Abstract
Hypercapnia, elevated partial pressure of carbon dioxide (PCO2) in blood and tissue, develops in many patients with chronic severe obstructive pulmonary disease and other advanced lung disorders. Patients with advanced disease frequently develop bacterial lung infections, and hypercapnia is a risk factor for mortality in such individuals. We previously demonstrated that hypercapnia suppresses induction of NF-κB-regulated innate immune response genes required for host defense in human, mouse and Drosophila cells, and increases mortality from bacterial infections in both mice and Drosophila. However, the molecular mediator(s) of hypercapnic immune suppression are undefined. Here, we report a genome-wide RNAi screen in Drosophila S2* cells stimulated with bacterial peptidoglycan (PGN). The screen identified 16 genes with human orthologs whose knockdown reduced hypercapnic suppression of the gene encoding the antimicrobial peptide (AMPs) Diptericin (Dipt), but did not increase Dipt mRNA levels in air. In vivo tests of one of the strongest screen hits, Zfh2 (mammalian orthologs ZFHX3/ATBF1 and ZFHX4), demonstrate that reducing zfh2 function using a mutation or RNAi improves survival of flies exposed to elevated CO2 and infected with S. aureus. Tissue-specific knockdown of zfh2 in the fat body, the major immune and metabolic organ of the fly, mitigates hypercapnia-induced reductions in Dipt and other AMPs and improves resistance of CO2-exposed flies to infection. Zfh2 mutations also partially rescue hypercapnia-induced delays in egg hatching, suggesting that Zfh2's role in mediating responses to hypercapnia extends beyond the immune system. Together, these results identify Zfh2 as the first in vivo mediator of hypercapnic immune suppression.
Keywords: Hypercapnia, immune suppression, Zfh2, Drosophila, genome-wide RNAi screen, CO2, infection, ZFHX3/ATBF1
Introduction
Hypercapnia, elevation of blood and tissue levels of CO2, develops in many patients with severe chronic obstructive pulmonary disease (COPD), currently the third leading cause of death in the U.S. (1), and other advanced lung diseases. Hypercapnia has long been recognized as a risk factor for mortality in patients with acute exacerbations of COPD (2-6), and recently it was shown that use of nocturnal ventilatory support to decrease CO2 levels improved survival of hypercapnic patients with COPD (7). Acute exacerbations of COPD, which are linked to mortality, are most commonly triggered by bacterial or viral respiratory infections (8-10). Hypercapnia is also a risk factor for mortality in patients hospitalized with community-acquired pneumonia (11, 12), children with adenoviral lung infections (13), and cystic fibrosis patients awaiting lung transplantation (14). These observations suggest that hypercapnia may contribute to poor clinical outcomes by increasing susceptibility to pulmonary infections.
Consistent with this hypothesis, we and others have shown that hypercapnia inhibits expression of IL-6, TNF and other cytokines important for host defense (15-17). Cummins and colleagues have shown that elevated CO2 inhibits activation of the canonical NF-κB pathway (18, 19) that drives expression of many host defense genes, while promoting activation of the non-canonical NF-κB component RelB (18, 19), whose function is largely anti-inflammatory and immunosuppressive. We also showed that hypercapnia suppresses phagocytosis, generation of reactive oxygen species and autophagy (15-17), all key phagocyte antimicrobial functions. Furthermore, we showed that hypercapnia reduces bacterial clearance and increases mortality in a mouse model of Pseudomonas pneumonia (16). Despite these observations, the molecular mechanisms by which elevated CO2 levels are sensed and transduced in immune cells are not yet understood.
One way that elevated CO2 might influence immune responses is by reducing extracellular and/or intracellular pH; indeed, there is evidence that acidosis may decrease the function of various immune cells (20). However, in our in vitro studies, suppression of cytokine gene expression, phagocytosis and autophagy by elevated CO2 levels was independent of acidosis (15-17). Further, hypercapnia increased the mortality of bacterial pneumonia in mice with acute and chronic hypercapnia to the same degree, despite more severe acidosis in animals with acute hypercapnia compared to chronic hypercapnia (due to renal compensation in the latter group) (16). Together, these results suggest that molecular CO2 has immune suppressive effects that are experimentally and physiologically distinguishable from the effects of acidosis.
To investigate the molecular mechanisms of hypercapnic immune suppression, we studied Drosophila as a tractable molecular genetic model system for non-neuronal responses to elevated CO2 (21). The response to hypercapnia in Drosophila and mammals has multiple parallels (15, 16): elevated CO2 levels suppress production of Drosophila immune response genes in vivo and in vitro at the transcriptional level via a mechanism that is downstream of NF-κB proteolytic activation. As in mammalian macrophages, down-regulation of immune response genes was independent of pH. Hypercapnia also significantly decreases resistance of adult flies to bacterial infections. The similarity of the hypercapnic immune suppression in flies and mammals, in combination with evidence that a conserved JNK-pathway controls Na,K-ATPase endocytosis during hypercapnia (22), suggest that elevated CO2 acts via specific evolutionarily conserved pathways to down-regulate host defenses.
Here we describe the results of a genome-wide RNAi screen in Drosophila S2* cells to identify mediators of CO2-induced immune suppression. The screen identified more than 16 genes with human orthologs whose knockdown attenuates hypercapnic suppression of the antimicrobial peptide (AMP) gene in PGN-stimulated S2* cells. In vivo characterization of one of these genes, the transcription factor zfh2 (mammalian orthologs ZFHX3/ATBF1 and ZFHX4), shows that reducing Zfh2 levels in the major immune organ of the fly, the fat body, improves resistance and survival to S. aureus infection during hypercapnia. This is the first description of a component of CO2 response pathways that mediate the in vivo effects of hypercapnia on immune responses.
Materials and Methods
CO2 treatment for cell lines and flies
Exposures to air (“normocapnia:” 0.039% CO2, 21% O2, 78% N2) and elevated CO2 were carried out in BioSpherix C-Chambers (BioSpherix Ltd.) fitted with ProCO2 regulators and CO2 and O2 sensors. 100% CO2 was injected into the chambers to raise the ambient CO2 levels to either 5% CO2 (“mild hypercapnia:” 5.0% CO2, 20% O2, 74% N2) or 13% CO2 (“hypercapnia:” 13.0% CO2, 18% O2, 68% N2). For fly experiments, humidity was maintained at ~60% using Drierite.
RNAi screening
Pilot screening was performed at Northwestern University's High Throughput Analysis Laboratory (NU HTA) using the S2* Diptericin-luciferase (Dipt-luc) cell line (gift from N. Silverman). The Dipt-luc reporter consists of 2.2kb of the Dipt promoter (23) driving firefly luciferase in the pGL3 vector (Promega) (24). A Renilla luciferase-Pol III (polIII-luc) reporter, in which a fragment of the promoter for the RNA polymerase 128 subunit drives Renilla luciferase (25) (gift of the Drosophila RNAi Screening Center [DRSC], Harvard Medical School, Boston, MA), was transfected into the Dipt-luc cell line using Effectene (Qiagen). Selected PCR amplicons were obtained from the DRSC and in vitro transcribed using the T7 MEGAScript Kit (#AMB1334, Applied Biosystems) into dsRNAs using DRSC protocols (26). Cells were maintained in 1 mg/mL G418 and 200 μg/mL hygromycin in Schneider's Insect Medium (Sigma) containing 10% FBS. All liquid handling was performed using a Matrix WellMate dispenser (Thermo Scientific). 500 cells per well in 384-well plates were bathed with pre-plated dsRNAs (~1-5 ug total dsRNA) for 3 days prior to CO2 exposure to ensure effective gene knockdown. 19 h prior to CO2 exposure cells were primed with 1μM 20-hydroxyecdysone (Sigma) to improve immune-responsiveness. Cells were exposed to 13% CO2 for 10 h in media neutralized with 25mM NaOH to maintain pH at 7.1 as previously described (21). Five h after start of CO2 exposure, cells were challenged with 100 ng/mL E. coli peptidoglycan (PGN, E. coli 0111:B4, InVivogen) to induce Dipt. Firefly and Renilla luminescence were measured sequentially using Dual-Glo luciferase reagent (Promega) with an Analyst GT plate reader (Molecular Devices). Data were analyzed using DRSC software and Microsoft Excel. Primary RNAi screening was carried out in duplicate in 13% CO2; these experiments were performed on-site at the DRSC. The raw data from the primary screen are available from the DRSC under project ID 128 at http://www.flyrnai.org/cgi-bin/DRSC_screen_csv.pl?project_id=128 and the normalized and formatted data are shown in supplementary Table S2.
dsRNAs of interest were identified based on their Z-score, which represents the number of standard deviations the signal from one well on a plate is above or below the plate mean. Z-scores were calculated by normalizing the Dipt-luc signal from each well to the polIII-luc signal from the same well, calculating the average Dipt-luc/polIII-luc ratio on a plate, and determining the deviation of each well from the plate average.
Secondary screening was performed in 13% CO2 in duplicate wells of duplicate 96-well plates filled using multichannel pipettes. A Z-score was calculated for each dsRNA by averaging the Z-scores for the dsRNAs on each of the two independent plates. Tertiary screening was performed as per secondary screening, except that duplicate plates with two dsRNAs on a plate were each screened in air and 13% CO2.
Fly stocks and maintenance
Drosophila stocks were kept on cornmeal food at room temperature or at 25°C. Stocks were obtained from the Bloomington Stock Center unless otherwise specified. zfh2MS209 (27), zfh22-M390.R (28) and zfh21-M707.R (28) were backcrossed five times to w1118 prior to infection experiments (zfh21-M707.R and zfh22-M390.R were provided by S. Elgin). The w1118 stock used for backcrossing was used as a control for zfh2 mutant experiments. For RNAi knockdown of zfh2, CG-GAL4 (29) and C754-GAL4 (30) were used to express UAS-zfh213305 (31) in the fat body. Control crosses for knockdown experiments used the w1118 isogenic strain V60000 from which UAS-zfh213305 was derived.
Western blotting
Western blotting was performed using ~106 S2* cells/sample, 20 fly heads, 20 dissected male abdominal fat bodies, or 20 carcasses after head and fat body removal. Fat bodies were dissected using the protocol of Krupp and Levine (32). For S. aureus challenge, samples were collected 4 h after infection. Samples were run on 4% acrylamide gels and transferred to nitrocellulose membrane by electroblotting at 100V for 80 min. Membranes were blocked with 5% skim milk in TBST (20Mm Tris-Cl pH7.5, 150 mM NaCl, 0.1% Tween20), incubated overnight with 1:500 rat anti-Zfh2 sera #205 (33), then 45 min with 1:10,000 goat anti-rat IgG-HRP (sc-2065, Santa Cruz Biotechnology). Actin (as a loading control) was detected using 1:200 mouse monoclonal antibody JLA20 (DSHB Hybridoma Bank) and 1:10,000 goat anti-mouse IgG-HRP (170-6516, Bio-Rad Laboratories). Amersham ECL Prime Western Blotting Detection Reagent (RPN2232, GE Healthcare Life Sciences) was used for detection, and chemiluminescence of bands was quantitated with the Odyssey Fc imaging system (LI-COR Biosciences).
Fly infection assays
Fly infection and mortality tests were performed on adult male flies as described in Helenius et al. (21). Bacterial load assays for Fig. 4E were performed as described in Helenius et al. (21), and for Fig. 4G were determined 16-18 h post-infection by washing single flies with 75% ethanol, rinsing with LB media and then homogenizing in 200μL fresh LB media. Homogenates were centrifuged on a bench-top centrifuge for 3 min at 2000 rpm, and 100μL of the bacterial supernatant transferred to 1.9mL of fresh LB media and then shaken at 37°C, for 8 h at which time OD600 was determined.
