Abstract
Studies of rad52 mutants in Saccharomyces cerevisiae have revealed a critical role of Rad52 protein in double-strand break repair and meiosis, and roles in both RAD51-dependent and -independent pathways of recombination. In vitro, both yeast and human Rad52 proteins play auxiliary roles with RPA in the action of Rad51. Rad52 also has annealing activity and promotes the formation of D-loops in superhelical DNA. The experiments described here show that Homo sapiens (Hs)Rad52 and yeast Rad52 proteins promote strand exchange as well. Strand exchange was promoted by the N-terminal domain of HsRad52 that contains residues 1–237, which includes the residues required to form rings of Rad52, whereas other truncated domains, both N-terminal and C-terminal, were inactive. For both yeast Rad52 and HsRad52, the yield of strand-exchange reactions was proportional to the fractional A·T content of the DNA substrates, but both enzymes catalyzed exchange with substrates that contained up to at least 50% G·C. Observations made on S. cerevisiae Rad52 protein from mutants with severe recombination deficiencies indicate that the strand-exchange activity measured in vitro reflects a biologically significant property of Rad52 protein.
Recombination, homologous and nonhomologous, comprises a set of mechanisms that maintain genome integrity by rejoining broken molecules of DNA. The term “nonhomologous end joining” characterizes the process that does not depend on matching DNA sequences, whereas homologous recombination requires the pairing of complementary sequences. This match can be accomplished by the Watson–Crick pairing of complementary single strands, termed “single-strand annealing,” or by the invasion of duplex DNA by a complementary single strand, termed “strand invasion.” The latter requires that an unpaired single strand recognize a complementary strand that is already paired in a stable duplex structure, the mechanism of which remains uncertain (see Discussion). Under suitable conditions, strand exchange follows strand invasion, leading to the production of heteroduplex DNA.
rad52, the eponymous member of the major epistasis group for homologous recombination in Saccharomyces cerevisiae, has homologs in human cells, chicken cells, and fission yeast. Studies of RAD52 mutants in S. cerevisiae, revealed a critical role of RAD52 in double-strand break repair and meiosis (1). In vitro, S. cerevisiae (Sc)Rad52 and Homo sapiens (Hs)Rad52 proteins perform auxiliary functions in reactions promoted by Rad51, a RecA homolog, but in S. cerevisiae it has been shown that Rad52 plays an important role as well in RAD51-independent pathways (1). In vitro, both ScRad52 and HsRad52 were shown to promote single-strand annealing (2, 3), and HsRad52 was shown to promote strand invasion (4).
We have found, as reported here, that both ScRad52 and HsRad52, acting on oligonucleotide substrates, promote strand exchange. This enlarged view of the biochemical activities of Rad52 may provide further insight into its critical roles in S. cerevisiae, and, taken together with other observations, it suggests a unifying view of the mechanisms underlying strand invasion and strand exchange.
Materials and Methods
Purification of HsRad52 Protein and Its Truncated Derivatives. The complete coding sequence of the HsRad52 gene was amplified by PCR from plasmid pEG2 (3) and inserted into bacterial expression vector pET 15b (Novagen) between NdeI and BamHI sites in frame with a 5′-end sequence coding for a series of six histidine residues, which function as a metal-binding domain in the translated fusion protein. The resulting plasmid was designated pEG161H1. The sequence of the cloned cDNA for HsRad52 was identical to the sequence published by Muris et al. (5). All truncated HsRad52 gene derivatives were made by PCR using plasmid pEG161H1 as a template. The fragments were inserted into bacterial expression vector pQE-31 (Qiagen, Valencia, CA) between BamH1 and KpnI sites in-frame with a 5′-terminal sequence coding for His6 residues. Truncated proteins EG174p, EG176p, EG181p, and EG182p carry amino acids 1–237, 1–184, 237–418, and 260–340, respectively. The plasmid was introduced into Escherichia coli strain BL21(DE3)pLys (Novagen). Bacteria were grown in TB medium (1.2% Bacto tryptone/2.4% Bacto yeast extract/17 mM KH2PO4/72 mM K2HPO4/0.4%, vol/vol, glycerol) (6) at 37°C to OD600 = 1.8, at which time synthesis of HsRad52 or its truncated derivative was induced by 1 mM isopropyl β-d-thiogalactoside. The proteins were isolated by using Ni-NTA (Qiagen) and SP-Sepharose (Amersham Biosciences) columns.
