Abstract
We previously identified an internal ribosome entry site (IRES) within the 5′ leader of the mRNA encoding the Gtx homeodomain protein and showed that shorter nonoverlapping segments of this 5′ leader could enhance the translation of a second cistron in a dicistronic mRNA. One of these segments was 9 nt in length, and when multiple copies of this IRES module were linked together, IRES activity was greatly enhanced. To further expand the potential uses of these synthetic constructs and facilitate analyses of the mechanism by which they affect translation, we show here that an IRES containing five linked copies of the 9-nt sequence can also enhance translation in the 5′ leader of a monocistronic mRNA. Moreover, a search for interactions of the IRES module with cellular factors revealed specific binding to 40S ribosomal subunits but not to other cellular components. Based on the results of earlier studies suggesting that this sequence could bind to a complementary segment of 18S rRNA, we tested various sequences for possible links between the length of the complementary match, their binding to ribosomes, and their influence on translational efficiency. We found that the length of the complementary match was directly correlated with the ability of RNA probes to bind to ribosomes. In addition, translation was maximally enhanced (≈8-fold) by a 7-nt segment of the 9-nt element; the enhancement declined progressively as the complementary stretches became progressively longer or shorter. The results suggest that the Gtx 9-nt sequence affects translation efficiency by a mechanism that involves base pairing to 18S rRNA.
In earlier studies, we identified cis-acting sequences within the 5′ leaders of the mRNAs for the homeodomain protein Gtx and the cold-stress-induced protein Rbm3 that appeared to function as internal ribosome entry sites (IRESs). When tested in isolation these relatively short sequences enhanced the translation of a second cistron in a dicistronic mRNA (1, 2). In addition, we found that RNA sequences with IRES activity could be selected from libraries of random 9- to 18-nt sequences (3, 4). Although in most cases, IRES activity mediated by an individual IRES element was relatively weak, it could be greatly enhanced when multiple copies of some of the individual IRES elements were linked together in tandem (1, 3).
Synthetic IRESs based on the 9-nt Gtx IRES module have proven to be particularly useful in various vector systems for generating high levels of expression from multicistronic mRNAs (e.g., refs. 5–7). Moreover, they are shorter and are more active than viral IRESs, such as the encephalomyocarditis virus IRES, that are typically used for these applications, and synthetic IRES activity can be enhanced or diminished by increasing or decreasing the number of IRES modules.
The question arises whether, in addition to enhancing translation in a dicistronic context, such constructs would also enhance translation in a monocistronic context, i.e., in the 5′ leader of a reporter mRNA. There are a number of reasons prompting an analysis of this sequence element in a monocistronic context. For one thing, a monocistronic context provides a better reference than dicistronic mRNAs for determining whether changes in protein production occur by mechanisms that alter the rate of translation or by a non-translational mechanism such as transcription, which can affect protein production by altering mRNA levels. In addition, in a monocistronic but not in dicistronic context, mutations that disrupt translational enhancement can be distinguished from those that actually block translation. A mutation that blocks translation in the monocistronic context will reduce translation to a level below that of the control. By contrast, in a dicistronic mRNA, a similar mutation will only reduce translation to the level of the control. Finally, for a similar degree of multimerization, protein production may be more efficient in monocistronic than in dicistronic constructs, and all of these properties may prove to be of practical significance.
In the present study, we showed that a synthetic IRES containing multiple copies of the Gtx 9-nt module enhanced translation efficiency in a monocistronic context by ≈3-fold. A search for trans-acting factors that bind to a single copy of the 9-nt element revealed a specific interaction with 40S ribosomal subunits but not to other cellular factors. Inasmuch as the 9-nt sequence is 100% complementary to a segment of 18S rRNA present in the 40S ribosomal subunit, we performed binding experiments by using 18-nt RNA probes in which the length of the 9-nt complementary match was either shortened or lengthened. The results showed that the relative binding affinities were directly correlated with the length of the complementary match, i.e., RNA probes with longer complementary matches bound to 40S ribosomal subunits with more affinity than RNA probes with shorter complementary matches.