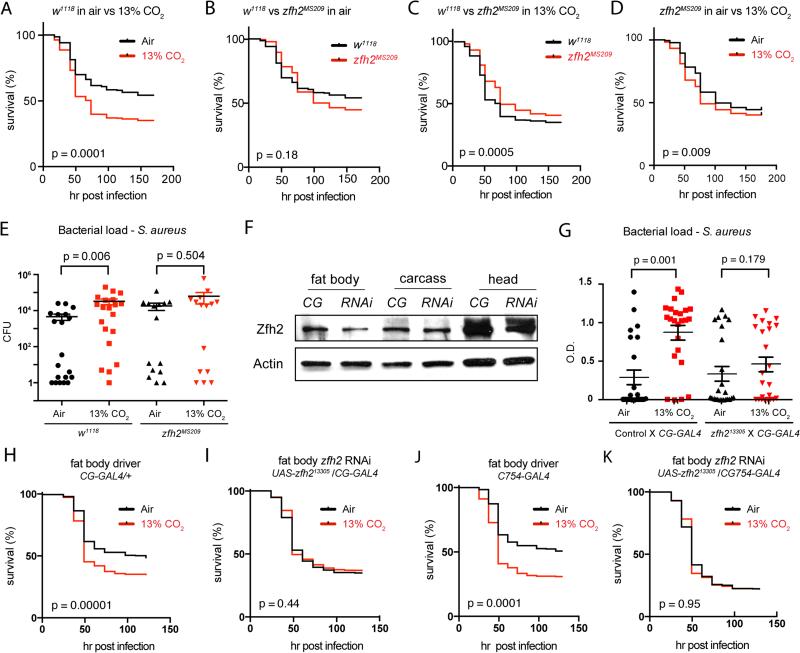
FIGURE 4. zfh2 mediates hypercapnic immune suppression in vivo.
(A) Control (w1118) flies exposed to hypercapnia (13% CO2) for 48 h, then inoculated with S. aureus, show reduced survival compared to S. aureus-infected flies exposed only to air (n= 636 for air, 622 for 13% CO2). For mortality experiments, p-values were calculated using the Gehan-Breslow-Wilcoxon test.
(B) In air, mortality of S. aureus-infection in zfh2MS209 mutant flies is not different than in w1118 control flies (n= 636 for w1118, 358 for w1118; zfh2MS209).
(C) When pre-exposed to 13% CO2, zfh2MS209 flies exhibit decreased mortality from S. aureus infection compared to w1118 control flies (n= 622 for w1118, 336 for w1118; zfh2MS209).
(D) zfh2MS209 flies are partially protected against the increase in mortality of S. aureus infection caused by exposure to elevated CO2, Genotype: w1118; zfh2MS209 (n=358 for air, 336 for 13% CO2). (Compare to w1118 control flies in Figure 4A.)
(E) Pre-exposure to 13% CO2 increases bacterial load in w1118 but not w1118; zfh2MS209 flies. Error bars show mean and SEM of the log10 load values (n= 20 for w1118 in air and 13% CO2, 16 for zfh2MS209 in air and CO2).
(F) Zfh2 protein levels are reduced in the fat body, but not the carcass or head, of flies with CG-GAL4 driven expression of the UAS-zfh213305 short hairpin RNAi construct (RNAi; genotype w1118;CG-GAL4/UAS-zfh213305). CG, control genotype w1118;CG-GAL4/+.
(G) Exposure to 13% CO2 for 48 h prior to infection increases bacterial load in control (w1118;CG-GAL4 /+) but not CG-GAL4/UAS-zfh213305 flies. Error bars show mean and SEM of the log10 load values (n= 24 for each condition).
(H) Exposure to 13% CO2 for 48 h prior to infection increases the mortality of S. aureus infection in control CG-GAL4 flies (w1118;CG-GAL4/+) (n= 428 for air, 465 for 13% CO2).
(I) Exposure to 13% CO2 for 48 h prior to infection does not increase mortality of S. aureus infection in flies in which zfh2 was knocked-down in the fat body (w1118;CG-GAL4/UAS - zfh213305) (n= 610 for air, 685 for 13% CO2).
(J,K) Results similar to those in H and I were obtained using another fat-body specific driver, C754-GAL4 (n= 314 for control-air; 357 for control-13% CO2, 459 for C754-GAL4/UAS-zfh213305-air; n = 488 for C754-GAL4/UAS-zfh213305-13% CO2).
Ex vivo fat body culture
For each experiment, three dissected fat bodies (see above) were placed in 1 ml of the S2* media (described above), and cultured in one well of a 24-well plate for 24 h in media equilibrated with air or 13% CO2. PGN treatment and media conditions were as for S2* cell induction (21). Total RNA was obtained from fat bodies using Trizol LS (#10296-028, Life Technologies). qPCR was performed per the manufacturer's protocol using the iScript™ cDNA Synthesis Kit (#170-8891, Bio-Rad Laboratories, Inc.), the iTaq™ Universal SYBR Green Supermix (#172-5124, Bio-Rad Laboratories, Inc.) and the Dipt primers 5’-ACCGCAGTACCCACTCAATC and 5’-ACTTTCCAGCTCGGTTCTGA. Primers for Att, Cec, Drs and Mtk were as previously described (21).
Statistical methods
Data are presented as means ± SEM. GraphPad Prism (version 5.04) and Sigmaplot (version 11.0) were used for statistical analysis. Differences between two groups were assessed using Student's t-test. Differences between multiple groups were assessed by ANOVA and the Tukey range test. For comparison of bacterial CFU data, the log10 values were used in the analysis. For mortality experiments, the Gehan-Breslow-Wilcoxon test was used. Significance was accepted at p<0.05. Symbols in figures: n.s., not significant (p>0.05); *, p<0.05, **, p<0.01; ***, p<0.001
Results
Diptericin-luciferase is a CO2-responsive reporter of innate immune responses
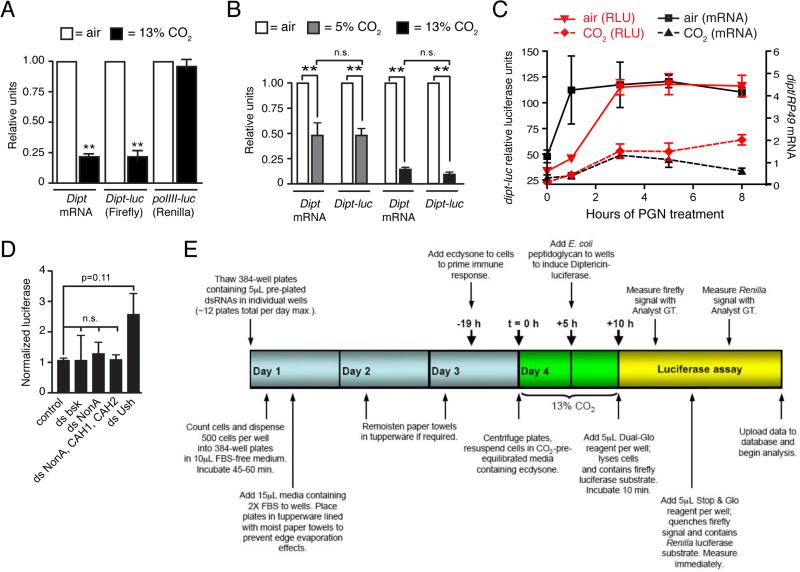
We had previously established Drosophila S2* cells as a model for investigating hypercapnic immune suppression by showing that E. coli PGN-stimulated induction of the mRNA for the antimicrobial peptide Diptericin (Dipt) was suppressed by hypercapnia in a concentration-dependent manner, without any cytotoxicity (21). In principle, this suppression could be the basis for a genome-wide screen to identify genes that mediate CO2-induced immune suppression. However, the previously used qPCR-based approach was poorly suited to high-throughput assays. We therefore tested whether the firefly Dipt-luc construct containing 2.2 kb of the Dipt promoter (24) would show the same CO2-induced suppression as the endogenous Dipt gene. We also tested whether expression of a Renilla luciferase driven by the promoter for the RNA polymerase III 128 subunit (polIII-luc, (25)) was independent of CO2 levels, which would enable it to be used as an internal control to account for treatment effects on cell growth and viability. In S2* cells stably transfected with the Dipt-luc and polIII-luc constructs, PGN-induced expression of the Dipt-luc reporter was suppressed ~5-fold in 13% CO2 compared to expression in air, whereas polIII-luc reporter expression was unaffected by elevated CO2 (Fig. 1A). Further testing established that the signal from the Dipt-luc construct also closely paralleled endogenous Dipt mRNA levels in mild hypercapnia (5% CO2 for a total of 10 h), sustained hypercapnia (13% CO2 for a total of 24 h), and over an 8-h time course following induction with PGN in air and 13% CO2 (Fig. 1B, 1C). Critically, signals from both Dipt-luc and polIII-luc were robustly detected from cells in 384-well plates. Thus, the Dipt-luc/polIII-luc combination appeared suitable for high-throughput screening.
FIGURE 1. A Diptericin-luciferase reporter construct enables a genome-wide screen for genes that mediate hypercapnic immune suppression.
(A) The Dipt-luc reporter containing 2.2 kb of the Dipt promoter region driving firefly luciferase (23) in S2* cells closely parallels expression of the endogenous Dipt locus in air and in neutral hypercapnia (13% CO2, pH 7.1: 5 h in 13% CO2, then 5 h PGN in 13% CO2). A reporter containing the promoter for the PolIII 128 subunit gene driving Renilla luciferase (25) is not responsive to CO2 and was used to calculate normalized activity of the Dipt-luc reporter. Dipt mRNA levels were assessed using qPCR normalized to RP49 mRNA levels.
(B, C) The Dipt-luc reporter closely parallels endogenous Dipt mRNA expression in PGN-stimulated S2* cells in (B) neutral mild hypercapnia (pH 7.1; 5 h in 5% CO2, then 5 h PGN in 5% CO2), in sustained neutral hypercapnia (13% CO2, pH 7.1: 19 h in 13% CO2, then 5 h PGN in 13% CO2) and (C) over an 8-h time course of PGN treatment (13% CO2, pH 7.1: 5 h pre-exposure in 13% CO2).
(D) dsRNAs targeting bsk (Drosophila JNK) or carbonic anhydrases, do not up-regulate Dipt-luc in elevated CO2 (results for additional candidate genes in pilot screening shown in Supplemental Table SI).
(E) Workflow for the genome-wide screen to identify genes that mediate hypercapnic immune suppression.
For all panels, **, p<0.01.
Hypercapnic immune suppression is not mediated by carbonic anhydrases or dJNK
Prior to performing a genome-wide RNAi screen, we conducted a pilot screen using S2* cells expressing Dipt-luc and polIII-luc at the Northwestern University's High Throughput Analysis Laboratory (HTA). We tested a panel of double-stranded RNAs (dsRNAs) targeting candidate genes that had previously been shown either to regulate AMP expression, to act in CO2-responsive pathways in other systems, or to interact biochemically with molecular CO2 and therefore plausibly mediate hypercapnic immune suppression (Supplemental Table SI). dsRNAs targeting the firefly and Renilla luciferases were used to confirm efficiency of RNAi knockdown. Knockdown of components of the Immune deficiency (Imd) pathway that regulates AMP expression either reduced (e.g. imd, rel) or increased (e.g. caspar) Dipt-luc expression as expected based on their known roles in regulation of Dipt expression, thus confirming the integrity of the assay (Supplemental Table S1). A dsRNA (DRSC Amplicon DRSC00843) targeting the gene u-shaped, a known regulator of AMP expression (34), typically up-regulated expression of the Dipt-luc reporter in both air and elevated CO2, and was used as a positive control in subsequent experiments.