Purification of ScRad52 Protein and Mutants. ScRad52 protein was expressed in E. coli JM109 by using plasmid pQE60-Rad52*-His6. The plasmid encodes a biologically competent recombinant ScRad52 protein, Rad52*-His6, which starts from the 34th N-terminal amino acid residue of the protein and has a His6-tag fused to its C terminus (2). Mutated genes ScRad52–1(N91A) (7) and ScRad52(F173A) (8) were made by PCR by using as a template ScRAD52 gene of plasmid pQE60-Rad52*-His6 (2). The sequences of all genes were confirmed by comparison with the published sequences.
For isolation of ScRad52 proteins or its mutants, E. coli JM109 carrying the relevant plasmid was grown in 2 liters of TB medium at 37°C until OD600 = 2.0, and synthesis of the protein was then induced by 1 mM isopropyl β-d-thiogalactoside. After 2.5 h, bacteria were harvested and lysed by sonication. Cell debris was removed by centrifugation at 27,000 × g for 20 min. ScRad52 was purified by chromatography through Ni-NTA and single-stranded (ss)DNA-cellulose columns.
The protein preparations described above produced a single band on gel electrophoresis and lacked detectable nuclease activity toward ssDNA or double-stranded DNA.
DNA Substrates. Sequences of 83-mer single-stranded oligonucleotides G16(-), G37(-), and G50(-) were described; the numbers in each name refer to the percentage of G·C content (9). The complementary strands are denoted as G16(+), G37(+), and G50(+), and the duplex forms as G16(+)/G16(-), etc.
EG673 was derived from G16(-) by making nine transversions, as indicated by underlined characters in the following sequences: AAATGAACATAAAGTAAATAAGTATAAGGTAAATACTAAATA and AGAAAATGAATAAACTTAGTAAATAAAGAAAAGGTAATAAA.
DNA Strand-Exchange Reactions. The 5′-32P-labeled 83-mers, G16(-), G37(-), or G50(-), were annealed with their complementary strands to form duplex DNA. HsRad52 protein, its truncated derivatives, or ScRad52 protein, were preincubated with the indicated single-stranded oligonucleotides at 37°C in a solution containing 25 mM Tris·HCl (pH 7.5), 1 mM MgCl2, 1 mM DTT, 0.1 mg/ml BSA, and 20 mM NaCl for 5 min at 37°C, after which duplex DNA was added at a final concentration of 2.5 μM. After 40 min, the reactions were stopped by stop solution (final concentrations: 0.5% SDS/50 mM EDTA/10 mg/ml proteinase K/0.5% glycerol/0.04% bromophenol blue/0.04% xylene cyanol), loaded onto a 15% polyacrylamide gel, and analyzed by electrophoresis at 20 V/cm for 3 h in 0.5× TBE (90 mM Tris·borate, pH 8.3/2 mM EDTA). The general procedure for RecA protein was the same except that the reaction mixture contained 50 mM Tris·HCl (pH 7.5), 20 mM MgCl2, 1 mM DTT, 2 mM ATP, 100 μg/ml BSA, and an ATP-regeneration system consisting of 10 units/ml pyruvate kinase and 2 mM phosphoenolpyruvate.
