Plants are host to diverse microbiome that might have co-evolved since millions of years. This resident microbiota can act as extended genome by contributing in plant growth, development and protection from biotic and abiotic stresses. Rice (Oryza Sativa) is a staple food consumed by more than 50% of the world's population. Herein we targeted the bacterial community associated with the healthy rice seeds. In this direction, we isolated and carried out whole genome sequencing of 100 bacterial isolates. These isolates belong to three major bacterial phyla Proteobacteria, Firmicutes, and Actinobacteria that spread over 15 distinct genus and 29 species. A phylogenetic tree based on a broad set of phylogenomic marker genes confirmed the evolutionary relationship amongst the strains and their phylogenetic grouping. Average Nucleotide Identity was also used to establish species identity of isolates that form a particular phylogenetic and taxonomic grouping. The data generated from the present study is one of the first major genomic resources in the field of phytobiome research. Whole genome sequence of the members will be invaluable in this era of big data driven research. Moreover, the majority of genus and species identified in this study are already known for plant probiotic properties. This genomic data with annotation will aid in comparative, evolutionary and ecological studies of bacteria associated with plants or multi-kingdom bacteria associated with nosocomial infections.
Methods
Isolation of bacteria from seed
Rice seeds were collected from farmer's field in Fazilka, Punjab, India practicing conventional farming and growing basmati variety. Bacterial isolations were done from a pool of seeds isolated from the same field grown and harvested in three successive years. First isolation was from the seeds harvested in the year 2011, next three isolations were from the crop harvested in 2012 and last from the year 2013. For bacterial extractions, 5 g of seeds were partially crushed (~80%) in normal saline (0.85% NaCl) using sterile pestle and mortar and suspended in 50 ml of the solution (10%; Cottyn et al., 2001). These solutions were incubated for 2 h at 4°C/28°C and then dilution plating was done up to 10−6. Samples from each dilution were plated in triplicates on six different media, Peptone Sucrose Agar (PSA), Glucose Yeast extract Calcium carbonate Agar (GYCA), Luria broth (LB) agar, King's B (KB) agar, Nutrient broth (NB) agar, and Potato Dextrose Agar (PDA). Plates were incubated at 28°C and growth was checked up to 6 days. Control plates with/without saline solution were also incubated to check for contamination up to 1 month. Bacterial colonies based on diverse morphology were selected and further processed, as the aim was to capture maximum diversity associated with rice seeds. Bacterial cultures were frozen in 15% glycerol at −80°C.
Identification by 16S rDNA sequencing
Bacterial isolates were streaked on nutrient broth (NB) agar to get single colonies and 3–4 colonies of each bacterium were suspended into 50 μl of water. Freeze-thaw shock was given to bacteria by freezing the vial at −80°C for 10 min and then incubation at 95°C for 5 min. After that samples were centrifuged at 10,000 rpm for 1 min to collect the supernatant and this step was repeated once again before proceeding further. Samples were quantified for DNA using NanoDrop (Thermo Scientific) and PCR was performed using universal 16S rRNA amplification primers 27F (AGAGTTTGATCMTGGCTCAG) and 1492R (GGTTACCTTGTTACGACTT). After checking for amplification on 1% agarose gel, samples were treated with Exo-Sap (USB, Affymetrix Inc. Cleveland, Ohio, USA) to remove single stranded DNA primers and unused dNTPs and samples were subjected to sanger sequencing using in-house facility ABI DNA sequencer. Data generated in ABI files were visualized in Finch TV v1.4.0 to select the sequences of high quality that were analyzed using Ez-BioCloud (Kim et al., 2012) to identify the closest bacterial species.
From five different isolations, 469 colonies were obtained as a pure culture. Further based on morphological characteristics on the agar plates, the sample size was reduced to 147 for identification of species by 16S rRNA sequencing. Out of these 147 cultures, further shortlisting was done to 100 isolates for genome sequencing, which consisted of minimum one representative of each species from each lot to represent the seed associated bacterial diversity.
