Abstract
We report on a microfluidic platform for culture of whole organs or tissue slices with the capability of point access reagent delivery to probe the transport of signaling events. Whole mice retina were maintained for multiple days with negative pressure applied to tightly but gently bind the bottom of the retina to a thin poly-(dimethylsiloxane) membrane, through which twelve 100 μm diameter through-holes served as fluidic access points. Staining with toluidine blue, transport of locally applied cholera toxin beta, and transient response to lipopolysaccharide in the retina demonstrated the capability of the microfluidic platform. The point access fluidic delivery capability could enable new assays in the study of various kinds of excised tissues, including retina.
Keywords: Microfluidic tissue culture, Retina, Microenvironment
1 Introduction
Microfluidic culture techniques have the potential to make transformative impacts on biological studies through their capability of implementing spatiotemporal control in local microenvironments (Meyvantsson and Beebe 2008; Squires and Quake 2005). In response to biological research needs, various kinds of microfluidic platforms were developed to culture single cells (Di Carlo et al. 2006; Lecault et al. 2011), populations of single cell types (Hung et al. 2005; Taylor et al. 2005), and to co-culture multiple cell types (Gao et al. 2011; Khetani and Bhatia 2008). To study more complex biological systems, recent efforts further expanded the capability to tissues or even organs (Berdichevsky et al. 2010; Borenstein et al. 2010; Choi et al. 2007; Günther et al. 2010; Huang et al. 2012; Khademhosseini et al. 2006; Kim et al. 2010; Passeraub et al. 2003; Queval et al. 2010; Vickerman et al. 2008). Assays based on microfluidic culture platforms not only promote fundamental understanding of complex biological processes but also address important practical issues, including testing the effectiveness of drugs.
Realizing the capability of microfluidics in dealing with single cells, various platforms were developed to catch and trap single cells, and probe their response to certain external stimuli (Di Carlo et al. 2006; Lecault et al. 2011). These platforms enable a variety of fundamental biological studies through observation of a single cell, without ambiguity from factors such as averaging over a heterogeneous ensemble. However, it is well-known that cell-cell interactions are crucial to cell functions and the behavior of a single cell often differs remarkably from cells in a larger population. As such, significant attention was paid to platforms that can culture and probe large groups of cells (Hung et al. 2005; Taylor et al. 2005). To study the interactions between different cell types, a number of microfluidic cell co-culture platforms were developed to enable the study of controlled cell-cell interactions (Gao et al. 2011; Khetani and Bhatia 2008). Recently, there was a great deal of interest in culturing different cell types in microfluidic platforms and organizing them to mimic the function of tissues or even organs.
Microfluidics provides opportunities for two types of tissue research: growing tissues and studying their functions. With respect to the former, significant efforts were made to organize cells into tissues (tissue engineering) (Borenstein et al. 2010; Choi et al. 2007; Khademhosseini et al. 2006; Vickerman et al. 2008). However, fewer microfluidic platforms were designed to study existing tissues or organs. To date, only a few microfluidic schemes have been reported discussing the culture, manipulation, and probing of breast tissue (Kim et al. 2010), arteries (Günther et al. 2010), and brain slices (Berdichevsky et al. 2010; Huang et al. 2012; Passeraub et al. 2003; Queval et al. 2010). In these studies, perfusion techniques were utilized to diagnose breast cancer, examine cardiovascular disease, and probe brain activities.
Most of the current platforms for tissue culture utilize laminar flow or focal perfusion over tissue slices to provide fluid to a relatively large and not well-defined area. This is suitable for studies that do not require high precision spatial control of media flow. To manipulate reagent delivery with a better resolution, some pioneering efforts have been made for point access to brain slices. For example, microfluidic platforms with microchannels underneath brain slices were constructed, in which the channels are separated from the brain slices by a layer of poly-(dimethylsiloxane) (PDMS) to act as a roof and allow interaction with the brain slices only via small through-holes (Huang et al. 2012). However, applying the reagents by positive pressure in the microchannel causes the molecules to spread over a much larger region than that defined by the small through-holes, leading to a degraded spatial resolution in reagent delivery.
Here we report on a new design of microfluidic tissue culture platform with multiple reagent access points suitable for probing tissue slices. Combined with a negative-pressure operational protocol, this platform allows for long-term culture and point access reagent delivery. We demonstrate this capability by sustaining and probing excised mouse retina. One significant challenge in implementing this scheme is the naturally curved shape of retina, which is not ideally compatible with the planar nature of the microfluidic platform. To address this issue, a fixture has been designed to facilitate the planar placing of the retina and to supply fresh culture media. The retina-on-a-chip scheme will enable studies of complex interactions between neurons and various kinds of glial cells that are often spatially regulated, in both healthy and pathological conditions, i.e. glaucoma (Echevarria et al. 2013; Formichella et al. 2014; Sims et al. 2012).