RNA sequences with longer or shorter complementary matches to 18S rRNA were then tested for activity in transiently transfected mouse neuroblastoma Neuro 2a (N2a) cells. The data showed that translational efficiencies were highest with a complementary matches of seven consecutive nucleotides and that enhancement of translation was reduced when either shorter or longer complementary stretches were used. The results of these studies indicate that multiple copies of the 9-nt Gtx element can enhance translation by a mechanism that involves base pairing to a segment of 18S rRNA within intact 40S ribosomal subunits.
Methods
DNA Constructs. Monocistronic reporter constructs contain the SV40 promoter, which drives transcription of the Photinus luciferase cistron. A multiple cloning site (MCS) containing SpeI and EcoRI restriction endonuclease sites was introduced 5′ of the Photinus luciferase cistron by using HindIII and NcoI restriction sites. Oligonucleotides containing five linked copies of nucleotides 133–141 of the Gtx 5′ leader or mutations of this sequence were cloned into the 5′ UTR of the Photinus luciferase cistron as described (1) by using either SpeI or EcoRI, and NcoI restriction sites. Mutated sequences were substituted into the full-length Gtx 5′ leader by using BseP1 and ApaI restriction endonuclease sites. Inserts containing mutated Gtx sequences were generated by using Pfu polymerase amplification and restricted with these same restriction endonucleases. The MCS oligonucleotides and 5′ leader sequences of the constructs used in this study are presented in Table 1, which is published as supporting information on the PNAS web site.
Transfection Analyses. Reporter constructs (0.5 μg) were transfected into mouse N2a cells (1 × 105) by using FuGENE 6 (Roche Applied Science). Transfection efficiencies were normalized by cotransfection with 0.2 μg of a LacZ reporter gene construct (pCMVβ, Clontech). Cells were harvested 24 h post-transfection, and reporter gene activities were determined (1). Translation efficiencies were calculated by normalizing the reporter gene activities to the corresponding mRNA levels, which were determined by using quantitative RT-PCR.
RNA Transcripts. RNA oligonucleotides obtained from Dharmacon (Lafayette, CO) were used as RNA probes. The sequences of the RNA oligonucleotides are listed in Table 2, which is published as supporting information on the PNAS web site. The 5′-end-radiolabeled RNA oligonucleotide probes were generated by using T4 polynucleotide kinase. Probes were purified away from free nucleotides by passage through two consecutive Microspin G25 columns (Pharmacia).
Electrophoretic Mobility Gel Shift and Nitrocellulose Filter Binding Assays. Radiolabeled RNA probes were incubated with cell extracts and electrophoresed on nondenaturing polyacrylamide gels as described (8, 9). For the filter binding experiments (see refs. 10 and 11), ≈15 fmol of radiolabeled RNA probes was denatured and incubated with puromycin dissociated ribosome subunits (12) or with purified 40S ribosomal subunits in binding buffer (13) and incubated at 37°C for 15 min. Samples were filtered through nitrocellulose membrane (0.45 μm) and washed several times with cold buffer. Ribosome-bound probe which was retained by the membrane was quantified by using a PhosphorImager (Molecular Dynamics). To determine binding specificity, competitor RNAs were added to the radiolabeled probe before the denaturation step.
Quantitative RT-PCR. Total RNA was extracted from transfected cells and 1 μg of RNA was reverse transcribed as described (14). PCR reactions were performed by using primer sets specific to the Photinus luciferase coding sequence (5′-TCAAAGAGGCGAACTGTGTG-3′ and 5′-GGTGTTGGAGCAAGATGGAT-3′) and lacZ (5′-GACGTCTCGTTGCTGCATAA-3′ and 5′-CAGCAGCAGACCATTTTCAA-3′) gene-specific primers. Fluorescent dNTPs were incorporated into PCR products by using SYBR Green PCR Master Mix (Applied Biosystems). Photinus luciferase and LacZ fluorescent PCR products were quantified after 15–18 and 17–19 cycles per sample, respectively, by using the GeneAmp 5700 sequence detector (Applied Biosystems). These cycle ranges reflect the cycle numbers at which these samples reached a predetermined threshold. The concentrations of cDNA at these cycle numbers were then determined by using a standard curve.