Genome sequencing, assembly, and analysis
Bacterial cultures were revived from −80°C stocks and ZR Fungal/Bacterial DNA isolation kit (Zymo Research) was used to isolate DNA from these. DNA quality check was done using NanoDrop (Thermo Scientific) and agarose gel electrophoresis and quantitation of DNA was performed using Qubit 2.0 Fluorometer (Life Technologies). Sequencing library preparation was performed using Nextera XT sample preparation kit (Illumina, Inc., San Diego, CA, USA) and loaded on to in-house Illumina MiSeq platform (Illumina, Inc., San Diego, CA, USA) using company supplied paired-end sequencing kits. Adapter trimming was done automatically by MiSeq Control Software (MCS) and additional adapter contamination identified by NCBI server was removed by manual trimming. De novo assembly of the sequences were done using CLC genomic workbench v7.5 (CLC bio, Aarhus, Denmark) with default settings. Sequences were annotated using NCBI Prokaryotic Genome Annotation Pipeline (http://www.ncbi.nlm.nih.gov/genome/annotation_prok/). RNAmmer 1.2 server was used to annotate the RNA sequences and Ez-BioCloud to identify the closest bacterial species. Protein sequences of 10 known phylogenomic marker genes (infC, rplB, rplC, rplD, rplE, rplF, rplM, rplN, and rplP) were extracted from the genomic sequences, aligned, and concatenated to obtain multi locus strain phylogeny. These are single copy and universally distributed genes with core housekeeping functions (Wu and Eisen, 2008) and importantly found to be relatively immune to horizontal gene transfer (Jain et al., 1999). Sequences were aligned using Mega v6.0 (Tamura et al., 2013) and a phylogenetic tree was constructed using the Neighbor-Joining method with 500 bootstrap replicates. JSpecies 1.2.1 software was used to calculate Average Nucleotide Identity (ANI) amongst different strains (Richter and Rosselló-Móra, 2009).
Nucleotide sequences accession numbers
The data has been submitted to NCBI GenBank under accession no. LDPZ00000000-LDTU00000000 and assembly statistics for the 100 bacterial genomes sequenced is provided in Table 1.
Table 1.
Assembly statistics and annotation features of bacterial isolates from rice and their accession numbers.
| S.No | Strain | Genome size (bp) | Coverage(x) | Contigs | N50(bp) | Coding Density (%) | Genes | tRNA | Accession No. |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Aureimonas ureilytica NS226 | 5119943 | 105 | 287 | 101071 | 80.2 | 4846 | 58 | LDPZ00000000 |
| 2 | Aureimonas ureilytica NS365 | 5013318 | 93 | 183 | 103973 | 81.0 | 4690 | 56 | LDQA00000000 |
| 3 | Curtobacterium citreum NS330 | 3454135 | 73 | 241 | 29468 | 81.4 | 3350 | 47 | LDQB00000000 |
