Abstract
Current nonviral genetic vaccine systems are less effective than viral vaccines, particularly in cancer systems where epitopes can be weakly immunogenic and antigen-presenting cell processing and presentation to T cells is down-regulated. A promising nonviral delivery method for genetic vaccines involves microencapsulation of antigen-encoding DNA, because such particles protect plasmid payloads and target them to phagocytic antigen-presenting cells. However, conventional microparticle formulations composed of poly lactic-co-glycolic acid take too long to release encapsulated payload and fail to induce high levels of target gene expression. Here, we describe a microparticle-based DNA delivery system composed of a degradable, pH-sensitive poly-β amino ester and poly lactic-co-glycolic acid. These formulations generate an increase of 3–5 orders of magnitude in transfection efficiency and are potent activators of dendritic cells in vitro. When used as vaccines in vivo, these microparticle formulations, unlike conventional formulations, induce antigen-specific rejection of transplanted syngenic tumor cells.
Genetic vaccination has tremendous potential for treating or preventing numerous diseases for which traditional vaccines are ineffective but can be limited by low immunogenicity in larger animals (1, 2). This deficiency is particularly pronounced in nonviral genetic vaccine cancer therapies where epitopes can be weakly recognized and tumors can down-regulate the ability of antigen-presenting cells (APC) to process and present antigen efficiently to T cells in an activated state (3). Current nonviral genetic vaccine systems are not designed to activate APCs (4) and lack the gene-delivery capacity of viral vectors. In an attempt to increase the effectiveness of nonviral systems, focus has shifted toward exploring the use of adjuvants, cytokines, and self-replicating RNA systems (5–8).
A promising method for nonviral delivery of genetic vaccines is microparticulate DNA delivery systems formulated with a degradable polymer such as poly lactic-co-glycolic acid (PLGA), because these particles facilitate targeting to phagocytic APCs (4, 9–11). Despite these advantages, even low-molecular-weight PLGA systems require up to 13 days to fully release encapsulated DNA after dendritic cell (DC) uptake in vitro (12). This period seems excessively long given evidence that most DCs die within 7 days after activation and migration to draining lymph nodes (13). Furthermore, PLGA microparticles can produce an extremely low pH microclimate (pH < 3.5) after only 3 days in an aqueous environment (14). This level of acidity has been shown to severely reduce the activity of plasmid DNA (15). PLGA microparticles also have been shown to remain confined to phagolysosomal vesicles and generate only low levels of gene expression in APCs (16). Improvements have been made to PLGA microparticles for delivery of plasmid DNA with promising results (4, 17–20), but novel delivery strategies are still needed to advance the potency of nonviral genetic vaccines for use in the clinic.
Recently, we described the synthesis of a degradable, pH-sensitive poly-β amino ester (PBAE) (21) and its application to microparticles capable of releasing fluorescently labeled payloads instantaneously upon pH changes in the physiological range (22). Here we report the encapsulation of plasmid DNA within hybrid PLGA/PBAE microparticles and demonstrate a resulting enhancement in intracellular delivery when compared with encapsulation by PLGA alone. We also report that these formulations are potent activators of primary DCs. To examine the resulting effect in vivo, we used a plasmid that contains a sequence for a fusion protein containing an octapeptide epitope (SIYRYYGL, henceforth called SIY), which associates with MHC class I (Kb) and can stimulate polyclonal CD8+ T cell responses in B6 mice (23). With this plasmid, we demonstrate the ability of hybrid PBAE/PLGA microparticles to induce an antigen-specific rejection of SIY-expressing tumor cells in vivo, unlike conventional PLGA microparticle and naked DNA formulations.
Materials and Methods
Materials. Poly(d,l-lactic-co-glycolic acid) polymer (RG502H Resomer 50:50) was purchased from Boehringer Ingelheim (Ingelheim, Germany). PBAE was synthesized as previously reported [number average molecular weight (Mn) ≈ 5 kDa] (22). Plasmid DNA encoding firefly luciferase (pCMV–Luc), β-galactosidase, or SIY/β-galactosidase fusion (pCMV-SIY) were obtained from Elim Biopharmaceuticals (Hayward, CA). Dextran-conjugated tetramethyl rhodamine (Mn ≈ 70 kDa) was purchased from Molecular Probes.
Mice. C57BL/6 (B6, H-2 Kb) mice (6–10 weeks old) were purchased from Taconic Farms.
Cells and Cell Lines. The P388D1 macrophage cell line was obtained from the American Type Culture Collection and was cultured as recommended. Leukopaks were obtained from Massachusetts General Hospital (Boston), and human peripheral mononuclear cells were isolated by adherence as described (24, 25). Human DCs were differentiated in Iscove's modified Dulbecco's medium (GIBCO) including 1% human serum (Valley Biomedical, Winchester, VA) along with 50 ng/ml granulocyte/macrophage colony-stimulating factor and 20 ng/ml IL-4 (R & D Systems). Primary bone marrow-derived DCs (BMDCs) were isolated from B6 mice and cultured as described (26) before purifying with magnetic beads (CD11c MACS, Miltenyi Biotec, Auburn, CA) (98% measured by anti-CD11c mAb in flow cytometry analysis). EL-4 murine thymoma cells (American Type Culture Collection) and a transduced, SIY-expressing EL-4 cell line were cultured in RPMI medium 1640 with 10% FCS with 1 mg/ml G418 (GIBCO). Previous studies have indicated that the SIY–Kb peptide MHC complex is expressed in the transfected EL-4 cells (27).