Results
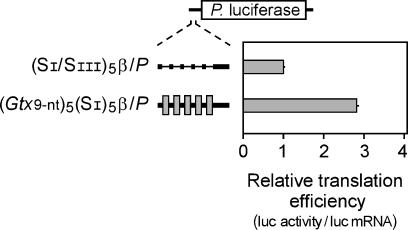
The 9-nt Gtx IRES Module Functions as a Translational Enhancer in a Monocistronic Context. A Photinus luciferase reporter construct with a synthetic 5′ leader sequence containing five linked copies of the 9-nt Gtx IRES module ((Gtx9-nt)5(SI)5β/P) was tested in transiently transfected N2a cells. Relative translation efficiencies were estimated by normalizing Photinus luciferase activities to Photinus luciferase mRNA levels (see Methods) and were compared to those obtained with control constructs lacking the Gtx sequences. In the Gtx construct, the individual 9-nt elements were separated from each other by segments of 9 nt taken from the β-globin 5′ UTR (designated SI; ref. 1). This assemblage of repeats was placed 25 nt upstream of the initiation codon by using another segment of the β-globin 5′ UTR as a spacer (Table 1). These same segments of the β-globin 5′ UTR were shown previously not to affect translation in an IRES assay (1). The results (Fig. 1) indicated that five linked copies of the Gtx IRES module in the 5′ UTR of a monocistronic mRNA enhanced the rate of translation by ≈3-fold when compared to the activities obtained with a control construct in which the 9-nt Gtx sequences were mutated to 9-nt stretches of poly(A).
Fig. 1.
Nine-nucleotide IRES module functions as a translational enhancer in a monocistronic context. A schematic representation of the Photinus luciferase monocistronic constructs is indicated. The 5′ leader sequences include five linked copies of the Gtx 9-nt IRES module, indicated as gray bars, and a control (SI/SIII) in which the Gtx sequences have been mutated to poly(A), indicated by thin black lines. Both constructs contain 25 nt of the mouse β-globin 5′ UTR sequence upstream of the Photinus luciferase initiation codon. These 25 nucleotides are represented by a thick black line. Constructs were assayed in transiently transfected N2a cells. Translation efficiencies were determined per unit mRNA and normalized to 1.0 for those obtained with the control construct containing the SI/SIII sequence. Horizontal lines indicate SEM.
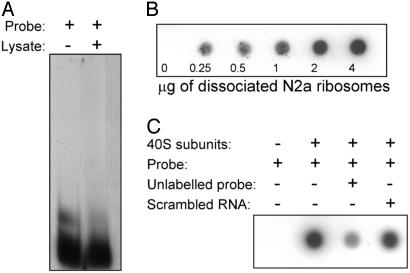
The 9-nt Element Binds Directly to Dissociated Ribosomal Subunits but Not to Other Cytoplasmic Factors. Electrophoretic mobility shift assays were performed by using radiolabeled RNA probes containing the 9-nt Gtx element. In these studies, a radiolabeled RNA probe containing a single copy of the 9-nt sequence was incubated with a cytoplasmic extract and electrophoresed on a nondenaturing gel (Fig. 2A). No difference in the mobility of this probe was found in the presence or absence of cytoplasmic factors, suggesting that this sequence does not bind specifically to cytoplasmic proteins contained within this extract.
Fig. 2.
Binding studies with the Gtx 9-nt sequence. (A) Gel mobility-shift assay. An 18-nt RNA probe containing the 9-nt Gtx sequence element (9-nt probe) was electrophoresed alone (probe +, extract -) or after incubation in an N2a cell extract (probe +, extract +), as indicated on the autoradiogram. (B) Nitrocellulose filter binding assays. The Gtx 9-nt probe was incubated with increasing concentrations of puromycin-dissociated N2a ribosomes and filtered through a nitrocellulose membrane. (C) Binding of the Gtx 9-nt probe to isolated 40S ribosomal subunits (4 μg). Binding specificity was tested by adding a 100-fold molar excess of the unlabeled probe as a specific competitor and confirmed with the same concentration of a nonspecific competitor, which was an unlabeled RNA of the same length and base composition as the Gtx probe but with a scrambled sequence.