One of the most interesting a priori candidate CO2 regulators was the c-Jun N-terminal kinase (JNK), an evolutionarily conserved modulator of the immune system that we had previously shown to be required for Na,K-ATPase endocytosis in response to elevated CO2 levels in both Drosophila S2 and mammalian lung epithelial cells (22). However, RNAi-mediated knockdown of Drosophila JNK (basket) did not mitigate hypercapnic suppression of Dipt-luc in S2* cells (Fig. 1D). Other candidates of particular interest were CO2-binding carbonic anhydrases. Flies have two conventional carbonic anhydrases, CAH1 and CAH2, and an ortholog of the non-canonical nuclear carbonic anhydrase nonO/p54, called nonA (35). However, RNAi knockdown of nonA alone or in combination with CAH1 and CAH2 did not abrogate the suppression of the Dipt-luc reporter by hypercapnia (Fig. 1D). Similar negative results were obtained with the other 34 candidate genes tested, including components of nitric oxide and hypoxia response pathways, components of the Imd pathway, and Gr63a/Gr21a that comprise the fly neuronal CO2 sensor (Supplemental Table SI). That none of the dsRNAs targeting candidate genes caused significant differential effects in air versus elevated CO2 suggests that hypercapnic immune suppression is mediated by novel mechanisms and underscores the need for discovery-based approaches to identify components of CO2 response pathways.
A genome-wide RNAi screen for components of CO2 response pathways
We next performed a genome-wide screen at the Harvard DRSC, using the DRSC 2.0 library that covered 13,900 of the ~14,000 annotated Drosophila genes (36), at an average of between 1 and 2 dsRNAs per gene. The dsRNA library was tested in duplicate in 384-well plates in 13% CO2 in media adjusted to pH 7.0 (see Fig. 1E for screen workflow, additional details in Materials and Methods). The primary genome-wide screen was performed only in 13% CO2. The full results of the screen have been deposited with the Harvard DRSC and are available at http://www.flyrnai.org/screensummary. As described below, top hits from the primary screen were further tested to identify those that differentially regulated Dipt-luc in elevated CO2 versus in air.
Genes of interest from the primary screen were identified as those whose knockdowns most increased Dipt-luc levels, after normalization with polIII-luc. This was quantified using the Z-score, which is the number of standard deviations that the normalized Dipt-luc level in a well treated with a given dsRNA is above or below the mean normalized Dipt-luc levels of all wells of the plate in which the dsRNA was arrayed (Materials and Methods). The primary screen identified a total of 126 dsRNAs targeting 123 genes that had an average Z-score from duplicate plates of ≥2.5 and 496 genes that had an average Z-score of ≥1.5 (Supplemental Table SII). The ability of the screen to identify biologically relevant regulators of immune function was confirmed by the identification of several genes previously described as negative regulators of the Imd/Dipt pathway, including falafel, kismet, cyclin D, enabled and Ras85D (37-39) (Supplemental Table SII). The robustness of the screen was further highlighted by the identification (Z-score ≤−1.5) of several known positive regulators of the Imd/Dipt pathway, including the peptidoglycan cell surface receptor PGRP-LC, Tak1 kinase, and the NF-κB transcription factor Relish that binds the Dipt promoter (Supplemental Table SII). The identification of known positive and negative regulators of the Imd/Dipt pathway, as well as genes far upstream (the PGRP-LC receptor) (40-42) and immediately regulating Dipt expression (the Rel transcription factor that drives Dipt expression) (43), suggested that the screen could also identify components of a CO2-responsive signaling pathway(s).
To begin assessing if the screen identified new pathways regulating Dipt expression, we analyzed the genes corresponding to the dsRNAs that had a Z-score of ≥1.5 in the primary screen for interactions using the STRING program (44). The resulting map contained one gene known to negatively regulate the Imd pathway (Ras85D) and showed connections between genes that positively regulate protein synthesis, but did not reveal an obvious new pathway that might mediate CO2-responses (data not shown). However, to our knowledge none of the data on the relationships between genes/proteins used by the STRING program has been generated under hypercapnia. Thus, interactions that occur specifically during conditions of elevated CO2 would not be expected to appear in the interaction map.
Functional classification, based on Gene Ontology (GO) biological processes, of the genes of interest identified in the primary screen revealed that the distribution of functions of genes with a Z-score of ≥+1.5 or ≤−1.5 from the screen is different than the fly genome as a whole (Supplementary Fig. 1). Further, the genes whose knockdown up-regulates the reporter fall into different functional groups than those whose knockdown down-regulates the reporter. The group of genes that negatively regulate Dipt expression in hypercapnia is enriched for genes that are involved in responding to external stimuli or are involved in movement, morphogenesis or differentiation, while the group of genes that positively regulate Dipt expression is enriched in spindle and centrosome functions (Supplementary Fig. 1). Notably, of the 126 primary hits, 51% have human orthologs based on analyses using HomoloGene (www.ncbi.nlm.nih.gov/homologene/) and InParanoid (45). Of those with orthologs, 25 have a Z-score >3 (Supplemental Table SII), suggesting the screen may have identified conserved genes that mediate the effects of hypercapnia on immune gene expression.
Secondary screening identifies 17 candidate mediators of CO2-induced immune suppression
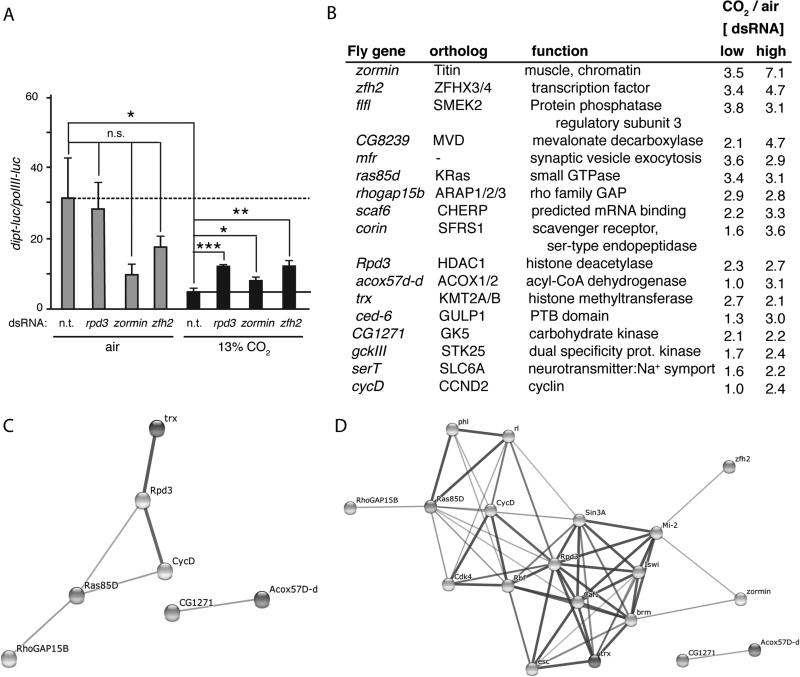
Based on strength of Z-score, presence of a human ortholog, and possible likelihood of functional significance as a CO2 pathway signal transducer (e.g. a kinase), 192 dsRNAs (representing 192 genes) were selected for further screening (Supplemental Table SII). Secondary screening was performed using the same protocols as the primary screen, except dsRNAs were assayed in 96-well plates in quadruplicate, rather than in duplicate (Materials and Methods). Of the 192 dsRNAs tested in secondary screening, 39 were found to up-regulate Dipt-luc by at least one standard deviation above the mean of all wells in at least one 96-well secondary screening plate (Supplemental Table SII). Thus, secondary screening delineated 39 dsRNAs that had the strongest effects in a head-to-head comparison of the 192 dsRNAs selected from the primary screen. These 39 dsRNAs were then further tested at two different RNA concentrations in 13% CO2 and ambient air to determine the CO2-specificity of the up-regulation of Dipt-luc (Fig. 2, Supplemental Table SIII). Candidate CO2-response pathway components were identified as those whose knockdown preferentially up-regulated Dipt-luc in hypercapnia but not in ambient air (Fig. 2). For 17 of the 39 candidate genes, induction of the Dipt-luc reporter was ≥2-fold higher in 13% CO2 than in air at one or both of the dsRNA concentrations tested (Fig. 2, Supplemental Table SIII). Thus, these 17 genes, 16 of which have human orthologs, were preferentially required for hypercapnia to suppress Dipt induction and were designated as candidate CO2-mediator genes.
FIGURE 2. Secondary screening identifies putative CO2-response mediator genes.
(A) Normalized Dipt-luc reporter expression in control S2* cells and in cells treated with dsRNAs targeting rpd3, zormin and zfh2. Dashed line, expression in control cells in air; solid line, expression in control cells in 13% CO2 in pH 7.1 media. n.t., control non-targeting dsRNA. Statistical significance determined using Student's t-test; *, p<0.05; **, p<0.01; ***, p<0.001; n.s., not significant p>0.05.
(B) The ratio of the expression of the normalized Dipt-luc reporter in CO2 versus air, for cells treated with dsRNAs at low and high concentrations. Shown are the 17 genes with the highest CO2/air ratios. The results for all 39 candidate genes tested in air and CO2 are shown in Supplemental Table SIII.
(C) An interaction network of genes in B predicted by the STRING program (44) using medium confidence settings. A subset of candidate CO2-mediators have previously identified interactions.
(D) An interaction network of genes in B predicted by the STRING program with extended connections that include known interactions that could relate zormin and zfh2 to other candidate CO2-mediators.
Putative functional categories of the 17 candidate CO2-mediators include a transcription factor (zfh2), chromatin associated proteins (zormin, flfl, rpd3, and trx), regulators of signal transduction (flfl, ras85D, rhogap15b), a cell surface protein (corin) and an RNA binding protein (scaf6). Interestingly, two of the candidate genes, flfl and ras85D (37), had previously been implicated in interacting with the Imd pathway, although they do not appear to act in linear order to control Imd signaling, and most proteins in the pathways these genes are associated with did not score as hits. GO and STRING analyses of the functions and interactions of these candidate mediators did not reveal an obvious CO2 sensor candidate or a common pathway or function among all of the genes. However, analysis of the top 17 candidate CO2-mediators using STRING revealed known connections between RhoGAP15B, Ras85D, Rpd3 and trx (Fig. 2C). Allowing the STRING program to search for potential connections by adding 10 nodes between the top 17 hits produced an interaction network connecting zormin and zfh2 to the Rpd3 group, however, only one of these new intervening nodes, brm, had a Z-score of ≤−1 in the primary screen. These results do not support the existence of a simple single linear pathway mediating the effects of CO2 on immune responses. Further characterization of the candidate CO2 mediators will be required to identify those that have instructive rather than permissive roles in responses to elevated CO2.
zfh2 encodes a large conserved transcription factor expressed in S2* cells and adult Drosophila
To begin validation of candidate effectors of CO2-induced immune suppression, we selected zinc finger homeodomain 2 (zfh2) for further analysis. zfh2 was a strong candidate because two separate dsRNAs targeting zfh2 in the primary screen had average Z-scores from duplicate plates in the top 10 of all dsRNAs (Amplicons DRSC17178 and DRSC28010, Supplemental Table SII), and further screening revealed robust up-regulation of the Dipt-luc reporter upon zfh2 knockdown in 13% CO2 (3.9- and 2-fold at low and high dsRNA concentrations, respectively) but not in air (1.1- and 0.4-fold) (Fig. 2A, Supplemental Table SIII). Further, as detailed below, zfh2 encodes a transcription factor, which we expect to have a more specific role in CO2 signal transduction than other candidate genes such as trx or rpd3 that encode chromatin-modifying proteins. Finally, previous work on zfh2 has generated important reagents, including mutant lines, UAS-RNAi lines, and antibodies.