Results
Experimental System. Previous observations on a small but diverse and widespread group of recombination enzymes have shown that the yields of pairing and strand-exchange reactions in vitro are proportional to the fractional content of A·T base pairs in the DNA substrates (9–12). By varying the A·T content of substrates in the experiments described here, we were able to find and characterize strand-exchange activities of yeast Rad52 and HsRad52.
Strand Exchange Promoted by HsRad52. In the following experiments, the substrates for strand exchange consisted of an unlabeled single-stranded oligonucleotide, called the minus strand, and a homologous duplex oligonucleotide in which the minus strand was labeled with 32P at its 5′ end. Strand exchange was indicated by the homology-dependent displacement of the labeled strand from a duplex to a single-stranded form, as detected by gel electrophoresis.
HsRad52 promoted strand exchange in a homology-dependent reaction when acting on 83-mer oligonucleotides with 16% G·C base pairs (Fig. 1). No exchange occurred when a heterologous single strand was added (Fig. 1, lane 2). Exchange was optimal at approximately one molecule of HsRad52 protein per 15- to 20-nt residues of ssDNA (Fig. 1 and data not shown). Strand exchange promoted by HsRad52 reached its maximum in ≈30 min (Fig. 2).
Fig. 1.
HsRad52 protein promotes strand exchange. Reactions were carried out as described in Materials and Methods. The oligonucleotide G16(-)(6 μM) was preincubated for 10 min with the indicated amounts of HsRad52 at 37°C, followed by addition of 2.5 μM homologous duplex oligonucleotide (5′-labeled with 32P on the minus strand). After 40 min, the reactions were stopped and analyzed by electrophoresis in 15% polyacrylamide and 0.5× TBE buffer (see Materials and Methods). The 32P-labeled ssDNA product was quantified by PhosphorImager analysis. Lane 1, no ssDNA; lane 2, heterologous ssDNA derived from the G16(-) oligonucleotide by making nine transversion mismatches; lane 3, no Rad52 protein; and lanes 4–7, increasing amounts of Rad52 as indicated. (At concentrations of Rad52 <0.4 μM, the yields decreased.)
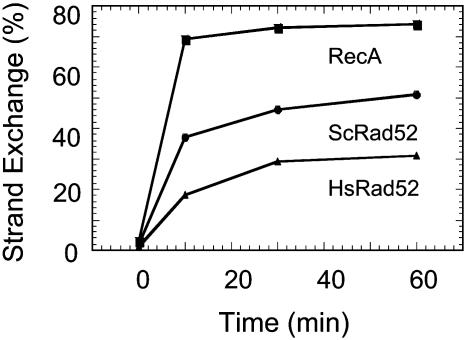
Fig. 2.
Time course of strand exchange promoted by HsRad52, ScRad52, and RecA proteins. Reactions of RecA protein contained 1 μM RecA, 3 μM ssDNA, and 2.5 μM duplex DNA. Reactions of HsRad52 contained 0.4 μM HsRad52, 6μM ssDNA, and 2.5 μM duplex DNA. Reactions of ScRad52 contained 1.5 μM ScRad52, 20 μM ssDNA, and 2.5 μM duplex DNA. Other conditions of reactions and gel electrophoresis were as described in Materials and Methods.
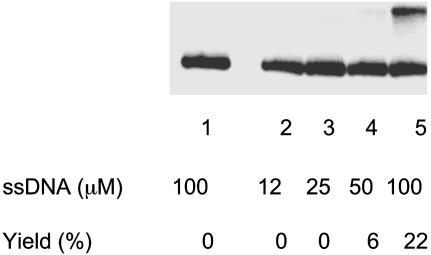
Under the conditions of the reaction that were just described (see Fig. 1 legend), HsRad52 did not promote strand exchange significantly when the content of G·C base pairs was approximately twice as great. We found, however, that a 5-fold increase in the concentration of protein and a 16-fold increase in the concentration ssDNA enabled HsRad52 to promote strand exchange with DNA that had 50% G·C base pairs (Fig. 3).
Fig. 3.