The RNA probe containing the single 9-nt element was also tested for its ability to bind to puromycin dissociated ribosomal subunits prepared from N2a cells. The radiolabeled RNA probe was incubated with ribosomes in binding buffer and filtered through a nitrocellulose membrane under salt conditions that retain protein but not free RNA. The results (Fig. 2B) showed that the Gtx probe was able to bind to ribosomes. When repeated with purified 40S ribosomal subunits, binding was again observed (Fig. 2C). This binding is specific because it was competed by a 100-fold molar excess of unlabeled probe but not by the same molar excess of an unlabeled probe containing a scrambled sequence (Fig. 2C).
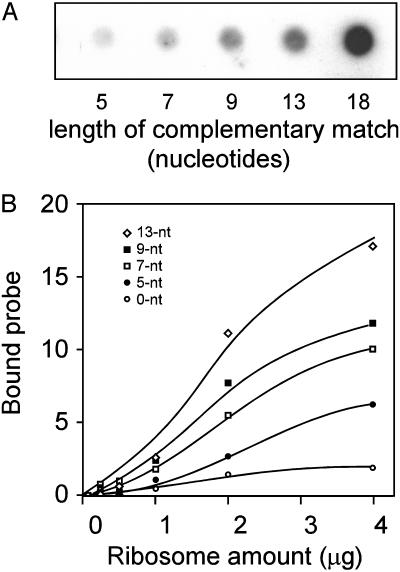
Ribosome Binding Is Directly Correlated with the Length of the Complementary Match to 18S rRNA. To assess the relationship between the length of the complementary match and ribosome binding, the binding affinity of an 18-nt radiolabeled RNA oligonucleotide probe containing a single copy of the 9-nt element was compared to the binding obtained with 18-nt probes that were mutated to increase or decrease the length of the complementary match to 18S rRNA (Fig. 3). The probe containing the 9-nt translational enhancer was flanked by β-globin 5′ leader sequences (SI; see Table 2). In constructs in which the length of the complementary match was decreased, nucleotides in the 9-nt Gtx element were mutated to A, whereas in constructs for which the length of the complementary match was increased, nucleotides in the flanking sequences were mutated accordingly. The binding of RNA probes containing longer complementary stretches showed progressively stronger relative binding affinities, whereas the RNA probes containing shorter complementary stretches showed progressively weaker relative binding affinities, indicating that the RNA probes were able to base pair to a complementary segment of the 18S rRNA within intact 40S subunits.
Fig. 3.
Relative binding affinity of six probes with varying lengths of complementarity to 18S rRNA. (A) Nitrocellulose filter binding assays of radiolabeled probes that differ in the length of the complementary match to 18S rRNA after mixing with isolated 40S ribosomal subunits (1 μg) from N2a cells. Filter-bound radiolabeled RNA oligonucleotides were quantified by exposure to a phosphor screen developed on a PhosphorImager. (B) Binding curves for the wild-type 9-nt Gtx element and mutated derivatives that increase or decrease the length of the complementary match to 18S rRNA. Binding curves were determined by using nitrocellulose filter binding assays performed with N2a puromycin dissociated ribosomal subunits.
