Abstract
Histone deacetylase (HDAC) activity, commonly correlated with transcriptional repression, was essential for transcriptional induction of IFN-stimulated genes (ISG). Inhibition of HDAC function led to global impairment of ISG expression, with little effect on basal expression. HDAC function was not required for signal transducer and activator of transcription tyrosine phosphorylation, nuclear translocation, or assembly on chromatin, but it was needed for full activity of the signal transducer and activator of transcription transactivation domain. HDAC function was also required for gene induction driven by the IFN regulatory factor 3 transcription factor activated by virus infection, and it was essential for establishment of an antiviral response against Flaviviridae, Rhabdoviridae, and Picornaviridae. Requirement for HDAC function in transcriptional activation may represent a general mechanism for rapid stimulation of ISG transcription.
Type I IFN is a family of related cytokines capable of inhibiting viral replication in sensitive target cells through induction of a large set of cellular antiviral proteins (1). Induced expression of antiviral proteins is regulated primarily through increased gene transcription, with expression of many genes increasing rapidly from nearly undetectable levels to robust levels of transcription (2, 3). It is likely that both the nearly silent preinduction levels of expression and the rapid and high postinduction expression are physiologically important. Antiviral functions can be deleterious to the host if expressed inappropriately but must respond rapidly in the event of viral attack. The combination of low basal but rapid and robust induced expression requires a sensitive molecular switch capable of responding to the presence of IFN.
It is hypothesized that gene regulation of this type requires a unique molecular machinery capable of rapidly transducing a cell surface signal into changes in gene expression (4). Part of the cellular solution to this problem was elucidated by the discovery of the Janus kinase–signal transducer and activator of transcription (Stat) signaling pathway in which engagement of a cell surface cytokine receptor results in rapid activation of a specific transcription factor complex by protein tyrosyl phosphorylation and nuclear translocation (5). In response to IFN stimulation, the transcription factors Stat1 and Stat2 are phosphorylated and assemble along with the DNA-binding IFN regulatory factor (IRF) 9 subunit into a multimeric transcription factor complex termed ISGF3 (6). Stat2 provides the majority of the transcriptional activation potential (7–10), whereas Stat1 and IRF9 contribute to DNA-binding specificity (8, 11).
Another aspect of the transcriptional induction of IFN-stimulated genes (ISGs) is the ability to be transcribed during cellular stress, for instance, during viral infection. This problem is solved, in part, by the ability of Stat2 to mediate gene induction through an alternative transcriptional initiation complex dependent on the histone acetyltransferase GCN5 and the general transcription factor TAF4 but lacking the TATA box-binding protein TBP (9). Such a complex allows transcription to continue in the face of the proteolytic degradation of TBP that occurs during some viral infections and inhibits the majority of cellular gene expression.
In exploring potential mechanisms for maintaining low basal levels of ISG expression in the absence of induction, we considered the possible involvement of histone deacetylases (HDACs). Gene repression is commonly associated with regions of deacetylated histones, presumably resulting in increased ionic interactions between histones and DNA to produce more compact chromatin and the absence of acetylation-dependent recruitment sites for necessary activation factors (12). Maintenance of deacetylated histones and silent chromatin is an active process mediated by HDAC enzymes countering the opposing action of histone acetyltransferases. Inhibition of HDAC function by pharmacological inhibitors, such as the microbial antibiotic trichostatin A (TSA), results in accumulation of acetylated histones, often accompanied by an increase in gene expression. Such changes in gene expression, presumably due to relief from HDAC-dependent repression, alter cellular behavior, inducing growth arrest, differentiation, or apoptosis, leading to successful use of HDAC inhibitors as chemotherapeutic anticancer drugs (13). To our surprise, however, treatment of cells with HDAC inhibitors did not increase the basal expression of most ISGs but rather prevented ISG transcription in response to IFN treatment. A similar requirement for HDAC activity was observed for virus-induced transcription mediated by the transcription factor IRF3. These results reveal another aspect of ISG transcriptional mechanisms and suggest that rapid induction of previously silent genes may exploit a unique transcriptional machinery.
Experimental Procedures
Cell Culture, Plasmids, Transfections, Viral Infections, and Drug Treatments. HeLa, U2OS, Cos1, and Huh-7.5 cells were maintained in DMEM supplemented with 10% calf serum and antibiotics. Cells were transfected by using the calcium phosphate method, and cellular extracts were collected for luminescent luciferase and β-galactosidase assays as described (8). Luciferase assays for ISG54-luc and Gal4-UAS-luc were performed in triplicate, and results were normalized to cotransfected β-galactosidase. Newcastle disease virus was a gift of A. García-Sastre (Mount Sinai School of Medicine, New York) and was added to cells at a multiplicity of infection of 10–30 (14). Where indicated, cells were also treated with IFN-α2a (Hoffman–La Roche) at 1,000 units/ml, IFNγ (Amgen Biologicals) at 1 ng/ml, TSA (Sigma–Aldrich) at 100 or 300 nM, valproic acid (VPA, Sigma–Aldrich) at 1 or 5 mM, cycloheximide (Sigma–Aldrich) at 50 μg/ml, or HC toxin (Biomol, Plymouth Meeting, PA) at 5 μM. Unless otherwise indicated, IFNα treatments were for 6 h, IFNγ treatments were for 15 h, and HDAC inhibitor treatments and cotreatments were for 6 h. Antiviral assays are described in Supporting Experimental Procedures, which is published as supporting information on the PNAS web site.