Promotion of strand exchange by HsRad52 with DNA containing 50% G·C base pairs. The reaction was carried out as described in Materials and Methods. Lane 1, no protein; lanes 2–5, 1.5 μM HsRad52 protein was incubated with the indicated concentrations of ssDNA, G50(-), followed by the addition of homologous duplex DNA (at 2.5 μM) and incubation for 40 min at 37°C.
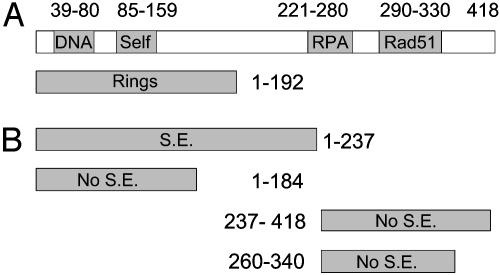
The N-Terminal Domain of HsRad52 Promotes Strand Exchange. The functions of HsRad52 have been mapped by various investigators, as diagrammed in Fig. 4A (4, 13–19). We used that map to validate further the strand-exchange activity of HsRad52. Rad52 forms several superstructures, including rings. N-terminal domains consisting respectively of amino acids 1–192, 1–209, 1–212, or 1–237 form rings and have variously been shown to bind ssDNA, anneal complementary single-strands, and promote the formation of D-loops. Two C-terminal moieties of Rad52 protein contain, respectively, the sites of interaction with RPA (221–280) and Rad51 (291–330).
Fig. 4.
Mapping the strand-exchange activity of HsRad52 protein. (A) A map of known domains of WT HsRad52 and truncated derivatives (see text). “DNA” identifies a site required for binding DNA; Self, one site required to form Rad52 multimers. (B) Summary of strand-exchange activity (S.E.) of tested fragments of HsRad52.
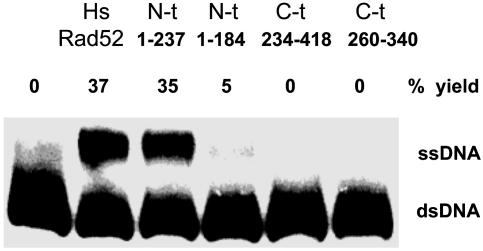
In a direct comparison, the domain comprising amino acids 1–237 promoted strand exchange as well as WT HsRad52, whereas an N-terminal fragment consisting of residues 1–184 did not catalyze exchange (Fig. 5). The latter Rad52 fragment is only 8 aa shorter than the shortest demonstrated ring-forming domain, 1–192. We also tested the C-terminal fragment from 237 to 418 and a shorter C-terminal fragment, neither of which promoted strand exchange. These observations, summarized in Fig. 4B, correlate with the published observations on the functionality of the N-terminal domain.
Fig. 5.
The N-terminal domain of HsRad52 promotes strand exchange. WT HsRad52 and truncated derivatives: Rad52 (1-237), Rad52 (1–184), Rad52 (234–418), or Rad52 (260–340), all at 0.4 μM, were incubated with 6 μM oligonucleotide G16(-), followed by addition of duplex oligonucleotide as described in Materials and Methods.
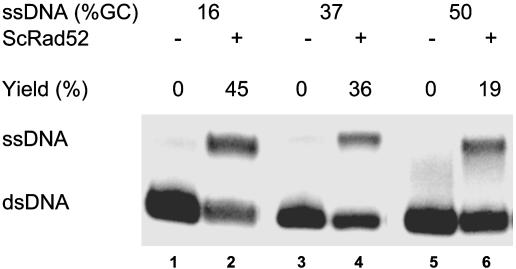
Promotion of Strand Exchange by ScRad52. Rad52 protein from S. cerevisiae also promoted strand exchange. Indeed, in direct comparisons, the yields of reactions promoted by ScRad52 were greater than those produced by HsRad52 (Fig. 2). The yeast enzyme was also sensitive to the G·C content of the DNA substrates (Fig. 6). However, even at 50% G·C content, ScRad52 yielded 19% strand exchange, but this yield was not increased by increasing the concentration of ssDNA and ScRad52 protein.