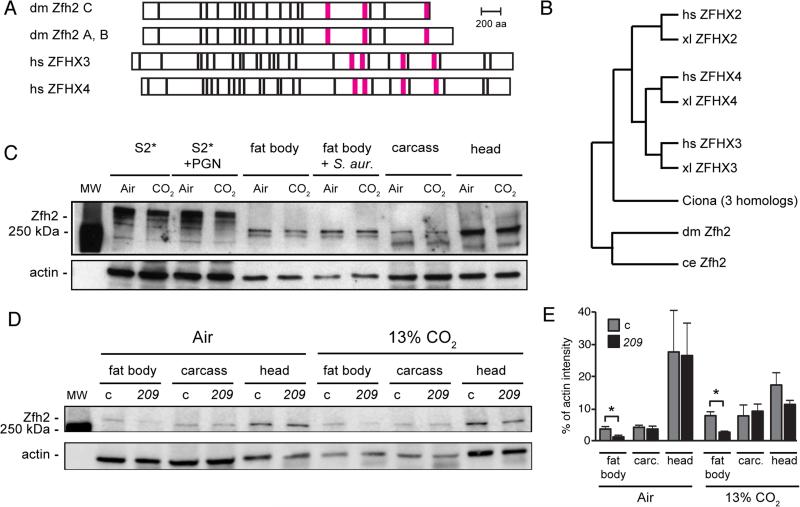
The zfh2 gene encodes three isoforms of a large, ~330 kDa protein containing 13 zinc fingers and 3 homeodomains. Two of the isoforms, Zfh2-PA and Zfh2-PB, are 3003 and 3005 aa long, respectively, and differ by only two amino acids. A third isoform, Zfh2-PC, is generated by an alternative splice near the 3’ end of the coding sequence that truncates the C-terminal 200 aa of the A and B isoforms, including part of the C-terminal homeodomain (Fig. 3A). The amplicons and UAS-RNAi constructs used in the RNAi screen and in vivo assays (see below) target all splice forms of zfh2. Zfh2 is part of a large family of zinc finger homeodomain proteins present in most metazoans (Fig. 3B and (46)), with the two clearest human orthologs of Zfh2 being ZFHX3 (also known as ATBF1, ATBT, ZNF927) (47-54) and ZFHX4 (55-57). Human ZFHX3 and ZFHX4 also encode large ~400 kDa proteins containing 23 zinc fingers and 4 homeodomains (Fig. 3A, Fig. 3B). Zfh2 is 25% and 24% identical to ZFHX3 and ZFHX4, respectively (58). The first three homeodomains of ZFHX3 share 77%, 69%, and 61% identity with the corresponding homeodomains of Zfh2, and several of the zinc finger motifs in ZFHX3 and Zfh2 share 52-71% identity (59). Notably, the 200 amino acids deleted in the Zfh2-PC isoform have 30% similarity to the C-termini of ZFHX3 and ZFHX4, suggesting that Zfh2-PC may have significant functional differences from Zfh2-PA and Zfh2–PB. A third potential human ortholog of Zfh2 is ZFHX2, but ZFHX2 appears to be further diverged from Zfh2 than either ZFHX3 or ZFHX4, sharing only 16% amino acid identity with Drosophila Zfh2 (58). Furthermore, using the DIOP ortholog search tool (60), ZFHX2 is identified as a Zfh2 ortholog by only 3 of 9 prediction programs (Homologene, Inparanoid, Isobase, OMA, OrthoDB, orthoMCL, Phylome, RoundUp, TreeFam), while ZFHX3 and ZFHX4 are identified by 7 and 6 databases, respectively. zfh2 has previously been shown to have an important role in neural and epithelial development and in regulating cell death (27, 61-65). ZFHX3 is involved in neural and epithelial development (47-50), implicated by GWAS in atrial fibrillation (51) and has roles in multiple types of cancer (52-54). Similarly, ZFHX4 is involved in epithelial and glial cancers (55, 56), and neuronal development (57). However, neither Zfh2 nor its human orthologs ZFHX3 and ZFHX4 had previously been shown to regulate immune gene transcription or gas sensing. Together, these observations suggest that Zfh2, ZFHX3 and ZFHX4 could define a family of conserved, but previously unidentified, proteins that mediate hypercapnic immune suppression.
FIGURE 3. zfh2 encodes a large conserved transcription factor expressed in immune tissues.
(A) Domain structure of Drosophila (dm) Zfh2 isoforms A, B and C, and human (hs) ZFHX3 and ZFHX4. Zfh2 isoforms A and B differ by only 2 aa and are shown together. Black bar, predicted zinc finger; magenta bar, predicted homeodomain.
(B) A phylogenetic tree showing the relationships between Drosophila (dm) Zfh2 and human (hs), xenopus (xl), Ciona and C. elegans (ce) zinc-finger homeodomain proteins. The tree was generated using Ensembl Release 81, June 2015 (76), which does not use branch length to represent evolutionary distance.
(C) Western blot using rat polyclonal anti-Zfh2 antiserum (33) shows that Zfh2 is expressed strongly in neural tissue (head), and at lower, but easily detectable, levels in the fat body and the S2* cell line. Zfh2 levels in S2* cells and all tissues appear similar following exposure to air or 13% CO2 and after immune challenge (PGN for S2* cells, S. aureus inoculation for adult flies; see Material and Methods). Fat body, fat body tissue dissected from the abdomens of adult flies (20/lane); carcass, remaining fly body after removal of the head and the abdominal fat body; actin, loading control.
(D) Western blot comparing Zfh2 expression in the fat bodies, carcasses and heads of control (c) and zfh2MS209 (209) mutant flies. Blot was performed using rat polyclonal anti-Zfh2 antiserum (33), with chemiluminescence detection imaged by an Odyssey Fc imaging system (LI-COR Biosciences). Shown blot is representative of triplicate experiments.
(E) Quantification of the bands from the Western blot in panel 3D and two replicates using the LI-COR Odyssey imaging system reveals that in zfh2MS209 flies (209), Zfh2 levels are reduced approximately two-fold in the fat body, but similar in the carcass and head, compared to control flies (c, w1118). carc., carcass; *, p<0.05.
To investigate a potential role for Zfh2 in immune regulation, we determined where Zfh2 was expressed and whether its levels were altered by immune stimulation or by hypercapnia. FlyBase listed Zfh2 as being enriched in the nervous system and not expressed significantly in other tissues or S2 cells (66). Western blotting (Fig. 3C) confirmed strong expression of Zfh2 in neural tissue (adult fly heads), and in addition revealed Zfh2 expression in both S2* cells and in the adult abdominal fat body, the major immune and metabolic organ in Drosophila. In adult flies, a predominant band of Zfh2 immunostaining at ~300 kDa is observed, however lower molecular weight bands are also present. In S2* cells, multiple higher molecular weight bands are observed that appear considerably larger than 300 kDa. The larger molecular weight bands may represent post-translationally modified Zfh2, since they are strongly reduced in RNAi and CRISPR experiments, but are not consistent with the predicted sizes of Zfh2 isoforms (Supplementary Fig. 1). PGN challenge of S2* cells and S. aureus infection of adult flies did not appear to alter Zfh2 protein levels or electrophoretic mobilities (Fig. 3C). Likewise, exposure to 13% CO2, as compared to air, did not dramatically alter Zfh2 protein levels in S2* cells or adult fly tissues (Fig. 3C, 3D, 3E).
Zfh2 is required for elevated CO2 levels to decrease resistance to bacterial infection in vivo
To determine if Zfh2 mediates hypercapnic immune suppression in vivo, we tested whether zfh2 mutant files were protected from the increase in mortality caused by elevated CO2 in an S. aureus infection assay (Fig. 4A and see Materials and Methods). We were unable to assess the impact of complete Zfh2 deficiency because flies homozygous for strong zfh2 mutations, such as zfh21-M707.R (27), do not survive to adulthood (data not shown). Trans-heterozygous combinations of zfh21-M707.R and weaker alleles, such as zfh2MS209 (27), are also not viable as adults (data not shown). However, we were able to analyze animals homozygous for the zfh2MS209 mutation, which is a hypomorphic allele resulting from a transposon insertion in the 3’ region of zfh2 (67). zfh2MS209 homozygotes have wing and leg developmental defects (67), but are viable for use in infection assays. zfh2MS209 mutant flies (w1118; zfh2MS209) and control flies (w1118) maintained in air experience approximately the same mortality after infection with S. aureus (Fig. 4B; red zfh2MS209 line above and below black control line, p=0.18). However when exposed to 13% CO2 for 48 h prior to infection, zfh2MS209 mutants exhibit significantly reduced mortality compared to w1118 controls (Fig. 4C; red zfh2MS209 line above black w1118 control line, p<0.0005). Note that flies were exposed to elevated CO2 before infection only, and placed in air after infection to avoid confounding effects of simultaneously exposing the pathogen as well as the host to hypercapnia. Consistent with the fact that zfh2MS209 is a partial loss-of-function mutation, Zfh2 levels in zfh2MS209 homozygotes are not different than control flies in the head and carcass, but are reduced approximately 2-fold in the fat body (Fig. 3D, 3E, 4F). zfh2MS209 mutants are not completely protected from the increased post-infection mortality caused by elevated CO2 (Fig. 4D, p=0.009), although the deleterious effect of hypercapnia is less than in control flies (compare Fig. 4A and 4D). Thus, mutation of zfh2 allows flies to better survive infection after exposure to hypercapnia.
We next investigated whether the improved survival of hypercapnia-exposed zfh2MS209 flies resulted from increased resistance (the ability to limit pathogen burden (68)), or increased tolerance (the ability to limit the health impact of a given pathogen burden (68)), of the bacterial infection. The zfh2MS209 mutation appears to increase resistance because hypercapnia leads to increased bacterial load in control animals, but not in zfh2MS209 homozygotes (Fig. 4E). Together, these results indicate that zfh2 is required for CO2 to suppress anti-bacterial host defense in vivo.
