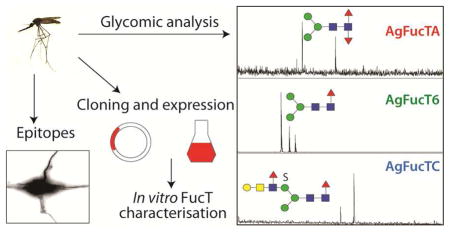
Abstract
Fucoconjugates are key mediators of protein-glycan interactions in prokaryotes and eukaryotes. As examples, N-glycans modified with the non-mammalian core α1,3-linked fucose have been detected in various organisms ranging from plants to insects and are immunogenic in mammals. The rabbit polyclonal antibody raised against plant horseradish peroxidase (anti-HRP) is able to recognize the α1,3-linked fucose epitope and is also known to specifically stain neural tissues in the fruit fly Drosophila melanogaster. In this study, we have detected and localized the anti-HRP cross-reactivity in another insect species, the malaria mosquito vector Anopheles gambiae. We were able to identify and structurally elucidate fucosylated N-glycans including core mono- and difucosylated structures (responsible for anti-HRP cross reactivity) as well as a Lewis-type antennal modification on mosquito anionic N-glycans by applying enzymatic and chemical treatments. The three mosquito fucosyltransferase open reading frames (FucT6, FucTA and FucTC) required for the in vivo biosynthesis of the fucosylated N-glycan epitopes were identified in the Anopheles gambiae genome, cloned and recombinantly expressed in Pichia pastoris. Using a robust MALDI-TOF MS approach, we characterised the activity of the three recombinant fucosyltransferases in vitro and demonstrate that they share similar enzymatic properties as compared to their homologues from D. melanogaster and Apis mellifera. Thus, not only do we confirm the neural reactivity of anti-HRP in a mosquito species, but also demonstrate enzymatic activity for all its α1,3- and α1,6-fucosyltransferase homologues, whose specificity matches the results of glycomic analyses.
Keywords: Anopheles gambiae, fucosyltransferases, anti-HRP, N-glycans, immunofluorescence, mass spectrometry
Graphical Abstract
1 INTRODUCTION
L-Fucose (6-deoxy-L-galactose) is a deoxyhexose and a common component of many glycolipids as well as of N- and O-linked glycoproteins occurring throughout nature. Fucose residues confer unique functional properties to oligosaccharides and mediate or regulate various biological processes such as cell adhesion, cell differentiation and cell growth. They are also involved in growth factor receptor modulation, microbial and viral infections, cancer and atherosclerosis (Staudacher et al., 1999, Becker and Lowe, 2003, Zhao et al., 2008). Among the most well-known fucosylated glycans, due to their biological and medical significance, are the ABO blood group antigens (Greenwell, 1997). Another fucosylated epitope of clinical relevance is the α1,3-linked fucose bound to the proximal N-acetylglucosamine of N-glycans (i.e. core α1,3 fucose). This non-mammalian epitope, found on plant and insect glycoproteins including pollen, food and venom allergens, is often a target of IgE in the sera of allergy patients (Paschinger et al., 2005a, Hoffmann-Sommergruber et al., 2011, Tretter et al., 1993). Furthermore, polyclonal antibodies raised against plant glycoproteins, especially horseradish peroxidase (HRP), have been extensively used to identify core α1,3-fucosylated and/or β1,2-xylosylated N-glycans in various non-vertebrate species. Anti-HRP was shown first some thirty years ago to specifically stain neural tissues in Drosophila melanogaster and grasshopper (Jan and Jan, 1982) and, to a lesser extent, in Caenorhabditis elegans (Siddiqui and Culotti, 1991). Since β1,2-xylose has not been found on N-glycans of either arthropods or nematodes, the major constituent of this cross-reactivity is due to the presence of core α1,3-linked fucose (Fabini et al., 2001). The fucosyltransferase responsible for the formation of this immunogenic epitope has been identified and characterised so far from several organisms including Arabidopsis thaliana, C. elegans, D. melanogaster, the honey bee A. mellifera and the silkworm Bombyx mori (Wilson et al., 2001, Paschinger et al., 2004, Fabini et al., 2001, Rendić et al., 2007, Minagawa et al., 2015). Analysis of honey bee venom revealed another fucosylated N-glycan epitope foreign to mammals, the Lewis-type fucosylated LacdiNAc [N-acetylgalactosaminyl-β1,4-N-acetylglucosamine (GalNAcβ1,4GlcNAc)] structure (Kubelka et al., 1993, Kubelka et al., 1995) also found in schistosome parasites (Smit et al., 2015).
Arthropods are not just model organisms, but a number are involved in transmission of diseases. The mosquito Anopheles gambiae is the most efficient vector for the malaria parasite Plasmodium falciparum in Sub-Saharan Africa (White et al., 2014). Two fundamental steps are required for the Plasmodium parasite development and transmission. Both the attachment of the parasite to the mosquito gut and salivary glands and of the Plasmodium sporozoites in the mammalian liver have been associated with the presence of chondroitin and heparin sulphate proteoglycans respectively (Dinglasan et al., 2007, Armistead et al., 2011, Pinzon-Ortiz et al., 2001).
In our recent study, we explored the N- and O-glycomes of An. gambiae larvae and revealed expected and novel epitopes including a large set of anionic glycans modified with sulphate, glucuronic acid or both which might represent additional glycan ligands, other than proteoglycans, involved in parasite-vector interactions (Kurz et al., 2015). Although, the detection of fucosylated N-glycans was somehow expected, considering that An. gambiae, like D. melanogaster, belongs to the order of the Diptera, we investigated whether anti-HRP IgG is localized predominantly to neural tissues in Anopheles larvae and adult mosquitoes by Western Blot analysis and immunofluorescence staining. By applying targeted chemical and enzymatic treatments, we were able to elucidate the mosquito N-glycan structures responsible for the in vivo anti-HRP cross-reactivity and also revealed antennae modified with the fucosylated LacdiNAc motif. Further, we identified three putative N-glycan processing fucosyltransferase open reading frames in the An. gambiae genome and describe their isolation, cloning and soluble expression in Pichia pastoris. Using a simple and robust MALDI-TOF MS based approach, we characterised the encoded enzymes which generate fucosylated glycans in vitro. Additionally, we demonstrate the activity and enzymatic properties of all three recombinant mosquito fucosyltransferases; these represent potential tools important for generating tailored epitopes suitable for glycan array analyses.
2 EXPERIMENTAL PROCEDURES
2.1 Biological material
Anopheles gambiae (Keele) were reared and maintained in an environmental chamber at 27 °C with a relative humidity ≥ 80% and a 12h light/dark cycle. Eggs were hatched in distilled water, transferred to plastic pans and larvae were fed on ground cat food (Purina cat chow). After 5 to 6 days of feeding, third (L3) or fourth (L4) instar larvae were either collected with a transfer pipette prior to centrifugation and storage at −80°C or allowed to pupate to collect adults. Adult female mosquitoes were fed on a 10% (w/v) sucrose solution ad libitum.
2.2 Mosquito dissections
L4 larvae and female adult An. gambiae (Keele) were cold-anesthetized prior to dissections. For subsequent Western Blot analysis, An. gambiae larvae (n=5) and adults (n=5) were partitioned into head, thorax and abdomen; tissues were collected into 1.5 mL microcentrifuge tubes containing 2x Laemmli sample buffer. For immunofluorescence staining experiments, adult An. gambiae were severed in the first and eighth abdominal segments, whereby the midgut was carefully removed at the cercus. The abdomens were further bisected in 1x phosphate buffered saline (PBS, pH 7.4; Gibco) along a coronal plane to obtain dorsal and ventral halves.
2.3 Western blotting
Collected larval and adult female An. gambiae tissues were homogenized in 100 μL 2x Laemmli sample buffer containing β-mercaptoethanol (Bio-Rad) using homogenization pestles (Bio-Rad) prior to boiling at 95°C for 10 min. Proteins were then separated by SDS-PAGE in a 4–20% polyacrylamide gel (Mini-PROTEAN TGX, Bio-Rad) and transferred to a nitrocellulose membrane for 2 hours at 100 V. Subsequently, the membrane was incubated in Odyssey blocking solution (LI-COR; mixed 1:1 with 1x PBS) overnight at 4°C prior to incubation with primary rabbit anti-HRP antibody (1:10000; Sigma-Aldrich) for 1 h at RT. After washing, the AlexaFluor 488 conjugated anti-rabbit IgG secondary antibody (1:20000; Life Technologies) was used for detection. The Western Blot was imaged using a LI-COR Odyssey near infrared imager (LI-COR) at an intensity setting between 3 to 5.
2.4 Immunofluorescence staining
Dissected adult An. gambiae abdomen halves were fixed in 4% (v/v) paraformaldehyde for 20 min at RT. Fixed abdomen halves were permeabilised by adding 0.5% (v/v) Triton-X 100 for 5 minutes and rinsed with PBS. Samples were blocked with 5% (v/v) foetal bovine serum (FBS; Gibco) prior to incubation with rabbit anti-HRP (1:500; Sigma-Aldrich) for 1 h at RT. After washing in PBS supplemented with 0.1% (v/v) Tween-20, the anti-rabbit DY-594 secondary antibody (1:400; Dyomics) was applied for one hour at RT. As a counterstain, samples were afterwards incubated with a mixture of Hoechst 33342 (1:1000; Invitrogen) and AlexaFluor 488 phalloidin (1:20; Life Technologies) for 10 minutes at RT. Dissected tissues were placed on glass slides and cover slips were mounted using Aqua Poly/Mount (Polysciences) and examined using a Nikon 90i microscope; images were analysed with Volocity 3D Image Analysis Software (Perkin-Elmer).
2.5 N-glycan release, purification and MALDI-TOF MS analysis
Preparation and subsequent analysis of L3/L4 instar An. gambiae larvae (Keele) N-glycans was performed as described recently (Kurz et al., 2015). Briefly, frozen larvae were boiled in deionised water before grinding and proteolysis using porcine pepsin (Sigma-Aldrich). After proteolysis, cation exchange and gel filtration chromatography, the N-glycans were released using PNGase F (Roche) followed by subsequent PNGase A (Roche) digestion of remaining glycopeptides to obtain core α1,3 fucosylated N-glycan structures, including core difucosylated N-glycans. PNGase F released N-glycans were then subjected to nonporous graphitized carbon (NPGC) chromatography for separation into neutral and anionic fractions. As required, neutral N-glycans were further purified by a second solid-phase extraction using a Lichroprep RP18 cartridge column (25–40 μm; Merck) and pyridylaminated. The fluorescently-labelled N-glycans were fractionated by HPLC using either an AscentisR Express RP-Amide column (150 × 4.6 mm, 2.7 μm; Supelco) with a gradient of 0.3% methanol per minute at a flow rate of 0.8 mL/min for neutral glycans or an IonPac AS11 column (HIAX, Dionex) for anionic glycans (Kurz et al., 2015). Collected HPLC fractions were lyophilized, reconstituted in water and analysed by MALDI-TOF MS in positive and negative ion modes (Bruker Autoflex Speed or Bruker UltrafleXtreme) with 6-aza-2-thiothymine (ATT) as matrix. Glycan spectra were initially processed using the manufacturer’s software (Bruker Daltonics FlexAnalysis 3.3.80) and then manually interpreted. Theoretical masses were calculated using the software GlycoWorkbench 2.0.