Fig. 6.
Promotion of strand exchange by ScRad52. Reactions were carried out as described in Materials and Methods with three different oligonucleotide substrates the G·C contents of which were 16%, 37%, and 50%, respectively. ScRad52 protein (1.5 μM) was preincubated with the three different single-stranded oligonucleotides at 20 μM for 5–10 min at 37°C. Double-stranded oligonucleotide (2.5 μM) containing a 5′-32P-labeled minus strand was added, and incubation was continued at 37°C for 40 min.
Strand exchange by ScRad52 required homology (data not shown), and the time course of exchange was similar to that for HsRad52 and E. coli RecA protein (Fig. 2).
Lack of Melting Activity of Rad52 Proteins. Lio et al. (20) recently reported that HsRad51C protein has an apparent strand-exchange activity that resulted from the protein-dependent melting of duplex oligonucleotides. After deproteinization of reaction mixtures, random spontaneous renaturation produced some heteroduplex DNA and displaced single strands, which was detected by adding an excess of homologous ssDNA together with the deproteinizing stop solution. We used a similar protocol to determine whether the activities of ScRad52 and HsRad52 described above are attributable to bona fide strand exchange activity.
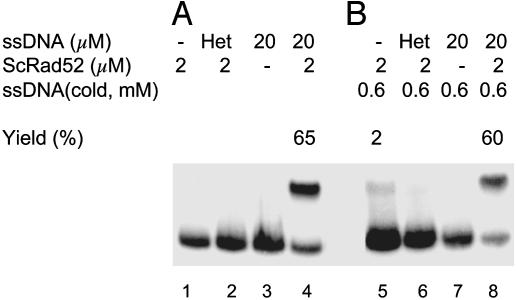
We examined reactions of yeast Rad52 in which we incubated terminally labeled duplex DNA, G16(-)32P/G16(+), for 40 min at 37°C either with or without heterologous single strands, and, upon deproteinization, we added unlabeled G16(-) strands at 600 mM, in 480-fold excess of the G16(-)32P strand with which it was intended to compete. If Rad52 had melted G16(-)32P/G16(+), spontaneous renaturation of G16(+) strands with unlabeled G16(-) strands that were present in large excess should have depleted all G16(+) strands and produced G16(-)32P single strands.
In Fig. 7A, as in reactions described above, ScRad52 yielded 65% displaced G16(-)32P single strands only when protein and unlabeled homologous G16(-) strands were present (lane 4). In Fig. 7B, a 480-fold excess of G16(-) strands was added together with stop-solution at the end of the 40-min incubation at 37°C. Fig. 7B, lane 8 was a control, a complete reaction, which showed that the addition of a large excess of unlabeled G16(-) strands added with the stop solution did not significantly alter the yield of G16(-)32P single strands displaced from the original duplex substrate. In lanes 5–7, reactions lacking any ssDNA, containing heterologous ssDNA, or lacking Rad52 there was no significant production of 32P-ssDNA, hence no evidence of melting of the duplex DNA, with or without Rad52. The experiment was repeated several times, and similar results were also obtained for HsRad52 (data not shown).
Fig. 7.
Rad52-promoted strand exchange is not mediated by a DNA-melting activity. The reaction was carried out as described in Materials and Methods. The concentration of double-stranded DNA was 2.5 μM. (A, lanes 1–4) Standard conditions, incubation at 37°C for 40 min. Contents of each reaction as indicated. Lane 2, (Het) nine transversion mismatches in G16(-) oligonucleotide as a heterologous control. (B, lanes 5–8) Contents of each reaction were the same as the corresponding lanes in A, except that a 480-fold excess of G16(-) strands, 600 μM, was added with stop solution after incubation of reaction mixtures at 37°C for 40 min.