Preparation of Microspheres. Plasmid-containing microparticles were prepared by double emulsion/solvent evaporation as described (22) by using varying amounts of PLGA blended with PBAE. Fluorescent microspheres were prepared similarly, but with dextran tetramethyl-rhodamine (200 μl, 1 mg/ml) in the primary emulsion. All in vitro cellular assays and in vivo tumor challenge experiments were performed by normalizing the microparticle amount to equalize plasmid DNA dosage.
Characterization of Microspheres. Loading of DNA microparticles was determined by dissolution in CH2Cl2 and extraction into 1× TAE buffer (40 mM Tris-acetate/1 mM EDTA, pH 8.0). DNA concentration was detected by using PicoGreen (Molecular Probes) and the Mithras plate reading fluorimeter (Berthold Technologies, Bad Wilbad, Germany). DNA integrity was determined by using gel electrophoresis (1% agarose) and nih image j software. Microsphere size distributions were measured by using a Multisizer 3 (Beckman Coulter). Zeta potentials were obtained by using a ZetaPALS analyzer (Brookhaven Instruments, Holtsville, NY). All microparticle formulations were certified to have a low endotoxin level (<0.50 endotoxin units per mg) by the Cambrex LAL testing service (Cambrex, Walkersville, MD).
3D Imaging of APCs. Human peripheral blood mononuclear cell-derived DCs were seeded on glass coverslips at 4 × 105 cells per well in six-well plates. Fluorescent microspheres were added (50 μg/ml of cell medium) and allowed to incubate for 4–6 h. Cells were then washed, fixed with 3.2% paraformaldehyde solution in PBS, and permeated by using 0.2% Triton X-100 (Sigma). Actin filaments and nuclear materials were labeled by using Alexa Fluor 488-conjugated phalloidin and Hoechst dye, respectively (Molecular Probes). Cells were imaged by using the Zeiss Axiovert fluorescent microscope with an Apochromat ×100 oil immersion lens, and vertical slices (0.2-μm separation) were deconvoluted with open-lab software (Improvision, Lexington, MA).
Reporter Gene Transfection. A modification of the six-well plate protocol used by Hedley et al. (10) was used to determine transfection efficiency in P388D1 macrophages in a 96-well-plate format. Briefly, P388D1 macrophages were seeded at 5 × 104 cells per well in fibronectin-coated, white, 96-well polystyrene plates and allowed to achieve 75% confluence. Medium was then replaced with a suspension of pCMV–Luc plasmid DNA-containing microspheres in cell medium. Lipofectamine 2000 (Invitrogen) was prepared with DNA as a positive control. At several time points, wells were washed with PBS and analyzed for luciferase protein content with the Bright Glo Luciferase Assay System (Promega) and a Mithras plate reading luminometer.
Total well protein content was determined with a Micro BCA Protein Assay (Pierce) in tandem with the bioluminescence assay. After lysis of the cells (Glo Lysis Buffer, Promega), bicinchoninic acid reagents were added, the cells were incubated for 3 h at 37°C, and absorption was read at 562 nm with the Spectra Max 384 Plus multiwell plate reader (Molecular Devices).
Flow Cytometry Analysis of Fluorescent Surface Markers. Primary BMDCs were plated at 1 × 106 cells per well of a six-well plate (BD Biosciences). Medium was then replaced with a suspension of microspheres (50 μg/ml) and incubated for 24 h. Untreated cells were used as negative controls. Positive controls were prepared by adding 100 ng/ml lipopolysaccharide (Sigma). At several time points, cells were harvested and stained with Abs for MHC class II (Pharmingen), F4/80 (Caltag, Burlingame, CA), mCD40, mCD86, mCD80, and m41BBL (e-Bioscience, San Diego) at 4°C for 30 min. Cells were then analyzed with a FACScan flow cytometer (Becton Dickinson) with propidium iodide gating (5 μg/ml) collecting a total of 30,000 total events.
Immunization and Tumor Challenge. Mice were immunized intradermally as described in ref. 28 twice at 2-week intervals with naked pCMV-SIY plasmid (10 μg); PLGA-encapsulated pCMV-SIY microspheres (10 μg plasmid total); PBAE/PLGA (15% and 25% wt/wt PBAE) hybrid-encapsulated pCMV-SIY microspheres (10 μg plasmid total); a PBS control group; PBAE/PLGA (25% wt/wt PBAE) microspheres with no encapsulated plasmid; and encapsulated pCMV-β-galactosidase control groups. One week after the last immunization, mice were challenged s.c. with a lethal number (3 × 106) of EL-4 cells on the right flank and an equal number of SIY-expressing EL-4 cells on the left flank. Beginning a week later, tumor size was measured with a caliper every other day in 2D for 9 days. Statistical analysis was performed by comparative ANOVA (samples to PBS-injected controls) by using a Dunnett's error confidence interval of 95%.