Transcription Measurements. Run-on transcription experiments (15) used ≈10 million cells per point. DNA probes were the following: Vector (pGEM1, Promega), γ-actin cDNA (16), ISG54 EcoRI–TaqI fragment from exon 2 (17), and ISG15 TaqI fragment from exon 2 (18). After hybridization, filter signals were quantified by PhosphorImager (Bio-Rad).
Protein Assays. Nuclear extracts were prepared and analyzed by immunoblotting as described (8) by using anti-IRF9, anti-Stat1, anti-Stat2, anti-phospho-Stat1 and Stat2 (Zymed, South San Francisco, CA), or anti-RPA-2 antibodies (NeoMarkers, Fremont, CA). ISGF3 levels were analyzed by electrophoretic mobility shift assay (19).
Microarray Expression Analysis. Total RNA was isolated with TRIzol reagent (Invitrogen) from untreated, IFN-treated, TSA-treated, and IFN/TSA-treated cells. RNA was fluorescently labeled by direct incorporation in a cDNA synthesis reaction. Unstimulated control RNA was labeled with Cy3-dUTP, and RNA from treated cells was labeled with Cy5–dUTP. Samples were hybridized overnight at 55°C in slide hyb no. 3 (Ambion, Austin, TX). Data were analyzed with genepix pro 4.0 and genespring 4.2.1. Low-intensity values were filtered out, and data were normalized on the basis of overall intensity.
Real-Time RT-PCR. RNA was isolated and converted to cDNA as described (14). Relative abundances of specific mRNA sequences were determined by real-time fluorescent PCR by using Syber green (Molecular Probes) by comparison with a standard curve generated by serial dilution of a cDNA sample containing abundant target sequences and normalized to the expression of GAPDH. All PCR reactions were performed in triplicate, and standard errors were <10% of mean values. Sequences of primers used are available on request. Hepatitis C virus (HCV) RNA abundance was estimated by Taqman-based real-time PCR (20).
Chromatin Immunoprecipitation (ChIP) PCR. ChIP assays were performed essentially as described (9). In brief, 1–2 million cells were treated with 1% formaldehyde for 30 min at 4°C and lysed in SDS-containing buffer. Cell extracts were sonicated to sheer DNA to ≈500 bp and immunoprecipitated with antibodies against Stat2 (Santa Cruz Biotechnology); the recovered chromatin was digested with proteinase K, treated at 65°C, and purified on ion-exchange spin columns. The presence of specific immunoprecipitated DNA sequences was quantified by real-time PCR compared with input genomic DNA.
Results
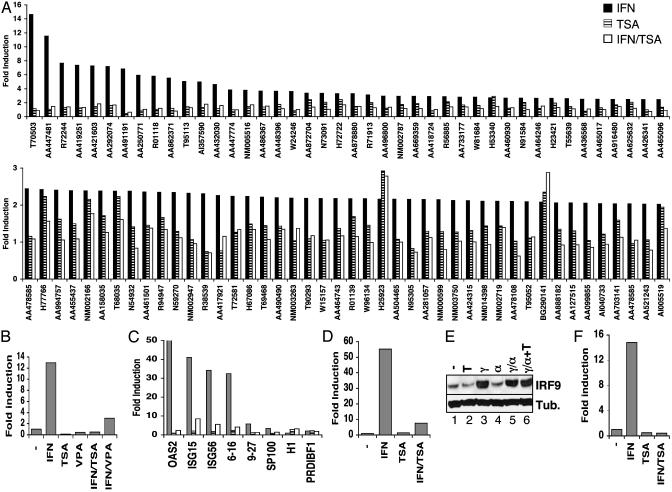
HDAC Inhibition Does Not Raise the Basal Expression of Most ISGs But Blocks Induced Expression in Response to IFN. We used a functional genomics approach to assess a role for HDAC activity during ISG regulation. U2OS cells were treated with TSA to inhibit HDAC activity, and mRNA was collected after 8 h. RNA was also prepared from cells treated with IFN, with a combination of IFN and TSA, or from control cells. RNA samples were used for microarray analysis with a custom ISG cDNA array containing probes for 850 unique putative ISGs and 100 control genes (21). Approximately 90 genes were induced at least 2-fold by IFN in these cells (Fig. 1A). Of these, a few were also induced at least 2-fold by TSA treatment, and only two gene sequences were induced by TSA to a greater extent than by IFN. Surprisingly, IFN-stimulated expression was almost universally impaired by combined treatment of IFN and TSA. These data do not support a general role for HDAC activity in maintaining the repressed state of ISG sequences, but they reveal an unexpected requirement for HDAC function during ISG expression.
Fig. 1.