To confirm the above results and to identify tissues in which zfh2 acts to mediate CO2-induced immune suppression, we used the GAL4/UAS system (69) to express a zfh2-targeted shRNA construct (31) under the control of tissue-specific drivers. Consistent with the lethal phenotype of strong zfh2 mutations, da-GAL4-driven ubiquitous expression of the zfh2 RNAi construct UAS-zfh213305 caused lethality during embryonic and larval stages, and no pupa were observed (data not shown). However, embryonic, larval and pupal development were not overtly perturbed by using the CG-GAL4 or C754-GAL4 drivers to express UAS-zfh213305 in the fat body, the major AMP-producing organ in the fly. Importantly, CG-GAL4-driven knockdown of zfh2 by UAS-zfh213305 reduces Zfh2 protein levels in the fat body, but not the rest of the body or head (Fig. 4F), and provides protection against hypercapnic immune suppression (Fig. 4I). Whereas exposure to 13% CO2 increases mortality of S. aureus infection in CG-GAL4 control adults (Fig. 4H; red CO2 line below the black air line, p=0.0001 for CG-GAL4/CG-GAL4; p=0.03 for CG-GAL4/+, data not shown), hypercapnia does not increase the mortality of infection in zfh213305/CG-GAL4 adults (Fig. 4I, red CO2 line superimposed on black air line, p=0.44). Expression of UAS-zfh213305 using another fat body driver, C754-GAL4, also improved survival of infected flies exposed to elevated CO2 (Fig. 4J, 4K). Improved survival of adult flies with fat body-specific knockdown of zfh2 is due at least in part to increased resistance to infection because bacterial load in UAS-zfh213305/CG-GAL4 was not increased in flies exposed to 13% CO2 compared to air (Fig. 4G). Importantly, no difference in bacterial loads was observed between control flies and flies with reduced Zfh2 levels in the fat body exposed to air alone (Fig. 4G), indicating that Zfh2 is not a major regulator of immune responses in ambient air. Together, these results identify Zfh2 as the first in vivo mediator of hypercapnic immune suppression. Further, the data provide strong evidence that the immunosuppressive effects of hypercapnia in Drosophila are mediated by a specific genetic pathway that functions in the fat body.
Zfh2 influences a non-immunological function affected by hypercapnia
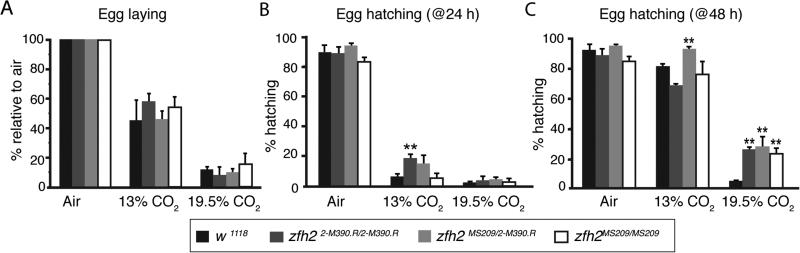
We next asked whether Zfh2 was a global mediator of hypercapnic responses, or if it acted specifically in the immune system. We assayed the ability of zfh2 mutations to prevent the hypercapnia-induced reductions in egg laying and delays in hatching that we had previously observed (21). The partial loss-of-function zfh2MS209 and zfh22-M390.R mutations do not significantly alter the suppression of egg laying by 13% or 19.5% CO2 (Fig. 5A), but can partially mitigate the delay in egg hatching by 24 h in 13% CO2 and by 48 h in 19.5% CO2 (Fig. 5B, 5C). These results provide evidence that zfh2 mediates additional deleterious effects of elevated CO2 on a process not directly related to immune function in tissues other than the fat body.
FIGURE 5. The partial loss-of-function mutations zfh2MS209 and zfh22-M390.R mitigate hypercapnia-induced reductions in egg hatching.
(A) Flies homozygous for the partial loss-of-function mutations zfh22-M390.R (dark grey) or zfh2MS209 (white), or the trans-heterozygous combination of zfh22-M390.R and zfh2MS209 (light grey), are not protected from the suppressive effect of 13% CO2 on egg laying seen in control animals (w1118, black). All zfh2 genotypes are w1118; zfh2. (n= 998 for w1118, 243 for zfh22-M390.R, 440 for zfh22-M390.R/MS209, 195 for zfh2MS209)
(B, C) Hypercapnic suppression of egg hatching is attenuated in zfh2 mutants flies. **, p<0.01 using Student's t-test compared to w1118 for each condition. (For B, n= 821 for w1118, 488 for zfh22-M390.R, 408 for zfh22-M390.R/MS209, 130 for zfh2MS209; for C, n= 845 for w1118, 682 for zfh22-M390.R, 350 for zfh22-M390.R/MS209, 207 for zfh2MS209)
Zfh2 mediates ex vivo hypercapnic suppression of multiple antimicrobial peptides
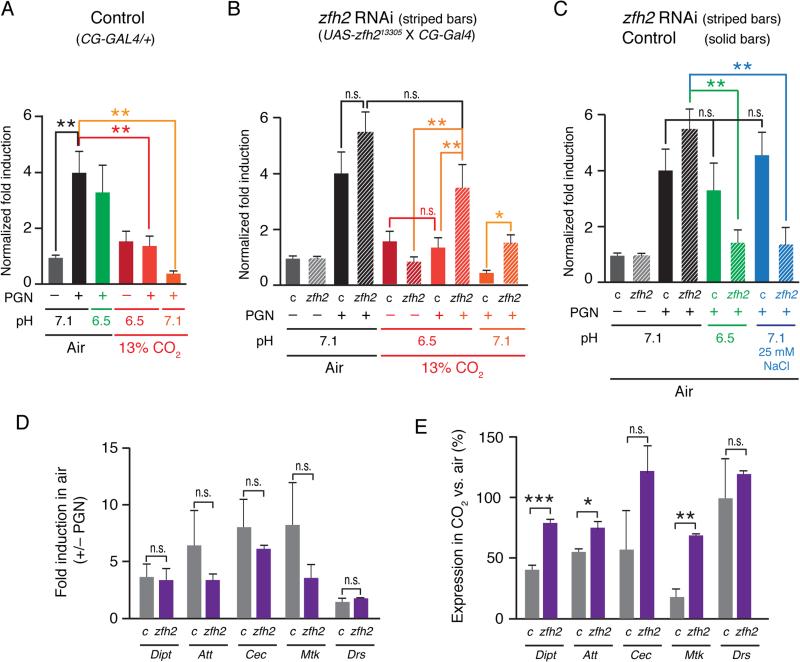
To determine whether increased expression of AMPs by the fat bodies of zfh2 knockdown flies could be at least partially responsible for the improved survival of zfh2 knockdown flies in elevated CO2, we developed an ex vivo system for culturing fat bodies (Materials and Methods). In this system, abdominal fat bodies are dissected from adult males and cultured in S2* media for 24 h in air or 13% CO2. PGN treatment and media conditions were the same as for S2* cell culture, using fat bodies from three flies per experimental condition. In this assay, PGN causes a 4-fold induction of endogenous Dipt mRNA levels that is not suppressed by culture in pH 6.5 media, but is suppressed 3-fold by culture in 13% CO2 (media pH 6.5), and 10-fold by culturing in 13% CO2 in pH-neutralized media (pH 7.1, Fig. 6A).
FIGURE 6. zfh2 mediates hypercapnic suppression of AMPs in adult fat bodies cultured ex vivo.
(A) Hypercapnia, but not normocapnic acidosis, suppresses Dipt induction in ex vivo cultured control fat bodies to a similar extent as the suppression of Dipt induction observed in S2* cells (21). Bars show Dipt mRNA levels (normalized to RP49 mRNA) in fat bodies from control adults (w1118; CG-Gal4/+, solid bars), that have been cultured ex vivo in media equilibrated with air or 13% CO2, un-induced and PGN-induced, at pH 7.1 (media without additions for air, or media + 25 mM NaOH for 13% CO2) or pH 6.5 (media + 19 mM MOPS for air, or media without additions for 13% CO2).
(B) Fat body-specific knockdown of zfh2 does not significantly increase Dipt induction when fat bodies are cultured in media equilibrated with air, but does prevent or reduce hypercapnia-induced suppression of Dipt induction in fat bodies cultured in media equilibrated with 13% CO2 at pH 6.5 or pH 7.1. Panel B shows data from the same experiment as in A, and in addition shows results obtained using fat bodies from adults in which zfh2 had been knocked down using a UAS-shRNA construct and the fat body driver, CG-Gal4 (genotype: w1118; UAS-zfh213305/CG-Gal4, stripped bars).
(C) Induction of Dipt is suppressed in zfh2 knockdown, but not control, fat bodies cultured in media equilibrated with air at pH 6.5 or supplemented with 25 mM NaCl.
(D) zfh2 knockdown (purple bars) does not alter induction of Dipt, Att, Cec or Mtk mRNAs in ex vivo–cultured fat bodies cultured in media equilibrated with air (grey bars – control (c), w1118; CG-Gal4/+; purple bars - zfh2 knockdown: w1118; UAS-zfh213305/CG-Gal4). n = 3.
(E) zfh2 knockdown attenuates hypercapnic suppression of PGN-induced mRNA for Dipt, Att and Mtk in ex vivo-cultured fat bodies. AMP mRNA levels in fat bodies cultured in 13% CO2 in neutral media (ph7.1) are expressed as a percentage of the corresponding mRNA level in fat bodies cultured in air. For example, Mtk was suppressed ~80% by hypercapnia in control fat bodies, but only ~25% in zfh2 knockdown fat bodies. Note that Drs, expression of which is not suppressed by 13% CO2, was not induced by PGN (panel 6D). Genotypes: same as 6D. n = 3. *, p<0.05; **, p<0.01; ***, p<0.001; n.s., p>0.05.
When cultured in air, Dipt induction by PGN was not different in zfh2 knockdown fat bodies than in control fat bodies (Fig. 6B). Notably, culture in 13% CO2 at pH 6.5 did not reduce induction of Dipt in zfh2 knockdown fat bodies (Fig. 6B), in marked contrast to results in control fat bodies (Fig. 6A). This finding corroborates the protective effect of zfh2 knockdown in the infection assays (Fig. 4I, 4K) and the CO2-specific increases in Dipt-luc expression observed in zfh2 RNAi-treated S2* cells (Fig. 2). Culture of zfh2 knockdown fat bodies in 13% CO2 in media buffered to pH 7.1 did reduce Dipt induction compared to culture in air, however, the reduction was significantly less than in control fat bodies, with zfh2 knockdown fat bodies inducing Dipt 3.6-fold more than control fat bodies under these culture conditions (Fig. 6B). Thus, Zfh2 mediates an immune suppressive effect of elevated CO2 on induction of Dipt transcription that is not dependent on extracellular acidosis.
To investigate whether Zfh2 regulates expression of AMPs other than Dipt, we determined the effects of zfh2 knockdown on Attacin (Att) and Cecropin (Cec) which can be induced by the Imd/Rel pathway (70), and Drosomycin (Drs), which is predominantly induced by the Toll pathway (71), and Metchnikowin (Mtk) which can be induced by both the Imd and Toll-family receptors (71). While knockdown of zfh2 had no effect on PGN-stimulated induction of these AMPs in fat bodies cultured ex vivo in air (Fig. 6D), zfh2 knockdown significantly attenuated hypercapnic suppression of Att and Mtk, in addition to Dipt (Fig. 6E) in fat bodies cultured in 13% CO2. Knockdown of zfh2 may have also attenuated hypercapnic suppression of Cec, however, Cec mRNA levels were more variable than those of the other AMPs, such that the increase in expression in CO2 relative to air did not reach statistical significance (Fig. 6E). Expression of Drs in elevated CO2 was not affected by zfh2 knockdown (Fig. 6E), but as Drs was not actually induced by PGN (Fig. 6D), this experiment does not address whether zfh2 modulates induction of genes by the Toll pathway. However, the results do indicate that Zfh2 does not control baseline (uninduced) levels of at least one Toll-regulated gene. Taken together, these findings demonstrate that zfh2 mediates hypercapnic suppression of multiple Imd/Rel-regulated immune genes in the fat body, and that zfh2's suppressive effect is specific to conditions of elevated CO2.