2.6 Chemical and exoglycosidase treatments of fucosylated N-glycans
Selected glycan fractions obtained from HPLC analysis of the recent study (Kurz et al., 2015) were subjected to chemical and enzymatic treatments. Thereby, hydrofluoric acid was used as it specifically releases α1,3-linked fucose residues (Haslam et al., 2000). Dried PA-labelled N-glycans were dissolved in 3 μL of 48% hydrofluoric acid (HF; Sigma-Aldrich) and incubated on ice in the cold room for 24 hours prior to repeated evaporation in the Speed Vac. Furthermore, glycans were incubated overnight with α1,6-specific fucosidase (from bovine kidney, Sigma-Aldrich) in 50 mM ammonium acetate, pH 5 at 37°C. Xanthomonas β1,3-specific galactosidase digestion (recombinant in E. coli from New England Biolabs, overnight at pH 5) with subsequent HF treatment for 24 hours was performed to elucidate the structural nature of the antennal modifications. All reaction mixtures were analysed without further purification by MALDI-TOF MS (Bruker Autoflex Speed) using 6-aza-2-thiothymine (ATT) as matrix.
2.7 Cloning of fucosyltransferase cDNAs
The full open reading frames of the FucTA α1,3-fucosyltransferase were first isolated by PCR using Expand polymerase (Roche; a mix of Tgo proof-reading DNA polymerase and Taq polymerase), An. gambiae cDNA (prepared by reverse transcription of midgut RNA) and the primers AgFucTA/1 (5′-gtctatgatgagactttcgaag-3′) and AgFucTA/2 (5′-cgcccctccacgctcatc-3′). The resulting full-length FucTA fragment from midgut cDNA was first ligated into pGEM-T (Promega) and then, based on sequencing, used to design new primers [AgFucTA/1/EcoRI (5′-cggaattcgtctatgatgagactttcgaa-3′) and AgFucTA/2/XbaI (5′-gctctagaccacgctcatcggctgtc-3′)]. The pGEM-T clone was then employed as a PCR template (using KOD proof-reading polymerase; Merck) in order to generate a partial FucTA reading frame which was digested with EcoRI and XbaI and ligated into pPICZαC3. As all FucTA clones were found to contain a missense mutation, inverse PCR with the 5′-phosphorylated primers AgFucTA_demut252_for, (5′-P-ctggaccgccacgtatagg-3′), and AgFucTA_demut252_rev, (5′-P-ttgaacacgtccggatacttc-3′), was performed on the vector using KOD polymerase in order to generate the final expression vector without the premature stop codon.
The partial and full open reading frames of FucTC were isolated by PCR using Expand polymerase, midgut cDNA and the respective primer pairs: AgFucTC/3/PstI (5′-aactgcaggacaagtgtctgctggcgac-3′) with AgFucTC/2/KpnI, (5′-ggggtaccaagatgcagctgaagcatgtg-3′) or AgFucTC/1/KpnI (5′-ggggtaccaagatgcagctgaagcatgtg-3′) with AgFucTC/2/XbaI (5′-gctctagaatcagaattcgatcttcgacct-3′). The full and partial reading frames of FucTC isolated using midgut cDNA were digested with the relevant enzymes and ligated into pIZTV5His or pPICZαC3 vectors. The partial FucT6 fragment from carcass cDNA was isolated by PCR using Expand polymerase and the primers AgFucT6/3/EcoRI (5′-cggaattctcgaagctgaacacgtcc-3′) and AgFucT6/2/XbaI (5′-gctctagaactacttcacgtacggatactgg-3′), before being cloned into pGEM-T; the resulting vector was cut with EcoRI and XbaI prior to ligation into pPICZαC3. For the pPICZαC3 vectors, the reading frames were also later cut prior to re-ligation into pPICZαHisFLAG (Nemčovičova et al., 2013).
2.8 Recombinant protein expression of An. gambiae fucosyltransferases
pPICZα vector constructs were linearised and used to transform Pichia pastoris GS115 competent cells followed by selection on YPDZeo plates [1% (w/v) yeast extract, 2% (w/v) peptone, 2% (w/v) agar, 2% (w/v) glucose, 100 μg/mL zeocin] plates. After selection, individual positive clones were cultivated either in 5 mL of glycerol-containing MGYC medium [1% (w/v) yeast extract, 2% (w/v) peptone, 1% (w/v) casamino acids, 1.34% (w/v) yeast nitrogen base, 4×10−5% (w/v) biotin and 1% (v/v) glycerol] or 20 mL YPD medium [1% (w/v) yeast extract, 2% (w/v) peptone, 2% (w/v) glucose]. Both pre-culture media were supplemented with 100 μg/mL of zeocin and cultivation was performed at 28°C, 200 rpm overnight. Cells were grown to OD600 ~3, collected by centrifugation at 4°C and 1500 × g, either washed once in 1.34% (w/v) yeast nitrogen base solution and resuspended in 50 mL methanol-containing MMYC medium [composition as for MGYC, except that 1% (v/v) methanol substitutes for glycerol] or directly resuspended in 200 mL YP medium containing 1% (v/v) methanol. Upon induction with methanol, cell cultures were grown at 25 °C, 200 rpm for 72 h and cultures were supplemented with additional 1% (v/v) methanol after 24 h and 48 h respectively.
2.9 Recombinant protein purification
After centrifugation at 4 °C (1500 × g, 30 min), Pichia supernatants were concentrated 25-fold using an Amicon ultrafiltration device (Millipore) with a 10 kDa NMWL (nominal molecular weight limit) filter, followed by buffer exchange to Ni-NTA column loading buffer (25 mM Tris-HCl, 300 mM NaCl, 7 mM imidazole, pH 8.5) or to Affi-Gel Blue column loading buffer (25 mM TRIS, pH 7.0). The concentrates were either mixed with pre-equilibrated 1 mL of Ni-NTA resin (Qiagen) in case of pPICZαHisFLAG constructs (AgFucT6 and AgFucTC) or with 6 mL of Affi-Gel Blue sepharose (Biorad) for the pPICZαFLAG construct (AgFucTA). After 1 h of incubation on a rotator at 4 °C, the protein-resin mixture was transferred to a column before washing and elution. For the Ni-NTA purification at room temperature, the mixture was loaded and, after washing with loading buffer (25 mM Tris-HCl, 300 mM NaCl, 7 mM imidazole, pH 8.5; six column volumes), the recombinant proteins were eluted in 500 mM imidazole (1/2 column volume per elution fraction) and collected into tubes kept on ice. For Affi-Gel Blue purification at 4 °C, the mixture was loaded and washed with 2 column volumes of loading buffer (25 mM TRIS, pH 7.0) while 2 mL fractions were collected. The recombinant enzyme (AgFucTA) was eluted in 3 column volumes of loading buffer containing a gradient of 0–3 M NaCl. Collected fractions were spectrophotometrically measured (A280) and protein-containing fractions were subjected to SDS-PAGE to assess purity. Pure fractions were pooled and imidazole was removed by exchanging into storage buffer (25 mM Tris-HCl, 150 mM NaCl, pH 7.0) using Vivaspin columns (30 kDa NMWL; Millipore). All three recombinant enzymes were aliquoted and kept at −20 °C for long-term storage; for characterisation, thawed portions were stable at 4 °C for at least one month.
2.10 MALDI-TOF MS based enzymatic activity assays
Standard enzymatic activity assays were performed using dabsyl-modified forms of GnGn/GalGal/βGNβGN glycopeptides as acceptor substrates. The dabsylated-GalGal [Galβ1,4-GlcNAcβ1,2-Manα1,6-(Galβ1,4-GlcNAcβ1,2-Manα1,3)Manβ1,4-GlcNAcβ1,4-GlcNAcβ1-Asn] glycopeptide, possessing the tetrapeptide sequence Gly-Glu-Asn-Arg (GENR), derived from Pronase E digestion of bovine fibrin, was remodelled to obtain dabsylated-GnGn and dabsylated-βGNβGN glycopeptides; the nomenclature (GnGn, GalGal, βGNβGN) is based on that of Schachter, as previously described (Mucha et al., 2004). The enzymatic activity assays were performed with 0.1 mM acceptor, 1 mM donor (GDP-Fuc), 10 mM MnCl2, 40 mM 2-morpholinoethanesulphonic acid (MES), pH 6.5 and 1 μL of crude P. pastoris supernatants or 0.5 μL of purified proteins in a final volume of 2.5 μL. All reactions have been supplemented with BSA to a final concentration of 400 μg/mL. Aliquots of the reaction were removed every hour up to 6 hours or overnight (generally at 25 °C) and analysed with an Autoflex Speed MALDI-TOF mass spectrometer (Bruker Daltonics) using α-cyano-4-hydroxycinnamic acid (ACH) as matrix.
To evaluate the generated fucose linkages, aliquots of fucosyltransferase assay mixtures were subject to chemical and enzymatic treatments as described above. Briefly, dabsylated products obtained from in vitro incubations were split into three parts, whereby one part remained untreated as control, the second part was treated with hydrofluoric acid and the third part incubated with bovine α1,6-fucosidase (Sigma-Aldrich) as described above for fucosylated N-glycans. All samples were then analysed without further purification by MALDI-TOF MS (Bruker Autoflex Speed) using α-cyano-4-hydroxycinnamic acid (ACH) as matrix.
2.11 Determination of the temperature optimum, pH optimum and ion susceptibility
In general, assays were performed within the linear range of the purified enzymes as described above in section 2.10 at 25 °C for 2 hours (AgFucT6 and AgFucTC) or 4 hours (AgFucTA). As acceptor substrates, dabsyl-modified GnGn glycopeptides were used for the putative core fucosyltransferases (AgFucTA and AgFucT6) and dabsyl-modified βGNβGN glycopeptides were used for the putative Lewis-type fucosyltransferase (AgFucTC). For determination of the temperature optimum, assays (n=4) were performed at different temperatures ranging from −20 °C to 70 °C in MES buffer pH 6.5. For the pH optima, MES buffers with a pH range from 5.0 to 7.0 and 2-amino-2-methyl-1,3-propanediol (AMPD) buffers with a range from 6.5 to 8.0 were used (n=2). For the determination of the ion susceptibility, assays (n=4) were supplemented with 10 mM divalent metal chlorides, 10 mM EDTA or without any divalent metal ions. All reaction mixtures were heat-inactivated for 5 min at 95 °C and analysed by an Autoflex Speed MALDI-TOF mass spectrometer using α-cyano-4-hydroxycinnamic acid (ACH) as matrix. The conversion into product ratio was calculated based on relative peak areas of acceptor substrate and product.