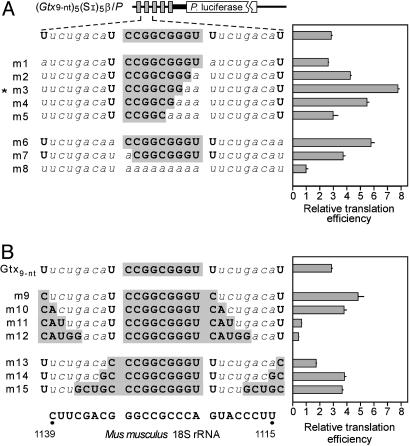
Translation Efficiency Is Correlated with an Optimal Size of the Sequence Complementary to 18S rRNA. The results of the nitrocellulose filter binding experiments suggested that binding affinities were directly related to the length of the complementary match. To assess whether these alterations in the relative binding affinity might affect translation efficiency, we determined the translation efficiency of constructs containing five linked copies of the Gtx 9-nt sequence and compared it to translation efficiencies obtained with constructs containing similar sequences that were mutated to increase or decrease the length of the complementary match to 18S rRNA (Fig. 4). The sequences were tested for activity in the 5′ UTR of a monocistronic Photinus luciferase cistron, and translational efficiencies were determined by normalizing reporter activities to mRNA levels (see Methods). The control mRNA for these studies (m8) is identical to the construct containing five copies of the 9-nt Gtx element ((Gtx9-nt)5(SI)5β/P), except that all of the Gtx nucleotides were mutated to A.
Fig. 4.
Defining the effective boundaries of a minimal IRES module in a monocistronic mRNA. Constructs are based on the (Gtx9-nt)5(SI)5β/P vector. A schematic representation of the constructs used in this analysis is indicated showing one Gtx 9-nt sequence (shaded in gray) flanked by two spacer sequences. Mutations of this sequence in constructs m1–m15 are indicated. The nucleotides complementary to 18S rRNA (shown below) are indicated in gray. Note that the same mutations were made to all of the repeated elements. Constructs were transfected into N2a cells, and relative translation efficiencies were determined (see Methods). The translation efficiency was maximally enhanced in construct m3 (indicated by *), which contains a stretch of seven complementary nucleotides. Numbers in parentheses represent SEM.
The 9-nt Gtx element in construct ((Gtx9-nt)5(SI)5β/P) is complementary to a 9-nt stretch of the 18S rRNA at nucleotides 1123–1131. The nucleotide sequence of the spacer contributes two additional G-U base pairs to this complementary stretch. In constructs m1–m15, the same mutations were made to all five IRES modules (Fig. 4). The results obtained with constructs in which the complementary stretch was shortened (Fig. 4A) showed that translation in transiently transfected N2a cells was maximally enhanced in construct m3 (indicated by *), which contains a stretch of seven complementary nucleotides. Translation was less efficiently enhanced when the complementary stretches were shorter or longer than seven consecutive nucleotides (see constructs m2 and m4–m8). When the length of the complementary match was increased at the 3′ end of the 9-nt sequence (Fig. 4B), the enhancement of translation was lost when complementarity was increased by three or four nucleotides (e.g., m11 and m12). Mutations that increased complementarity 5′ of the 9-nt sequence had little or no effect on translation efficiency (m13–m15).
One of the mutations that increased the length of the complementary match to 18S rRNA (m12) introduced potential AUG initiation codons into the 5′ UTR. These AUGs generate ORFs that overlap the Photinus luciferase reading frame. Interestingly, the presence of these upstream AUGs did not appear to inhibit translation as might be expected if they were used as an initiation codons because construct m11 lacks the upstream AUGs and construct m12 contains them, yet the translation efficiencies of these constructs are not significantly different from each other. Note that although the activities of these constructs is relatively low compared to the other constructs in Fig. 4, these constructs were translated with an efficiency that is ≈50% of that obtained with construct m8. Thus, they are actually translated relatively efficiently.
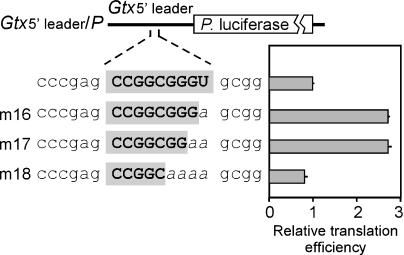
Activity of the Gtx 9-nt Element in Its Native Context. To determine whether alteration of the 9-nt element affects translation when present in the Gtx 5′ leader, we mutated this sequence in this context (Fig. 5). In constructs m16 to m18, the 9-nt sequence was mutated to decrease the length of the complementary match as in constructs m2, m3, and m5 (Fig. 4). The results showed that disruption of the 9-nt sequence actually enhanced translation by ≈3-fold compared to the translation of a construct containing the wild-type sequence. These data are consistent with those in Fig. 4A (m1, m2, m3, and m5). When the length of the complementary match was increased at the 5′ or 3′ ends, the results were more variable, perhaps reflecting the disruption of other cisacting sequences or structures in the 5′ leader (data not shown).