2 Device design/fabrication
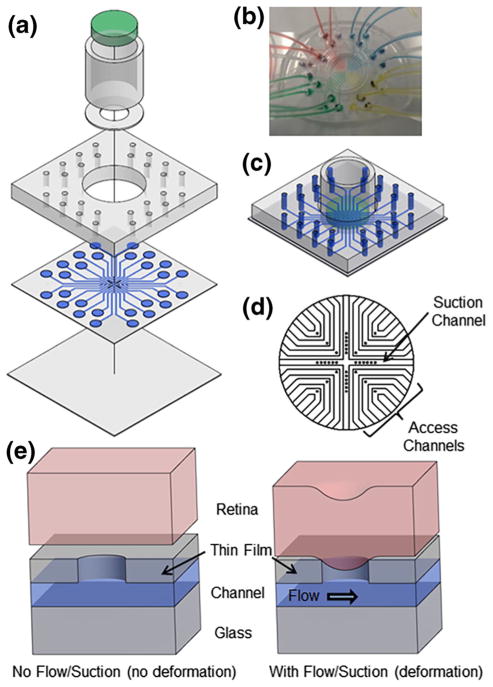
Other than the glass coverslip as the substrate, the device is composed of three main components: the PDMS thin-film layer with molded microchannels and access through-holes, tubing support layer, and media cylinder (as shown in Fig. 1). Each component is fabricated separately using soft lithography techniques, and then assembled to construct the full device. The overall function is to bring signaling molecules into point contact with the retina and observe their transport as well as the cellular response within the retina. This requires holding the retina flat, in-place, and keeping it healthy throughout the process of imaging the downstream events resulting from the point-delivery of molecules to the retina.
Fig. 1.
Retina-on-a-chip microfluidic platform is shown in (a) exploded view with media cylinder broken into the retina well, agar gel (green, at the top), and glass cylinder, (b) top view with a PDMS disk substituting for the retina and media cylinder removed to show the path of fluid flow within the device, and (c) assembled view with media in channels. d Close-up schematic of the layout of access channels and suction channel in the thin film PDMS layer. All channels are 300 μm wide and all through-holes are 100 μm in diameter. e Schematics of deflection of retina into through-holes in PDMS thin film layer. Without flow or suction, the retina does not deform and seal the through-hole and channel. When negative pressure is applied to the outlet tubing via a syringe, the retina deforms into the through-hole sealing the access channel and allowing for fluid flow. Note that the retina deforms in both the access and suction channels to seal the through-holes due to flow through the channel or suction pressure in the channel, respectively
Each of the three components of the device contributes to the overall function in a specific way. The 100-μm thick PDMS thin-film layer allows for point contact of media with the retina by pulling the tissue into contact with the media at a set of through-holes. The tubing support layer allows for tubing connections to the thin film layer along with an alignment guide for the media cylinder. The media cylinder holds the retina flat on the thin film layer and also supplies fresh nutrient media to the retina. The tubing support layer and the media cylinder are included to address the challenge that retina is naturally curved while point access fluidic delivery and confocal imaging need to keep the retina flat on the thin-film layer. With the small thickness of the PDMS thin-film layer, the device is also designed to be compatible with confocal microscopy, which limits the allowable distance from the bottom of the device to the retina tissue to about 210 μm.
The first component, the PDMS thin-film layer, is designed to bring signaling molecules into point contact with the retina. The design is composed of twelve access channels and a single suction channel all with dimensions of 300 μm wide and 50 μm tall. The suction channel, which is used to pull the retina into contact with the through-holes, forms an X-shape through the middle of the device separating the twelve access channels into four quadrants of three access points each, as shown in Fig. 1d. The access channels, arranged in a grid array at right angles, allow for drugs or signaling molecules flowing underneath the retina to come into point contacts with the retina at a variety of locations. Since each access channel has its own inlet, outlet, and through-hole, different drugs or signaling molecules may be applied at the twelve different access points via through-holes of 100 μm diameter and 50 μm depth. Note that using positive pressure to deliver fluid to an access point would endanger the integrity of the seal between the through-hole and tissue thus causing excess fluid delivery (Huang et al. 2012). Therefore, we chose to withdraw fluid from the outlet, which prevents excess fluid delivery and ensures that only the tissue section above the through-hole receives the signaling molecules.
The second PDMS piece of the device is composed of a much simpler design but is required for the structural stability and handiness of the device. The tubing support layer provides tubing connections for each of the inlets and outlets on the thin film layer and a large hole in the center (11 mm diameter) for the tissue sample to be positioned using the media cylinder.
The media cylinder is composed of three subcomponents: a tissue well, a glass cylinder, and a slice of agar gel. These are combined to create a tissue placement fixture and media supplier. A thin washer of PDMS that defines the tissue well is used to hold the retina in place within its horizontal plane on top of the PDMS thin-film layer. The thickness of the tissue well is approximately the same as the retina to ensure that it stays in a flattened form without rolling or folding into the naturally curved state during assays. The tissue well is attached to the bottom of a glass cylinder. A slice of agar gel is then placed inside the glass cylinder and set flush with the top of the tissue well as shown in Fig. 1c. The agar gel serves as a slightly sticky substrate to hold the retina in place as well as a porous filter that allows fresh media to slowly perfuse through to supply nutrients to the retina from the cylinder. With the media cylinder filled, the weight of the media on top of the agar gel holds the cylinder in place on top of the retina to restrict curling.
The fabrication of the device is based on soft lithography methods using PDMS and assembly by plasma-assisted bonding. In this study, even though the fabrication process follows the general soft-lithography procedure, it differs from the standard approach in its use of plastic master molding (Desai et al. 2009) and the PDMS thin-film technique (Hsu et al. 2004; Jo et al. 2000; Zhang et al. 2010) as described in Methods.