Supporting Information. For more details, see Figs. 6 and 7 and Movies 1 and 2, which are published as supporting information on the PNAS web site.
Results
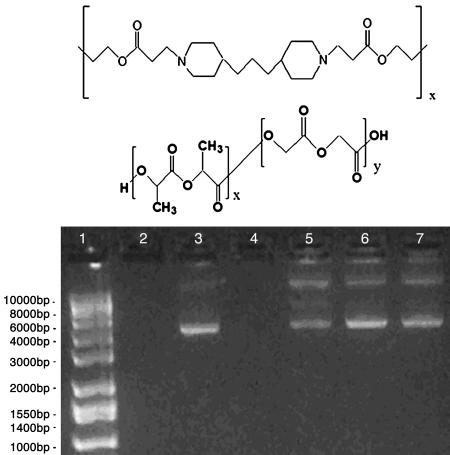
Hybrid Polymeric Microparticles Have Properties Well Suited for Genetic Vaccine Delivery. Plasmid DNA was encapsulated into polymeric microparticles as described in Materials and Methods. We initially fabricated hybrid PBAE/PLGA (molecular structures shown in Fig. 1 Top and Middle, respectively) microparticles ranging from 0% to 100% PBAE to investigate the effects of PBAE content on particle physical properties and function. Formulations containing larger amounts of this cationic polymer (e.g., 35–100%) generally did not release encapsulated DNA as efficiently as those containing lesser amounts of PBAE by delaying release of DNA for several days after administration (data not shown). Formulations containing 15% and 25% PBAE showed rapid release and were therefore used in all subsequent studies. Scanning electron microscopy analysis of microsphere preparations reveals a smooth, spherical surface on all microsphere preparations (see supporting information), and all formulations ranged between 1 and 10 μm in average diameter (Table 1), allowing for a passive APC-targeting mechanism by phagocytosis.
Fig. 1.
Encapsulation of plasmid DNA using PLGA and PBAE. Molecular formulas of PBAE (Top) and PLGA (Middle). (Bottom) One percent agarose gel demonstrating DNA extracted from microparticles prepared by double emulsion. Lane 1, ladder; lanes 2 and 4, empty; lane 3, unprocessed control (88% supercoiled); lanes 5–7, aqueous extract from PLGA (lane 5), 15% PBAE (lane 6), and 25% PBAE (lane 7) microparticles after lyophilization. Percent super-coiled content for lanes 5–7 is reported in Table 1.
Table 1. Characteristics of microparticles made from PBAE and PLGA.
| Formulation* | Volume % diameter, μm | Encapsulation efficiency, % | Supercoiled content, % | Zeta potential, mV |
|---|---|---|---|---|
| 100% PLGA | 4.35 ± 2.34 | ≈69 | ≈45 | -3.76 ± 0.40 |
| 15% PBAE/85% PLGA | 6.01 ± 2.06 | ≈68 | ≈72 | -0.86 ± 0.62 |
| 25% PBAE/75% PLGA | 5.53 ± 2.31 | ≈78 | ≈64 | 0.46 ± 0.38 |
| 25% PBAE/75% PLGA/No DNA | 5.12 ± 2.20 | 0.41 ± 0.36 |
Percentage by weight
Microspheres incorporating PBAE (Fig. 1 Bottom, lanes 6 and 7) had similar or greater encapsulation efficiencies when compared with PLGA microparticles (Table 1) but exhibited higher supercoiled plasmid content than those prepared with PLGA alone (Fig. 1 Bottom, lane 5). Zeta potential analysis of microspheres indicated a negative surface charge on PLGA micoparticles similar to those previously reported (29). In contrast, 15% and 25% PBAE preparations showed slightly more positive zeta potentials than PLGA formulations (Table 1).
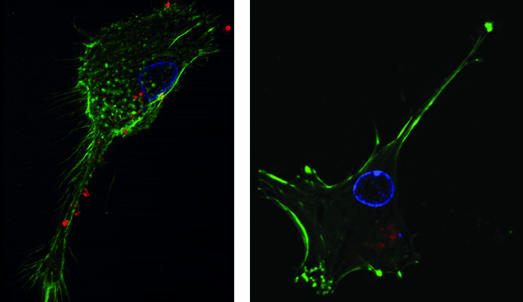
Uptake of PBAE/PLGA Microparticles by DCs in Vitro. To examine the effects of PBAE on APC phagocytosis, primary human DCs derived from peripheral blood mononuclear cells were incubated with particle formulations containing rhodamine-conjugated dextran, fluorescently stained, and examined by visual fluorescence microscopy. 3D overhead views of treated cells are shown in Fig. 2.