Fig. 5.
Evaluation of alterations to the 9-nt element in the context of the full-length Gtx 5′ leader. A schematic representation of the Photinus luciferase constructs is indicated. Constructs include the full-length wild-type Gtx 5′ leader, replacement of the Gtx module in mutation m16, in which the complementary match is shortened, and replacement by mutation m17, in which the complementary match is lengthened. The nucleotides complementary to 18S rRNA are indicated in gray. Constructs were assayed in transiently transfected N2a cells. Translation efficiencies were determined per unit mRNA and indicated as a % change (+/-) relative to the translation efficiency obtained with the construct containing the wild-type 5′ leader sequence. Horizontal lines indicate SEM.
Discussion
In the present study, we found that a synthetic construct containing five linked copies of the 9-nt Gtx IRES module enhanced translation when tested in the 5′ leader of a monocistronic mRNA. These nine nucleotides are contained within a larger segment of the 5′ leader that was previously shown to be inhibitory to translation (15). In an earlier study, we showed that a 40-nt segment of the Gtx 5′ leader containing the 9-nt sequence could be cross-linked to 40S ribosomal subunits via 18S rRNA (15). However, we did not localize binding sites within the mRNA sequence. We now show that the 9-nt sequence itself can bind specifically to 40S ribosomal subunits and demonstrate that the length of the complementary match is directly correlated with the ability to bind 40S ribosomal subunits. We found that the ability of this sequence to enhance translation increases as the length of the complementary match is decreased to 7 nt, and then declines with shorter or longer complementary matches. Mutations of the 9-nt sequence in the context of the full-length Gtx 5′ leader also suggested that this element affects the translation of the Gtx mRNA by base pairing to 18S rRNA. Mutations that shortened the complementary match increased translation efficiency (Fig. 5, m16 and m17). This enhancement was lost when the match was decreased to 5 nt (m18).
The translation data indicate that the strength of the binding interaction obtained with seven complementary nucleotides is optimal for enhancing translation efficiency, suggesting that shorter complementary matches may fail to efficiently recruit ribosomes, whereas some longer complementary matches may bind 40S subunits too strongly (m11 and m12). However, increased matches at the 5′ end did not block translation (m14 and m15). These differences may be due to variables, such as RNA secondary structures, that differentially affect the presentation of this sequence in the synthetic IRES.
Direct binding to ribosomes has now been shown to occur for two eukaryotic messages, Gtx and Rbm3 (8, 15). Some viral IRESs, including those contained within RNAs from the hepatitis C virus, coxsackievirus, classic swine fever virus, bovine viral diarrhea virus, and cricket paralysis virus also appear to facilitate translation initiation by mechanisms that involve binding to 40S subunits (16–19). It has been suggested that various picornaviral IRESs base pair to 18S rRNA, e.g., the coxsackievirus B3 IRES is thought to bind to 40S subunits via a complementary segment at the 3′ end of the 18S rRNA (19), and the hepatitis C virus IRES appears to enhance translation by binding to ribosomal proteins (20).
In addition to the Gtx synthetic IRES reported here, several cellular and viral IRESs have been tested in a monocistronic context, and their rates of translation have been compared to those obtained with control constructs lacking the IRES sequences; some IRESs enhanced the rate of translation, whereas others inhibited it (see, e.g., refs. 21 and 22). The ability of an IRES to enhance or inhibit the rate of translation relative to that of a capped monocistronic mRNA may be determined by whether translation is derived from the combined contributions of the IRES and other elements such as the cap, or whether it is derived solely from the IRES. In the latter case, the efficiency of the IRES alone might determine the rate of translation.