| 4 | Curtobacterium luteum NS184 | 3586193 | 100 | 209 | 44951 | 80.2 | 3460 | 45 | LDQC00000000 |
| 5 | Curtobacterium oceanosedimentum NS263 | 3370512 | 97 | 164 | 44334 | 79.7 | 3206 | 46 | LDRB00000000 |
| 6 | Curtobacterium oceanosedimentum NS359 | 3421352 | 94 | 188 | 39494 | 79.4 | 3258 | 45 | LDRC00000000 |
| 7 | Enterobacter asburiae NS23 | 4716160 | 71 | 102 | 154724 | 89.0 | 4472 | 71 | LDQD00000000 |
| 8 | Enterobacter asburiae NS34 | 4722850 | 64 | 72 | 165694 | 88.9 | 4462 | 70 | LDQE00000000 |
| 9 | Enterobacter asburiae NS7 | 4728317 | 166 | 70 | 235927 | 89.0 | 4464 | 70 | LDQF00000000 |
| 10 | Enterobacter cancerogenus NS104 | 4884126 | 117 | 52 | 261552 | 87.7 | 4630 | 76 | LDQG00000000 |
| 11 | Enterobacter cancerogenus NS111 | 4874699 | 99 | 49 | 268762 | 87.7 | 4622 | 79 | LDQH00000000 |
| 12 | Enterobacter cancerogenus NS188 | 4889753 | 117 | 62 | 254143 | 87.8 | 4644 | 75 | LDQI00000000 |
| 13 | Enterobacter cancerogenus NS31 | 4869574 | 96 | 45 | 261547 | 87.8 | 4620 | 76 | LDQJ00000000 |
| 14 | Enterobacter xiangfangensis NS19 | 4731803 | 91 | 75 | 347372 | 88.9 | 4436 | 69 | LDQK00000000 |
| 15 | Enterobacter xiangfangensis NS24 | 4705572 | 98 | 187 | 109643 | 88.7 | 4444 | 67 | LDQL00000000 |
| 16 | Enterobacter xiangfangensis NS28 | 4736807 | 146 | 38 | 431385 | 88.9 | 4418 | 73 | LDQM00000000 |
| 17 | Enterobacter xiangfangensis NS29 | 4712167 | 82 | 129 | 160476 | 88.8 | 4449 | 73 | LDQN00000000 |
| 18 | Enterobacter xiangfangensis NS371 | 4725300 | 125 | 37 | 345357 | 88.8 | 4402 | 71 | LDQO00000000 |
| 19 | Enterobacter xiangfangensis NS49 | 4719851 | 128 | 33 | 371391 | 88.9 | 4401 | 70 | LDQP00000000 |
| 20 | Enterobacter xiangfangensis NS57 | 4730229 | 129 | 44 | 265382 | 88.9 | 4417 | 74 | LDQQ00000000 |
| 21 | Enterobacter xiangfangensis NS64 | 4723970 | 102 | 76 | 277091 | 88.8 | 4434 | 71 | LDQR00000000 |
| 22 | Enterobacter xiangfangensis NS75 | 4120285 | 92 | 60 | 131492 | 88.9 | 4408 | 72 | LDQS00000000 |
| 23 | Enterobacter xiangfangensis NS80 | 4722881 | 85 | 32 | 282477 | 88.9 | 4405 | 73 | LDQT00000000 |
| 24 | Enterobacter xiangfangensis RSA8 | 4707136 | 72 | 172 | 134845 | 88.7 | 4467 | 71 | LDQU00000000 |
| 25 | Exiguobacterium indicum RSA11 | 3083121 | 89 | 53 | 109567 | 88.0 | 3213 | 57 | LDQV00000000 |
| 26 | Exiguobacterium indicum RSA42 | 3095093 | 79 | 126 | 69291 | 87.6 | 3233 | 65 | LDQW00000000 |
| 27 | Kocuria kristinae RSA28 | 2302499 | 205 | 159 | 25292 | 76.4 | 2072 | 47 | LDRD00000000 |
| 28 | Kocuria kristinae RSA5 | 2282497 | 236 | 204 | 21282 | 75.9 | 2074 | 47 | LDRE00000000 |