3 RESULTS
3.1 Detection and in vivo localisation of core α1,3 fucosylated N-glycan epitopes in An. gambiae
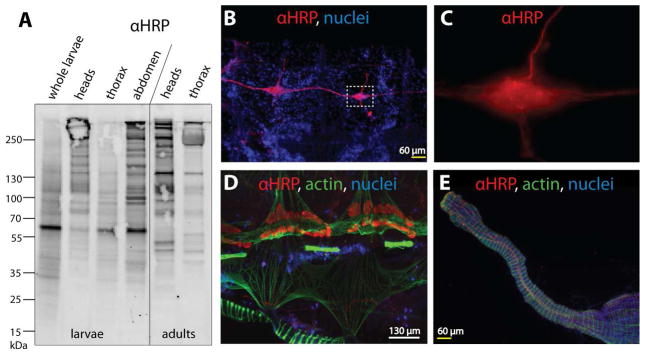
In previous studies, we have used anti-HRP (see also introduction) to specifically stain neural tissues in D. melanogaster embryos (Fabini et al., 2001, Rendic et al., 2010) and further demonstrated anti-HRP cross-reactivity with glycoproteins from other sources as diverse as nematodes, plants, amoeba and trichomonads by Western Blot analysis (Paschinger et al., 2004, Poltl et al., 2007, Schiller et al., 2009, Schiller et al., 2012, Paschinger et al., 2012). As an initial screen to elucidate the anti-HRP cross-reactivity in mosquitoes, we dissected An. gambiae larvae and female adults prior to tissue disruption and Western Blot analysis. Anti-HRP binding was observed with glycoproteins in the molecular range between 55 and 250 kDa throughout the Anopheles larval and adult tissues (Figure 1A). In all larval preparations, a major band of 65 kDa was observed, which was absent in the adult tissues. In general, anti-HRP cross-reactivity was significantly enriched in larval abdomens and adult heads suggesting changes in glycosylation of tissue-specific glycoproteins during development; this is compatible with previous findings in D. melanogaster, in which both neural-specific proteins and male reproductive tissue display anti-HRP reactivity (Wang et al., 1994).
Figure 1. In vivo detection of core α1,3-fucosylated N-glycan epitopes using anti-HRP antibody.
(A) Dissected tissues of Anopheles gambiae larvae and female adults as well as whole larvae were subject to Western Blot analysis using anti-HRP antibody (αHRP) which demonstrates a particular concentration of the α1,3-fucose epitope in the adult head; Coomassie staining (not shown) indicated comparable amounts of protein for the samples shown. (B–E) Female adult abdomens were further dissected into dorsal and ventral halves prior to immunofluorescence staining with anti-HRP. (B) Anti-HRP staining of the ventral nerve cord and ganglia from two abdominal segments; nuclei of surrounding fat body, muscle cells and haemocytes can be seen in the blue channel (Hoechst 33342). (C) High magnification image of inset from panel B, showing the increased magnitude and seemingly punctate anti-HRP reactivity in the concentrated nervous tissue of the ganglion. (D) Anti-HRP staining of pericardial cells (nephrocytes) along with muscular staining of the attached mosquito heart (at top) and some of the ventral abdominal muscles (at bottom); actin stained with phalloidin (in green). (E) Anti-HRP reactivity with nervous tissue attached to the mosquito midgut (muscles shown in green); in this instance anti-HRP reactivity was observed on a finer scale than seen in panels A–C.
To further elucidate neuron-specific binding in An. gambiae, we investigated the anti-HRP cross-reactivity by immunofluorescence staining of the abdomen dissected from adult, female Anopheles; the sections were also stained with Hoechst 33342 and phalloidin as appropriate. On the ventral half of the abdomen, anti-HRP cross-reactivity was associated with the ventral nerve cord (in red, Figure 1B); the magnified image (Figure 1C) shows seemingly punctate anti-HRP staining generally reminiscent of that observed in adult Drosophila for the nerve-tissue specific sodium pump Nervana2, although this has not been well characterized in adult tissues (Sun et al., 1999). In the dorsal half, the anopheline heart, a horizontally-extending phalloidin-stained tubular structure, is flanked by pericardial cells, which also demonstrated anti-HRP reactivity (Figure 1D); examination of the midgut showed very fine anti-HRP staining (Figure 1E). The observation of neural staining are in agreement with previous findings in D. melanogaster and grasshopper embryos (Jan and Jan, 1982).
3.2 An. gambiae larvae bear known and novel fucosylated N-glycan epitopes
In our recent study (Kurz et al., 2015), we examined the N-glycome of An. gambiae by using PNGase F and subsequently PNGase A (post PNGase F) to release N-glycans. Following enrichment, the pyridylamino (PA)-labelled N-glycans from the neutral and anionic pools were fractionated respectively using RP-Amide or HIAX columns, prior to MALDI-TOF MS. Using this approach, we were able to identify both novel and familiar N-glycan epitopes as compared to other glycomic studies in insects. A number of the glycans, not characterised in that study focussing on anionic structures (Kurz et al., 2015), were core monofucosylated and core difucosylated as indicated by diagnostic fragments of m/z 446 (single core fucose; inset in Figure 2C) and m/z 592 (core difucose; inset in Figure 2A) in MS/MS analysis. Core difucosylation was only detected on mosquito N-glycans released by PNGase A (post PNGase F) and indicate α1,3- and α1,6-fucosylation of the same reducing terminal GlcNAc residue, compatible with the anti-HRP reactivity observed in larvae. Using fused core RP-HPLC, we were able to separate N-glycans based on their isomeric status and compare the basic trends of the elution profiles in terms of glucose units (g.u.) to previous studies with both classical and fused core RP-HPLC columns (Tomiya et al., 1988, Tomiya et al., 1991)(Yan et al., 2015). A well-known example of isomeric separation by RP-HPLC is the ability to distinguish core α1,3- from core α1,6-fucosylated N-glycans (core α1,3- elute earlier than core α1,6-fucosylated structures respectively; see Kurz et al., 2015 for the RP-HPLC chromatograms).
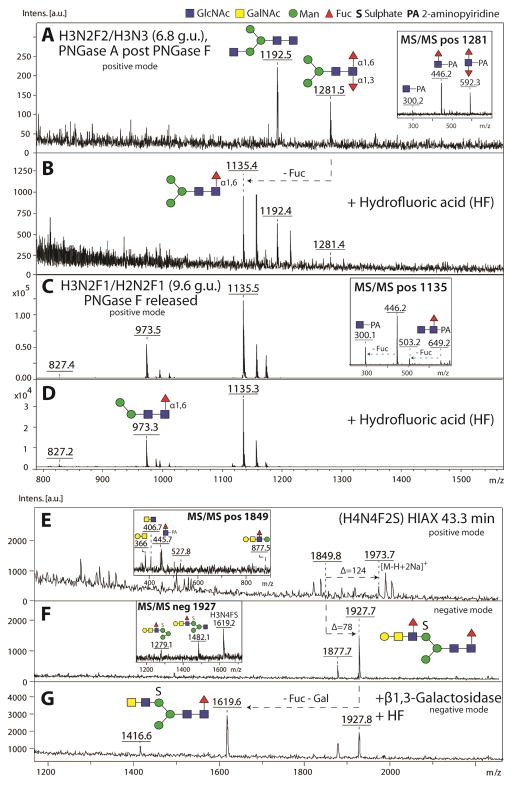
Figure 2. Structure elucidation of fucosylated N-glycans from Anopheles gambiae larvae.
N-glycans from Anopheles larvae were released by PNGase F prior to PNGase A (post PNGase F), enriched in neutral and anionic pools, prior to pyridylamino-labelling, MALDI-TOF MS and HPLC analysis. The neutral pools were fractionated using an RP-Amide column and the anionic pool using a HIAX column (see Kurz et. al, 2015). MALDI-TOF MS spectra of selected HPLC fractions containing fucosylated N-glycans from neutral (A–D) and anionic pools (E–G) are shown before (A, C, E) and after either hydrofluoric acid (B, D, G) or enzymatic treatment (G). After HF treatment, loss of α1,3-linked fucose (Δm/z 146) could be detected for the core-difucosylated N-glycan species (m/z 1281; compare panel A and B), whereas α1,6-linked fucose residues remained HF resistant (m/z 1135; B and D). The anionic N-glycan species of Hex4HexNAc4Fuc2(SO3)1 was sensitive to a combined treatment with β1,3-galactosidase and HF indicating the presence of an antennal α1,3-linked Lewis-type fucose (G). MS/MS key fragments of untreated parent glycans are depicted in insets. N-glycan structures are assigned using the symbolic nomenclature of the Consortium of Functional Glycomics (circles, hexose; squares, N-acetylhexosamine; triangles, deoxyhexose; S, sulphate) and glycan compositions are abbreviated in the form HxNyFzS (HexxHexNAcyFuczS0-1).
To further elucidate the structural nature of these fucosylated structures, we performed chemical and enzymatic treatments of selected HPLC fractions followed by MALDI-TOF MS and MS/MS analysis. Using hydrofluoric acid, known to specifically release α1,3-linked fucose, we were able to detect a loss of one fucose residue (indicated by the loss of m/z 146) from a difucosylated N-glycan species of the composition Hex3HexNAc2Fuc2 (m/z 1281; compare panels A and B in Figure 2). However, the major monofucosylated N-glycan species of Hex2-3HexNAc2Fuc1 released by PNGase F eluting at 9.6 g.u. (m/z 1135) remained resistant to this chemical treatment (Figure 2D) and were solely susceptible to bovine α1,6-specific fucosidase treatment and therefore concluded to be in α1,6-linkage (data not shown). In contrast, a minor monofucosylated N-glycan species of Hex3HexNAc2Fuc1 released by PNGase A (post PNGase F) eluting at 5.2 g.u. demonstrated a m/z 446 fragment in MS/MS analysis but was resistant to bovine α1,6 fucosidase treatment (data not shown); thus, we assumed a core α1,3-linked fucose epitope for this glycan species due to its resistance towards bovine α1,6 fucosidase and the early RP-HPLC elution. This is in keeping with data on nematode monofucosylated N-glycans of the same retention time (Yan et al., 2015).