Fig. 2.
DCs phagocytose microparticle formulations of PLGA and PBAE in vitro. Peripheral blood mononuclear cell-derived DCs were incubated with rhodamine-conjugated dextran-encapsulated microparticles (red) for 5 h, fixed, and then stained with Hoechst dye for nucleus (blue) and phalloidin–Alexa Fluor 488 for actin (green). 3D fluorescent microscopy images indicate uptake of both PLGA microsphere formulations (Left) and 25% PBAE/75% PLGA microsphere formulations (Right). Intracellular rhodamine signals were seen as bright, localized spheres in 100% PLGA-treated DCs (Left). In 25% PBAE microsphere-treated cells, rhodamine distributions were sometimes seen as dim and dispersed, as though in the cell cytoplasm (Right).
Imaging of DCs incubated for 4–5 h with PLGA and 25% PBAE particle formulations revealed substantial uptake of all microparticle formulations, even at microparticle concentrations as low as 1 μg/ml. In general, PLGA microparticles containing labeled dextran remained sharp, bright, spherical objects, as though restricted to phagosomal compartments (Fig. 2 Left). In contrast, a population of cells treated with 25% PBAE formulations demonstrated a dimmer, diffuse fluorescent signal, suggesting release from phagosomes (Fig. 2 Right).
PBAE/PLGA Blends Induce Higher Levels of Reporter Gene Expression in P388D1 Macrophages than Conventional PLGA Microparticles. To compare the transfection efficiency of formulations composed of PBAE and PLGA, we used an established APC system and reporter gene that previously exhibited positive transfection using PLGA microparticles (10). Lipofectamine 2000/pCMV–Luc plasmid complexes were chosen as a positive control.
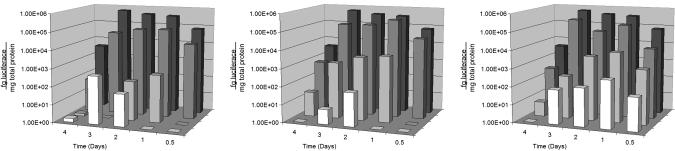
Microsphere formulations composed of 25% PBAE demonstrated optimal reporter gene expression, comparable to that of an optimal formulation of Lipofectamine 2000 (Fig. 3). However, Lipofectamine required 25 times more DNA than the 25% PBAE microparticles to achieve the same level of transfection (300 ng per well vs. 12 ng per well). An optimal formulation of 25% PBAE microparticles generated ≈3 orders of magnitude higher transfection efficiency than an optimal formulation of PLGA (Fig. 2), and at some time points 25% PBAE microparticles exhibited up to 5 orders of magnitude higher luciferase expression than those containing only PLGA. Fifteen percent PBAE microspheres were not as effective as 25% PBAE formulations but still maintained a significant increase of 1–3 orders of magnitude in transfection efficiency relative to PLGA microspheres at most time points (Fig. 3). Naked plasmid DNA at concentrations equal to that administered at the maximum dosage used in Lipofectamine controls yielded no detectable expression of luciferase (data not shown). Formulation groups were compared by using ANOVA and t test analysis for significance (α = 0.05).
Fig. 3.
Transfection of P388D1 macrophages with PBAE/PLGA microspheres. Results are displayed as femptograms of luciferase (luminescence assay) per mg of total protein (bicinchoninic acid assay) vs. time and formulation. Microparticle concentrations incubated with the cells were 10 μg/ml (Left)30 μg/ml (Center), and 100 μg/ml (Right). An optimal formulation of Lipofectamine 2000 (0.8:1 Lipofectamine:DNA) is displayed as a positive control in each plot (black). Twenty-five percent PBAE formulations (dark gray) consistently performed at par to 1 log unit lower than positive controls despite using 25 times less plasmid DNA, whereas 15% PBAE formulations (light gray), although lower than the 25% formulations, consistently transfected at a higher level than PLGA microparticles (white).
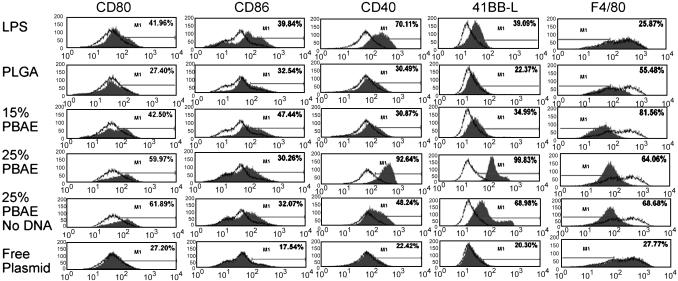
PBAE-Containing Microparticles Activate Primary DCs. Primary BMDCs were analyzed for surface expression of costimulatory molecules, CD80 (B7-1), CD86 (B7-2), CD40, and 41BB ligand (CD137L). F4/80 surface expression was also examined to demonstrate the absence of nonspecific binding of Ab to PBAE microparticles on the surface of cells due to its characteristic down-regulation upon DC maturation (30, 31). Lipopolysaccharide (100 ng/ml) treatment was used as a positive control.