Global impairment of IFN-stimulated gene expression by HDAC inhibition. (A) U2OS cells were stimulated with IFNα in the absence (filled bars) or presence of TSA (open bars) or were treated with TSA alone (hatched bars) for 8 h. Isolated RNA was analyzed by two-color microarray hybridization in comparison with RNA isolated from untreated cells. A Upper represents genes induced by IFNα >2.5-fold; A Lower includes genes induced >2-fold. (B) HeLa cells were treated with IFNα, TSA, and VPA, as indicated, for 6 h before isolation of RNA. Expression of ISG54 was quantified by real-time RT-PCR and represented as fold induction over untreated cells. (C) Expression of additional ISG transcripts was analyzed as in B, except that HeLa cells were treated overnight with IFNγ; cells were treated with IFNα (filled bars), TSA (hatched bars), or IFNα plus TSA (open bars) for 4.5 h before isolation of RNA and analysis by quantitative RT-PCR with primers specific for the indicated genes. (D) As in B, except human fibroblast FS2 cells were treated for 1 h with IFNα and/or TSA, as indicated, before quantitation of ISG54 transcript levels. (E) Western blot analysis of IRF9 and β-tubulin expression in HeLa cells treated with TSA (T), IFNγ (γ), and IFNα (α), as indicated. (F) As in C, except expression of ISG54 nuclear RNA precursors rather than mature mRNA was quantified by using an intron/exon primer pair.
To confirm and extend these observations, we performed quantitative real-time RT-PCR for selected ISG sequences. Initially, we examined the expression of ISG54, a gene highly responsive to IFN stimulation (17). ISG54 mRNA was induced ≈13-fold by IFN, but induced expression was completely suppressed by cotreatment with TSA (Fig. 1B). To confirm that the effect of TSA was through inhibition of HDAC activity rather than through any uncharacterized effects of this drug, additional HDAC inhibitors were tested for their effect on ISG54 expression. VPA (Fig. 1B) and HC toxin (data not shown), HDAC inhibitors structurally unrelated to TSA (22, 23), both inhibited ISG induction in the presence of IFN. Therefore, it is very likely that the effects reported here are mechanism-based, indicating that HDAC catalytic function is required for ISG expression.
The induced expression of additional ISG species tested was also impaired in the presence of TSA, often by greater than a factor of 10 (Fig. 1C). Inhibition of ISG expression was observed not only in U2OS cells but also in other human and mouse IFN-responsive cell lines, such as HeLa (Fig. 1 B and C), normal human fibroblasts (Fig. 1D), mouse embryonic fibroblasts, and L929 cells (data not shown). ISG promoters have been recently classified into two groups based on their requirement for BRG1-dependent chromatin remodeling for induced expression (24, 25). Genes from both classes were equally sensitive to TSA inhibition. For instance, expression of ISG54, ISG15, and genes 6–16 (BRG1-independent genes) was inhibited in the presence of TSA. Likewise, induction of the BRG1-dependent genes 9–27 and oligoadenylate synthetase was blocked by treatment with TSA (Fig. 1C). In contrast, genes that were induced by TSA alone, such as histone H1 and PRDI-BFI, were still equally induced by combined IFN and TSA treatment (Fig. 1C).
ISG induction in HeLa cells largely depends on prior induction of IRF9, the DNA-binding subunit of ISGF3 (26). We considered whether TSA inhibition of ISG expression was due to prevention of IRF9 induction. Indeed, IRF9 protein levels did not accumulate in cells treated with IFN and TSA (data not shown). However, IRF9 is also inducible by IFNγ (27), and cells pretreated with IFNγ remained sensitive to the effects of TSA on subsequent treatment with IFNα (Fig. 1F), despite abundant levels of IRF9 protein (Fig. 1E). Moreover, human FS2 fibroblasts that constitutively express high levels of IRF9 were similarly sensitive to TSA, even after exposure to drug and cytokine for as short as 1 h (Fig. 1D). Therefore, TSA inhibited ISG expression by mechanisms independent of or in addition to impaired accumulation of IRF9.
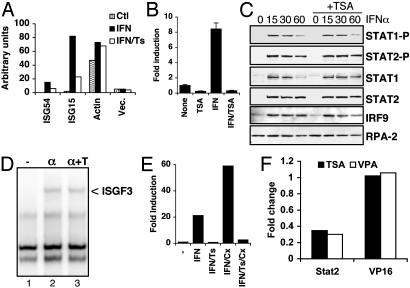
HDAC Activity Is Required for Transcription but Does Not Affect Stat Protein Activation. The data shown in Fig. 1F scored the abundance of ISG54 primary nuclear transcripts by using PCR primers specific for unspliced mRNA retaining the intron between exons 1 and 2. This measurement closely approximates transcription rates (17). Inhibition of ISG54 primary transcript abundance in the presence of TSA suggests that HDAC functions at the transcriptional level. To confirm a transcriptional role, the effect of TSA on ISG transcription was analyzed by in vitro nuclear run-on assays. Nuclei were isolated from HeLa cells after 1-h treatments with IFN and TSA, RNA was transcribed in the presence of [32P]UTP, and transcription rates were estimated by filter hybridization (Fig. 2A). ISG54 and ISG15 transcription increased substantially in response to IFN, whereas actin transcription remained approximately uniform, as expected (18). However, addition of TSA together with IFN impaired transcriptional induction of ISG54 and ISG15 by 60% and 75%, respectively, while having no effect on actin transcription.