The majority of the anionic glycans, modified with sulphate and glucuronic acid, were also core α1,6-fucosylated. Amongst these, minor sulphated forms with additional antennal N-acetylhexosamine, hexose and fucose residues were found, which suggested the presence of fucosylated Lewis-type epitopes. Using HIAX-HPLC, we separated anionic N-glycans according to size and charge (for the full repertoire of anionic mosquito N-glycans see Kurz et al., 2015). In a HIAX fraction collected at 43.3 minutes, a glycan species of the composition Hex4HexNAc4Fuc2(SO3)1 was detected in both positive and negative ion modes (respectively at m/z 1973 as [M-H+2Na]+ and m/z 1927 as [M-H]−317 ; Figure 2E and F). This mass shift is indicative for the presence of sulphate or phosphate residues with the former modification easily dissociating from the N-glycan backbone during MALDI-TOF MS ionization in positive ion mode. Positive ion mode MS/MS fragmentation of this glycan revealed the presence of the key fragments m/z 366 (Hex1HexNAc1), m/z 407 (HexNAc2) and m/z 877 (Hex2HexNAc2Fuc1 corresponding to the entire α1,6-arm) indicative for a fucosylated LacdiNAc motif (GalNAcβ1,4[Fucα1,3]GlcNAc) putatively capped with an additional β1,3-linked galactose (inset in Figure 2E). Furthermore, negative ion mode MS/MS resulted in key fragments of m/z 1279, 1482 and 1619 compatible with the presence of a sulphate group (inset in Figure 2F), which is as in other mosquito glycans presumably linked to the α1,6-mannose residue (Kurz et al., 2015).
To verify the capping with β1,3-linked galactose, the glycan species of m/z 1927 was subjected to serial β1,3 galactosidase and HF treatment to remove the putative Lewis-type α1,3-linked fucose residue attached to the galactosylated LacdiNAc motif. Negative ion mode MS analysis demonstrated the partial loss of one galactose and one fucose residue as judged by the presence of a product of m/z 1619 (Figure 2G). As the anionic modification remained unaltered by the HF treatment, we can confirm the presence of sulphate, rather than HF-sensitive phosphate. Thus, we demonstrate the presence of a sulphated N-glycan with an antennal β1,3-galactose capping the α1,3-fucosylated LacdiNAc motif. Interestingly, we were not able to detect the non-sulphated, neutral structure in the N-glycome analysis of this organism, but a non-fucosylated non-sulphated variant (a T-antigen-like structure) is known from honeybee royal jelly glycoproteins (Kimura et al., 2006), whereas fucosylated LacdiNAc is known from honeybee venom phospholipase (Kubelka et al., 1993).
3.3 Cloning and sequence analysis of fucosyltransferase cDNAs
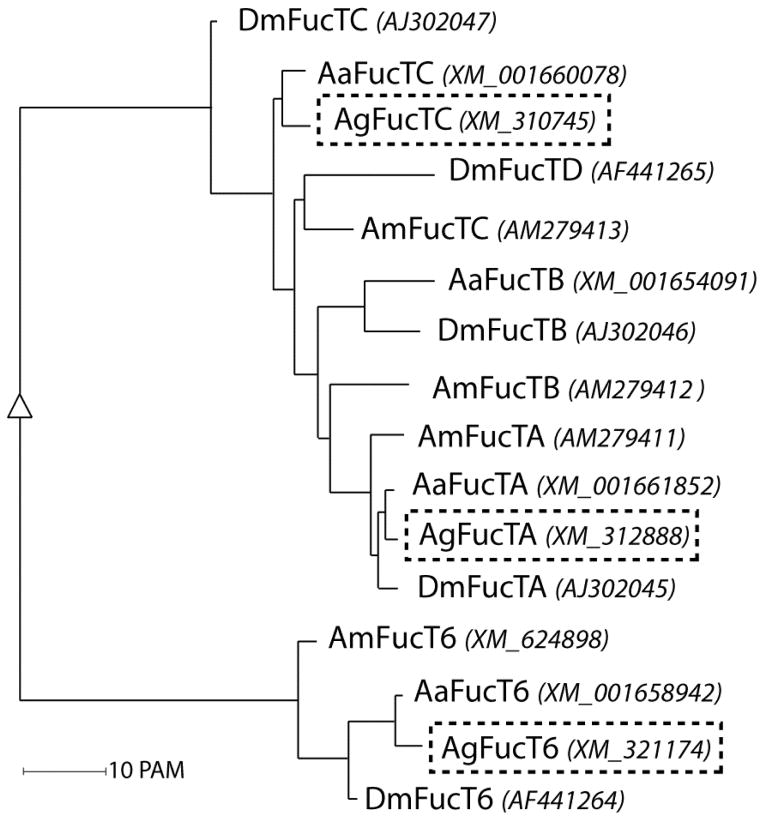
All three fucosylated N-glycan motifs examined in this study are indicative for the in vivo activity of multiple mosquito fucosyltransferases. As the genome of An. gambiae is known (Holt et al., 2002), we identified potential fucosyltransferase genes responsible for the biosynthesis of these epitopes and then characterised the cognate recombinant enzymes in vitro. Using tBLASTn, potential N-glycan modifying fucosyltransferase genes were identified in the genome of An. gambiae with homology to the previously-described A. mellifera and D. melanogaster FucTA and FucT6 (respectively AGAP003191-PA and AGAP001888-PA); also a third fucosyltransferase gene, closest to A. mellifera FucTC, was predicted (AGAP000365-PA; see Figure 3 for a phylogenetic comparison). However, the predicted FucTA reading frame lacked a region encoding a cytosolic and transmembrane domain, which would be typical for a Golgi-localised glycosyltransferase. Thus, the genomic DNA sequence upstream of the predicted FucTA gene was visually analysed in all three reading frames; various primer combinations and PCR conditions were tried before a sequence-verified FucTA clone was isolated; as FucTA clones from two independent PCR reactions had a potential missense mutation in the 252nd codon as compared to the genomic sequence (TGG to TAG), inverse PCR was performed in order to obtain a Pichia expression vector with the expected sequence. The corrected FucTA reading frame encodes a protein of ~61 kDa, as compared to ~67 kDa and ~46 kDa for AgFucT6 and AgFucTC respectively. Sequence identities to the closest A. mellifera and D. melanogaster homologues are 51% and 60% for FucTA, 52% and 58% for FucT6, and 37% and 46% for FucTC.
Figure 3. Phylogenetic comparison of fucosyltransferases.
Selected insect fucosyltransferases were analysed using the Multiple Sequence Alignment server (http://multalin.toulouse.inra.fr/multalin/ (Corpet, 1988). The resulting phylogenetic data were visualised using the Alignment and tree description (rfd) software on the same webpage. The abbreviations for the respective organisms used are as followed: Aa, Aedes aegypti; Am, Apis mellifera; Ag, Anopheles gambiae; and Dm, Drosophila melanogaster. A phylogenetic tree of predicted insect fucosyltransferase amino acid sequences with corresponding gene bank accession numbers in brackets and italic are shown. The three putative fucosyltransferases from An. gambiae examined in this study are indicated by dashed boxes.
3.4 Substrate specificities of recombinant Anopheles fucosyltransferases in vitro
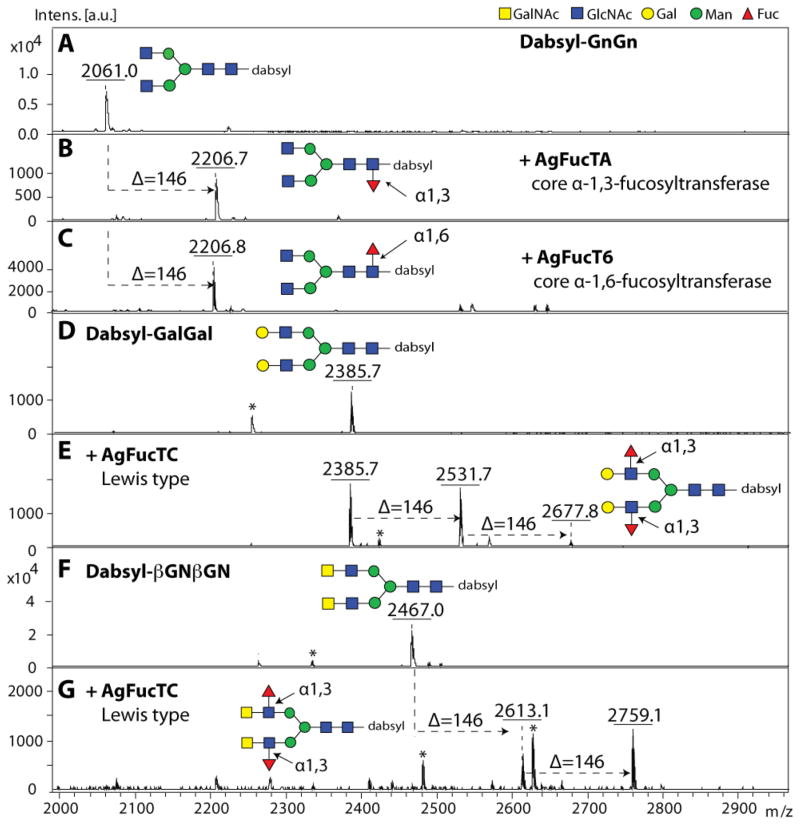
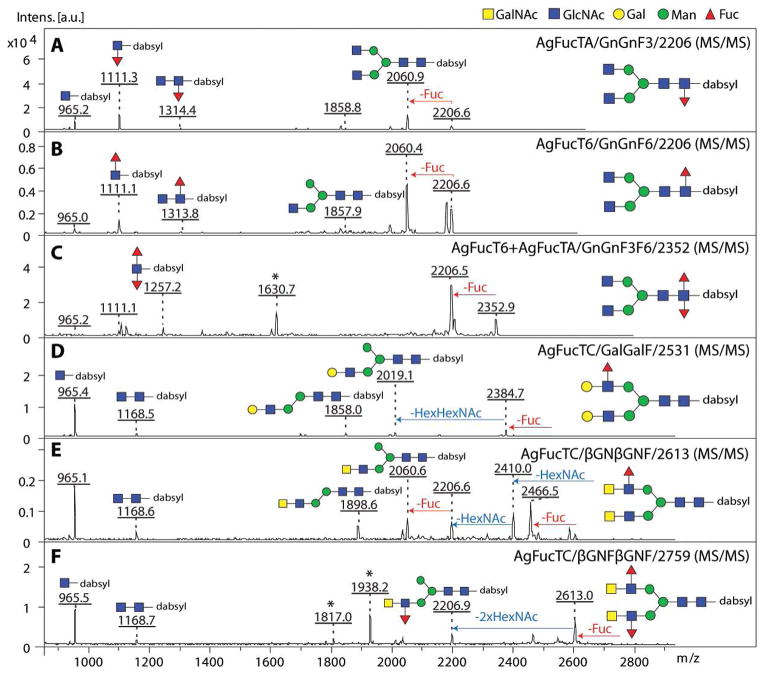
Initially P. pastoris supernatants were assayed for fucosyltransferase activity by MALDI-TOF MS using GDP-Fuc as donor substrate and various dabsylated glycopeptides (GENR; Gly-Glu-Asn-Arg) carrying either GnGn, GalGal or βGNβGN N-glycans as acceptor substrates. In general, GnGn serves as an acceptor substrate specific for core fucosyltransferases, GalGal (i.e. GnGn with two non-reducing terminal β1,4-galactose residues) is suitable for Lewis-X generating α1,3-fucosyltransferases, and βGNβGN (i.e. GnGn with two non-reducing terminal β1,4 N-acetyl-D-galactosamine residues) for LacdiNAc (GalNAcβ1,4GlcNAc) preferring Lewis-type α1,3/4-fucosyltransferases. In this study, all P. pastoris supernatants demonstrated fucosyltransferase activity with selected dabsylated glycopeptides after overnight incubation in the presence of GDP-Fuc as indicated by a mass shift of 146 or 292 (2 × 146) units higher than the dabsylated glycopeptide controls (Figure 4). As judged by MALDI-TOF MS, both recombinant core fucosyltransferases (AgFucTA and AgFucT6) were able to transfer one fucose residue to the dabsylated-GnGn acceptor substrate as indicated by the presence of products of m/z 2206 (Figure 4B–C), whereas the recombinant Lewis-type AgFucTC showed activity to either dabsylated glycopeptides carrying GalGal (m/z 2385) or βGNβGN (m/z 2467) N-glycans as acceptor substrate with a transfer of up to two fucose residues to form either GalFGalF (m/z 2677; Figure 4E) or, preferentially, βGNFβGNF (m/z 2759; Figure 4G). Supernatants of Pichia transformed with vector lacking inserts displayed no fucosyltransferase activity with any of the acceptor substrates tested (data not shown).