After incubation with conventional PLGA microparticle formulations, the costimulatory profile changed slightly across the spectrum (Fig. 4). However, with 15% and 25% PBAE formulations containing plasmid DNA, the amount of F4/80 greatly decreased and the surface expression of costimulatory molecules was markedly increased, indicating an activated, mature phenotype (Fig. 4). Twenty-five percent PBAE formulations with no encapsulated DNA also activated the DCs but to a lower extent in CD40 and 41BBL than 25% PBAE particles that included plasmid DNA. In addition, BMDC incubated with naked plasmid DNA in quantities equivalent to the total amount of DNA in microparticles (assuming 100% loading during encapsulation and instantaneous release) demonstrated costimulatory profiles similar to that of untreated cells (Fig. 4).
Fig. 4.
Activation of primary APCs by incubation with PBAE microsphere formulations. Histograms show expression levels of the indicated costimulatory molecules after 18 h of incubation with 100 ng/ml lipopolysaccharide, PLGA, 15% PBAE, and 25% PBAE microparticles encapsulating plasmid DNA (50 μg/ml), 25% PBAE microparticles without encapsulated plasmid, and an amount of free plasmid equivalent to 100% theoretical loading of microparticle treatments and instantaneous release into the supernatant. Untreated controls are shown as the open background trace in each histogram. Cells incubated with lipopolysaccharide demonstrated higher expression of costimulatory molecules but unchanged F4/80 expression. PLGA microsphere-treated cells appear to have a slightly activated phenotype, whereas cells incubated with 15% PBAE and also 25% PBAE microsphere formulations with and without plasmid DNA are activated as indicated by both down-regulation of F4/80 and up-regulation of costimulatory molecules. Results are representative of three independent experiments.
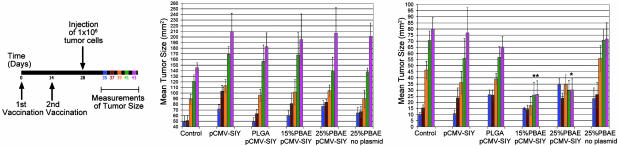
Vaccination with PBAE Microparticle Formulations Containing pCMV-SIY Results in Antigen-Specific Rejection of SIY-Expressing Tumor Cells in B6 Mice. To compare the immunogenic efficacy of PBAE-containing microparticle formulations, B6 mice (five per group) were vaccinated with plasmid-containing microparticles, naked DNA, empty microparticles, or PBS following the schedule represented in Fig. 5 Left and described in Materials and Methods. The SIY–Kb complex is presented on the surface of EL-4 tumor cells transfected with SIY plasmid (administered left flank) but not presented on the surface of untransfected EL-4 tumor cells used as a control (administered right flank) (23, 27). SIY-expressing EL-4 tumor cells on the left flank grew at similar rates in mice injected with PBS controls, naked DNA, PLGA/DNA microparticles, and blank 25% PBAE microparticle formulations. Conversely, in mice injected with formulations composed of 15% and 25% PBAE-containing pCMV-SIY, the average rate of growth of tumors expressing SIY was distinctly reduced (Fig. 5 Right). Also, in two of the five mice in the 15% PBAE formulation group and in one of the five mice in the 25% PBAE formulation group, the SIY-expressing tumors decreased in size and completely disappeared on the days indicated (Fig. 5 Right, *). On the right flank, control, untransfected EL4 tumors grew progressively in all groups (Fig. 5 Center). Moreover, vaccination with a plasmid that exclusively expresses β-galactosidase (without the added SIY sequence) did not inhibit growth of the SIY-expressing tumor cells (data not shown).
Fig. 5.
In vivo tumor rejection in B6 mice after treatment with genetic vaccine formulations. Mice were vaccinated and challenged by using the schedule shown in Left, and mean tumor size was measured by using a caliper in 2D 7 (blue), 9 (red), 11 (orange), 13 (green), and 15 (pink) days after s.c. injection of 3 × 106 normal EL4 thymoma cells (Center) or EL4 cells that are transfected with SIY and express it on their surface (Right). Standard error bars are shown for comparison. * indicates one mouse in which the tumor expressing SIY had completely regressed by this time point.
Statistical analysis using comparative ANOVA showed that the 15% PBAE formulation was significantly different from the PBS-injected control after day 11 and that formulations containing 25% PBAE were significantly different after day 13. No other group showed significantly reduced tumor size when compared with the control group at any time point.
Discussion
Polymeric microparticles that physically encapsulate antigen-encoding plasmid offer several potential benefits to genetic vaccine formulations, including protection of the encapsulated plasmid (32) and size-based adjuvancy and targeting to phagocytic APCs (9). Furthermore, unlike viral delivery, microparticle delivery systems possess the capacity to hold extremely large payloads, allowing for vaccines with multiple antigen expression constructs (multivalent) and coencapsulation of immunomodulating cytokines. Despite these advantages, current microparticle systems prepared from PLGA exhibit extremely low levels of gene expression in APCs. Although such low amounts of antigen expression may be sufficient to induce some immune responses, it is likely that increasing levels of gene expression will lead to a corresponding increase in vaccine potency. We hypothesized that the incorporation of a degradable, pH-sensitive polymer in conventional PLGA microparticle formulations would increase gene-delivery capacity by facilitating intracellular release of plasmid payload upon phagosomal acidification.