Once the device is fabricated and the retina is excised, the tissue is placed on the bottom of the agar gel inside the tissue well, unfolded, and flattened. The media cylinder is then inverted and inserted into the large hole in the middle of the tubing support layer such that the retina comes into contact with the top of the thin-film layer. In our studies, the inner retina is placed opposite the agar gel such that this side will be in contact with the fluid delivery from the through-holes once inverted. Culture media is then added into the glass cylinder on top of the agar gel to keep the retina healthy. The placement of the flattened retina into the device as shown in Fig. 2 demonstrates that the retina is held flat and remains close to the thin-film layer. Visibility of the nerve head and vasculature shows that there was no gross deformation of the major anatomical structure of the tissue during explant and placement onto the device.
Fig. 2.
Once the retina is placed onto the device, the nerve head can be found with an array of surrounding vessels, vasculature, and RGC axons. The visibility of such anatomy within the retina shows there was no gross deformation of the major anatomical structures after explant on the device. a Image taken at the focal plane of the retina to show the detailed retina components. b Image taken at a focal plane closer to the thin film PDMS layer to demonstrate placement on the device. (Inset) Retina stained with toluidine blue. Note that the vessels and RGC axons do not take up the dye, but the surrounding cells are stained. Scale bar =100 μm
3 Retina-on-a-chip assay results and discussion
To demonstrate the ability of the microfluidic platform through a retina-on-a-chip assay, retina from CX3CR1-GFP mice with microglia expressing GFP under the CX3CR1 promoter were carefully excised as described in Methods and placed in the device. Using a needle and syringe attached to the suction channel, a vacuum pressure of approximately 7 kPa was applied to pull the retina into tight contact with the thin film layer. When the vacuum pressure was applied to the device, the retina deformed slightly into the through-holes, as shown schematically in Fig. 1e. This seals the edge of the through-holes and confines the tissue contact to the access point only. Note that negative pressure was used to both bring the retina into contact with the thin-film layer using the suction channel and to pull culture media through the access channels, such that the reagent contacted the tissue at the through-hole access point only. Fluorescent imaging revealed the capability of the device to isolate a single microglial cell (inset of Fig. 3c), while maintaining its relationship to the surrounding retinal milieu.
Fig. 3.
Demonstration of suction pressure and fluid flow through the device. a Retina is at rest with no suction applied to the channels. b Negative pressure is gently applied in the suction channel. c When the retina deforms into the through-holes in the suction channel, GFP+ glial cells are seen in the focal plane of the thin film PDMS layer. d Toluidine blue is applied to one of the access channels by pulling fluid through the device via a needle and syringe attached by tubing. Note there is no leakage from the through-hole due to the retina deformation creating a seal. e The toluidine blue in the access channel is replaced by media leaving a small and focused stain on the retina. f If the device is not used properly and negative pressure is not used to create a seal between the tissue and through-hole, dye will leak throughout the device staining a much larger area than needed. Scale bar =100 μm
To demonstrate delivery of reagents to well-defined and specific points in intact tissue, we delivered the histological stain toluidine blue (Sigma, St. Louis, MO) to through-hole access points. To add the dye to an access point, a suction pressure of approximately 6 kPa was applied from the outlet via a needle and syringe attached by tubing to the access channel, which caused the retina to deform into and seal the through-hole. The pressure in the channel to induce fluid flow from the reservoir was recorded using a pressure gauge attached via a T-connector to the syringe. Once the access point was in tight contact with the retina, toluidine blue was supplied from the inlet reservoir through the tubing and channel, which comes into contact with the retina at the through-hole area of that channel (Fig. 3d). The access channel was then flushed with culture media to remove excess toluidine blue from the channel (Fig. 3e). Imaging results show that the remaining toluidine blue was associated with and restricted to tissue area accessed by the through-hole (Fig. 3e). In contrast to our negative pressure protocol, delivery of toluidine via the access channels by positive pressure resulted in significant spreading of the dye from the through-hole into the surrounding tissue (Fig. 3f). A large suction pressure of approximately 23 kPa also caused leakage at the through-hole due to rupture of the RGC layer. Note that deflection of the retina via negative pressure occurred at through-holes in both the access channels and the suction channel. In our experiment, we also found that by clamping the inlet and outlet tubing after fluid delivery, the seal between the through-hole and tissue can be held for a period of more than 24 h to allow for the media to stain the tissue. This operation mode could be very useful for assays involving expensive reagents such that only a small quantity, approximately 20–30 μL, is needed.
As with all ex vivo assays, maintenance of tissue health is a pre-requisite. To demonstrate: 1) healthy retina maintained over an extended time period, and 2) the ability to assess long-term biological endpoints in spatially-isolated microenvironments of intact tissue, we treated retina with the neural tracer cholera toxin beta subunit (CTB; Invitrogen) via the access points. CTB is an active uptake, active transport neural tracer that is specific to retinal ganglion cells (RGCs) in retina (Crish et al. 2010). RGCs and their axons, which form the optic nerve, reside in the innermost layers of the retina and actively uptake CTB, via the GM1 receptor, in the cell soma and transport it to their axon terminals in the brain (Crish et al. 2010). We elected to examine RGCs specifically because these neurons experience the most cellular stress during and after explanting of the retina, which requires complete transection of their axons in the optic nerve. Since CTB uptake and transport are both active processes, deficits in the uptake and transport of CTB are indicators of RGC degeneration (Crish et al. 2010). Accordingly, deficits in CTB uptake and transport are well-described in neurodegenerative disease, including those in the retina (Calkins 2012; Chidlow et al. 2012; Chidlow et al. 2011; Crish et al. 2013; Crish et al. 2010; Echevarria et al. 2013; Formichella et al. 2014).