Figure 4. MALDI-TOF MS based activity assays of recombinant fucosyltransferases.
After 3 days of expression, P. pastoris supernatants were tested in MALDI-TOF MS based assays for fucosyltransferase activities. (A–G) Incubation of selected dabsylated glycopeptides (dabsyl-GENR; Gly-Glu-Asn-Arg) with the recombinant enzyme preparations was performed overnight in absence (A, D, F) or presence (B, C, E, G) of GDP-Fuc as donor substrate. In all cases, the recombinant fucosyltransferases transferred fucose (indicated by the Δm/z = 146; one in the case of dabysl-GENR carrying GnGn, and up to two in the case of dabsyl-GENR carrying either GalGal or βGNβGN). Asterisks indicate ions resulting from partial laser-induced breakdown of dabsyl moiety of the substrates and products (peaks of 132 mass units smaller than the parent glycopeptides).
Extended substrate preference tests were performed using purified forms of the recombinant enzymes incubated with either dabsylated GnGn, GnGnF3, GnGnF6, GalGal, βGNβGN or βGNFβGNF glycopeptide acceptors (Table I) and the assays were analysed by MALDI-TOF MS; the results with the purified enzymes were compatible with those for the supernatants. These tests verified that both putative core fucosyltransferases (AgFucTA and AgFucT6) have a significant preference towards dabsylated-GnGn. Furthermore, subsequent incubation of the AgFucT6 product (dabsyl-GnGnF6) with the putative core α1,3-fucosyltransferase (AgFucTA) resulted in a difucosylated product (dabsyl-GnGnF3F6; see also MS/MS fragmentation indicating presence of the diagnostic fragment at m/z 1257; Figure 5C). In contrast, the AgFucTA product (dabsyl-GnGnF3) was not a substrate for AgFucT6; therefore, the α1,6- before α1,3-fucosylation rule, previously determined for D. melanogaster core fucosyltransferases (Paschinger et al., 2005b), also applies in the case of the mosquito.
Table I.
Substrate specificity of recombinant FucTs
| Substrate | AgFucTA | AgFucT6 | AgFucTC |
|---|---|---|---|
| GnGn | 96% | 100% | ND |
| GnGnF3 | - | ND | ND |
| GnGnF6 | 94% | - | ND |
| GalGal | ND | 1% | 59% (1F) |
| βGNβGN | 1% | 1% | 70% (2F) |
| βGNFβGNF | ND | ND | - |
Dabsylated glycopeptides were used for fucosyltransferase assays. GnGnF3, GnGnF6 and βGNFβGNF were obtained from prior incubation with purified AgFucTA, AgFucT6 and AgFucTC respectively. Relative conversion to product in % was determined by calculating peak areas obtained from MALDI-TOF MS spectra. In case of AgFucTC, either transfer of one or two fucoses residues was detected (indicated by 1F or 2F respectively). ND, not detectable; -, not performed.
Figure 5. MALDI-TOF MS/MS fragmentation of recombinant FucT products.
MS/MS fragmentation patterns of dabsylated glycopeptide products in the range of m/z 900–2800 are shown. In the lower mass range, presence of the diagnostic fragment ion at m/z 1111 in AgFucTA and AgFucT6 products are indicating the presence of core fucose (A–C), which is absent in Lewis-type AgFucTC products (D–F). In case of the dabsylated GnGnF3F6 product, a diagnostic fragment of m/z 1257 was observed indicating a difucosylated N-glycan core (C). Terminal loss of one fucose residue from the parent ion (loss of m/z 146) was observed for all products, whereby in AgFucTC products loss of either ‘en-block’ Hex1HexNAc1, indicated by the loss of m/z 365 (D) or ‘en-block’ loss of two HexNAc residues, indicated by the loss of m/z 407 (E, F) were detected. The assumed fragmentation pattern with loss of the α1,6 mannose arm is based on previous observations for N-glycans. Asterisks represent unidentified signals.
In the case of the purified Lewis-type fucosyltransferase (AgFucTC), the stronger preference for the LacdiNAc (GalNAcβ1,4GlcNAc) over the LacNAc (Galβ1,4GlcNAc) motif was verified and no activity was determined towards GnGn, GnGnF3 or GnGnF6. Transfer of fucose by either recombinant core fucosyltransferase to GalGal and βGNβGN substrates was only at marginally-detectable levels and, therefore, no subsequent substrate specificity tests with the putative Lewis-type fucosyltransferase using core fucosylated GalGal or βGNβGN could be performed. Also, due to a lack of the relevant sulphotransferase, we were also not able to investigate the recombinant AgFucTC activity towards acceptor substrates carrying anionic N-glycans in vitro in order to mimic the in vivo products observed in the glycomic analysis. Thus, conclusions for the biosynthetic pathway of the putative Lewis-type fucosyltransferases can only be hypothesized, but not be proven.
3.5 Characterisation of the in vitro fucosylation products
To address the position of the enzymatically added fucose residues, MS/MS fragmentation of the dabsylated products was performed first (Figure 5), followed by chemical and exoglycosidase treatments and subsequent MALDI-TOF MS analysis (Table II). MS/MS fragmentation of all products resulted in terminal loss of one fucose residue; in the case of the AgFucTA and AgFucT6 core-modified products, diagnostic fragment ions of m/z 1111 and 1313 were found which correspond to HexNAc1-2Fuc1 bound to dabsylated-GENR (Figure 5A and B). These fragments were absent in the case of AgFucTC products and were replaced by a fragment of m/z 1168 indicative for dabsyl-GENR carrying only HexNAc2 (Figure 5D–F), which verifies that the Lewis-type enzyme does not modify the core region. Unfortunately, MS/MS fragments specifically supporting the presence of fucose residues linked to LacNAc and LacdiNAc motifs (e.g. Hex1HexNAc1Fuc1, m/z 512 or HexNAc2Fuc1, m/z 553) were not detectable in either of the dabsylated AgFucTC products even though the precursor structures lacking the fucose moiety (m/z 366 and 407) were observed (data not shown), possibly due to the lability of the Lewis-type linkages during MALDI-TOF MS/MS.
Table II.
Chemical and enzymatic characterisation of recombinant FucT products
| dabsylated-product (recombinant enzyme; product; m/z) | Hydrofluoric acid (HF) | α1,6-fucosidase (bovine kidney) |
|---|---|---|
| AgFucTA/GnGnF3/2206 | + | − |
| AgFucT6/GnGnF6/2206 | − | + |
| AgFucTC/GalGalF/2531 | + | − |
| AgFucTC/βGNβGNF/2613 | + | − |
Dabsylated products (GnGnF3, GnGnF6, GalGalF, βGNβGNF) obtained from incubation with recombinant fucosyltransferases (AgFucTA, AgFucT6, AgFucTC) were split and incubated with hydrofluoric acid and α1,6-fucosidase from bovine kidney respectively. Core α1,3- and Lewis-type fucose residues demonstrated sensitivity towards hydrofluoric acid treatment, whereas core α1,6-fucose was only susceptible to bovine α1,6-fucosidase. Sensitivity towards a treatment is indicated by (+).
For further analysis, the dabsylated fucosyltransferase products were split into three parts and used for chemical and enzymatic treatments (Table II). Based on previous studies in our and other laboratories (Yan et al., 2012, Haslam et al., 2000), an α1,3-linked fucose – regardless if linked to the proximal GlcNAc or to the non-reducing terminal GlcNAc – shows sensitivity towards chemical treatment with hydrofluoric acid. In contrast, an α1,6-linked fucose will remain resistant, but can be released using α1,6-fucosidase. As summarised in Table II, the dabsyl-GnGnF6 product generated by the putative core α1,6-fucosyltransferase (AgFucT6) showed sensitivity towards the fucosidase treatment by the loss of one fucose residue and was the only enzymatic product resistant to the hydrofluoric acid treatment; on the other hand, the fucose residues in the GnGnF3, GalGalF and βGNβGNF products of AgFucTA and AgFucTC are α1,3-linked as judged by their sensitivity to hydrofluoric acid. Therefore the recombinant mosquito fucosyltransferases are generating the fucose linkages expected from their homologies.
3.6 Enzymatic properties of recombinant mosquito fucosyltransferases
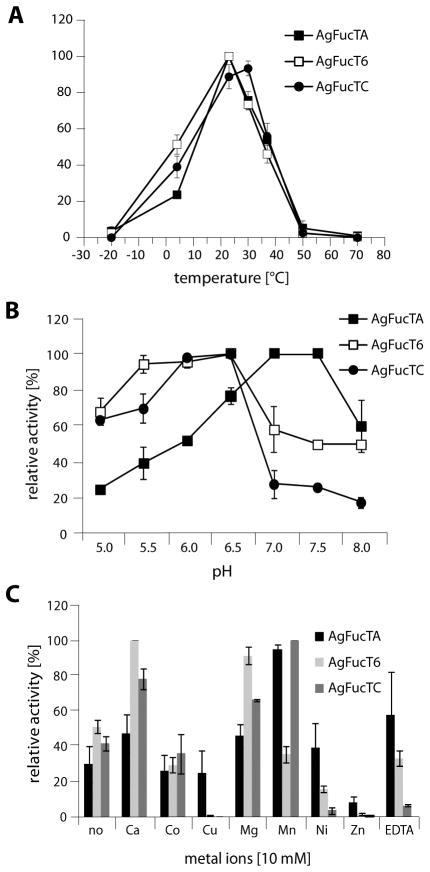
Further parameters including the temperature, pH and ion dependence of all three recombinant fucosyltransferases were investigated using the relevant preferred dabsyl-glycopeptide in MALDI-TOF MS based assays. In experiments at various temperatures, at pH 6.5 and in the presence of Mn2+ ions, optima at 25–30 °C were determined for all enzymes with activity being detected for a broad temperature range from 4 °C to 37 °C (Figure 6A). For the determination of the pH optima, two different buffer systems (MES and AMPD) were used to cover a broad pH range from 5.0 to 8.0. A feature of many Golgi enzymes includes a tendency of maximum enzymatic activity around pH 6; this was demonstrated to be the case for AgFucT6 and AgFucTC with a pH optimum around pH 6 (between 5.5 and 6.5). In contrast, AgFucTA showed maximal activity in neutral to mild alkaline conditions with an optimal pH of 7–7.5 (Figure 6B). Amongst the different cations tested, highest activity was found in presence of manganese (AgFucTA and AgFucTC) and calcium (AgFucT6) ions, whereas copper and zinc ions nearly inhibited all the studied enzymes. On the other hand, activity was demonstrated also in the absence of cations as well as, other than AgFucTC, in the presence of the chelating agent EDTA; this indicates that the three enzymes have no absolute requirement for any divalent cation (Figure 6C).