The ex vivo fat body experiments also provided insight into the detrimental effects of zfh2 knockdown in flies maintained in air, both in the absence and presence of infection. Flies in which zfh2 has been knocked down in the fat body take approximately four days longer to reach adulthood than control animals (data not shown), and mortality of zfh2 fat body knockdown flies after infection in air is increased compared to controls (Fig. 4J, 4K). This may be explained by the finding that fat bodies from zfh2 knockdown flies raised in air had smaller volumes than fat bodies from control flies. Also, induction of Dipt was reduced in zfh2 knockdown fat bodies compared to control fat bodies, when cultured in air at pH 6.5 or at pH 7.1 with hypertonic media (25 mM NaCl) (Fig. 6C). Given the delayed growth of the zfh2 knockdown flies and the adverse effects of culture in low pH or hypertonic media in air on Dipt induction in cultured fat bodies from these animals, it is striking that induction of Dipt in cultured zfh2 knockdown fat bodies is not suppressed by elevated CO2 and that zfh2 knockdown in adult flies prevents 13% CO2 from increasing the mortality of S. aureus infection. Together, these results show that Zfh2 has multiple functions in the adult fat body, one of which is to mediate the immunosuppressive effects of elevated CO2.
Discussion
Although several CO2 sensing pathways have been defined in neuronal tissues, it has largely been assumed that the non-neuronal physiological effects of CO2 are exerted indirectly, either by H+ and HCO3− or by neuronal signaling. However, a growing body of evidence indicates that multiple non-neuronal cell types respond to elevated CO2 levels under neutral pH conditions, and responses to CO2 can be distinguished from responses to acidosis or bicarbonate (reviewed in (72)). The molecular basis of these responses is a focus of current research, however, to date the mechanisms by which CO2 influences immune systems have not been elucidated. Here, we report a genome-wide RNAi screen that used Drosophila S2* cells to identify genes that mediate CO2-induced suppression of Diptericin (Dipt), an important antimicrobial peptide. We identified 16 genes with human orthologs whose knockdown could cause the PGN-induced levels of a Dipt reporter to increase ≥2-fold more in 13% CO2 than in air. In vivo analysis of one of these genes, which encodes the transcription factor Zfh2, demonstrates that reducing zfh2 function in adult fat bodies by RNAi does not alter PGN-induced expression of AMP mRNAs during culture in air, but increases expression of Dipt and several other AMPs when cultured in elevated CO2. Correspondingly, reducing zfh2 function in whole flies by mutation, or more specifically in the fat body by RNAi knockdown, improves the ability of adult flies to clear and survive infection with S. aureus.
Implications of Zfh2 as a CO2 mediator
The identification of Zfh2 as a CO2 mediator is important for guiding further investigation of hypercapnic immune suppression in Drosophila. In particular, the ability of fat body-directed knockdown of zfh2 to prevent hypercapnic immune suppression identifies a site of action for this CO2 response pathway. Since the fat body is the major immune organ of the fly, it is not surprising that it is an important target site of action for immunomodulatory effects of CO2, however, it was nonetheless critical to establish this fact experimentally. Moreover, when combined with conditional knockdown tools (31, 69), identification of a site of action enables the study of other putative CO2 response genes that have essential roles in embryonic development and thus are difficult or impossible to study in simple homozygous mutants.
Identification of Zfh2 is also a critical step in understanding the molecular mechanisms of hypercapnic immune suppression. zfh2 knockdown in vivo almost fully rescues resistance to bacterial infection during hypercapnia, thus identifying zfh2 as a major mediator of hypercapnic immune suppression. In S2* cells and in ex vivo fat bodies, knockdown of zfh2 can eliminate up to 50% of the effect of elevated CO2 on Dipt induction, suggesting that hypercapnia suppresses Dipt by acting on one or a small number of signaling pathways rather than by non-specific mechanisms affecting the activity of many targets. This hypothesis is bolstered by our finding that increases in Dipt expression resulting from zfh2 knockdown are specific for hypercapnia, and do not occur during culture in air or in acidified media without hypercapnia. zfh2 knockdown in fat bodies not only fails to mitigate the effects of non-hypercapnic acidosis, it makes fat bodies more sensitive to acidosis and less sensitive to elevated CO2 levels. An important implication of the suggestion that the immune suppressive effects of hypercapnia are mediated by one or a small number of pathways is that, in principle, it should be possible to therapeutically block the effects of elevated CO2 using small molecules.
Perhaps the most important aspect of the identification of Zfh2 as a mediator of hypercapnic immune suppression is that it may provide an entry point to understanding the molecular mechanism of hypercapnic immune suppression in humans. Zfh2 has established orthologs in mammals, and we have previously shown that there are strong parallels between hypercapnic immune suppression in Drosophila and mammals (15, 16, 21). While it is possible that Zfh2 acquired a role as a CO2 mediator after flies and mammals diverged, strong conservation of the innate immune pathways, other gas-sensing pathways, as well as most other signaling systems, suggests that mammalian ZFHX3 and/or ZFHX4 could also mediate immunomodulatory effects of CO2. In addition, given that zfh2 mutations partially blocked the effects of elevated CO2 on egg hatching, it is also possible that ZFHX3 and ZFHX4 could act outside the immune system to mediate non-immunological effects of elevated CO2, such as hypercapnia-induced muscle wasting (73).
Possible models for how Zfh2 acts
How does Zfh2 mediate hypercapnic immune suppression? Zfh2 has 13 zinc fingers and three homeodomains, and has been demonstrated to bind DNA. While it is therefore likely that Zfh2 acts as a transcription factor in transducing CO2 responses, zinc fingers can also mediate protein-protein interactions, raising the possibility that a non-transcriptional function of Zfh2 mediates CO2 responses. In either case, the genetic data are consistent with Zfh2 acting as a CO2-specific negative regulator of AMP induction, either by directly binding AMP promoters or by binding and inhibiting a positive regulators of AMP induction. At this point it is unclear if Zfh2 is the sole mediator of the immune-suppressive effects of CO2 because the lethality of Zfh2 mutants made it impossible to completely remove Zfh2 function. However, it seems likely that additional CO2-responsive factors may suppress innate immune function in flies because the limited role of Zfh2 in CO2-induced suppression of egg laying indicates other CO2-responsive pathways exist, and few immunological responses are regulated by a single pathway.
Zinc-finger homeodomain proteins: a family of immune regulators?
In addition to zfh2/ZFHX3/ZFHX4, the Drosophila and human genomes encode a second family of zinc-finger homeodomain containing proteins (46). Members of this family, which includes Drosophila zfh1 and human ZEB1 and ZEB2, are about 1/3 the size of the Zfh2/ZFHX3/ZFHX4 protein family (~1,100 aa) and each contains one homeodomain and 7 to 9 zinc-finger domains. Notably, despite the considerable evolutionary divergence between these families, both Zfh1 and ZEB1 have been shown to also modulate NF-κB-regulated innate immune responses (74). Like Zfh2, Zfh1 is a negative regulator of Dipt and other Imd-responsive AMPs including Att and Cec. Interestingly, epistasis experiments indicated that Zfh1 acts downstream of or in parallel to Rel, similar to our findings that elevated CO2, apparently via Zfh2, suppresses AMPs downstream of Rel proteolytic activation (21). However, a critical difference between Zfh1 and Zfh2 is that while knockdown of zfh1 in normocapnia increases expression of multiple AMPs, knockdown of zfh2 only increases AMP expression in hypercapnia. These findings raise the possibility that an ancestral zinc-finger homeodomain protein may have regulated immune responses, with the Zfh1/ZEB1/ZEB2 family evolving as general immune regulators, while Zfh2, and potentially other members of the Zfh2 family, evolved as CO2-responsive regulators of innate immune responses. This specialization also suggests that the ability to separately regulate immune responses in normocapnia and hypercapnia is biologically important.
Was an immunomodulatory CO2 sensor identified by the screen?
An interesting question is whether the screen identified a CO2 sensor that controls hypercapnic immune suppression. The variety in known CO2 sensing mechanisms, particularly the recent identification of a connexin subunit as a CO2 sensor by carbamylation (75), indicates that, in principle, almost any protein could act as a sensor, thus making it difficult to predict CO2 sensing ability based on known function or sequence. Thus, the screen may well have identified the sensor, but we are unable to recognize it as such at this time. While it is possible that Zfh2 is carbamylated in elevated CO2 levels and thus is both a sensor and effector, another likely scenario is that CO2 levels are sensed by an as yet unidentified upstream pathway component, leading to post-translational modification of Zfh2 that alters its capacity for DNA binding or protein-protein interactions, thereby altering transcription of AMPs and possibly other immune genes. Alternatively, secondary screening may not have identified the CO2 sensor because we focused on dsRNAs whose knockdown in air had minimal effects, but in hypercapnia prevented suppression of Dipt-luc. If the sensor, or any part of a hypercapnia response pathway, is essential for Imd pathway signaling in air, it would not have scored as a positive in the screen. Further analysis of the hits from this screen, and the use of confirmed CO2 mediators in assays of pathway function, will ultimately define the pathway of hypercapnic immune suppression and the CO2 sensor that regulates it.
Concluding remarks
The goal of this work was to identify genes that mediate immune suppressive effects of hypercapnia. Our in vitro screen identified 16 candidate Drosophila CO2 mediator genes that have human orthologs, one of which was tested for immunomodulatory effects in adult flies. These experiments defined Zfh2 as the first known mediator of hypercapnic immune suppression. The screen was also expected to identify components of the Imd pathway that regulates the Dipt product, and given that 12 previously-known positive and negative regulators of the Imd pathway were identified, it is likely that several of the candidate CO2-mediators will be in vivo mediators of hypercapnic immune suppression. Thus, this screen is an important starting point in the quest to fully define the components of the signaling pathway(s) that mediate hypercapnic immune suppression.
Supplementary Material
Acknowledgements
We thank the staff of the Northwestern High Throughput Analysis Laboratory and Harvard DRSC, in particular Sara Fernandez Dunne and Chi-Hao Luan (NU HTA) and Stephanie Mohr and Quentin Gilly (Harvard DRSC), for technical assistance. We also thank Emilia Lecuona and Neal Silverman for advice, Chris Doe for anti-Zfh2 antisera, and the Bloomington Drosophila Stock Center, the Transgenic RNAi Project at Harvard Medical School and Sarah Elgin for fly stocks.