For the CTB studies, retina from CX3CR1-GFP mice were placed in the microfluidic platform and a solution of 1 % CTB in culture media was applied to the access points, as described above. Retina was then cultured for 24 h in the microfluidic platform under standard culture conditions prior to real-time confocal imaging of GFP labeling in microglia and CTB labeling in the retinal ganglion cell and nerve fiber layers of the retina. After culture for 24 h, the access channels were rinsed with media and the retina was imaged. As demonstrated in Fig. 4, CTB labeling remained restricted to RGCs located at the access point and the presence of GFP+/CTB− microglia were also apparent in and around the through-hole (Fig. 4a). To demonstrate the ability to keep the retina healthy for extended periods, the retina was cultured for 5 days on the device. Four days after explanting retina from C57BL/6 mice, CTB conjugated to Alexa Fluor 594 (CTB-594) was added to the media cylinder and CTB conjugated to Alexa Fluor 488 (CTB-488) was added only to the access channels. As such, the entire retina, including that associated with the through-holes, was exposed to CTB-594, and only retina associated with the through-holes was exposed to CTB-488. After culture overnight (16 h) with CTB-488, the channels were rinsed with media and the retina was imaged. Figure 5 demonstrates that RGCs in retina remained healthy enough after 4 days in the device to actively uptake CTB. Furthermore, the seal in the suction channel was maintained for a minimum of 4 days, as CTB-594 labeling was noted in the entire retina while CTB-488 labeling was noted only in retina associated with through-holes (Fig. 5a–d). Note that although CTB-488 uptake to RGCs is restricted to the immediate vicinity of the access points, there was some diffusion of CTB-488 into the tissue surrounding the access points during the overnight culture. However, this diffusion beyond the access point boundary extended an average of only 39 μm with a standard deviation of 19 μm (Fig. 5e). Note that using Fick’s law and a reported value of 0.26 μm2/s as the diffusion coefficient for CTB-488 in cells (Kang et al. 2012), a quick estimation yields a diffusion length of about 120 μm over 16 h, which is three-fold of the observed diffusion range. One possible reason for the difference is that in our set-up, media perfuse from the media cylinder to the access channel due to a negative pressure in the access channel, which might offset the diffusion process and lead to smaller affected range, as shown in Fig. 5e. Together, these data demonstrate the capacity of this system to retain tissue health and integrity while also maintaining localized delivery of reagents over a period of days.
Fig. 4.

a Cholera toxin beta subunit (CTB), a protein related to cholera infection, was added to one of the access channels and the retina was cultured for 24 h. The CTB in the channel was then flushed with culture media and imaged. b Note some excess CTB remains in the channel in the vicinity of the through-hole after flushing. c Close up image of the RGC staining at the through-hole in (a). RGC somas (bold arrow) that were stained demonstrate axonal transport of the CTB
Fig. 5.
a Red CTB was added to the media cylinder to stain the entire retina while green CTB was added to access channels 4 days after explanting to demonstrate successful culture on the microfluidic platform. After culture for 24 h, the green CTB in the channels was replaced with media. Note that CTB can only be absorbed by living cells, thus the retina was shown to be healthy after 4 days in culture. (b–d) Shown in larger images are the three through-holes where green CTB has stained the healthy retinal ganglion cells in the retinal area within and immediately surrounding the access point. e The mean distance of green CTB diffusion and labeling from edge of access point is restricted to an average of 40 μm over 16 h
To demonstrate the ability of our microfluidic platform to examine real-time cellular activities and interactions in discrete cellular microenvironments of whole tissue, we examined the behavioral response of microglia in retina to lipopolysaccharide (LPS). LPS is a bacterial endotoxin that is well-known for inducing robust inflammation via activation of glial cells, particularly microglia (Beurel and Jope 2009; Lee et al. 1993; Sébire et al. 1993). Past research using LPS showed that microglia respond by activation and migration between 6 and 48 h after application (Buttini et al. 1996; Cui et al. 2002; Horvath et al. 2008; Kloss et al. 2001; Lee et al. 2005). Many of these studies used typical methods of LPS application such as a bath for tissue slices or intravenous injection into a live animal. These methods result in delivery of LPS to an unconfined large area of tissue in contrast to our current method. One study, with some similarity to our method, microinjected LPS in vivo into the corpus callosum of a mouse (Lee et al. 2005). This microinjection may give a much smaller affected region of tissue but the authors did not report an estimation of the region size, and the microglia reactivity was not evaluated until six hours post-injection. Although past results have shown that microglia are activated and migrate due to LPS application, there is little to no research demonstrating the early spatiotemporal attributes of microglia response to LPS. Given the prevalence of LPS as a global stimulator of inflammation and more specifically, a generic activator of microglia, identification of immediate and early response to directed application of this noxious stimulus provides the opportunity to examine previously uncharacterized behavior of microglia.