Fig. 2.
HDAC function is required for transcription without affecting Janus kinase–Stat signaling. (A) HeLa cells were untreated or treated with IFNα with or without TSA for 1 h, before isolation of nuclei for in vitro run-on transcription reactions. Labeled RNA was hybridized with filter-bound cDNA for the indicated sequences and quantified by PhosphorImager analysis. (B) HeLa cells transfected with ISG54-luciferase together with CMV-LacZ were treated with TSA and IFNα, as indicated, and cell extracts were analyzed for luciferase activity, normalized to β-galactosidase activity, and reported as fold induction over untreated cells. (C) HeLa cells were untreated or treated with IFNα in the absence or presence of TSA, as indicated, before isolation of nuclear extracts. Western blots were probed with antibodies against phospho-Stat1 and Stat2, total Stat1 and Stat2, IRF9, and RPA-2, as indicated. (D) Extracts from HeLa cells treated with IFNα with or without TSA were analyzed for the presence of ISGF3 by electrophoretic mobility-shift assay. (E) ISG54 transcript levels were quantified from HeLa cells treated with IFNα, TSA, and Cx, as indicated. (F) HeLa cells were transfected with UAS-luciferase along with Gal4 DNA-binding domain alone or fused with the transactivation domain of Stat2 or VP16. Fold change in luciferase activity in the presence of TSA or VPA relative to untreated samples is shown.
The effect of TSA on ISG promoter activity was measured by using reporter assays (28). Cells transfected with ISG54-luciferase showed that IFN-stimulated expression was substantially blocked by the presence of TSA (Fig. 2B). Neither ectopic expression of IRF9 nor pretreatment of cells with IFNγ, two methods for increasing cellular responsiveness to IFN (29), prevented transcriptional inhibition by TSA (data not shown). The combination of nuclear run-on assays, quantitation of nuclear RNA precursors, and reporter assays provides compelling evidence that ISG induction by IFN is impaired at the transcriptional level in the presence of TSA.
ISG transcription depends on activation of the Janus kinase–Stat pathway, resulting in tyrosyl phosphorylation of Stat1 and Stat2 and nuclear accumulation of ISGF3. To determine the effect of TSA on these steps, we examined Stat tyrosyl phosphorylation and the ability of ISGF3 to bind DNA. Both Stat1 and Stat2 became phosphorylated on tyrosine in response to IFN, regardless of the presence of TSA (Fig. 2C). Similarly, the ability of these proteins to be recovered from nuclear extracts of IFN-treated cells was unaffected by TSA treatment. ISGF3 assembly and function was directly assayed by gel mobility shift experiments, but no differences were noted when extracts from IFN-treated cells were compared with those from cells treated simultaneously with IFN and TSA (Fig. 2D). These data indicate that TSA is not a direct inhibitor of the Janus kinase–Stat pathway and suggests that the requirement for HDAC activity is downstream of ISGF3 formation.
Two potential mechanisms of TSA-mediated inhibition of ISG expression were considered. It was possible that TSA inhibited a critical HDAC function directly required for transcription from ISG promoters. Alternatively, HDAC function might be required indirectly, for example, to prevent accumulation of a protein repressor that blocks ISG expression. To investigate the latter hypothesis, we treated cells with TSA in the presence of the translation inhibitor cycloheximide (Cx). Blocking ongoing protein synthesis impairs ISG induction in HeLa cells due to prevention of IRF9 accumulation, but this effect can be overcome by preinduction of IRF9 by IFNγ treatment (26). Cx did not prevent expression of ISG mRNA in response to IFN treatment in cells previously exposed to IFNγ, as expected (Fig. 2E), but rather led to enhanced accumulation, most likely because of stabilization of short-lived mRNA (30, 31). However, treatment with Cx did not reverse the effect of TSA, which remained capable of blocking ISG induction. These data indicate that the HDAC activity required for induction of ISG expression that is blocked by TSA does not act indirectly through an intermediate protein but rather directly inhibits ISG transcription.
We used a mammalian one-hybrid approach to test a direct HDAC role in ISG transcription. The majority of ISGF3 transactivation function resides in the carboxyl terminus of Stat2, and this region functions as a transactivation domain when fused to the yeast Gal4 DNA-binding domain (8, 11). Significantly, Stat2-driven gene expression was reduced 3- to 4-fold by treatment with TSA or VPA (Fig. 2F). In contrast, expression driven by VP16 was not affected by HDAC inhibitors. These data indicate that inhibition of transcription by HDAC inactivation occurred at least in part by inhibition of Stat2 function.