Incorporation of PBAE into the microsphere matrix did not alter the structure or loading of the particles significantly. It was possible to encapsulate relatively high quantities of supercoiled plasmid in our formulations using standard double emulsion techniques, despite previous indications that encapsulating supercoiled plasmid is difficult (33).
We demonstrated previously that microspheres fabricated from PBAE have the ability to rapidly release contents when exposed to acidic endosomal pH (22). Consistent with this finding, a population of DCs treated with PBAE/PLGA micro-particles exhibited internally dispersed rhodamine fluorescence (Fig. 2), suggesting that PBAE may facilitate release from phagosomes. In a solid state, PBAE in the microparticle structure should remain mostly unprotonated (Fig. 1 Top) and subsequently absorb protons during phagosomal acidification. PBAE may thus provide a means of phagosomal escape by osmotic membrane disruption using a proton sponge-like mechanism (34). Although the exact intracellular delivery mechanism is unclear, we observed an increase of 3–5 orders of magnitude in reporter gene transfection upon addition of PBAE to the microparticle formulations, consistent with a corresponding increase in intracellular delivery of plasmid DNA (Fig. 3).
In addition to sufficient gene expression by APCs, upregulation of costimulatory molecules on these cells during epitope presentation is crucial to vaccine potency. Costimulatory molecules are particularly important, because, in their absence, antigen presentation by immature/inactivated DCs may induce tolerance to that antigen (35, 36). It was thus notable that primary BMDCs were strikingly stimulated by microparticle formulations to up-regulate expression of several costimulatory molecules (Fig. 4). Interestingly, 25% PBAE microparticle formulations with no encapsulated DNA still activated DCs to a greater extent than PLGA microparticles but not as fully as 25% PBAE microparticles with encapsulated plasmid, suggesting that the PBAE polymer microparticle on its own can activate DCs. The mechanisms behind this effect on DCs are not clear and warrant further investigation. One possible mechanism stems from the observation by Thiele et al. (37) that the addition of cationic polymer (polyl-lysine) to the surface of polystyrene beads up-regulates CD83 on primary human DCs. This finding introduces the possibility that the increasingly positive surface charge of the cationic PBAE-containing formulations may be partially responsible for the observed effect.
To determine whether antigen-encoding plasmid DNA encapsulated within PBAE-containing microparticles can generate CD8+ T cell response to a model antigen, we used an antigen expression system based on a particular peptide–MHC complex in which SIY is associated with Kb, a class I MHC protein (38). Cho et al. (23) showed that a fusion protein containing the SIY sequence can stimulate mice to produce polyclonal CD8+ T cells that react specifically to the SIY–Kb complex.
Accordingly, we compared the ability of the pCMV-SIY plasmid in various microparticle formulations and as naked DNA to stimulate SIY–Kb-specific rejection of EL-4 (express SIY and Kb+) (27) tumor cells in B6 mice. Only in mice immunized with pCMV-SIY DNA in PBAE-containing micro-particles was growth of SIY-producing EL-4 cells reduced. This effect was antigen-specific, because EL-4 cells not expressing SIY grew unhindered in the same mice in which SIY+ EL-4 cells were affected. In contrast, the same plasmid in PLGA micro-particles, or as unencapsulated naked plasmid, had no apparent effects on the SIY+ EL-4 cells. In addition, preliminary experiments demonstrated naïve anti-SIY–Kb cells (2C T cells) (39) adoptively transferred into B6 mice persisted and up-regulated a T cell activation marker (CD69) after i.p. injection of the SIY peptide in three of four mice only if the mice had been previously vaccinated with microparticles containing PBAE (or with the naked pCMV-SIY DNA) but not with those made exclusively with PLGA (data not shown). The antigen-specific tumor regression observed was likely due to a polyclonal anti-SIY–Kb CD8+ T cell response, but CD4+ T cells that recognize class II MHC–peptide complexes derived from the fusion protein encoded by the plasmid could have contributed to the tumor regression. Nevertheless, the responses seen were antigen-specific, reinforcing and extending in vitro evidence that micro-spheres containing PBAE are far more effective than those made exclusively from PLGA in their transfection ability and effect on primary DCs.