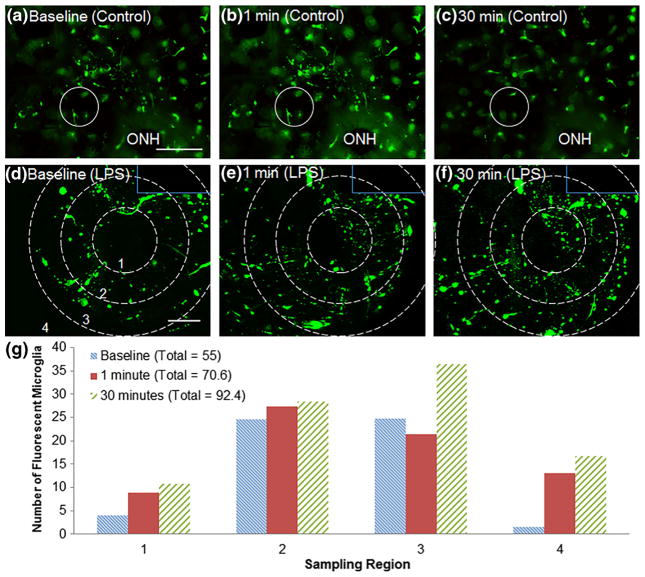
For our study, we delivered 2.5 ng/μL LPS via the through-holes to retina from CX3CR1-GFP mice. Within one minute of LPS delivery, microglia began migrating toward the access point (Fig. 6d,e). Over the course of 30 min post-LPS delivery, microglia began to migrate into the area of retina that occupies the access point (Fig. 6f). Similarly, microglia that originated from outside the field of view also migrated into the retinal area surrounding the access point (Fig. 6f). This phenomenon was not observed in retina exposed to only culture media, in which microglia moved in and out of the plane of focus without regard for location of the through-hole (Fig. 6a–c). These data indicate that the microglia migration observed in Fig. 6d–f is attributable to LPS application rather than other factors. To quantify LPS-induced migration of microglia, cells were hand-counted in circular regions surrounding the access point. Details of this analysis can be found in the Methods section. As shown in Fig. 6g, the total number of microglia within the image frame increased after one minute and thirty minutes of LPS application. One minute after application, microglia are activated and show more processes in the entire image, but only show immediate migration in the first two regions. After 30 min, microglia from outside the image boundaries were activated and began migrating toward the site of application.
Fig. 6.
a–c GFP+ microglia in retina from CX3CR1-GFP mice with exposure to culture media only are imaged over 30 min. Note the microglia in the circle, which indicates the through-hole, do not migrate directionally throughout the 30 min of imaging. ONH denotes the optical nerve head of the retina tissue. d GFP+ microglia surrounding an access point (Region 1) in retina from CX3CR1-GFP mice prior to LPS delivery. e Activation and migration of microglia to access point at 1 min and f 30 min post-LPS delivery to access point. Solid lines delineate edge of microfluidic channel. Dotted lines indicate the boundaries of the four sampling regions. g Five hand-counted sample sets were taken at each time point and averaged to give the number of fluorescently labeled microglia in each region. Evidence of immediate microglia migration was observed at 1 min in regions closest to LPS application and delayed migration toward the point of application was observed in regions farther from LPS exposure at 30 min. The total number of microglia at each time point is displayed in the legend. Scale bar (a–c) = 200 μm. Scale bar (d–f) = 50 μm
It would be interesting to consider whether the microglia migration is due to cell-cell signaling or LPS diffusion. LPS is a very small molecule and could be highly diffusive, and the reported LPS diffusion coefficient spans a large range. Most studies gave a diffusion coefficient ranging from 0.001 to 10 μm2/s (Morrison and Leive 1975; Schindler et al. 1980; Shawkat et al. 2008). Using the upper limit, 10 μm2/s, a diffusion length of 24.5 μm is obtained for a time period of 1 min. Considering the size of the view field in Fig. 6 (300 μm long) and the fact that media perfusion from the pressure bias would offset molecule diffusion as shown in Fig. 5 for the case of CTB, we speculate that the initial activation of microglia as seen in Fig. 6e is due to signaling instead of diffusion through the tissue and media. However, Triantafilou et al. (2001) suggested even higher diffusion coefficients ranging from 14.4 to 121 μm2/s, which could yield a diffusion length as large as 85 μm over 1 min. As such, achieving a more solid conclusion of whether the microglia response is due to cell signaling or LPS diffusion would require a measurement of the LPS diffusion coefficient in retina first. Nonetheless, the LPS assay results further confirm that the retina-on-a-chip platform can provide restricted access to intact tissue and its utility for the real-time manipulation and documentation of cell activity in a highly localized and controlled manner.
Together, all the assay results indicate that the design of our microfluidic platform permits real-time imaging of biological processes in discrete microenvironments through directed delivery of pharmacological agents to spatially confined locations in intact tissue.
4 Summary
In summary, we developed a microfluidic device for tissue slice culture with point access capability for localized and controlled application of drugs or signaling molecules. We verified the platform through long-term culture of retina, controlled and highly localized delivery of pharmacological agents, staining, tracing, and real-time examination of localized glial cell behavior. We expect that the platform could enable novel assays in ex vivo tissue studies.