Figure 6. Enzymatic properties of recombinant mosquito FucTs.
In MALDI-TOF MS-based assays, activity of recombinant, purified fucosyltransferases was measured at various temperatures (A), pH values (B) and in presence of various ions (C). Reactions (A–C) were performed with selected dabsylated glycopeptides as acceptor substrates (dabsyl-GnGn for core fucosyltransferases; dabsyl-βGNβGN for Lewis-type fucosyltransferase) and GDP-Fuc as enzyme donor. Relative activity [%] was normalised based on the condition giving the highest rate of fucose transfer and all values represent averages +/− standard error.
4 DISCUSSION
4.1 In vivo localisation and characterisation of fucosylated epitopes in insects
Since the earlier discovery in 1982 by Jan and Jan, antibodies raised against horseradish peroxidase (HRP) have been extensively used as a tool to stain neural tissues in arthropods and nematodes (Jan and Jan, 1982, Katz et al., 1988, Wang et al., 1994, Snow et al., 1987, Siddiqui and Culotti, 1991). In D. melanogaster, the neural anti-HRP staining occurs very early in embryogenesis and can be detected throughout the development of the fly nervous system. Using dissected abdomen tissues, we confirm the neural anti-HRP cross-reactivity of the ventral nerve cord in An. gambiae and further detected staining of the anopheline pericardial cells, which are nephrocytes involved in haemolymph recycling. This is in agreement with previous findings as limited distribution of anti-HRP staining was observed for non-neural tissues including larval garland cells, pericardial cells, adult rectal papillae and adult male accessory glands in D. melanogaster (Jan and Jan, 1982). Using Western Blot analysis to correlate anti-HRP reactive components with tissue staining, several candidates for neural and non-neural anti-HRP reactivity have been identified in the fly. Among the neural specific candidates, surface glycoproteins such as Fascilin I, II and III were characterised (Snow et al., 1987, Patel et al., 1987), whereas the non-membrane associated protein esterase-6 was identified to be a putative candidate responsible for the non-neural anti-HRP staining of adult fruit fly male reproductive tissues (Wang et al., 1994). The relevant glycoproteins associated with neural and non-neural anti-HRP staining in An. gambiae will need to be identified by glycoproteomic analyses in order to show, e.g., whether the pericardial reactivity is due to proteins endogenous to these cells or to uptake of α1,3-fucosylated proteins from the haemolymph.
It took many years after the initial observation of anti-HRP staining to prove that the epitopes in insects were indeed based on fucose. Key to this were the definition of a relevant core α1,3-fucosyltransferase (FucTA) in D. melanogaster, RNAi knock-down of FucTA in an insect neural cell line, overexpression of FucTA in D. melanogaster embyros and the proof that the anti-HRP-defective nac mutant was due to a point mutation in the Golgi GDP-Fuc transporter (Fabini et al., 2001, Rendić et al., 2006, Rendic et al., 2010, Geisler et al., 2012). In comparison to the fruit fly, there has been no data to date regarding anti-HRP epitopes in mosquitoes; previous studies were limited primarily to lectin blots and histochemistry with actual mass spectrometric N-glycan analyses performed only on a couple of glycoproteins from mosquitoes or mosquito cell lines (Wilkins and Billingsley, 2001, Rudin and Hecker, 1989, Andrews et al., 1997, Li and Li, 2005, Crispin et al., 2014). Thus, our recent paper on the analysis of neutral and anionic glycans from An. gambiae and Aedes aegypti was the first comprehensive glycomic analyses of these two mosquito species. As rather mixed and contradictory data can be obtained by use of lectins alone, only the direct mass spectrometric analysis of fucosylated glycans can prove the nature of the linkages of fucose to the N-linked oligosaccharides. The MS/MS and digestion data shown here prove the presence of core α1,3-fucose, core α1,6-fucose and antennal α1,3-fucose on N-glycans of An. gambiae. Whereas the modifications of fucose on the core are also known from D. melanogaster (Fabini et al., 2001), the antennal fucose has not been observed previously in a dipteran species. In insects, this ‘Lewis-like’ epitope is previously only known from the venom of honeybee (Kubelka et al., 1993), a hymenopteran.
4.2 Genomic basis for N-glycan fucosylation
Following the histochemical and glycomic analyses, we identified three putative fucosyltransferases in the Anopheles genome possibly involved in the biosynthesis of fucosylated glycan epitopes detected in N-glycan analysis. As previously observed in vertebrates and other invertebrates, the Anopheles genome encodes only one homologue (designated FucT6) of the mammalian core α1,6 fucosyltransferase (FUT8). The high abundance of core α1,6-fucosylated N-glycans examined in neutral and anionic PNGase F released N-glycan pools, emphasises the in vivo activity of mosquito core α1,6-fucosyltransferases. Other insect homologues have been identified and characterised from D. melanogaster (Paschinger et al., 2005b) as well as lepidopteran sources (Sf9 cells, Bombyx mori) (Juliant et al., 2014, Ihara et al., 2014). The excellent activity of the An. gambiae FucT6 has already been exploited for preparing glycans for microarray printing (Beloqui et al., 2013). Only two putative α1,3-fucosyltransferases (designated FucTA and FucTC) are predicted in the Anopheles genome. In general, α1,3-fucosyltransferases can either transfer fucose moieties to the N-glycan core region or to the antennae (Lewis-type α1,3- or α1,4-fucosyltransferases). The presence of difucosylated N-glycans revealed by N-glycomic analysis of An. gambiae and anti-HRP reactivity in Western blot analysis indicates an in vivo activity of the core α1,3-fucosyltransferase, which correlates with the in vitro data on the recombinant AgFucTA. The Lewis-like activity of the second mosquito α1,3-fucosyltransferase (AgFucTC), on the other hand, correlates with the detection of N-glycan species with antennal fucose. However, in contrast to the Lewis-type fucosylated LacdiNAc modifications of neutral N-glycans on honeybee venom glycoproteins (Kubelka et al., 1993), in Anopheles this motif was only detected on sulphated N-glycans. Thus one may ask: Why were fucosylated LacdiNAc motifs only detected on sulphated N-glycans for which the sulphate moieties are putatively associated with core α1,6-mannose? This, of course, may depend on the tissue-specificity of the expression of AgFucTC and the relevant sulphotransferase. Interestingly, three fruit fly α1,3-fucosyltransferase genes (FucTB, FucTC and FucTD) have not yet been proven to encode active enzymes, whereas only two of three α1,3-fucosyltransferase homologues from the honeybee (the core-modifying FucTA and the Lewis-like FucTC) have detectable biochemical function (Fabini et al., 2001, Rendić et al., 2007). Thus, An. gambiae is the first insect for which all α1,3- and α1,6-fucosyltransferase homologues can be assigned a biochemical function.
4.3 Characterisation of insect fucosyltransferases
The actual biochemical function of the three fucosyltransferases identified in the An. gambiae genome was examined by using recombinant forms expressed in Pichia pastoris. All three have proven to be active in the presence of GDP-Fuc as donor substrate and dabsylated glycopeptides. In comparison to other techniques for testing enzymatic fucosyltransferase activities in vitro, we opted for a robust MALDI-TOF MS approach, which is fast, simple and minimally work-intensive. In the past, radioactive-based assays were predominantly performed for enzymatic activity assays using free oligosaccharides as acceptor substrates and GDP-[14C]Fuc as donor. Besides the hazards and costs associated with radioisotopes, the mere measurement of counts does not identify the nature of the products of either transfer or breakdown. A similar conclusion can be obtained when using fluorescently labelled oligosaccharides as acceptor substrates with subsequent HPLC analysis; shifts in retention time merely are suggestive for the transfer of fucose moieties. In contrast, in our MALDI-TOF MS approach, we used glycopeptides carrying tailored N-glycan structures as acceptor substrates thereby mimicking the in vivo situation for the recombinant enzymes in an in vitro experimental setup and allowing product characterisation by MS/MS.
As previously described for D. melanogaster and for insect cell lines, the in vitro reconstitution of core fucosylation events in An. gambiae follows a strict order whereby α1,6-fucosylation takes place before α1,3 fucosylation (Paschinger et al., 2005b). Both recombinant core fucosyltransferases (AgFucTA and AgFucT6) demonstrated a substrate preference towards dabsylated-GnGn. Subsequent incubation of the AgFucT6 product (dabsyl-GnGnF6) with the core α1,3-fucosyltransferase (AgFucTA) promoted further addition of one fucose residue resulting in a difucosylated product (dabsyl-GnGnF3F6). On the contrary, incubation of the AgFucTA product (dabsyl-GnGnF3) with AgFucT6 prevented any further addition of fucose following the strict rule of α1,6 before α1,3-fucosylation. In addition, both core fucosyltransferases were not able to use dabsylated GalGal or βGNβGN as substrates, whereas the recombinant Lewis-type fucosyltransferase (AgFucTC) could only utilise GalGal and βGNβGN; the seeming preference of AgFucTC for the latter is in accordance with the detection of fucosylated LacdiNAc on An. gambiae N-glycans. The positions and linkages of the transferred fucose residues were assessed by MS/MS fragmentation and chemical or enzymatic treatments. Only the GnGnF3, GnGnF6 or GnGnF3F6 products revealed key fragments indicative for core mono- and di-fucosylated reducing GlcNAc residues linked to the dabsylated glycopeptide. On the other hand, solely the GnGnF6 product generated by AgFucT6 was sensitive to bovine α1,6-fucosidase but remained resistant to HF treatment. In keeping with the homology-predicted enzyme specificities, only α1,3-fucosylated glycoforms generated by the core and Lewis-type α1,3-fucosyltransferases showed sensitivity towards HF treatment.