Because of their inherent adjuvancy, the PBAE-containing microparticles may be widely applicable as a platform for delivery in circumstances in which the antigen of interest is not immunogenic enough for plasmid DNA vaccination alone, as in the case of B cell malignancies or in individuals with weakened or tolerized immune capacity (40). The presence of strong adjuvancy as a innate property of the delivery system also bypasses adverse systemic effects from using cytokines or conventional adjuvants to augment the immune reaction (8). We are currently exploring combinations of PBAE microparticles with complementary technologies such as plasmid-encoded cytokine and immunogenic fusion constructs along with targeting moieties on the microparticle surface, which may even further enhance vaccine potency. Finally, the intracellular delivery capacity of PBAE microparticles may have implications for delivery of other drugs to APCs, such as in the case of lysosomal storage disorders, where targeted, effective delivery to macrophages could lead to enhancements in enzyme replacement therapy.
Supplementary Material
Acknowledgments
We thank Daniel S. Kohane for advice on statistical analysis. This work was supported by the Center for Innovative Minimally Invasive Therapies, the National Science Foundation (through the Massachusetts Institute of Technology Biotechnology Processing and Engineering Center), and the National Institutes of Health (Grants EB00244 and CA60686). S.R.L. is the recipient of a graduate fellowship from the National Science Foundation.
Abbreviations: APC, antigen-presenting cell; DC, dendritic cell; BMDC, bone marrow-derived DC; PLGA, poly lactic-co-glycolic acid; PBAE, poly-β amino ester; SIY, SIYRYYGL octapeptide.
References
- 1.Gurunathan, S., Klinman, D. M. & Seder, R. A. (2000) Annu. Rev. Immunol. 18, 927-974. [DOI] [PubMed] [Google Scholar]
- 2.McKenzie, B. S., Corbett, A. J., Brady, J. L., Dyer, C. M., Strugnell, R. A., Kent, S. J., Kramer, D. R., Boyle, J. S. & Lew, A. M. (2001) Immunol. Res. 24, 225-244. [DOI] [PubMed] [Google Scholar]
- 3.Pardoll, D. M. (2002) Nat. Rev. Immunol. 2, 227-238. [DOI] [PubMed] [Google Scholar]
- 4.McKeever, U., Barman, S., Hao, T., Chambers, P., Song, S., Lunsford, L., Hsu, Y. Y., Roy, K. & Hedley, M. L. (2002) Vaccine 20, 1524-1531. [DOI] [PubMed] [Google Scholar]
- 5.Leitner, W. W., Hwang, L. N., deVeer, M. J., Zhou, A. M., Silverman, R. H., Williams, B. R. G., Dubensky, T. W., Ying, H. & Restifo, N. P. (2003) Nat. Med. 9, 33-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pachuk, C. J., McCallus, D. E., Weiner, D. B. & Satishchandran, C. (2000) Curr. Opin. Mol. Ther. 2, 188-198. [PubMed] [Google Scholar]
- 7.Leitner, W. W., Hammerl, P. & Thalhamer, J. (2001) Curr. Pharm. Des. 7, 1641-1667. [DOI] [PubMed] [Google Scholar]
- 8.O'Hagan, D. T., MacKichan, M. L. & Singh, M. (2001) Biomol. Eng. 18, 69-85. [DOI] [PubMed] [Google Scholar]
- 9.O'Hagan, D. T., Singh, M. & Gupta, R. K. (1998) Adv. Drug Delivery Rev. 32, 225-246. [PubMed] [Google Scholar]
- 10.Hedley, M. L., Curley, J. & Urban, R. (1998) Nat. Med. 4, 365-368. [DOI] [PubMed] [Google Scholar]
- 11.O'Hagan, D., Singh, M., Ugozzoli, M., Wild, C., Barnett, S., Chen, M. C., Schaefer, M., Doe, B., Otten, G. R. & Ulmer, J. B. (2001) J. Virol. 75, 9037-9043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walter, E., Dreher, D., Kok, M., Thiele, L., Kiama, S. G., Gehr, P. & Merkle, H. P. (2001) J. Controlled Release 76, 149-168. [DOI] [PubMed] [Google Scholar]
- 13.Garg, S., Oran, A., Wajchman, J., Sasaki, S., Maris, C. H., Kapp, J. A. & Jacob, J. (2003) Nat. Immunol. 4, 907-912. [DOI] [PubMed] [Google Scholar]
- 14.Fu, K., Pack, D. W., Klibanov, A. M. & Langer, R. (2000) Pharm. Res. 17, 100-106. [DOI] [PubMed] [Google Scholar]
- 15.Walter, E., Moelling, K., Pavlovic, J. & Merkle, H. P. (1999) J. Controlled Release 61, 361-374. [DOI] [PubMed] [Google Scholar]