If melting had occurred but conditions were not favorable for renaturation, then ssDNA should have appeared in control reactions lacking the 480-fold excess of unlabeled G16(-) strands, but none was present (Fig. 7A, lanes 1–3). In addition, to determine directly whether conditions were suitable for renaturation, we incubated unlabeled G16(-)/G16(+) with and without Rad52 and, upon deproteinization, added G16(-)32P and a 25-fold excess of G16(+), in which case all of the labeled strands were recovered in double-stranded form as a result of spontaneous renaturation (data not shown).
We conclude that under the conditions of strand-exchange reactions, our most A·T-rich, most unstable DNA substrate did not melt, either spontaneously or as a result of the presence of yeast Rad52 or HsRad52 protein.
Effect of ScRad52 Mutants on Strand Exchange in Vitro. Among the genes of the RAD52 epistasis group in yeast, mutations of RAD52 itself have the strongest effect on recombination (8). To assess the biological relevance of the strand-exchange activity of Rad52 protein described here, we purified and tested two mutant proteins, rad52–1 (N91A) (7), and rad52-F173A (8), both of which are located in the N-terminal portion of the protein and have severe recombination phenotypes. The mutant protein Rad52(F173A) was strongly defective in strand-exchange activity in the presence of both 10 and 50 mM NaCl, whereas Rad52(N91A) was defective in the presence of 50 mM salt (Table 1). These observations support the conclusion that the activity measured in vitro reflects a biologically significant property of Rad52 protein.
Table 1. Effect of ScRad52 mutants on strand exchange in vitro.
| Strand exchange, %
|
|||
|---|---|---|---|
| NaCl | WT | N91A | F173A |
| 10 mM | 23 | 26 | 3 |
| 50 mM | 29 | 0 | 3 |
Wild-type and mutant proteins (1.5 μM) were preincubated with 20 μM 83-mer (G16-) for 5-10 min at 37°C in the presence of different concentrations of NaCl. Labeled homologous duplex DNA was added at 2.5 μM, the reaction was continued for 40 min at 37°C, and strand exchange was measured as described in Materials and Methods.
Discussion
The experiments described here show that HsRad52 protein and Rad52 from budding yeast, acting on 83-mer oligonucleotides can promote strand exchange. This exchange is favored by substrates that contain a large fraction of A·T base pairs, but conditions were readily found under which yeast Rad52 and HsRad52 catalyzed strand exchange with DNA substrates that contained 50% G·C base pairs. Experiments with our most A·T-rich substrate revealed no significant melting activity of yeast Rad52. Thus, any hypothetical melting of A·T-rich substrates occurs too slowly to account for the observed strand-exchange activities of Rad52.
Oligonucleotides, whose DNA sequence and composition can be modified readily, played an important role in uncovering the strand-exchange activities of the human and yeast Rad52 proteins, but oligonucleotides do not provide information about the propagation of strand exchange, which in the case of RecA protein and its homologs can produce heteroduplex DNA products that are kilobases in length. The extent to which Rad52 can propagate strand exchange remains to be determined.
Other studies have established that, in vitro and in vivo, Rad52 interacts with Rad51 and RPA to play an auxiliary or mediator function that potentiates the action of Rad51. On the basis of in vitro studies, it has been proposed that this mediator function governs the loading of Rad51 on DNA. The annealing activity of Rad52 in vitro has been invoked as the basis for the single-strand annealing pathway of recombination in vivo (1). The demonstration that Rad52 can promote the formation of D-loops opened the possibility of another more direct action of Rad52 in strand invasion, independent of Rad51 (4). The present studies add further evidence of the multifunctionality of Rad52 and its potential for initiating recombination in the absence of Rad51.