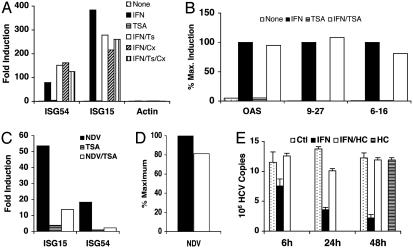
HDAC Activity Is Required for a Transcriptional Step Downstream of ISGF3 Assembly on Chromatin. The preceding results indicated that TSA blocks HDAC activity required for ISG transcriptional induction at a step downstream of ISGF3 formation, because Stat1 and Stat2 tyrosine phosphorylation, nuclear translocation, and DNA binding ability were unaffected. However, because an important target for HDAC activity is histone modification, we examined whether ISGF3 recruitment to chromatin required active deacetylation. To this end, we compared the recruitment of ISGF3 to target promoters in vivo by ChIP. As shown previously (9), the ability of antibodies against Stat2 to recover ISG54 promoter sequences was greatly enhanced by IFN treatment (Fig. 3A), demonstrating that Stat2 is recruited to this promoter in response to IFN. Similarly, Stat2 ChIP recovered other ISG promoter sequences in an IFN-dependent manner, including the promoters for ISG15 (Fig. 3A), oligoadenylate synthetase, gene 9–27, and gene 6–16 (Fig. 3B). Cotreatment of cells with IFN and TSA did not prevent the interaction of Stat2 with these target promoters, demonstrating that HDAC is not required for Stat2 recruitment to chromatin. Neither Cx treatment nor combined Cx and TSA treatment prevented Stat2 recruitment in response to IFN (Fig. 3A). As expected, antibodies against Stat1 and IRF9 also precipitated ISG promoter sequences after IFN treatment, and this outcome was not prevented by TSA (not shown). The background recovery of control sequences (e.g., β-actin) was unaffected by either IFN or TSA treatment and was near the limits of detectability (Fig. 3A). These results demonstrate that assembly of ISGF3 on chromatin in vivo does not require HDAC activity despite the severe impairment of transcriptional activation of target genes.
Fig. 3.
HDAC function is not required for ISGF3 assembly on chromatin but is essential for IFN suppression of HCV replication. (A) ChIP assays were performed on HeLa cells treated with IFNα, TSA, and Cx, as indicated, with antibodies against Stat2. Amounts of ISG54, ISG15, and actin promoter sequences recovered in immunoprecipitates relative to input levels were quantified by real-time PCR and reported as fold induction relative to untreated cells. (B) As in A, except promoter sequences of OAS2, gene 9–27, and gene 6–16 were quantified and reported relative to maximal amount recovered from IFNα-treated cells. (C) Induction of ISG15 and ISG54 transcript levels was quantified in Vero cells infected with NDV in the absence or presence of TSA. (D) NDV transcripts from Vero cells infected in the absence (filled bar) or presence of TSA (open bar) were quantified by RT-PCR. (E) Inhibition of HCV replication by IFNα treatment in the absence or presence of HC toxin was monitored by RT-PCR.
Many ISG promoters are transcriptionally induced not only in response to IFN treatment but also after virus infection through recruitment of transcription factor IRF3 (32). To determine whether induction of the same ISG promoters by an alternative transcription factor also required HDAC activity, we measured ISG expression in virus-infected cells. To preclude virus-induced autocrine IFN, we infected Vero cells that had sustained a chromosomal deletion removing the type I IFN locus (33). Newcastle disease virus (NDV)-infected Vero cells displayed a robust induction of ISG54 and ISG15, and this induction was severely impaired by TSA (Fig. 3C). Expression of genes 6–16 was largely unaffected by virus infection (not shown), consistent with its promoter being a poor target for IRF3 (34). TSA did not inhibit virus infection as judged by nearly equal production of viral RNA (Fig. 3D) and viral proteins (data not shown). Similar to results obtained after IFN treatment, the block in promoter activity in response to activated IRF3 in the presence of TSA was downstream of transcription factor assembly, as judged by ChIP for IRF3 (not shown). These data provide compelling evidence that transcriptional induction of ISG promoters, whether by Stat2 or IRF3, requires HDAC function. HDAC activity is required subsequent to transcription factor assembly on chromatin.
HDAC Activity Is Required for Establishment of an Antiviral State. HCV replication is inhibited by IFN, making IFN the preferred treatment for HCV infections in vivo (35). To determine whether the block in ISG induction observed in the absence of HDAC activity would affect IFN antiviral action, we treated Huh-7.5 cells containing a HCV replicon (20) with IFN in the presence or absence of the HDAC inhibitor, HC toxin (Fig. 3E). IFN inhibited HCV in this system, resulting in a progressive loss of HCV RNA with >80% inhibition of viral RNA after 48 h of IFN treatment. This antiviral response required HDAC activity because little or no inhibition of viral RNA synthesis was observed in IFN-treated cells exposed to HC toxin. HC toxin produced no nonspecific effects on viral replication, because no change in viral RNA abundance was observed in cells incubated in the presence of HC toxin alone (Fig. 3E). The antiviral response against two other viruses, encephalomyocarditis virus (EMCV) and vesicular stomatitis virus (VSV), was also impaired in the presence of HDAC inhibitors. IFN treatment of HeLa cells inhibited the cytopathic effect of EMCV (IC50, 4 units/ml) and VSV (IC50, 125 units/ml); however, in the presence of HC toxin, TSA, or VPA, IFN failed to inhibit the cytopathic effect of EMCV or VSV, even at 1,000 units/ml (Figs. 4–7, which are published as supporting information on the PNAS web site). In fact, treatment with HDAC inhibitors increased the viral cytopathic effect, likely because of inhibition of autocrine IFN.