5 Methods
5.1 Device fabrication
SU8 molds were created by photolithography techniques to generate PDMS microchannel structures. Using typical soft lithography techniques, liquid PDMS mixed at a 10:1 ratio of base polymer to curing agent is poured over the SU8 molds, degassed under vacuum pressure, and cured in a 70 °C oven for at least 4 h. The PDMS pieces were then cut and removed from the mold and assembled via irreversible oxygen plasma-assisted bonding.
For the thin film layer fabrication, instead of an SU8 mold, a plastic mold (Desai et al. 2009) was utilized to increase the mold durability. To create the mold, a two-layer SU8 pre-master mold was fabricated following the standard photolithography approach. The first layer of SU8 contained the 50 μm tall access and suction channels while the second layer contained the 50 μm tall through-holes and tubing connection holes. Once the SU8 mold was prepared, a PDMS pre-mold was generated from the SU8 mold. To create a plastic master, Smooth-Cast 300 Parts A and B were poured into separate containers and degassed under vacuum for 20 min. The two parts were then thoroughly mixed at a 1:1 ratio in a third container and poured over the PDMS pre-mold. The plastic was allowed to cure at room temperature for about 6 h. The PDMS was then removed and the plastic was placed in an oven at 70 °C to hard-cure for about 4 h. Once cooled, the plastic was ready to be used for fabrication of the first layer of the device with open through-holes using a thin-film fabrication technique. To do so, a small drop of liquid PDMS was placed at the center of the plastic mold. A fluoro-polymer liner (Hsu et al. 2004) (3 M Scotchpak 1022) was cut to size and placed active side down onto the liquid PDMS. The PDMS was then degassed under vacuum pressure till most air bubbles were removed. Any remaining air bubbles were further removed by pushing them toward the edge of the mold using a flat edge. The PDMS and the mold were then placed under normal pressure using weights to squeeze out excess PDMS from between the liner and features on the mold. Once an acceptable amount of weight was applied, the whole fixture was placed in an oven at 70 °C to cure for at least 4 h. After the PDMS had cooled, the thin film layer was removed from the mold and bonded to a thin glass coverslip. The fluoro-polymer liner was then gently removed from the PDMS layer to expose the through-holes and tubing connection holes of the PDMS thin-film piece.
Similarly, another plastic mold was used to fabricate the tubing support layer. Unlike the thin-film layer, here the plastic mold was directly prepared with a PDMS pre-mold. This was fabricated by punching holes from a 2–3 mm thick slab of PDMS, a small hole for each tubing connection and one large hole in the center as the cylinder alignment guide. The two parts of the plastic were again degassed, mixed, and poured over the PDMS pre-mold. Care was taken as to remove any air bubbles from the smaller holes immediately after pouring to ensure solid posts remained. After curing at room temperature for 6 h, the PDMS was removed and the plastic mold was hard-cured in a 70 °C oven for 4 h. Liquid PDMS was then poured over the plastic mold, cured, and removed to create the tubing support layer.
Once the first two pieces were fabricated separately, the thin film and tubing support layer were exposed to oxygen plasma, aligned and bonded. Tubing was then inserted into the appropriate holes. The third piece of the device was fabricated in two steps. First a 200–300 μm thick layer of liquid PDMS was spin-coated onto a piece of the fluoro-polymer liner and cured. Two holes were punched to create a PDMS washer which was then attached to the bottom of a 10 mm glass cylinder (Pyrex 3166–10) using liquid PDMS. Once the liquid PDMS had cured, the fluoro-polymer liner was removed. To create the agar gel piece which fits inside the glass cylinder on top of the PDMS washer, agar powder was dissolved in boiling water at a w/v ratio of 3 % and poured into a flat dish to about 2 mm thickness. Once cooled, the agar gel was removed by punching a plug from the dish and placed into the cylinder. The flattened retina was then placed on the agar gel inside the PDMS washer on the glass cylinder, which was then inverted and placed into the large hole on the device for culture and examination.
5.2 Animal procurement and ethics statement
This study was conducted in accordance with regulations set forth in the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. Animal protocols were approved by the Institutional Animal Care and Use Committee of Vanderbilt University Medical Center. C57BL/6 and CX3CR1-GFP mice were originally obtained from Jackson Laboratories (Bar Harbor, ME) and colonies were established in the animal facility at Vanderbilt University Medical Center. Mice utilized in this study were obtained from these in-house colonies.
5.3 Organotypic retina culture
Adult mice (2–4 months) were sacrificed by cervical dislocation and decapitation. Eyes were enucleated and live retina were dissected for mounting in the microfluidic platform. Whole retina cultures were maintained for 1 h to 4 days in the microfluidic device with serum-free, B27-supplemented medium (NeuroBasal; Gibco, Carlsbad, CA), as previously described (Sappington et al. 2009; Sappington et al. 2006). Media also contained 2 mM glutamine, 0.1 % gentamicin, 1 % N2 supplement (500 μg/mL insulin, 10 mg/mL transferrin, 630 ng/mL progesterone, 1.6 mg/mL putrescine, 520 mg/mL selenite; Gibco), 50 ng/mL brain-derived neurotrophic factor (Invitrogen, Carlsbad, CA), 20 ng/mL ciliary neurotrophic factor (Invitrogen) and 100 μM inosine (Sigma, St. Louis, MO). For culture intervals lasting longer than one hour, the entire microfluidic device was maintained in a standard cell culture incubator with 5 % CO2 at 37 °C.