The recombinant enzymes were characterised to assess any cation dependency as well as the temperature and pH optima. The temperature optima were in the range between 25 – 30 °C, which is compatible with the habitat of the mosquito. Comparable to many Golgi enzymes, a broad pH range between 5.5 and 6.5 was observed for core α1,6- and Lewis type α1,3-fucosyltransferases, whereby the optimal pH of 7.5 was examined for the core α1,3 fucosyltransferase. Rather broad pH optima were also observed for the honeybee α1,3-fucosyltransferases (Rendić et al., 2007). Some glycosyltransferases, including fucosyltransferases with a GT-B fold, require divalent cations for their optimal transferase activity. The enzymatic activity of the α1,3-fucosyltransferases was boosted by the addition of Mn(II), and for the core α1,6-fucosyltransferase by the addition of Mg(II). An alternative divalent metal cofactor was Ca(II), whereby Cu(II) and Zn(II) ions demonstrated to be inhibitory for all three recombinant fucosyltransferases. Interestingly, the enzymes demonstrated activity in the absence of divalent cofactors and in the presence of the chelating agent EDTA, thereby indicating that the three recombinant fucosyltransferases tested have no absolute requirement for any special divalent cation, which has also been described for C. elegans core α1,3- and α1,6-fucosyltransferases FUT-1 and FUT-8 (Paschinger et al., 2004, Paschinger et al., 2005b). In the case of the two honeybee α1,3-fucosyltransferases, FucTA was inhibited by use of EDTA (Rendić et al., 2007).
4.4 Conclusion
By using histochemical, glycomic and enzymatic analyses we reveal the neural specificity of anti-HRP in An. gambiae; we identified genes required for the biosynthesis of fucosylated N-glycans and characterised the resulting in vivo N-glycan products of the mosquito fucosyltransferases using a combined approach of state-of-the-art mass spectrometry, HPLC separation as well as exoglycosidase and chemical treatments. We demonstrate the enzymatic activity and properties for all α1,3- and α1,6-fucosyltransferase homologues from this insect species by applying a robust MALDI-TOF MS based approach. Not only can these enzymes serve as potential tools in generating epitopes for glycan array analyses, but their demonstrated activity corroborates the glycogenomic capacity of the organism with its proven glycomic potential.
Highlights.
We revealed the neural specificity of anti-HRP in Anopheles gambiae
Identification of the relevant genes involved in biosynthesis of fucosylated N-glycans in the mosquito
Native fucosylated N-glycans of the malaria vector were characterised
Robust MALDI-TOF MS based assays demonstrated in vitro enzymatic activity of three fucosyltransferases
Acknowledgments
This work was supported by the FWF-funded doctoral programme “Biomolecular Technology of Proteins” [W1224] (SK), FWF grant P21946 (KP) and NIH grant 5R01AI082587-05 (RRD). The authors thank Ajda Flašker (visiting student) and Dr. Shi Yan for help in cloning as well as Daniel Gregorich for help in initial trials on the recombinant expression and purification of AgFucTA and AgFucTC. The authors also thank Paul Eggleston and Hilary Hurd for providing the An. gambiae KEELE strain.
Footnotes
Authors’ contributions
SK performed the Western Blotting, immunofluorescence staining, N-glycan preparation, N-glycan analysis by HPLC and MALDI-TOF MS, recombinant expression, purification and characterisation experiments. JGK assisted in preparing the mosquito sections and immunofluorescence staining experiments. IBHW performed cloning and transformation into P. pastoris. RRD, KP and IBHW assisted in the experimental design and data evaluation. SK, KP and IBHW wrote the manuscript. KP and IBHW conceived the study.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- ANDREWS L, LAUGHINGHOUSE A, SINA BJ. Lectin binding characteristics of male and female salivary gland proteins of Anopheles gambiae: Identification and characterization of female specific glycoproteins. Insect Biochem Mol Biol. 1997;27:159–166. [Google Scholar]
- ARMISTEAD JS, WILSON IBH, VAN KUPPEVELT TH, DINGLASAN RR. A role for heparan sulfate proteoglycans in Plasmodium falciparum sporozoite invasion of anopheline mosquito salivary glands. Biochem J. 2011;438:475–83. doi: 10.1042/BJ20110694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BECKER DJ, LOWE JB. Fucose: biosynthesis and biological function in mammals. Glycobiology. 2003;13:41R–53R. doi: 10.1093/glycob/cwg054. [DOI] [PubMed] [Google Scholar]
- BELOQUI A, CALVO J, SERNA S, YAN S, WILSON IBH, MARTIN-LOMAS M, REICHARDT NC. Analysis of Microarrays by MALDI-TOF MS. Angew Chem Int Ed Engl. 2013;52:7477–81. doi: 10.1002/anie.201302455. [DOI] [PubMed] [Google Scholar]
- CORPET F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–90. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CRISPIN M, HARVEY DJ, BITTO D, BONOMELLI C, EDGEWORTH M, SCRIVENS JH, HUISKONEN JT, BOWDEN TA. Structural plasticity of the Semliki Forest virus glycome upon interspecies transmission. J Proteome Res. 2014;13:1702–12. doi: 10.1021/pr401162k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DINGLASAN RR, ALAGANAN A, GHOSH AK, SAITO A, VAN KUPPEVELT TH, JACOBS-LORENA M. Plasmodium falciparum ookinetes require mosquito midgut chondroitin sulfate proteoglycans for cell invasion. Proc Natl Acad Sci U S A. 2007;104:15882–7. doi: 10.1073/pnas.0706340104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FABINI G, FREILINGER A, ALTMANN F, WILSON IBH. Identification of core a1,3-fucosylated glycans and the requisite fucosyltransferase in Drosophila melanogaster. Potential basis of the neural anti-horseradish peroxidase epitope. J Biol Chem. 2001;276:28058–28067. doi: 10.1074/jbc.M100573200. [DOI] [PubMed] [Google Scholar]
- GEISLER C, KOTU V, SHARROW M, RENDIC D, POLTL G, TIEMEYER M, WILSON IBH, JARVIS DL. The Drosophila neurally altered carbohydrate mutant has a defective Golgi GDP-fucose transporter. J Biol Chem. 2012;287:29599–609. doi: 10.1074/jbc.M112.379313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GREENWELL P. Blood group antigens: molecules seeking a function? Glycoconj J. 1997;14:159–73. doi: 10.1023/a:1018581503164. [DOI] [PubMed] [Google Scholar]
- HASLAM SM, COLES GC, MORRIS HR, DELL A. Structural characterisation of the N-glycans of Dictyocaulus viviparus: discovery of the Lewisx structure in a nematode. Glycobiology. 2000;10:223–229. doi: 10.1093/glycob/10.2.223. [DOI] [PubMed] [Google Scholar]
- HOFFMANN-SOMMERGRUBER K, PASCHINGER K, WILSON IBH. Glycomarkers in parasitic infections and allergy. Biochem Soc Trans. 2011;39:360–4. doi: 10.1042/BST0390360. [DOI] [PubMed] [Google Scholar]
- HOLT RA, SUBRAMANIAN GM, HALPERN A, SUTTON GG, CHARLAB R, NUSSKERN DR, WINCKER P, CLARK AG, RIBEIRO JM, WIDES R, SALZBERG SL, LOFTUS B, YANDELL M, MAJOROS WH, RUSCH DB, LAI Z, KRAFT CL, ABRIL JF, ANTHOUARD V, ARENSBURGER P, ATKINSON PW, BADEN H, DE BERARDINIS V, BALDWIN D, BENES V, BIEDLER J, BLASS C, BOLANOS R, BOSCUS D, BARNSTEAD M, CAI S, CENTER A, CHATURVERDI K, CHRISTOPHIDES GK, CHRYSTAL MA, CLAMP M, CRAVCHIK A, CURWEN V, DANA A, DELCHER A, DEW I, EVANS CA, FLANIGAN M, GRUNDSCHOBER-FREIMOSER A, FRIEDLI L, GU Z, GUAN P, GUIGO R, HILLENMEYER ME, HLADUN SL, HOGAN JR, HONG YS, HOOVER J, JAILLON O, KE Z, KODIRA C, KOKOZA E, KOUTSOS A, LETUNIC I, LEVITSKY A, LIANG Y, LIN JJ, LOBO NF, LOPEZ JR, MALEK JA, MCINTOSH TC, MEISTER S, MILLER J, MOBARRY C, MONGIN E, MURPHY SD, O’BROCHTA DA, PFANNKOCH C, QI R, REGIER MA, REMINGTON K, SHAO H, SHARAKHOVA MV, SITTER CD, SHETTY J, SMITH TJ, STRONG R, SUN J, THOMASOVA D, TON LQ, TOPALIS P, TU Z, UNGER MF, WALENZ B, WANG A, WANG J, WANG M, WANG X, WOODFORD KJ, WORTMAN JR, WU M, YAO A, ZDOBNOV EM, ZHANG H, ZHAO Q, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–49. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- IHARA H, OKADA T, IKEDA Y. Cloning, expression and characterization of Bombyx mori α1,6-fucosyltransferase. Biochem Biophys Res Commun. 2014;450:953–60. doi: 10.1016/j.bbrc.2014.06.087. [DOI] [PubMed] [Google Scholar]
- JAN LY, JAN YN. Antibodies to horseradish peroxidase as specific neuronal markers in Drosophila and in grasshopper embryos. Proc Natl Acad Sci USA. 1982;79:2700–2704. doi: 10.1073/pnas.79.8.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JULIANT S, HARDUIN-LEPERS A, MONJARET F, CATIEAU B, VIOLET ML, CERUTTI P, OZIL A, DUONOR-CERUTTI M. The α1,6-fucosyltransferase gene (fut8) from the Sf9 lepidopteran insect cell line: insights into fut8 evolution. PLoS One. 2014;9:e110422. doi: 10.1371/journal.pone.0110422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KATZ F, MOATS W, JAN YN. A carbohydrate epitope expressed uniquely on the cell surface of Drosophila neurons is altered in the mutant nac (neurally altered carbohydrate) EMBO J. 1988;7:3471–3477. doi: 10.1002/j.1460-2075.1988.tb03222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIMURA Y, USHIJIMA T, MAEDA M, HAMA Y, KIMURA M, OKIHARA K, SUGIMOTO H, YAMADA H. Tumor antigen occurs in N-glycan of royal jelly glycoproteins: honeybee cells synthesize T-antigen unit in N-glycan moiety. Biosci Biotechnol Biochem. 2006;70:2583–7. doi: 10.1271/bbb.60331. [DOI] [PubMed] [Google Scholar]
- KUBELKA V, ALTMANN F, MARZ L. The asparagine-linked carbohydrate of honeybee venom hyaluronidase. Glycoconjugate J. 1995;12:77–83. doi: 10.1007/BF00731872. [DOI] [PubMed] [Google Scholar]