Discussion
HDAC activity has been commonly associated with transcriptional repression, and several HDAC-containing repressor complexes have been identified. In these situations, HDAC proteins act as corepressor molecules that prevent transcription of target genes once recruited to promoter regions. It is assumed that HDAC-mediated transcriptional repression involves erasure of acetylation marks of active transcription by deacetylation of histones, resulting in repressed chromatin inaccessible to transcription factor binding (12). Inhibition of HDAC activity results in loss of repression, leading to increased transcription of repressed genes.
Evidence for other transcriptional roles for HDAC activity has also been reported, including a role in transcriptional activation. For instance, expression profiling of yeast strains lacking HDAC genes or of wild-type yeast treated with TSA revealed a complex pattern of both induced and repressed genes (36). A similar result was observed in TSA-treated human cells (37), suggesting that HDAC activity can be both inhibitory and stimulatory for gene expression. However, mechanisms by which HDAC activity would mediate transcriptional induction rather than repression remain largely unknown. Possible mechanisms for HDAC function in transcription include direct effects through modification of histones, modification of non-histone chromatin proteins, or direct or indirect effects through alternative substrates. For instance, acetylation of lysine 12 of histone H4 is associated with gene silencing rather than activation (38), and HDAC-mediated deacetylation of this lysine may therefore derepress these silenced genes. Histones are not the only targets for acetyltransferases, because many transcriptional regulators and other cellular proteins can be modified by acetylation (39). Acetylation of such proteins can be inhibitory, implying that reversal by deacetylation would be stimulatory for transcription. For instance, acetylation of coactivator proteins can disrupt transcription complex assembly, leading to transcriptional repression (40). Deacetylation would therefore favor transcription. Similarly, acetylation of the activator C/EBPβ inhibits its DNA-binding ability and its ability to induce gene expression. Deacetylation of C/EBPβ by a recruited HDAC contributes to transcriptional activation (41). Other cellular proteins that do not have direct roles in transcription but may nonetheless indirectly affect gene expression are also modulated by acetylation and deacetylation.
The results reported here predict a HDAC requirement for ISG transcription. We have previously shown that these genes are regulated by a distinct mechanism dependent on the transactivation domain of Stat2 that involves GCN5 and TAF4 (9). Our data demonstrate that the HDAC requirement lies down-stream of Stat activation and assembly on chromatin but before initiation of transcription. However, the HDAC requirement may be more general than the GCN5-dependent initiation mechanism used by Stat2, because virus-induced gene expression was also blocked by TSA. Thus, ISG54 induction by IFN, mediated by a GCN5/TAF4 mechanism recruited by Stat2, and induction by virus, mediated by an IRF3/p300 mechanism (42), were both impaired when HDAC function was blocked. HDAC involvement also appears to be independent of the requirement for ATP-dependent chromatin remodeling, because both BRG1-dependent and independent gene induction (24, 25) were impaired in the absence of HDAC function. Therefore, the requirement for HDAC function in gene induction, as opposed to its more commonly observed role in gene repression, may represent a general regulatory mechanism for acutely inducible genes. In addition, it was recently reported that inducible gene expression mediated by cytokine activation of Stat5 requires HDAC function (43). However, not all inducible genes require HDAC function for activation. In fact, although it was recently shown that some IFNγ-responsive genes are inhibited by TSA (44), some genes induced by IFNγ exhibit the more traditional HDAC-dependent repression. For instance, IFNγ induction of major histocompatibility II genes is enhanced by inhibition of HDAC function (45), and we found that inhibition of HCV by IFNγ was not impaired in the absence of HDAC activity, unlike the antiviral action of IFNα (M.P., unpublished observations). In addition, expression of TGFβ-responsive genes is augmented rather than inhibited by cotreatment with HDAC inhibitors such as TSA (46).
The molecular mechanism of HDAC action during ISG induction remains undefined. The finding that three unrelated HDAC inhibitors produce equivalent effects on ISG expression argues that this inhibition is mechanism-based through impaired HDAC catalytic function. The absence of any HDAC requirement for ISGF3 formation and assembly on chromatin implies that deacetylation of a target protein is required during transcriptional initiation, possibly during coactivator recruitment or action. Indeed, the impaired activity of a Gal4-Stat2 fusion transactivator in the presence of TSA (Fig. 2F) supports this notion. A potential target could be the transactivation domain of Stat2 itself; however, we have failed to detect any significant acetylation of Stat2 (H.-M.C., unpublished data). Recently, Civas and colleagues (47) suggested that deacetylation affects Stat2 nuclear accumulation in response to IFN. Their observations are in contrast to those reported here or reported recently by Nusinzon and Horvath (44). It should be noted that the conclusions drawn by Civas and colleagues were based on extended treatments with TSA for 24–40 h, which are likely to result in additional indirect effects, including impaired positive feedback and possibly cellular toxicity (48).