Abbreviations used in this paper
- AMP
antimicrobial peptide
- COPD
chronic obstructive pulmonary disease
- Dipt
Diptericin
- Dipt-luc
firefly Diptericin-luciferase
- Att
Attacin
- Cec
Cecropin
- Drs
Drosomycin
- Mtk
Metchnikowin
- DRSC
Drosophila RNAi screening center
- dsRNA
double-stranded RNA
- NU HTA
Northwestern University High Throughput Analysis Laboratory
- PGN
peptidoglycan
- polIII-luc
Renilla polIII-luciferase
- qPCR
quantitative PCR
- SEM
standard error of the mean
Footnotes
Author contributions: ITH, RJH, PHSS and GJB designed research; ITH, RJH, YJK, JAH, TK and SMCM performed research; all authors contributed to data analysis; ITH, RJH, PHSS and GJB wrote the paper
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Minino AM. Death in the United States, 2011. NCHS Data Brief. 2013:1–8. [PubMed] [Google Scholar]
- 2.Sethi S, Murphy TF. Infection in the Pathogenesis and Course of Chronic Obstructive Pulmonary Disease. N. Engl. J. Med. 2008;359:2355–2365. doi: 10.1056/NEJMra0800353. [DOI] [PubMed] [Google Scholar]
- 3.Moser KM, Shibel EM, Beamon AJ. Acute respiratory failure in obstructive lung disease. Long-term survival after treatment in an intensive care unit. JAMA. 1973;225:705–707. [PubMed] [Google Scholar]
- 4.Martin TR, Lewis SW, Albert RK. The prognosis of patients with chronic obstructive pulmonary disease after hospitalization for acute respiratory failure. Chest. 1982;82:310–314. doi: 10.1378/chest.82.3.310. [DOI] [PubMed] [Google Scholar]
- 5.Goel A, Pinckney RG, Littenberg B. APACHE II Predicts Long-term Survival in COPD Patients Admitted to a General Medical Ward. J. Gen. Intern. Med. 2003;18:824–830. doi: 10.1046/j.1525-1497.2003.20615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Groenewegen KH, Schols AM, Wouters EF. Mortality and mortality-related factors after hospitalization for acute exacerbation of COPD. Chest. 2003;124:459–467. doi: 10.1378/chest.124.2.459. [DOI] [PubMed] [Google Scholar]
- 7.Kohnlein T, Windisch W, Kohler D, Drabik A, Geiseler J, Hartl S, Karg O, Laier-Groeneveld G, Nava S, Schonhofer B, Schucher B, Wegscheider K, Criee CP, Welte T. Non-invasive positive pressure ventilation for the treatment of severe stable chronic obstructive pulmonary disease: a prospective, multicentre, randomised, controlled clinical trial. Lancet Respir Med. 2014;2:698–705. doi: 10.1016/S2213-2600(14)70153-5. [DOI] [PubMed] [Google Scholar]
- 8.Wedzicha JA, Seemungal TA. COPD exacerbations: defining their cause and prevention. Lancet. 2007;370:786–796. doi: 10.1016/S0140-6736(07)61382-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bafadhel M, McKenna S, Terry S, Mistry V, Reid C, Haldar P, McCormick M, Haldar K, Kebadze T, Duvoix A, Lindblad K, Patel H, Rugman P, Dodson P, Jenkins M, Saunders M, Newbold P, Green RH, Venge P, Lomas DA, Barer MR, Johnston SL, Pavord ID, Brightling CE. Acute exacerbations of chronic obstructive pulmonary disease: identification of biologic clusters and their biomarkers. Am J Respir Crit Care Med. 2011;184:662–671. doi: 10.1164/rccm.201104-0597OC. [DOI] [PubMed] [Google Scholar]
- 10.Beasley V, Joshi PV, Singanayagam A, Molyneaux PL, Johnston SL, Mallia P. Lung microbiology and exacerbations in COPD. Int J Chron Obstruct Pulmon Dis. 2012;7:555–569. doi: 10.2147/COPD.S28286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sin DD, Man SF, Marrie TJ. Arterial carbon dioxide tension on admission as a marker of in-hospital mortality in community-acquired pneumonia. The American journal of medicine. 2005;118:145–150. doi: 10.1016/j.amjmed.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 12.Laserna E, Sibila O, Aguilar PR, Mortensen EM, Anzueto A, Blanquer JM, Sanz F, Rello J, Marcos PJ, Velez MI, Aziz N, Restrepo MI. Hypocapnia and hypercapnia are predictors for ICU admission and mortality in hospitalized patients with community-acquired pneumonia. Chest. 2012;142:1193–1199. doi: 10.1378/chest.12-0576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murtagh P, Giubergia V, Viale D, Bauer G, Pena HG. Lower respiratory infections by adenovirus in children. Clinical features and risk factors for bronchiolitis obliterans and mortality. Pediatr Pulmonol. 2009;44:450–456. doi: 10.1002/ppul.20984. [DOI] [PubMed] [Google Scholar]
- 14.Belkin RA, Henig NR, Singer LG, Chaparro C, Rubenstein RC, Xie SX, Yee JY, Kotloff RM, Lipson DA, Bunin GR. Risk factors for death of patients with cystic fibrosis awaiting lung transplantation. Am J Respir Crit Care Med. 2006;173:659–666. doi: 10.1164/rccm.200410-1369OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang N, Gates KL, Trejo H, Favoreto S, Jr., Schleimer RP, Sznajder JI, Beitel GJ, Sporn PH. Elevated CO2 selectively inhibits interleukin-6 and tumor necrosis factor expression and decreases phagocytosis in the macrophage. FASEB J. 2010;24:2178–2190. doi: 10.1096/fj.09-136895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gates KL, Howell HA, Nair A, Vohwinkel CU, Welch LC, Beitel GJ, Hauser AR, Sznajder JI, Sporn PH. Hypercapnia impairs lung neutrophil function and increases mortality in murine pseudomonas pneumonia. Am J Respir Cell Mol Biol. 2013;49:821–828. doi: 10.1165/rcmb.2012-0487OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casalino-Matsuda SM, Nair A, Beitel GJ, Gates KL, Sporn PH. Hypercapnia Inhibits Autophagy and Bacterial Killing in Human Macrophages by Increasing Expression of Bcl-2 and Bcl-xL. J Immunol. 2015;194:5388–5396. doi: 10.4049/jimmunol.1500150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cummins EP, Oliver KM, Lenihan CR, Fitzpatrick SF, Bruning U, Scholz CC, Slattery C, Leonard MO, McLoughlin P, Taylor CT. NF-kappaB links CO2 sensing to innate immunity and inflammation in mammalian cells. J Immunol. 2010;185:4439–4445. doi: 10.4049/jimmunol.1000701. [DOI] [PubMed] [Google Scholar]
- 19.Oliver KM, Lenihan CR, Bruning U, Cheong A, Laffey JG, McLoughlin P, Taylor CT, Cummins EP. Hypercapnia induces cleavage and nuclear localization of RelB protein, giving insight into CO2 sensing and signaling. J Biol Chem. 2012;287:14004–14011. doi: 10.1074/jbc.M112.347971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lardner A. The effects of extracellular pH on immune function. J Leukoc Biol. 2001;69:522–530. [PubMed] [Google Scholar]
- 21.Helenius IT, Krupinski T, Turnbull DW, Gruenbaum Y, Silverman N, Johnson EA, Sporn PHS, Sznajder JI, Beitel GJ. Elevated CO2 suppresses specific Drosophila innate immune responses and resistance to bacterial infection. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0905925106. Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vadasz I, Dada LA, Briva A, Helenius IT, Sharabi K, Welch LC, Kelly AM, Grzesik BA, Budinger GR, Liu J, Seeger W, Beitel GJ, Gruenbaum Y, Sznajder JI. Evolutionary conserved role of c-Jun-N-terminal kinase in CO2-induced epithelial dysfunction. PLoS One. 2012;7:e46696. doi: 10.1371/journal.pone.0046696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reichhart JM, Meister M, Dimarcq JL, Zachary D, Hoffmann D, Ruiz C, Richards G, Hoffmann JA. Insect immunity: developmental and inducible activity of the Drosophila diptericin promoter. EMBO J. 1992;11:1469–1477. doi: 10.1002/j.1460-2075.1992.tb05191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tauszig S, Jouanguy E, Hoffmann JA, Imler JL. Toll-related receptors and the control of antimicrobial peptide expression in Drosophila. Proc Natl Acad Sci U S A. 2000;97:10520–10525. doi: 10.1073/pnas.180130797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nybakken K, Vokes SA, Lin TY, McMahon AP, Perrimon N. A genome-wide RNA interference screen in Drosophila melanogaster cells for new components of the Hh signaling pathway. Nat Genet. 2005;37:1323–1332. doi: 10.1038/ng1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramadan N, Flockhart I, Booker M, Perrimon N, Mathey-Prevot B. Design and implementation of high-throughput RNAi screens in cultured Drosophila cells. Nat Protoc. 2007;2:2245–2264. doi: 10.1038/nprot.2007.250. [DOI] [PubMed] [Google Scholar]
- 27.Whitworth AJ, Russell S. Temporally dynamic response to Wingless directs the sequential elaboration of the proximodistal axis of the Drosophila wing. Dev Biol. 2003;254:277–288. doi: 10.1016/s0012-1606(02)00036-2. [DOI] [PubMed] [Google Scholar]
- 28.Sun FL, Cuaycong MH, Craig CA, Wallrath LL, Locke J, Elgin SC. The fourth chromosome of Drosophila melanogaster: interspersed euchromatic and heterochromatic domains. Proc Natl Acad Sci U S A. 2000;97:5340–5345. doi: 10.1073/pnas.090530797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hennig KM, Colombani J, Neufeld TP. TOR coordinates bulk and targeted endocytosis in the Drosophila melanogaster fat body to regulate cell growth. J Cell Biol. 2006;173:963–974. doi: 10.1083/jcb.200511140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hrdlicka L, Gibson M, Kiger A, Micchelli C, Schober M, Schock F, Perrimon N. Analysis of twenty-four Gal4 lines in Drosophila melanogaster. Genesis. 2002;34:51–57. doi: 10.1002/gene.10125. [DOI] [PubMed] [Google Scholar]
- 31.Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, Shim HS, Tao R, Handler D, Karpowicz P, Binari R, Booker M, Brennecke J, Perkins LA, Hannon GJ, Perrimon N. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods. 2011;8:405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krupp JJ, Levine JD. Dissection of oenocytes from adult Drosophila melanogaster. J Vis Exp. 2010 doi: 10.3791/2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tran KD, Miller MR, Doe CQ. Recombineering Hunchback identifies two conserved domains required to maintain neuroblast competence and specify early-born neuronal identity. Development. 2010;137:1421–1430. doi: 10.1242/dev.048678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valanne S, Myllymaki H, Kallio J, Schmid MR, Kleino A, Murumagi A, Airaksinen L, Kotipelto T, Kaustio M, Ulvila J, Esfahani SS, Engstrom Y, Silvennoinen O, Hultmark D, Parikka M, Ramet M. Genome-wide RNA interference in Drosophila cells identifies G protein-coupled receptor kinase 2 as a conserved regulator of NF-kappaB signaling. J Immunol. 184:6188–6198. doi: 10.4049/jimmunol.1000261. [DOI] [PubMed] [Google Scholar]
- 35.Karhumaa P, Parkkila S, Waheed A, Parkkila AK, Kaunisto K, Tucker PW, Huang CJ, Sly WS, Rajaniemi H. Nuclear NonO/p54(nrb) protein is a nonclassical carbonic anhydrase. J Biol Chem. 2000;275:16044–16049. doi: 10.1074/jbc.275.21.16044. [DOI] [PubMed] [Google Scholar]