- KUBELKA V, ALTMANN F, STAUDACHER E, TRETTER V, MARZ L, HARD K, KAMERLING JP, VLIEGENTHART JFG. Primary structures of the N-linked carbohydrate chains from honeybee venom phospholipase A2. Eur J Biochem. 1993;213:1193–204. doi: 10.1111/j.1432-1033.1993.tb17870.x. [DOI] [PubMed] [Google Scholar]
- KURZ S, AOKI K, JIN C, KARLSSON NG, TIEMEYER M, WILSON IB, PASCHINGER K. Targeted release and fractionation reveal glucuronylated and sulphated N- and O-glycans in larvae of dipteran insects. J Proteomics. 2015;126:172–188. doi: 10.1016/j.jprot.2015.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI JS, LI J. Characterization of N-linked oligosaccharides in chorion peroxidase of Aedes aegypti mosquito. Protein Sci. 2005;14:2370–86. doi: 10.1110/ps.051419105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MINAGAWA S, SEKIGUCHI S, NAKASO Y, TOMITA M, TAKAHISA M, YASUDA H. Identification of Core α1,3-Fucosyltransferase Gene From Silkworm: An Insect Popularly Used to Express Mammalian Proteins. J Insect Sci. 2015:15. doi: 10.1093/jisesa/iev088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MUCHA J, DOMLATIL J, LOCHNIT G, RENDIĆ D, PASCHINGER K, HINTERKORNER G, HOFINGER A, KOSMA P, WILSON IBH. The Drosophila melanogaster homologue of the humna histo-blood group Pk gene encodes a glycolipid-modifying α1,4-N-acetylgalactosaminyltransferase. Biochem J. 2004;382:67–74. doi: 10.1042/BJ20040535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEMČOVIČOVA I, SESTAK S, RENDIC D, PLSKOVA M, MUCHA J, WILSON IBH. Characterisation of class I and II α-mannosidases from Drosophila melanogaster. Glycoconj J. 2013;30:899–909. doi: 10.1007/s10719-013-9495-5. [DOI] [PubMed] [Google Scholar]
- PASCHINGER K, FABINI G, SCHUSTER D, RENDIĆ D, WILSON IBH. Definition of immunogenic carbohydrate epitopes. Acta Biochim Pol. 2005a;52:629–632. [PubMed] [Google Scholar]
- PASCHINGER K, HYKOLLARI A, RAZZAZI-FAZELI E, GREENWELL P, LEITSCH D, WALOCHNIK J, WILSON IBH. The N-glycans of Trichomonas vaginalis contain variable core and antennal modifications. Glycobiology. 2012;22:300–313. doi: 10.1093/glycob/cwr149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PASCHINGER K, RENDIĆ D, LOCHNIT G, JANTSCH V, WILSON IBH. Molecular basis of anti-horseradish peroxidase staining in Caenorhabditis elegans. J Biol Chem. 2004;279:49588–49598. doi: 10.1074/jbc.M408978200. [DOI] [PubMed] [Google Scholar]
- PASCHINGER K, STAUDACHER E, STEMMER U, FABINI G, WILSON IBH. Fucosyltransferase substrate specificity and the order of fucosylation in invertebrates. Glycobiology. 2005b;15:463–474. doi: 10.1093/glycob/cwi028. [DOI] [PubMed] [Google Scholar]
- PATEL NH, SNOW PM, GOODMAN CS. Characterization and cloning of fasciclin III: a glycoprotein expressed on a subset of neurons and axon pathways in Drosophila. Cell. 1987;48:975–88. doi: 10.1016/0092-8674(87)90706-9. [DOI] [PubMed] [Google Scholar]
- PINZON-ORTIZ C, FRIEDMAN J, ESKO J, SINNIS P. The binding of the circumsporozoite protein to cell surface heparan sulfate proteoglycans is required for plasmodium sporozoite attachment to target cells. J Biol Chem. 2001;276:26784–91. doi: 10.1074/jbc.M104038200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POLTL G, AHRAZEM O, PASCHINGER K, IBANEZ MD, SALCEDO G, WILSON IBH. Molecular and immunological characterization of the glycosylated orange allergen Cit s 1. Glycobiology. 2007;17:220–30. doi: 10.1093/glycob/cwl068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RENDIĆ D, KLAUDINY J, STEMMER U, SCHMIDT J, PASCHINGER K, WILSON IBH. Towards abolition of immunogenic structures in insect cells: characterization of a honey-bee (Apis mellifera) multi-gene family reveals both an allergy-related core α1,3-fucosyltransferase and the first insect Lewis-histo-blood-group-related antigen-synthesizing enzyme. Biochem J. 2007;402:105–15. doi: 10.1042/BJ20060964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RENDIĆ D, LINDER A, PASCHINGER K, BORTH N, WILSON IBH, FABINI G. Modulation of neural carbohydrate epitope expression in Drosophila melanogaster cells. J Biol Chem. 2006;281:3343–3353. doi: 10.1074/jbc.M508334200. [DOI] [PubMed] [Google Scholar]
- RENDIC D, SHARROW M, KATOH T, OVERCARSH B, NGUYEN K, KAPURCH J, AOKI K, WILSON IB, TIEMEYER M. Neural-specific alpha3-fucosylation of N-linked glycans in the Drosophila embryo requires fucosyltransferase A and influences developmental signaling associated with O-glycosylation. Glycobiology. 2010;20:1353–65. doi: 10.1093/glycob/cwq119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RUDIN W, HECKER H. Lectin-binding sites in the midgut of the mosquitoes Anopheles stephensi Liston and Aedes aegypti L. (Diptera: Culicidae) Parasitol Res. 1989;75:268–79. doi: 10.1007/BF00931811. [DOI] [PubMed] [Google Scholar]
- SCHILLER B, HYKOLLARI A, VOGLMEIR J, POLTL G, HUMMEL K, RAZZAZI-FAZELI E, GEYER R, WILSON IBH. Development of Dictyostelium discoideum is associated with alteration of fucosylated N678 glycan structures. Biochem J. 2009;423:41–52. doi: 10.1042/BJ20090786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHILLER B, HYKOLLARI A, YAN S, PASCHINGER K, WILSON IBH. Complicated N-linked glycans in simple organisms. Biol Chem (Hoppe Seyler) 2012;393:661–673. doi: 10.1515/hsz-2012-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIDDIQUI SS, CULOTTI JG. Examination of neurons in wild type and mutants of Caenorhabditis elegans using antibodies to horseradish peroxidase. J Neurogenet. 1991;7:193–211. doi: 10.3109/01677069109167433. [DOI] [PubMed] [Google Scholar]
- SMIT CH, VAN DIEPEN A, NGUYEN DL, WUHRER M, HOFFMANN KF, DEELDER AM, HOKKE CH. Glycomic Analysis of Life Stages of the Human Parasite Schistosoma mansoni Reveals Developmental Expression Profiles of Functional and Antigenic Glycan Motifs. Mol Cell Proteomics. 2015;14:1750–69. doi: 10.1074/mcp.M115.048280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SNOW PM, PATEL NH, HARRELSON AL, GOODMAN CS. Neural-specific carbohydrate moiety shared by many surface glycoproteins in Drosophila and grasshopper embyros. J Neurosci. 1987;7:4137–4144. doi: 10.1523/JNEUROSCI.07-12-04137.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STAUDACHER E, ALTMANN F, WILSON IBH, MARZ L. Fucose in N-glycans: from plant to man. Biochim Biophys Acta. 1999;1473:216–36. doi: 10.1016/s0304-4165(99)00181-6. [DOI] [PubMed] [Google Scholar]
- SUN B, XU P, SALVATERRA PM. Dynamic visualization of nervous system in live Drosophila. Proc Natl Acad Sci U S A. 1999;96:10438–43. doi: 10.1073/pnas.96.18.10438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TOMIYA N, AWAYA J, KURONO M, ENDO S, ARATA Y, TAKAHASHI N. Analyses of N-linked oligosaccharides using a two-dimensional mapping technique. Anal Biochem. 1988;171:73–90. doi: 10.1016/0003-2697(88)90126-1. [DOI] [PubMed] [Google Scholar]
- TOMIYA N, LEE YC, YOSHIDA T, WADA Y, AWAYA J, KURONO M, TAKAHASHI N. Calculated two-dimensional sugar map of pyridylaminated oligosaccharides: Elucidation of the jack bean a-mannosidase digestion pathway of Man9GlcNAc2. Analytical Biochemistry. 1991;193:90–100. doi: 10.1016/0003-2697(91)90047-w. [DOI] [PubMed] [Google Scholar]
- TRETTER V, ALTMANN F, KUBELKA V, MARZ L, BECKER WM. Fucose α1,3-linked to the core region of glycoprotein N-glycans creates an important epitope for IgE from honeybee venom allergic individuals. Int Arch Allergy Immunol. 1993;102:259–66. doi: 10.1159/000236534. [DOI] [PubMed] [Google Scholar]
- WANG X, SUN B, YASUYAMA K, SALVATERRA PM. Biochemical analysis of proteins recognised by anti-HRP antibodies in Drosophila melanogaster: Identification of neuron specific and male specific glycoproteins. Insect Biochem Mol Biol. 1994;24:233–242. doi: 10.1016/0965-1748(94)90002-7. [DOI] [PubMed] [Google Scholar]
- WHITE NJ, PUKRITTAYAKAMEE S, HIEN TT, FAIZ MA, MOKUOLU OA, DONDORP AM. Malaria. Lancet. 2014;383:723–35. doi: 10.1016/S0140-6736(13)60024-0. [DOI] [PubMed] [Google Scholar]
- WILKINS S, BILLINGSLEY PF. Partial characterization of oligosaccharides expressed on midgut microvillar glycoproteins of the mosquito, Anopheles stephensi Liston. Insect Biochem Mol Biol. 2001;31:937–48. doi: 10.1016/s0965-1748(01)00040-6. [DOI] [PubMed] [Google Scholar]
- WILSON IBH, RENDIĆ D, DUMIĆ J, FREILINGER A, ALTMANN F, MUCHA J, MULLER S, HAUSER MT. Cloning and expression of α1,3-fucosyltransferase homologues from Arabidopsis thaliana. Biochim Biophys Acta. 2001;1527:88–96. doi: 10.1016/s0304-4165(01)00151-9. [DOI] [PubMed] [Google Scholar]
- YAN S, BLEULER-MARTINEZ S, PLAZA GUTIERREZ DF, KUENZLER M, AEBI M, JOACHIM A, RAZZAZI-FAZELI E, JANTSCH V, GEYER R, WILSON IBH, PASCHINGER K. Galactosylated fucose epitopes in nematodes: increased expression in a Caenorhabditis mutant associated with altered lectin sensitivity and occurrence in parasitic species. J Biol Chem. 2012;287:28276–28290. doi: 10.1074/jbc.M112.353128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAN S, WILSON IBH, PASCHINGER K. Comparison of RP-HPLC modes to analyse the N-glycome of the free-living nematode Pristionchus pacificus. Electrophoresis. 2015;36:1314–29. doi: 10.1002/elps.201400528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHAO Y, SATO Y, ISAJI T, FUKUDA T, MATSUMOTO A, MIYOSHI E, GU J, TANIGUCHI N. Branched N-glycans regulate the biological functions of integrins and cadherins. Febs J. 2008;275:1939–48. doi: 10.1111/j.1742-4658.2008.06346.x. [DOI] [PubMed] [Google Scholar]