5.4 Live retina imaging
Live imaging of retina organotypic cultures was performed through the Vanderbilt University Medical Center Cell Imaging Shared Resource Core on an inverted confocal microscope (Olympus FV-1000; Center Valley, PA) equipped with laser scanning fluorescence (blue/green, green/red, red/far-red). Three dimensional z-series images of the retina through the ganglion cell and nerve fiber layers were acquired using a digital camera and image analysis software (FV-10 ASW; Olympus).
5.5 Cell migration analysis
To quantify microglia migration, images from three time points during LPS delivery to one of the access points were analyzed by hand-counting cells in each of four regions surrounding the initial application site. Region 1 was defined as the area within the access point. Region 2 and 3 were defined as concentric circles about region 1 with diameters of 200 and 300 μm respectively. Region 4 contained the remainder of the view field. GFP+ microglia in each region at each time point were hand-counted five times and averaged. The total number of cells at each time point is also given in the legend in Fig. 6.
Acknowledgments
This material is based upon work supported by the following grants 1) National Eye Institute - RO1EY020496-01 (RMS) and P30EY08126 (Vanderbilt Vision Research Center), 2) Research to Prevent Blindness, Inc. - Unrestricted Grant (Vanderbilt Eye Institute) and Career Development Award (RMS), and 3) National Science Foundation Graduate Research Fellowship Program under Grant No. 0909667 and 1445197.
Contributor Information
Deyu Li, Email: deyu.li@vanderbilt.edu.
Rebecca M. Sappington, Email: rebecca.m.sappington@vanderbilt.edu.
Jon F. Edd, Email: jonedd2@gmail.com.
References
- Berdichevsky Y, Staley KJ, Yarmush ML. Lab Chip. 2010;10:999. doi: 10.1039/b922365g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beurel E, Jope RS. J Neuroinflammation. 2009;6:9. doi: 10.1186/1742-2094-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borenstein JT, Tupper MM, Mack PJ, Weinberg EJ, Khalil AS, Hsiao J, García-Cardeña G. Biomed Microdevices. 2010;12:71. doi: 10.1007/s10544-009-9361-1. [DOI] [PubMed] [Google Scholar]
- Buttini M, Limonta S, Boddeke HWGM. Neurochem Int. 1996;29:25. doi: 10.1016/0197-0186(95)00141-7. [DOI] [PubMed] [Google Scholar]
- Calkins DJ. Prog Retin Eye Res. 2012;31:702. doi: 10.1016/j.preteyeres.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chidlow G, Ebneter A, Wood JPM, Casson RJ. Acta Neuropathol. 2011;121:737. doi: 10.1007/s00401-011-0807-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chidlow G, Wood JPM, Ebneter A, Casson RJ. Neurobiol Dis. 2012;48:568. doi: 10.1016/j.nbd.2012.07.026. [DOI] [PubMed] [Google Scholar]
- Choi NW, Cabodi M, Held B, Gleghorn JP, Bonassar LJ, Stroock AD. Nat Mater. 2007;6:908. doi: 10.1038/nmat2022. [DOI] [PubMed] [Google Scholar]
- Crish SD, Sappington RM, Inman DM, Horner PJ, Calkins DJ. Proc Natl Acad Sci U S A. 2010;107:5196. doi: 10.1073/pnas.0913141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crish SD, Dapper JD, MacNamee SE, Balaram P, Sidorova TN, Lambert WS, Calkins DJ. Neuroscience. 2013;229:55. doi: 10.1016/j.neuroscience.2012.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui YH, Le Y, Gong W, Proost P, Van Damme J, Murphy WJ, Wang JM. J Immunol. 2002;168:434. doi: 10.4049/jimmunol.168.1.434. [DOI] [PubMed] [Google Scholar]
- Desai SP, Freeman DM, Voldman J. Lab Chip. 2009;9:1631. doi: 10.1039/b822081f. [DOI] [PubMed] [Google Scholar]
- Di Carlo D, Wu LY, Lee LP. Lab Chip. 2006;6:1445. doi: 10.1039/b605937f. [DOI] [PubMed] [Google Scholar]