- 16.Walter, E., Thiele, L. & Merkle, H. P. (2001) STP Pharma. Sci. 11, 45-56. [Google Scholar]
- 17.Singh, M., Briones, M., Ott, G. & O'Hagan, D. (2000) Proc. Natl. Acad. Sci. USA 97, 811-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang, C., Ge, Q., Ting, D., Nguyen, D., Shen, H. R., Chen, J., Eisen, H. N., Heller, J., Langer, R. & Putnam, D. (2004) Nat. Mater. 3, 190-196. [DOI] [PubMed] [Google Scholar]
- 19.Walter, E. & Merkle, H. P. (2002) J. Drug Targeting 10, 11-21. [DOI] [PubMed] [Google Scholar]
- 20.Luo, Y. P., O'Hagan, D., Zhou, H., Singh, M., Ulmer, J., Reisfeld, R. A., Primus, F. J. & Xiang, R. (2003) Vaccine 21, 1938-1947. [DOI] [PubMed] [Google Scholar]
- 21.Lynn, D. M. & Langer, R. (2000) J. Am. Chem. Soc. 122, 10761-10768. [Google Scholar]
- 22.Lynn, D. M., Amiji, M. M. & Langer, R. (2001) Angew. Chem. Int. Ed. 40, 1707-1710. [PubMed] [Google Scholar]
- 23.Cho, B. K., Palliser, D., Guillen, E., Wisniewski, J., Young, R. A., Chen, J. Z. & Eisen, H. N. (2000) Immunity 12, 263-272. [DOI] [PubMed] [Google Scholar]
- 24.Romani, N., Gruner, S., Brang, D., Kampgen, E., Lenz, A., Trockenbacher, B., Konwalinka, G., Fritsch, P. O., Steinman, R. M. & Schuler, G. (1994) J. Exp. Med. 180, 83-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thurner, B., Roder, C., Dieckmann, D., Heuer, H., Kruse, M., Glaser, A., Keikavoussi, P., Kampgen, E., Bender, A. & Schuler, G. (1999) J. Immunol. Methods 223, 1-15. [DOI] [PubMed] [Google Scholar]
- 26.Inaba, K., Inaba, M., Romani, N., Aya, H., Deguchi, M., Ikehara, S., Muramatsu, S. & Steinman, R. M. (1992) J. Exp. Med. 176, 1693-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho, B. K., Rao, V. P., Ge, Q., Eisen, H. N. & Chen, J. Z. (2000) J. Exp. Med. 192, 549-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis, H. L. (1999) in DNA Vaccines: Methods and Protocols, eds. Lowrie, D. B. & Whalen, R. G. (Humana, Totowa, NJ), pp. 71-78.
- 29.Sahoo, S. K., Panyam, J., Prabha, S. & Labhasetwar, V. (2002) J. Controlled Release 82, 105-114. [DOI] [PubMed] [Google Scholar]
- 30.McKnight, A. J., Macfarlane, A. J., Dri, P., Turley, L., Willis, A. C. & Gordon, S. (1996) J. Biol. Chem. 271, 486-489. [DOI] [PubMed] [Google Scholar]
- 31.Caminschi, I., Lucas, K. M., O'Keeffe, M. A., Hochrein, H., Laabi, Y., Kontgen, F., Lew, A. M., Shortman, K. & Wright, M. D. (2001) J. Immunol. 167, 3570-3576. [DOI] [PubMed] [Google Scholar]
- 32.Capan, Y., Woo, B. H., Gebrekidan, S., Ahmed, S. & DeLuca, P. P. (1999) Pharm. Res. 16, 509-513. [DOI] [PubMed] [Google Scholar]
- 33.Singh, M., Ott, G., Kazzaz, J., Ugozzoli, M., Briones, M., Donnelly, J. & O'Hagan, D. T. (2001) Pharm. Res. 18, 1476-1479. [DOI] [PubMed] [Google Scholar]
- 34.Demeneix, B. & Behr, J. P. (1996) in Artificial Self-Assembling Systems for Gene Delivery, eds. Felgner, P. L., Heller, M. J., Lehn, P., Behr, J. P. & Szoka, F. C., Jr. (Oxford Univ. Press, Washington, DC), pp. 146-151.
- 35.Steinman, R. M., Hawiger, D., Liu, K., Bonifaz, L., Bonnyay, D., Mahnke, K., Iyoda, T., Ravetch, J., Dhodapkar, M., Inaba, K. & Nussenzweig, M. (2003) Ann. N.Y. Acad. Sci. 987, 15-25. [DOI] [PubMed] [Google Scholar]
- 36.Steinman, R. M., Hawiger, D. & Nussenzweig, M. C. (2003) Annu. Rev. Immunol. 21, 685-711. [DOI] [PubMed] [Google Scholar]
- 37.Thiele, L., Rothen-Rutishauser, B., Jilek, S., Wunderli-Allenspach, H., Merkle, H. P. & Walter, E. (2001) J. Controlled Release 76, 59-71. [DOI] [PubMed] [Google Scholar]
- 38.Udaka, K., Wiesmuller, K. H., Kienle, S., Jung, G. & Walden, P. (1996) J. Immunol. 157, 670-678. [PubMed] [Google Scholar]
- 39.Sykulev, Y., Vugmeyster, Y., Brunmark, A., Ploegh, H. L. & Eisen, H. N. (1998) Immunity 9, 475-483. [DOI] [PubMed] [Google Scholar]
- 40.Savelyeva, N., Munday, R., Spellerberg, M. B., Lomonossoff, G. P. & Stevenson, F. K. (2001) Nat. Biotechnol. 19, 760-764. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.