A potential implication of these results relates to the clinical use of HDAC inhibitors, where inhibition of ISG expression and the subsequent impaired antiviral response could be a consideration. HDAC inhibitors are chemotherapeutic agents because of their ability to induce differentiation, growth arrest, and apoptosis of tumor cells (13). Acute promyelocytic leukemia responds to both HDAC inhibitor and IFN therapy (49). However, the inhibition of IFN responsiveness by HDAC inhibitors would suggest caution in the possible combined use of these agents. Similarly, attention to the possibility of unexpected susceptibility to viral infections during HDAC inhibitor therapy may be appropriate. The HDAC inhibitor VPA is a standard antiepileptic treatment. Given the impaired ISG responses observed in cells exposed to VPA, a similar effect is possible in patients receiving this drug. For instance, VPA treatment has been found to exacerbate chronic HCV infections (50), a disease that is often responsive to IFN therapy in a HDAC-dependent manner (Fig. 3E). Investigation of possible impaired IFN responsiveness in other biological consequences of HDAC inhibitors is warranted.
Supplementary Material
Acknowledgments
We thank Gregory David for helpful discussion and the gift of reagents, Annick Gauthier for discussions, and Wei Zhu and Esther Casas for expert technical assistance. This work was supported by the National Institutes of Health, the American Heart Association, the Dr. Louis A. Schneider Professorship in Molecular Pathology, and the Greenberg Medical Research Institute.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ChIP, chromatin immunoprecipitation; Cx, cycloheximide; HDAC, histone deacetylase; HCV, hepatitis C virus; IRF, IFN regulatory factor; ISG, IFN-stimulated gene(s); NDV, Newcastle disease virus; Stat, signal transducer and activator of transcription; TSA, trichostatin A; VPA, valproic acid; VSV, vesicular stomatitis virus.
References
- 1.Stark, G. R., Kerr, I. M., Williams, B. R., Silverman, R. H. & Schreiber, R. D. (1998) Annu. Rev. Biochem. 67, 227-264. [DOI] [PubMed] [Google Scholar]
- 2.Friedman, R. L., Manly, S. P., McMahon, M., Kerr, I. M. & Stark, G. R. (1984) Cell 38, 745-755. [DOI] [PubMed] [Google Scholar]
- 3.Larner, A. C., Jonak, G., Cheng, Y. S., Korant, B. D., Knight, E. & Darnell, J. E. (1984) Proc. Natl. Acad. Sci. USA 81, 6733-6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levy, D. E. & Darnell, J. E. (1990) New Biol. 2, 923-928. [PubMed] [Google Scholar]
- 5.Levy, D. E. & Darnell, J. E., Jr. (2002) Nat. Rev. Mol. Cell Biol. 3, 651-662. [DOI] [PubMed] [Google Scholar]
- 6.Bluyssen, H. A. R., Durbin, J. E. & Levy, D. E. (1996) Cytokine Growth Factor Rev. 7, 11-17. [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharya, S., Eckner, R., Grossman, S., Oldread, E., Arany, Z., D'Andrea, A. & Livingston, D. M. (1996) Nature 383, 344-347. [DOI] [PubMed] [Google Scholar]
- 8.Paulson, M., Pisharody, S., Pan, L., Guadagno, S., Mui, A. L. & Levy, D. E. (1999) J. Biol. Chem. 274, 25343-25349. [DOI] [PubMed] [Google Scholar]
- 9.Paulson, M., Press, C., Smith, E., Tanese, N. & Levy, D. E. (2002) Nat. Cell Biol. 4, 140-147. [DOI] [PubMed] [Google Scholar]
- 10.Lau, J. F., Nusinzon, I., Burakov, D., Freedman, L. P. & Horvath, C. M. (2003) Mol. Cell. Biol. 23, 620-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qureshi, S. A., Leung, S., Kerr, I. M., Stark, G. R. & Darnell, J. E. (1996) Mol. Cell. Biol. 16, 288-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischle, W., Wang, Y. & Allis, C. D. (2003) Curr. Opin. Cell Biol. 15, 172-183. [DOI] [PubMed] [Google Scholar]
- 13.Marks, P., Rifkind, R. A., Richon, V. M., Breslow, R., Miller, T. & Kelly, W. K. (2001) Nat. Rev. Cancer 1, 194-202. [DOI] [PubMed] [Google Scholar]
- 14.Marié, I., Durbin, J. E. & Levy, D. E. (1998) EMBO J. 17, 6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Decker, T., Lew, D. J., Cheng, Y. S., Levy, D. E. & Darnell, J. E. (1989) EMBO J. 8, 2009-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cleveland, D. W., Lopata, M. A., MacDonald, R. J., Cowan, N. J., Rutter, W. J. & Kirschner, M. W. (1980) Cell 20, 95-105. [DOI] [PubMed] [Google Scholar]
- 17.Levy, D. E., Larner, A. C., Chaudhuri, A., Babiss, L. E. & Darnell, J. E. (1986) Proc. Natl. Acad. Sci. USA 83, 8929-8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reich, N., Evans, B., Levy, D. E., Fahey, D., Knight, E. & Darnell, J. E. (1987) Proc. Natl. Acad. Sci. USA 84, 6394-6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levy, D. E. (1998) Methods 15, 167-174. [DOI] [PubMed] [Google Scholar]