- 36.Misra S, Crosby MA, Mungall CJ, Matthews BB, Campbell KS, Hradecky P, Huang Y, Kaminker JS, Millburn GH, Prochnik SE, Smith CD, Tupy JL, Whitfied EJ, Bayraktaroglu L, Berman BP, Bettencourt BR, Celniker SE, de Grey AD, Drysdale RA, Harris NL, Richter J, Russo S, Schroeder AJ, Shu SQ, Stapleton M, Yamada C, Ashburner M, Gelbart WM, Rubin GM, Lewis SE. Annotation of the Drosophila melanogaster euchromatic genome: a systematic review. Genome Biol. 2002;3:RESEARCH0083. doi: 10.1186/gb-2002-3-12-research0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Foley E, O'Farrell PH. Functional dissection of an innate immune response by a genome-wide RNAi screen. PLoS Biol. 2004;2:E203. doi: 10.1371/journal.pbio.0020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleino A, Valanne S, Ulvila J, Kallio J, Myllymaki H, Enwald H, Stoven S, Poidevin M, Ueda R, Hultmark D, Lemaitre B, Ramet M. Inhibitor of apoptosis 2 and TAK1-binding protein are components of the Drosophila Imd pathway. EMBO J. 2005;24:3423–3434. doi: 10.1038/sj.emboj.7600807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bond D, Foley E. A quantitative RNAi screen for JNK modifiers identifies Pvr as a novel regulator of Drosophila immune signaling. PLoS Pathog. 2009;5:e1000655. doi: 10.1371/journal.ppat.1000655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature. 2002;416:644–648. doi: 10.1038/nature735. [DOI] [PubMed] [Google Scholar]
- 41.Choe KM, Werner T, Stoven S, Hultmark D, Anderson KV. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science. 2002;296:359–362. doi: 10.1126/science.1070216. [DOI] [PubMed] [Google Scholar]
- 42.Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, Ferrandon D, Royet J. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature. 2002;416:640–644. doi: 10.1038/nature734. [DOI] [PubMed] [Google Scholar]
- 43.Hedengren M, Asling B, Dushay MS, Ando I, Ekengren S, Wihlborg M, Hultmark D. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol Cell. 1999;4:827–837. doi: 10.1016/s1097-2765(00)80392-5. [DOI] [PubMed] [Google Scholar]
- 44.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Remm M, Storm CE, Sonnhammer EL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 46.Seetharam A, Bai Y, Stuart GW. A survey of well conserved families of C2H2 zinc-finger genes in Daphnia. BMC Genomics. 2010;11:276. doi: 10.1186/1471-2164-11-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jung CG, Kim HJ, Kawaguchi M, Khanna KK, Hida H, Asai K, Nishino H, Miura Y. Homeotic factor ATBF1 induces the cell cycle arrest associated with neuronal differentiation. Development. 2005;132:5137–5145. doi: 10.1242/dev.02098. [DOI] [PubMed] [Google Scholar]
- 48.Ishii Y, Kawaguchi M, Takagawa K, Oya T, Nogami S, Tamura A, Miura Y, Ido A, Sakata N, Hashimoto-Tamaoki T, Kimura T, Saito T, Tamaoki T, Sasahara M. ATBF1-A protein, but not ATBF1-B, is preferentially expressed in developing rat brain. J Comp Neurol. 2003;465:57–71. doi: 10.1002/cne.10807. [DOI] [PubMed] [Google Scholar]
- 49.Miura Y, Tam T, Ido A, Morinaga T, Miki T, Hashimoto T, Tamaoki T. Cloning and characterization of an ATBF1 isoform that expresses in a neuronal differentiation-dependent manner. J Biol Chem. 1995;270:26840–26848. doi: 10.1074/jbc.270.45.26840. [DOI] [PubMed] [Google Scholar]
- 50.Li M, Fu X, Ma G, Sun X, Dong X, Nagy T, Xing C, Li J, Dong JT. Atbf1 regulates pubertal mammary gland development likely by inhibiting the pro proliferative function of estrogen-ER signaling. PLoS One. 2012;7:e51283. doi: 10.1371/journal.pone.0051283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou M, Liao Y, Tu X. The role of transcription factors in atrial fibrillation. J Thorac Dis. 2015;7:152–158. doi: 10.3978/j.issn.2072-1439.2015.01.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mori Y, Kataoka H, Miura Y, Kawaguchi M, Kubota E, Ogasawara N, Oshima T, Tanida S, Sasaki M, Ohara H, Mizoshita T, Tatematsu M, Asai K, Joh T. Subcellular localization of ATBF1 regulates MUC5AC transcription in gastric cancer. Int J Cancer. 2007;121:241–247. doi: 10.1002/ijc.22654. [DOI] [PubMed] [Google Scholar]
- 53.Sun X, Fu X, Li J, Xing C, Frierson HF, Wu H, Ding X, Ju T, Cummings RD, Dong JT. Deletion of atbf1/zfhx3 in mouse prostate causes neoplastic lesions, likely by attenuation of membrane and secretory proteins and multiple signaling pathways. Neoplasia. 2014;16:377–389. doi: 10.1016/j.neo.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen DB, Yang HJ. Comparison of gene regulatory networks of benign and malignant breast cancer samples with normal samples. Genet Mol Res. 2014;13:9453–9462. doi: 10.4238/2014.November.11.10. [DOI] [PubMed] [Google Scholar]
- 55.Stacey SN, Helgason H, Gudjonsson SA, Thorleifsson G, Zink F, Sigurdsson A, Kehr B, Gudmundsson J, Sulem P, Sigurgeirsson B, Benediktsdottir KR, Thorisdottir K, Ragnarsson R, Fuentelsaz V, Corredera C, Gilaberte Y, Grasa M, Planelles D, Sanmartin O, Rudnai P, Gurzau E, Koppova K, Nexo BA, Tjonneland A, Overvad K, Jonasson JG, Tryggvadottir L, Johannsdottir H, Kristinsdottir AM, Stefansson H, Masson G, Magnusson OT, Halldorsson BV, Kong A, Rafnar T, Thorsteinsdottir U, Vogel U, Kumar R, Nagore E, Mayordomo JI, Gudbjartsson DF, Olafsson JH, Stefansson K. New basal cell carcinoma susceptibility loci. Nat Commun. 2015;6:6825. doi: 10.1038/ncomms7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chudnovsky Y, Kim D, Zheng S, Whyte WA, Bansal M, Bray MA, Gopal S, Theisen MA, Bilodeau S, Thiru P, Muffat J, Yilmaz OH, Mitalipova M, Woolard K, Lee J, Nishimura R, Sakata N, Fine HA, Carpenter AE, Silver SJ, Verhaak RG, Califano A, Young RA, Ligon KL, Mellinghoff IK, Root DE, Sabatini DM, Hahn WC, Chheda MG. ZFHX4 interacts with the NuRD core member CHD4 and regulates the glioblastoma tumor-initiating cell state. Cell Rep. 2014;6:313–324. doi: 10.1016/j.celrep.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hemmi K, Ma D, Miura Y, Kawaguchi M, Sasahara M, Hashimoto-Tamaoki T, Tamaoki T, Sakata N, Tsuchiya K. A homeodomain-zinc finger protein, ZFHX4, is expressed in neuronal differentiation manner and suppressed in muscle differentiation manner. Biol Pharm Bull. 2006;29:1830–1835. doi: 10.1248/bpb.29.1830. [DOI] [PubMed] [Google Scholar]
- 58.Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, Gil L, Giron CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Johnson N, Juettemann T, Kahari AK, Keenan S, Martin FJ, Maurel T, McLaren W, Murphy DN, Nag R, Overduin B, Parker A, Patricio M, Perry E, Pignatelli M, Riat HS, Sheppard D, Taylor K, Thormann A, Vullo A, Wilder SP, Zadissa A, Aken BL, Birney E, Harrow J, Kinsella R, Muffato M, Ruffier M, Searle SM, Spudich G, Trevanion SJ, Yates A, Zerbino DR, Flicek P. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hashimoto T, Nakano Y, Morinaga T, Tamaoki T. A new family of homeobox genes encoding multiple homeodomain and zinc finger motifs. Mech Dev. 1992;39:125–126. doi: 10.1016/0925-4773(92)90031-e. [DOI] [PubMed] [Google Scholar]
- 60.Hu Y, Flockhart I, Vinayagam A, Bergwitz C, Berger B, Perrimon N, Mohr SE. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics. 2011;12:357. doi: 10.1186/1471-2105-12-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gabilondo H, Losada-Perez M, del Saz D, Molina I, Leon Y, Canal I, Torroja L, Benito-Sipos J. A targeted genetic screen identifies crucial players in the specification of the Drosophila abdominal Capaergic neurons. Mech Dev. 2011;128:208–221. doi: 10.1016/j.mod.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 62.Perea D, Molohon K, Edwards K, Diaz-Benjumea FJ. Multiple roles of the gene zinc finger homeodomain-2 in the development of the Drosophila wing. Mech Dev. 2013;130:467–481. doi: 10.1016/j.mod.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 63.Terriente J, Perea D, Suzanne M, Diaz-Benjumea FJ. The Drosophila gene zfh2 is required to establish proximal-distal domains in the wing disc. Dev Biol. 2008;320:102–112. doi: 10.1016/j.ydbio.2008.04.028. [DOI] [PubMed] [Google Scholar]
- 64.Lundell MJ, Hirsh J. The zfh-2 gene product is a potential regulator of neuron-specific dopa decarboxylase gene expression in Drosophila. Dev Biol. 1992;154:84–94. doi: 10.1016/0012-1606(92)90050-q. [DOI] [PubMed] [Google Scholar]
- 65.Guarner A, Manjon C, Edwards K, Steller H, Suzanne M, Sanchez-Herrero E. The zinc finger homeodomain-2 gene of Drosophila controls Notch targets and regulates apoptosis in the tarsal segments. Dev Biol. 2014;385:350–365. doi: 10.1016/j.ydbio.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 66.dos Santos G, Schroeder AJ, Goodman JL, Strelets VB, Crosby MA, Thurmond J, Emmert DB, Gelbart WM, FlyBase C. FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res. 2015;43:D690–697. doi: 10.1093/nar/gku1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Capdevila J, Guerrero I. Targeted expression of the signaling molecule decapentaplegic induces pattern duplications and growth alterations in Drosophila wings. EMBO J. 1994;13:4459–4468. doi: 10.1002/j.1460-2075.1994.tb06768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schneider DS, Ayres JS. Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nat Rev Immunol. 2008;8:889–895. doi: 10.1038/nri2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 70.Tanji T, Hu X, Weber AN, Ip YT. Toll and IMD pathways synergistically activate an innate immune response in Drosophila melanogaster. Mol Cell Biol. 2007;27:4578–4588. doi: 10.1128/MCB.01814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Valanne S, Wang JH, Ramet M. The Drosophila Toll signaling pathway. J Immunol. 2011;186:649–656. doi: 10.4049/jimmunol.1002302. [DOI] [PubMed] [Google Scholar]
- 72.Cummins EP, Selfridge AC, Sporn PH, Sznajder JI, Taylor CT. Carbon dioxide-sensing in organisms and its implications for human disease. Cell Mol Life Sci. 2014;71:831–845. doi: 10.1007/s00018-013-1470-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jaitovich A, Angulo M, Lecuona E, Dada LA, Welch LC, Cheng Y, Gusarova G, Ceco E, Liu C, Shigemura M, Barreiro E, Patterson C, Nader GA, Sznajder JI. High CO2 levels cause skeletal muscle atrophy via AMP-activated kinase (AMPK), FoxO3a protein, and muscle-specific Ring finger protein 1 (MuRF1). J Biol Chem. 2015;290:9183–9194. doi: 10.1074/jbc.M114.625715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Myllymaki H, Ramet M. Transcription factor zfh1 downregulates Drosophila Imd pathway. Dev Comp Immunol. 2013;39:188–197. doi: 10.1016/j.dci.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 75.Meigh L, Greenhalgh SA, Rodgers TL, Cann MJ, Roper DI, Dale N. CO(2)directly modulates connexin 26 by formation of carbamate bridges between subunits. Elife. 2013;2:e01213. doi: 10.7554/eLife.01213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.