The present observations add to a list of diverse enzymes that are capable of promoting the full set of enzymatic functions that are central to homologous recombination, namely, single-strand annealing, strand invasion, and strand exchange. The latter two functions, strand invasion and strand exchange, are common to a set comprising prokaryotic, eukaryotic, ATP-dependent, and ATP-independent recombination enzymes, including HsRad51, Rad52, and Dmc1, budding yeast Rad52, E. coli RecA and RecT, β protein of phage λ, and UvsX of phage T4 (10, 12, 21–26, and unpublished observations). Single-strand annealing has been demonstrated for all of these except, to our knowledge, Dmc1 and UvsX. Is there a common mechanism?
In a study on the effect of base composition on HsRad51, Gupta et al. (9) provided evidence that recognition of homology, or formation of synaptic complexes, which occurs before strand exchange, requires the switching of base pairs, mediated preferentially by A·T base pairs. More limited evidence indicated the same to be true of human Dmc1 and RecT (11, 12). In one set of experiments on HsRad51, as the A·T content of the substrates was raised, the kinetic difference between recognition of homology and strand exchange progressively diminished until, at 84% A·T, no kinetic difference was detectable (9). This observation suggests that recognition of homology and initiation of strand exchange are mechanistically the same process. Both recognition and exchange are accomplished by the switching of base pairs, but when the content of DNA is not strongly biased toward A·T base pairs, strand exchange is slower than the formation of three-stranded synaptic complexes because G·C base pairs switch more slowly. Thus, if recognition of homology and strand exchange are mechanistically the same process separated kinetically only by the slower exchange of G·C base pairs, it is not surprising that any enzyme that can promote strand invasion can promote strand exchange. Such is the case for the set of enzymes cited above. According to this hypothesis, the common mechanism that links this diverse group of enzymes is the switching of base pairs. In the case of E. coli RecA protein and HsRad51 protein, Nishinaka and colleagues (27, 28) solved the structure of DNA within the nucleoprotein filament and suggested how that structure promotes the switching of base pairs. The preferential switching of A·T base pairs that has been proposed as the basis for the recognition of homology (9) is consistent with a fundamental property of DNA in solution, namely the breathing of base pairs, which occurs more rapidly for A·T than G·C base pairs (29).
Acknowledgments
We thank Rodney Rothstein for plasmid pQE60-Rad52*-His6, Kavitha Kabaleeswaran for technical assistance, Jin Li Liu for assistance with computer graphics, Ewa Folta-Stogniew for scientific critiques, and Janice Zulkeski for administrative assistance. This work was supported by National Institute of General Medical Sciences Grant R37-GM33504.
Abbreviations: ssDNA, single-stranded DNA; Sc, Saccharomyces cerevisiae; Hs, Homo sapiens.
References
- 1.Symington, L. S. (2002) Microbiol. Mol. Biol. Rev. 66, 630-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mortensen, U. H., Bendixen, C., Sunjevaric, I. & Rothstein, R. (1996) Proc. Natl. Acad. Sci. USA 93, 10729-10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reddy, G., Golub, E. I. & Radding, C. M. (1997) Mutat. Res. 377, 53-59. [DOI] [PubMed] [Google Scholar]