- Echevarria F, Walker C, Abella S, Won M, Sappington R. J Clin Exp Ophthalmol. 2013;4:286. doi: 10.4172/2155-9570.1000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formichella CR, Abella SK, Sims SM, Cathcart HM, Sappington RM. J Clin Cell Immunol. 2014;5:1. doi: 10.4172/2155-9899.1000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Majumdar D, Jovanovic B, Shaifer C, Lin PC, Zijlstra A, Webb DJ, Li D. Biomed Microdevices. 2011;13:539. doi: 10.1007/s10544-011-9523-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther A, Yasotharan S, Vagaon A, Lochovsky C, Pinto S, Yang J, Lau C, Voigtlaender-Bolz J, Bolz SS. Lab Chip. 2010;10:2341. doi: 10.1039/c004675b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath RJ, Nutile-McMenemy N, Alkaitis MS, DeLeo JA. J Neurochem. 2008;107:557. doi: 10.1111/j.1471-4159.2008.05633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CH, Chen C, Folch A. Lab Chip. 2004;4:420. doi: 10.1039/b404956j. [DOI] [PubMed] [Google Scholar]
- Huang Y, Williams JC, Johnson SM. Lab Chip. 2012;12:2103. doi: 10.1039/c2lc21142d. [DOI] [PubMed] [Google Scholar]
- Hung PJ, Lee PJ, Sabounchi P, Aghdam N, Lin R, Lee LP. Lab Chip. 2005;5:44. doi: 10.1039/b410743h. [DOI] [PubMed] [Google Scholar]
- Jo B, Van Lerberghe LM, Motsegood KM, Beebe DJ. J Microelectromech Syst. 2000;9:76. [Google Scholar]
- Kang M, Day CA, Kenworthy AK, DiBenedetto E. Traffic. 2012;13:1589. doi: 10.1111/tra.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khademhosseini A, Langer R, Borenstein J, Vacanti JP. Proc Natl Acad Sci U S A. 2006;103:2480. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khetani SR, Bhatia SN. Nat Biotechnol. 2008;26:120. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- Kim MS, Kim T, Kong SY, Kwon S, Bae CY, Choi J, Kim CH, Lee ES, Park JK. PLoS One. 2010;5:e10441. doi: 10.1371/journal.pone.0010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss CU, Bohatschek M, Kreutzberg GW, Raivich G. Exp Neurol. 2001;168:32. doi: 10.1006/exnr.2000.7575. [DOI] [PubMed] [Google Scholar]
- Lecault V, Vaninsberghe M, Sekulovic S, Knapp DJHF, Wohrer S, Bowden W, Viel F, McLaughlin T, Jarandehei A, Miller M, Falconnet D, White AK, Kent DG, Copley MR, Taghipour F, Eaves CJ, Humphries RK, Piret JM, Hansen CL. Nat Methods. 2011;8:581. doi: 10.1038/nmeth.1614. [DOI] [PubMed] [Google Scholar]
- Lee SC, Liu W, Dickson DW, Brosnan CF, Berman JW. J Immunol. 1993;150:2659. [PubMed] [Google Scholar]
- Lee JC, Cho GS, Hye JK, Lim JH, Oh YK, Nam W, Chung JH, Kim WK. Glia. 2005;50:168. doi: 10.1002/glia.20164. [DOI] [PubMed] [Google Scholar]
- Meyvantsson I, Beebe DJ. Annu Rev Anal Chem (Palo Alto, Calif) 2008;1:423. doi: 10.1146/annurev.anchem.1.031207.113042. [DOI] [PubMed] [Google Scholar]
- Morrison DC, Leive L. J Biol Chem. 1975;250:2911. [PubMed] [Google Scholar]
- Passeraub P, Almeida A, Thakor N. Biomed Microdevices. 2003;5:147. [Google Scholar]
- Queval A, Ghattamaneni NR, Perrault CM, Gill R, Mirzaei M, McKinney RA, Juncker D. Lab Chip. 2010;10:326. doi: 10.1039/b916669f. [DOI] [PubMed] [Google Scholar]
- Sappington RM, Chan M, Calkins DJ. Investig Ophthalmol Vis Sci. 2006;47:2932. doi: 10.1167/iovs.05-1407. [DOI] [PubMed] [Google Scholar]
- Sappington RM, Sidorova T, Long DJ, Calkins DJ. Investig Ophthalmol Vis Sci. 2009;50:717. doi: 10.1167/iovs.08-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler M, Osborn MJ, Koppel DE. Nature. 1980;285:261. doi: 10.1038/285261a0. [DOI] [PubMed] [Google Scholar]
- Sébire G, Emilie D, Wallon C, Héry C, Devergne O, Delfraissy JF, Galanaud P, Tardieu M. J Immunol. 1993;150:1517. [PubMed] [Google Scholar]
- Shawkat S, Karima R, Tojo T, Tadakuma H, Saitoh SI, Akashi-Takamura S, Miyake K, Funatsu T, Matsushima K. J Biol Chem. 2008;283:22962. doi: 10.1074/jbc.M801366200. [DOI] [PubMed] [Google Scholar]
- Sims SM, Holmgren L, Cathcart HM, Sappington RM. Am J Neurodegener Dis. 2012;1:168. [PMC free article] [PubMed] [Google Scholar]
- Squires TM, Quake SR. Rev Mod Phys. 2005;77:977. [Google Scholar]
- Taylor AM, Blurton-Jones M, Rhee SW, Cribbs DH, Cotman CW, Jeon NL. Nat Methods. 2005;2:599. doi: 10.1038/nmeth777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantafilou K, Triantafilou M, Ladha S, Mackie A, Dedrick RL, Fernandez N, Cherry R. J Cell Sci. 2001;114:2535. doi: 10.1242/jcs.114.13.2535. [DOI] [PubMed] [Google Scholar]
- Vickerman V, Blundo J, Chung S, Kamm R. Lab Chip. 2008;8:1468. doi: 10.1039/b802395f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Wu J, Wang L, Xiao K, Wen W. Lab Chip. 2010;10:1199. doi: 10.1039/b923101c. [DOI] [PubMed] [Google Scholar]