron KM, et al. RASA3 is a critical inhibitor of RAP1-dependent platelet activation. J Clin Invest. 2015;125:1419–1432. doi: 10.1172/JCI77993. doi: 10.1172/JCI77993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yarwood S, Bouyoucef-Cherchalli D, Cullen PJ, Kupzig S. The GAP1 family of GTPase-activating proteins: spatial and temporal regulators of small GTPase signalling. Biochem Soc Trans. 2006;34:846–850. doi: 10.1042/BST0340846. doi: 10.1042/BST0340846. [DOI] [PubMed] [Google Scholar]
- 40.Cullen PJ, Hsuan JJ, Truong O, Letcher AJ, Jackson TR, Dawson AP, Irvine RF. Identification of a specific Ins(1,3,4,5)P4-binding protein as a member of the GAP1 family. Nature. 1995;376:527–530. doi: 10.1038/376527a0. doi: 10.1038/376527a0. [DOI] [PubMed] [Google Scholar]
- 41.Kupzig S, Bouyoucef-Cherchalli D, Yarwood S, Sessions R, Cullen PJ. The Ability of GAP1IP4BP To Function as a Rap1 GTPase-Activating Protein (GAP) Requires Its Ras GAP-Related Domain and an Arginine Finger Rather than an Asparagine Thumb. Molecular and Cellular Biology. 2009;29:3929–3940. doi: 10.1128/MCB.00427-09. doi: 10.1128/MCB.00427-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sot B, Kötting C, Deaconescu D, Suveyzdis Y, Gerwert K, Wittinghofer A. Unravelling the mechanism of dual-specificity GAPs. The EMBO Journal. 2010;29:1205–1214. doi: 10.1038/emboj.2010.20. doi: 10.1038/emboj.2010.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lockyer PJ, Bottomley JR, Reynolds JS, McNulty TJ, Venkateswarlu K, Potter BV, Dempsey CE, Cullen PJ. Distinct subcellular localisations of the putative inositol 1,3,4,5-tetrakisphosphate receptors GAP1IP4BP and GAP1m result from the GAP1IP4BP PH domain directing plasma membrane targeting. Current Biology. 1997;7:1007–1010. doi: 10.1016/s0960-9822(06)00423-4. [DOI] [PubMed] [Google Scholar]
- 44.Cozier GE, Lockyer PJ, Reynolds JS, Kupzig S, Bottomley JR, Millard TH, Banting G, Cullen PJ. GAP1IP4BP contains a novel group I pleckstrin homology domain that directs constitutive plasma membrane association. J Biol Chem. 2000;275:28261–28268. doi: 10.1074/jbc.M000469200. doi: 10.1074/jbc.M000469200. [DOI] [PubMed] [Google Scholar]
- 45.Cozier GE, Bouyoucef D, Cullen PJ. Engineering the phosphoinositide-binding profile of a class I pleckstrin homology domain. J Biol Chem. 2003;278:39489–39496. doi: 10.1074/jbc.M307785200. doi: 10.1074/jbc.M307785200. [DOI] [PubMed] [Google Scholar]
- 46.Cullen PJ. Bridging the GAP in inositol 1,3,4,5-tetrakisphosphate signalling. Biochim Biophys Acta. 1998;1436:35–47. doi: 10.1016/s0005-2760(98)00149-0. [DOI] [PubMed] [Google Scholar]
- 47.Wang J, Richards DA. Segregation of PIP2 and PIP3 into distinct nanoscale regions within the plasma membrane. Biology Open. 2012;1:857–862. doi: 10.1242/bio.20122071. doi: 10.1242/bio.20122071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iwashita S, Kobayashi M, Kubo Y, Hinohara Y, Sezaki M, Nakamura K, Suzuki-Migishima R, Yokoyama M, Sato S, Fukuda M, et al. Versatile Roles of R-Ras GAP in Neurite Formation of PC12 Cells and Embryonic Vascular Development. Journal of Biological Chemistry. 2006;282:3413–3417. doi: 10.1074/jbc.C600293200. doi: 10.1074/jbc.C600293200. [DOI] [PubMed] [Google Scholar]
- 49.Boulaftali Y, Hess PR, Kahn ML, Bergmeier W. Platelet immunoreceptor tyrosine-based activation motif (ITAM) signaling and vascular integrity. Circ Res. 2014;114:1174–1184. doi: 10.1161/CIRCRESAHA.114.301611. doi: 10.1161/CIRCRESAHA.114.301611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Molina-Ortiz P, Polizzi S, Ramery E, Gayral S, Delierneux C, Oury C, Iwashita S, Schurmans S. Rasa3 controls megakaryocyte Rap1 activation, integrin signaling and differentiation into proplatelet. PLoS Genet. 2014;10:e1004420. doi: 10.1371/journal.pgen.1004420. doi: 10.1371/journal.pgen.1004420. [DOI] [PMC free article] [PubMed] [Google Scholar]