The ability of the Gtx 9-nt sequence to affect translation efficiency may depend on the accessibility of the complementary sequence within 18S rRNA. This notion is supported by the location of this site within helix 26, which appears to be accessible based on current structural models of prokaryotic ribosomes (23, 24). In an earlier study (1), we noted that the activity of a synthetic IRES containing 10 linked copies of the 9-nt element varied dramatically depending on the cell type. This raises the possibility that the accessibility of the complementary nucleotides within the 40S subunit differs in ribosomes from different cell types, either because of ribosome heterogeneity (see ref. 25) or because of ribosome binding factors that may include proteins or other RNAs that mask the binding site.
Although the present results are consistent with the conclusion that the 9-nt Gtx sequence can enhance translation by a mechanism that involves base pairing interaction to 18S rRNA, a critical test of this mechanism will require studies showing that the activity of the Gtx element actually depends on the integrity of the complementary sequences within the 18S rRNA. This will involve assessment of the activity of the Gtx sequence in cells in which the complementary site within the 18S rRNA has been mutated and comparison with the activity obtained in cells containing the unmutated RNA.
Multiple copies of the 9-nt Gtx element may enhance translation initiation by increasing the local concentration of ribosomes in the vicinity of the mRNA. This notion is also supported by the results of an analysis of complementarity in which the 5′ leader sequences from Fig. 4 were tested in the intercistronic region of a dicistronic vector (data not shown). The results obtained in dicistronic constructs mirrored those obtained in the monocistronic vector with one difference: translation was enhanced in monocistronic constructs with as few as five complementary matches; in the dicistronic mRNA, however, elements shorter than 6 nt did not function as IRESs.
Our analysis of the synthetic construct containing the 9-nt Gtx element suggests that short sequence elements contained within natural mRNAs can affect translation efficiency. However, the 9-nt element was only one of several cis-acting sequences shown to affect the translation of the Gtx mRNA (1). Our examination of the 5′ leader of the Rbm3 mRNA revealed a similar modular composition (8), as have deletion analyses of many other mRNA 5′ leaders (1). These observations are consistent with the notion that the translation efficiencies of many mRNAs are determined by the cumulative effects of numerous sequence elements in the 5′ leaders that individually have only small effects on translation.
We have found that, in addition to Gtx, a large number of other eukaryotic mRNAs contain rRNA-like sequences that either resemble rRNA or are complementary to it (26). These rRNA-like sequences have been identified both in coding and noncoding segments of mRNAs. We previously suggested that some of these sequences might affect the translation efficiency of their host mRNAs by mediating interactions with RNA or protein ribosomal components. This notion was further elaborated in the ribosome filter hypothesis (25). This hypothesis suggests that these interactions may be regulated by factors affecting the accessibility of the binding sites on the ribosomes, including ribosome heterogeneity and ribosome binding factors.
Supplementary Material
Acknowledgments
We thank Luke Burman and Rachel Eachus for excellent technical assistance; Drs. Anthony Stonehouse and Megumi Adachi for valuable advice regarding the quantitative PCR analyses; Dr. George Rogers for providing 40S ribosomal subunits from N2a cells; Dr. John Dresios for helpful discussions; and Drs. Bruce Cunningham and Ralph Greenspan for critical reading of the manuscript. This work was supported by the G. Harold and Leila Y. Mathers Foundation and National Institutes of Health Grants GM61725 (to V.P.M.) and HD09635 (to G.M.E.). Support for S.A.C. was provided by a postdoctoral fellowship from The Skaggs Institute for Chemical Biology.
Abbreviations: IRES, internal ribosome entry site; N2a, Neuro 2a.