| 29 | Kocuria kristinae SA11 | 2243566 | 194 | 202 | 19601 | 76.3 | 2043 | 44 | LDRF00000000 |
| 30 | Kocuria kristinae SA12 | 2217668 | 129 | 249 | 17427 | 76.0 | 2049 | 46 | LDRG00000000 |
| 31 | Kocuria kristinae SA13 | 2252040 | 158 | 233 | 15889 | 76.0 | 2077 | 44 | LDRH00000000 |
| 32 | Kocuria kristinae SA14 | 2245805 | 270 | 175 | 23072 | 76.0 | 1996 | 43 | LDRI00000000 |
| 33 | Kocuria kristinae SA15 | 2265963 | 210 | 211 | 19130 | 76.1 | 2061 | 47 | LDRJ00000000 |
| 34 | Leucobacter chromiiresistens NS354 | 2841040 | 101 | 194 | 37446 | 84.6 | 2618 | 44 | LDRK00000000 |
| 35 | Methylobacterium aquaticum NS228 | 6323349 | 110 | 487 | 29045 | 80.0 | 5861 | 67 | LDRL00000000 |
| 36 | Methylobacterium aquaticum NS229 | 6377294 | 110 | 443 | 30018 | 80.1 | 5907 | 69 | LDRM00000000 |
| 37 | Methylobacterium aquaticum NS230 | 6353803 | 127 | 384 | 32708 | 79.9 | 5835 | 65 | LDRN00000000 |
| 38 | Methylobacterium radiotolerans SB2 | 6647570 | 116 | 334 | 55436 | 81.8 | 6254 | 47 | LDRO00000000 |
| 39 | Methylobacterium radiotolerans SB3 | 6693841 | 110 | 351 | 55393 | 81.8 | 6361 | 53 | LDRP00000000 |
| 40 | Microbacterium oxydans NS234 | 4013326 | 114 | 136 | 89218 | 87.8 | 3976 | 41 | LDRQ00000000 |
| 41 | Microbacterium testaceum NS183 | 3942620 | 94 | 305 | 37174 | 84.7 | 3760 | 45 | LDRR00000000 |
| 42 | Microbacterium testaceum NS206 | 3881914 | 113 | 300 | 44670 | 84.7 | 3687 | 44 | LDRS00000000 |
| 43 | Microbacterium testaceum NS220 | 4023911 | 118 | 291 | 32198 | 82.9 | 3829 | 43 | LDRT00000000 |
| 44 | Microbacterium testaceum NS283 | 3903457 | 140 | 177 | 50419 | 86.3 | 3655 | 45 | LDRU00000000 |
| 45 | Microbacterium testaceum RSA3 | 3914164 | 102 | 222 | 42959 | 84.9 | 3712 | 45 | LDRV00000000 |
| 46 | Paenibacillus jamilae NS115 | 5586901 | 49 | 311 | 58412 | 84.5 | 4997 | 58 | LDRX00000000 |
| 47 | Novosphingobium barchaimii NS277 | 3553442 | 135 | 75 | 193132 | 84.3 | 3332 | 45 | LDRW00000000 |
| 48 | Pantoea ananatis NS296 | 4734861 | 69 | 54 | 193582 | 85.2 | 4439 | 65 | LDQX00000000 |
| 49 | Pantoea ananatis NS303 | 4733153 | 80 | 69 | 182398 | 85.3 | 4421 | 59 | LDQY00000000 |
| 50 | Pantoea ananatis NS311 | 4721638 | 73 | 174 | 118598 | 85.1 | 4449 | 64 | LDQZ00000000 |
| 51 | Pantoea ananatis RSA47 | 4738039 | 91 | 46 | 229877 | 85.2 | 4439 | 65 | LDRA00000000 |
| 52 | Pantoea dispersa NS215 | 4843865 | 84 | 35 | 271354 | 86.9 | 4425 | 67 | LDRY00000000 |
| 53 | Pantoea dispersa NS375 | 4887220 | 81 | 42 | 318516 | 86.5 | 4510 | 73 | LDRZ00000000 |
| 54 | Pantoea dispersa NS380 | 4801064 | 74 | 123 | 195710 | 86.3 | 4454 | 70 | LDSA00000000 |
| 55 | Pantoea dispersa NS389 | 4819785 | 73 | 54 | 228728 | 86.5 | 4420 | 61 | LDSB00000000 |