- 4.Kagawa, W., Kurumizaka, H., Ikawa, S., Yokoyama, S. & Shibata, T. (2001) J. Biol. Chem. 276, 35201-35208. [DOI] [PubMed] [Google Scholar]
- 5.Muris, D. F. R., Bezzubova, O., Buerstedde, J.-M., Vreeken, K., Baljee, A. S., Osgood, C. J., Troelstra, C., Hoeijmakers, J. H. J., Osterman, K., Schmidt, H., et al. (1994) Mutat. Res. DNA Repair 315, 295-305. [DOI] [PubMed] [Google Scholar]
- 6.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 7.Adzuma, K., Ogawa, T. & Ogawa, H. (1984) Mol. Cell. Biol. 4, 2735-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mortensen, U., H., Erdeniz, N., Feng, Q. & Rothstein, R. (2002) Genetics 161, 549-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta, R. C., Folta-Stogniew, E., O'Malley, S. O., Takahashi, M. & Radding, C. M. (1999) Mol. Cell 4, 705-714. [DOI] [PubMed] [Google Scholar]
- 10.Gupta, R. C., Folta-Stogniew, E. & Radding, C. M. (1999) J. Biol. Chem. 274, 1248-1256. [DOI] [PubMed] [Google Scholar]
- 11.Gupta, R. C., Golub, E., Bi, B. & Radding, C. M. (2001) Proc. Natl. Acad. Sci. USA 98, 8433-8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noirot, P., Gupta, R. C., Radding, C. M. & Kolodner, R. D. (2003) EMBO J. 22, 324-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen, Z., Cloud, K. G., Chen, D. J. & Park, M. S. (1996) J. Biol. Chem. 271, 148-152. [DOI] [PubMed] [Google Scholar]
- 14.Park, M. S., Ludwig, D. L., Stigger, E. & Lee, S.-H. (1996) J. Biol. Chem. 271, 18996-19000. [DOI] [PubMed] [Google Scholar]
- 15.Kurumizaka, H., Aihara, H., Kagawa, W., Shibata, T. & Yokoyama, S. (1999) J. Mol. Biol. 291, 537-548. [DOI] [PubMed] [Google Scholar]
- 16.Ranatunga, W., Jackson, D., Lloyd, J., Forget, A. & Knight, K. (2001) J. Biol. Chem. 276, 15, 876-15880. [DOI] [PubMed] [Google Scholar]
- 17.Liu, J., Meng, X. & Shen, Z. (2002) Biochem. Biophys. Res. Comm. 297, 1191-1198. [DOI] [PubMed] [Google Scholar]
- 18.Kagawa, W., Kurumizaka, H., Ishitani, R., Fukai, S., Nureki, O., Shibata, T. & Yokoyama, S. (2002) Mol. Cell 10, 359-371. [DOI] [PubMed] [Google Scholar]
- 19.Singleton, M., Wentzell, L., Liu, Y., West, S. & Wigley, D. (2002) Proc. Natl. Acad. Sci. USA 99, 13492-13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lio, Y.-C., Mazin, A. V., Kowalczykowski, S. C. & Chen, D. J. (2003) J. Biol. Chem. 278, 2469-2478. [DOI] [PubMed] [Google Scholar]
- 21.Roca, A. I. & Cox, M. M. (1997) Prog. Nucleic Acid Res. 56, 129-223. [DOI] [PubMed] [Google Scholar]
- 22.Bianco, P. R., Tracy, R. B. & Kowalczykowski, S. C. (1998) Front. Biosci. 3, 570-603. [DOI] [PubMed] [Google Scholar]
- 23.Li, Z., Karakousis, G., Chiu, S. K. & Radding, C. M. (1998) J. Mol. Biol. 276, 733-744. [DOI] [PubMed] [Google Scholar]
- 24.Li, Z., Golub, E. I., Gupta, R. & Radding, C. M. (1997) Proc. Natl. Acad. Sci. USA 94, 11221-11226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, H.-K., Morimatsu, K., Norden, B., Ardhammer, M. & Takahashi, M. (2002) Genes Cells 7, 1125-1134. [DOI] [PubMed] [Google Scholar]
- 26.Formosa, T. & Alberts, B. M. (1986) J. Biol. Chem. 261, 6107-6118. [PubMed] [Google Scholar]
- 27.Nishinaka, T., A., S., Ito, Y., Yokoyama, S. & Shibata, T. (1998) Proc. Natl. Acad. Sci. USA 95, 11071-11076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishinaka, T., Ito, Y., Yokoyama, S. & Shibata, T. (1997) Proc. Natl. Acad. Sci. USA 94, 6623-6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frank-Kamenetskii, M. (1987) Nature 328, 17-18. [DOI] [PubMed] [Google Scholar]