References
- 1.Chappell, S. A., Edelman, G. M. & Mauro, V. P. (2000) Proc. Natl. Acad. Sci. USA 97, 1536-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chappell, S. A., Owens, G. C. & Mauro, V. P. (2001) J. Biol. Chem. 276, 36917-36922. [DOI] [PubMed] [Google Scholar]
- 3.Owens, G. C., Chappell, S. A., Mauro, V. P. & Edelman, G. M. (2001) Proc. Natl. Acad. Sci. USA 98, 1471-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou, W., Edelman, G. M. & Mauro, V. P. (2003) Proc. Natl. Acad. Sci. USA 100, 4457-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reiser, J., Lai, Z., Zhang, X. & Brady, R. (2000) J. Virol. 74, 10589-10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li, T. & Zhang, J. (2004) J. Virol. Methods 115, 137-144. [DOI] [PubMed] [Google Scholar]
- 7.Zhang, X., La Russa, V., Bao, L., Kolls, J., Schwarzenberger, P. & Reiser, J. (2002) Mol. Ther. 5, 555-565. [DOI] [PubMed] [Google Scholar]
- 8.Chappell, S. A. & Mauro, V. P. (2003) J. Biol. Chem. 278, 33793-33800. [DOI] [PubMed] [Google Scholar]
- 9.Park, Y. W. & Katze, M. G. (1995) J. Biol. Chem. 270, 28433-28439. [DOI] [PubMed] [Google Scholar]
- 10.Lasater, L. S., Olson, H. M., Cann, P. A. & Glitz, D. G. (1988) Biochemistry 27, 4687-4695. [DOI] [PubMed] [Google Scholar]
- 11.Draper, D. E., Deckman, I. C. & Vartikar, J. V. (1988) Methods Enzymol. 164, 203-219. [DOI] [PubMed] [Google Scholar]
- 12.Blobel, G. & Sabatini, D. (1971) Proc. Natl. Acad. Sci. USA 68, 390-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tranque, P., Hu, M. C.-Y., Edelman, G. M. & Mauro, V. P. (1998) Proc. Natl. Acad. Sci. USA 95, 12238-12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stonehouse, A. H., Adachi, M., Walcott, E. C. & Jones, F. S. (2003) Mol. Pharmacol. 64, 1463-1473. [DOI] [PubMed] [Google Scholar]
- 15.Hu, M. C.-Y., Tranque, P., Edelman, G. M. & Mauro, V. P. (1999) Proc. Natl. Acad. Sci. USA 96, 1339-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pestova, T. V., Shatsky, I. N., Fletcher, S. P., Jackson, R. J. & Hellen, C. U. T. (1998) Genes Dev. 12, 67-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pestova, T. V. & Hellen, C. U. T. (1999) Virology 258, 249-256. [DOI] [PubMed] [Google Scholar]
- 18.Kolupaeva, V. G., Pestova, T. V. & Hellen, C. U. (2000) RNA 6, 1791-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang, D., Cheung, P., Sun, Y., Yuan, J., Zhang, H., Carthy, C. M., Anderson, D. R., Bohunek, L., Wilson, J. E. & McManus, B. M. (2003) Virology 305, 31-43. [DOI] [PubMed] [Google Scholar]
- 20.Otto, G. A., Lukavsky, P. J., Lancaster, A. M., Sarnow, P. & Puglisi, J. D. (2002) RNA 8, 913-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oumard, A., Hennecke, M., Hauser, H. & Nourbakhsh, M. (2000) Mol. Cell. Biol. 20, 2755-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Attal, J., Theron, M. C., Rival, S., Puissant, C. & Houdebine, L. M. (2000) Mol. Biol. Rep. 27, 21-26. [DOI] [PubMed] [Google Scholar]
- 23.Wimberly, B., Brodersen, D., Clemons, W. J., Morgan-Warren, R., Carter, A., Vonrhein, C., Hartsch, T. & Ramakrishnan, V. (2000) Nature 407, 327-339. [DOI] [PubMed] [Google Scholar]
- 24.Brodersen, D., Clemons, W., Carter, A., Wimberly, B. & Ramakrishnan, V. (2002) J. Mol. Biol. 316, 725-768. [DOI] [PubMed] [Google Scholar]
- 25.Mauro, V. P. & Edelman, G. M. (2002) Proc. Natl. Acad. Sci. USA 99, 12031-12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mauro, V. P. & Edelman, G. M. (1997) Proc. Natl. Acad. Sci. USA 94, 422-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.