| 56 | Pantoea dispersa RSA31 | 4949525 | 90 | 194 | 266158 | 86.7 | 4607 | 64 | LDSC00000000 |
| 57 | Pantoea dispersa SA2 | 4906145 | 52 | 52 | 255271 | 85.8 | 4488 | 66 | LDSD00000000 |
| 58 | Pantoea dispersa SA3 | 4905368 | 72 | 83 | 185303 | 85.9 | 4494 | 65 | LDSE00000000 |
| 59 | Pantoea dispersa SA4 | 4911078 | 55 | 58 | 156533 | 85.8 | 4503 | 68 | LDSF00000000 |
| 60 | Pantoea dispersa SA5 | 4897186 | 77 | 98 | 190714 | 85.9 | 4500 | 64 | LDSG00000000 |
| 61 | Pantoea stewartii NS381 | 4693026 | 82 | 51 | 268280 | 84.7 | 4338 | 68 | LDSH00000000 |
| 62 | Pantoea stewartii RSA13 | 4766695 | 96 | 48 | 167594 | 84.4 | 4423 | 67 | LDSI00000000 |
| 63 | Pantoea stewartii RSA30 | 4760407 | 65 | 84 | 211952 | 84.3 | 4420 | 65 | LDSJ00000000 |
| 64 | Pantoea stewartii RSA36 | 4791295 | 139 | 76 | 285765 | 84.2 | 4451 | 64 | LDSK00000000 |
| 65 | Pseudacidovorax intermedius NS331 | 5547045 | 97 | 295 | 34362 | 84.5 | 5076 | 49 | LDSL00000000 |
| 66 | Pseudomonas parafulva NS212 | 4823858 | 147 | 147 | 107693 | 86.7 | 4359 | 65 | LDSM00000000 |
| 67 | Pseudomonas parafulva NS96 | 4672265 | 177 | 162 | 108451 | 87.2 | 4283 | 64 | LDSN00000000 |
| 68 | Pseudomonas psychrotolerans NS2 | 5271410 | 146 | 130 | 143537 | 86.4 | 4687 | 59 | LDSO00000000 |
| 69 | Pseudomonas psychrotolerans NS201 | 5262847 | 139 | 172 | 119605 | 86.7 | 4783 | 55 | LDSP00000000 |
| 70 | Pseudomonas psychrotolerans NS274 | 5418049 | 135 | 103 | 95239 | 86.2 | 4848 | 60 | LDSQ00000000 |
| 71 | Pseudomonas psychrotolerans NS337 | 4837497 | 62 | 192 | 118956 | 88.0 | 4559 | 59 | LDSR00000000 |
| 72 | Pseudomonas psychrotolerans NS376 | 5326810 | 95 | 136 | 99843 | 86.0 | 4790 | 60 | LDSS00000000 |
| 73 | Pseudomonas psychrotolerans NS383 | 5413173 | 113 | 174 | 94742 | 86.0 | 4877 | 55 | LDST00000000 |
| 74 | Pseudomonas psychrotolerans RSA46 | 5455836 | 120 | 197 | 97624 | 86.0 | 4934 | 59 | LDSU00000000 |
| 75 | Pseudomonas psychrotolerans SB11 | 5202479 | 113 | 232 | 134485 | 86.7 | 4817 | 59 | LDSV00000000 |
| 76 | Pseudomonas psychrotolerans SB14 | 5345440 | 124 | 169 | 129021 | 86.0 | 4823 | 61 | LDSW00000000 |
| 77 | Pseudomonas psychrotolerans SB18 | 5339762 | 147 | 173 | 104148 | 86.4 | 4889 | 59 | LDSX00000000 |
| 78 | Pseudomonas psychrotolerans SB5 | 5435284 | 113 | 171 | 92521 | 86.1 | 4884 | 60 | LDSY00000000 |
| 79 | Pseudomonas psychrotolerans SB8 | 5130436 | 138 | 116 | 137111 | 86.5 | 4598 | 60 | LDSZ00000000 |
| 80 | Pseudomonas psychrotolerans SB9 | 5283903 | 104 | 121 | 91801 | 86.7 | 4803 | 57 | LDTA00000000 |
| 81 | Sphingomonas endophytica NS334 | 3642958 | 164 | 330 | 58696 | 85.2 | 3506 | 47 | LDTB00000000 |