- 20.Blight, K. J., McKeating, J. A. & Rice, C. M. (2002) J. Virol. 76, 13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sledz, C. A., Holko, M., De Veer, M. J., Silverman, R. H. & Williams, B. R. (2003) Nat. Cell Biol. 5, 834-839. [DOI] [PubMed] [Google Scholar]
- 22.Gottlicher, M., Minucci, S., Zhu, P., Kramer, O. H., Schimpf, A., Giavara, S., Sleeman, J. P., Lo Coco, F., Nervi, C., Pelicci, P. G. & Heinzel, T. (2001) EMBO J. 20, 6969-6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brosch, G., Ransom, R., Lechner, T., Walton, J. D. & Loidl, P. (1995) Plant Cell 7, 1941-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, H., Kang, H., Liu, R., Chen, X. & Zhao, K. (2002) Mol. Cell. Biol. 22, 6471-6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang, M., Qian, F., Hu, Y., Ang, C., Li, Z. & Wen, Z. (2002) Nat. Cell Biol. 4, 774-781. [DOI] [PubMed] [Google Scholar]
- 26.Levy, D. E., Lew, D. J., Decker, T., Kessler, D. S. & Darnell, J. E. (1990) EMBO J. 9, 1105-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Veals, S. A., Kessler, D. S., Leonard, D., Josiah, S. & Levy, D. E. (1991) J. Interferon Res. 11, S82. [DOI] [PubMed] [Google Scholar]
- 28.Bluyssen, H. A. R. & Levy, D. E. (1997) J. Biol. Chem. 272, 4600-4605. [DOI] [PubMed] [Google Scholar]
- 29.Bluyssen, H. A. R., Muzaffar, R., Vlieststra, R. J., van der Made, A. C. J., Leung, S., Stark, G. R., Kerr, I. M., Trapman, J. & Levy, D. E. (1995) Proc. Natl. Acad. Sci. USA 92, 5645-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larner, A. C., Chaudhuri, A. & Darnell, J. E. (1986) J. Biol. Chem. 261, 453-459. [PubMed] [Google Scholar]
- 31.Kusari, J. & Sen, G. C. (1987) Mol. Cell. Biol. 7, 528-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wathelet, M. G., Lin, C. H., Parekh, B. S., Ronco, L. V., Howley, P. M. & Maniatis, T. (1998) Mol. Cell 1, 507-518. [DOI] [PubMed] [Google Scholar]
- 33.Desmyter, J., Melnick, J. L. & Rawls, W. E. (1968) J. Virol. 2, 955-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wathelet, M. G., Berr, P. M. & Huez, G. A. (1992) Eur. J. Biochem. 206, 901-910. [DOI] [PubMed] [Google Scholar]
- 35.McHutchison, J. G. & Fried, M. W. (2003) Clin. Liver Dis. 7, 149-161. [DOI] [PubMed] [Google Scholar]
- 36.Bernstein, B. E., Tong, J. K. & Schreiber, S. L. (2000) Proc. Natl. Acad. Sci. USA 97, 13708-13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eickhoff, B., Ruller, S., Laue, T., Kohler, G., Stahl, C., Schlaak, M. & van der Bosch, J. (2000) Biol. Chem. 381, 107-112. [DOI] [PubMed] [Google Scholar]
- 38.Braunstein, M., Sobel, R. E., Allis, C. D., Turner, B. M. & Broach, J. R. (1996) Mol. Cell. Biol. 16, 4349-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sterner, D. E. & Berger, S. L. (2000) Microbiol. Mol. Biol. Rev. 64, 435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen, H., Lin, R. J., Xie, W., Wilpitz, D. & Evans, R. M. (1999) Cell 98, 675-686. [DOI] [PubMed] [Google Scholar]
- 41.Xu, M., Nie, L., Kim, S.-H. & Sun, X.-H. (2003) EMBO J. 22, 893-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoneyama, M., Suhara, W., Fukuhara, Y., Fukuda, M., Nishida, E. & Fujita, T. (1998) EMBO J. 17, 1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rascle, A., Johnston, J. A. & Amati, B. (2003) Mol. Cell. Biol. 23, 4162-4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nusinzon, I. & Horvath, C. M. (2003) Proc. Natl. Acad. Sci. USA 100, 14742-14747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zika, E., Greer, S. F., Zhu, X. S. & Ting, J. P. (2003) Mol. Cell. Biol. 23, 3091-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Su, G. H., Sohn, T. A., Ryu, B. & Kern, S. E. (2000) Cancer Res. 60, 3137-3142. [PubMed] [Google Scholar]
- 47.Genin, P., Morin, P. & Civas, A. (2003) J. Virol. 77, 7113-7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henderson, C., Mizzau, M., Paroni, G., Maestro, R., Schneider, C. & Brancolini, C. (2003) J. Biol. Chem. 278, 12579-12589. [DOI] [PubMed] [Google Scholar]
- 49.de The, H. & Chelbi-Alix, M. K. (2001) Oncogene 20, 7136-7139. [DOI] [PubMed] [Google Scholar]
- 50.Felker, B. L., Sloan, K. L., Dominitz, J. A. & Barnes, R. F. (2003) Am. J. Psychiatry 160, 174-178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.