| 82 | Sphingomonas sanguinis NS258 | 3868819 | 129 | 346 | 36442 | 85.0 | 3708 | 45 | LDTC00000000 |
| 83 | Sphingomonas sanguinis NS319 | 4169594 | 110 | 280 | 33184 | 84.9 | 3912 | 48 | LDTD00000000 |
| 84 | Sphingomonas sanguinis SB4 | 4019921 | 97 | 226 | 40305 | 84.5 | 3772 | 49 | LDTE00000000 |
| 85 | Sphingomonas yabuuchiae NS355 | 3910679 | 133 | 190 | 45578 | 85.0 | 3612 | 49 | LDTF00000000 |
| 86 | Staphylococcus epidermidis SA6 | 2540147 | 111 | 66 | 227679 | 82.1 | 2443 | 62 | LDTG00000000 |
| 87 | Staphylococcus epidermidis SA8 | 2505031 | 84 | 181 | 30972 | 82.4 | 2419 | 49 | LDTH00000000 |
| 88 | Staphylococcus epidermidis SB7b | 2506997 | 86 | 148 | 44745 | 82.5 | 2423 | 55 | LDTI00000000 |
| 89 | Staphylococcus epidermidis SB7c | 2559409 | 118 | 96 | 109441 | 82.3 | 2485 | 58 | LDTJ00000000 |
| 90 | Staphylococcus sciuri NS1 | 2918429 | 80 | 127 | 70746 | 85.4 | 2972 | 54 | LDTK00000000 |
| 91 | Staphylococcus sciuri NS112 | 2878717 | 73 | 136 | 49833 | 85.3 | 2926 | 52 | LDTL00000000 |
| 92 | Staphylococcus sciuri NS202 | 2743697 | 84 | 56 | 93562 | 86.8 | 2757 | 56 | LDTM00000000 |
| 93 | Staphylococcus sciuri NS36 | 2881364 | 90 | 117 | 52335 | 85.3 | 2907 | 54 | LDTN00000000 |
| 94 | Staphylococcus sciuri NS44 | 2979824 | 160 | 164 | 142946 | 84.5 | 3038 | 55 | LDTO00000000 |
| 95 | Staphylococcus sciuri NS53 | 2897114 | 85 | 155 | 53334 | 85.3 | 2951 | 51 | LDTP00000000 |
| 96 | Staphylococcus sciuri RSA37 | 2890453 | 130 | 72 | 81742 | 85.5 | 2911 | 54 | LDTQ00000000 |
| 97 | Staphylococcus warneri NS346 | 2563869 | 96 | 87 | 84728 | 83.6 | 2485 | 50 | LDTR00000000 |
| 98 | Staphylococcus warneri SA10 | 2549551 | 84 | 145 | 36722 | 82.4 | 2497 | 53 | LDTS00000000 |
| 99 | Staphylococcus warneri SA9 | 2541660 | 96 | 77 | 114371 | 82.0 | 2482 | 56 | LDTT00000000 |
| 100 | Staphylococcus xylosus NS341 | 2842280 | 89 | 111 | 59876 | 82.2 | 2702 | 54 | LDTU00000000 |
Interpretation of data set
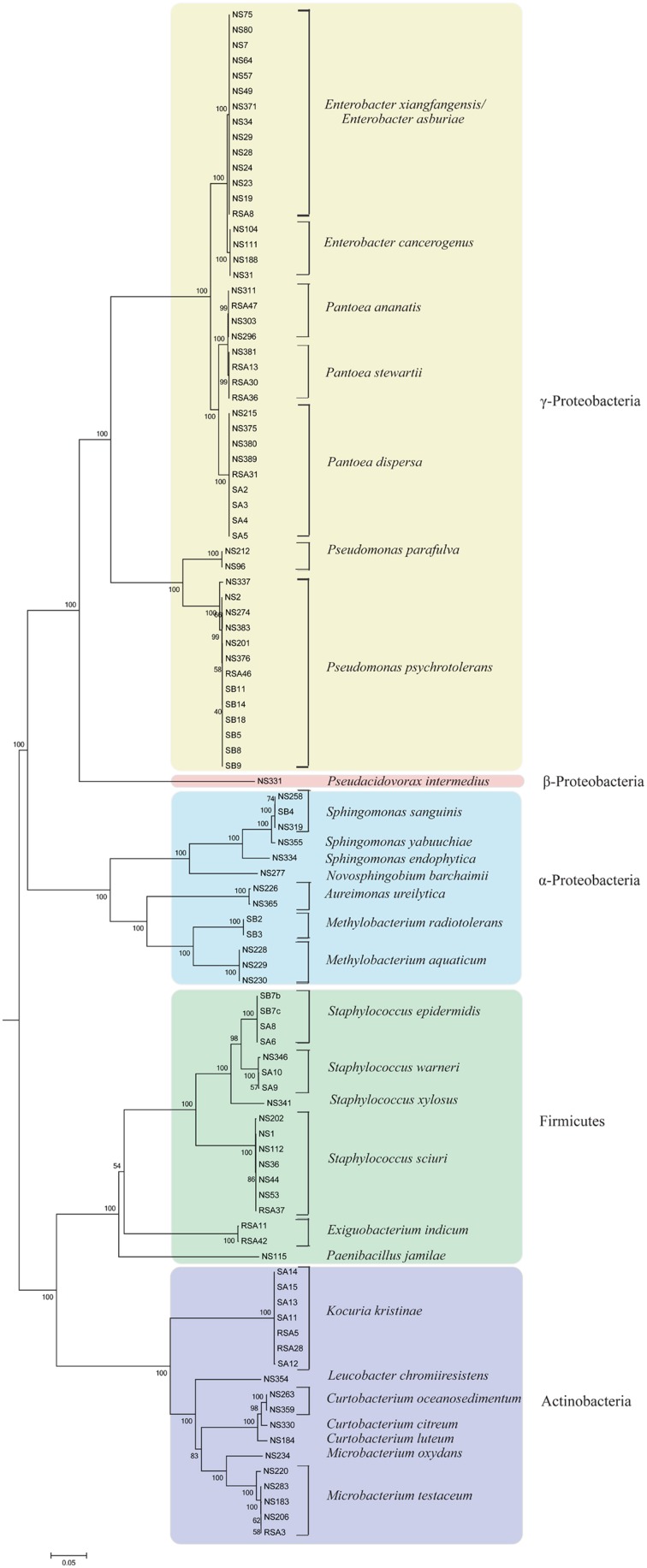
High-quality sequencing data generated for each strain (Table 1) was de novo assembled with coverage ranging from 49x to 270x. Analysis based on complete 16S rRNA sequence extracted from the whole genome sequences assigned them to 15 distinct genus and 29 species. It is also pertinent to note that genus/species assignment has been validated by a new QA protocol of NCBI during submission process. Here, the “input fasta sequences are BLASTed against a collection of 23 bacterial ribosomal protein COG families during submission.” Multilocus phylogenetic tree based on marker genes further supports the distinction between different groups of bacteria (Figure 1) to strain level. Description of the strains assigned to 15 different genera is provided below:
Figure 1.
Multi locus sequence analysis of rice seed associated bacterial isolates constructed using 10 phylogenomic marker genes with Neighbor-Joining method and 500 bootstrap replications. Bacterial strains belonging to different phylum; Proteobacteria (α, β, and γ), Firmicutes, and Actinobacteria, are highlighted with different background colors. Strains belonging to each species are grouped together with high boot strap values.
Genus: Kocuria
Kocuria is a gram-positive bacterium, belonging to phylum Actinobacteria. Seven isolates (SA11, SA12, SA13, SA14, SA15, RSA5, and RSA28) belonging to this genus were sequenced from two different libraries and two different year lots. Complete 16S rRNA typing has assigned the seven isolates to same species Kocuria kristinae. Further ANI analysis showed the seven strains have genome level identity >99.8%, much above the cut-off of 94–96% for delineation of species (Konstantinidis and Tiedje, 2005; Richter and Rosselló-Móra, 2009) and suggests their monophyletic/clonal nature. Interestingly only in case of Kocuria, a single species was detected even after having multiple strains.
Genus: Curtobacterium
Curtobacterium is also a gram-positive bacterium belonging to phylum Actinobacteria and class Microbacteriaceae. Four isolates of Curtobacterium were obtained from seed microbiome that belong to three different species on the basis of 16S rRNA sequences i.e. Curtobacterium luteum (NS184), Curtobacterium citreum (NS330), and Curtobacterium oceanosedimentum (NS263, NS359), sequenced from two different libraries. ANI values amongst the genomes of four strains also support the presence of three species.
Genus: Leucobacter
Leucobacter is another gram-positive Actinobacteria, belonging to class Microbacteriaceae. One isolate NS354 belonging to this genus was isolated from rice seeds and was assigned to Leucobacter chromiiresistens on the basis of 16S rRNA sequences.
Genus: Microbacterium
Microbacterium is another gram-positive Actinobacteria and six isolates from rice seeds were assigned to genus Microbacterium. These isolates were extracted from three different libraries and two different rice lots. 16S rRNA sequences assigned them into two different species Microbacterium testaceum (NS183, NS206, NS220, NS283, and RSA3) and Microbacterium oxydans (NS234), while ANI values suggest NS220 to be a different species as the values are less than 87.5% with all the other strains.
Genus: Exiguobacterium
Exiguobacterium is a gram-positive bacterium that is assigned to phylum Firmicutes. Two isolates (RSA11 and RSA42) belonging to this genus were isolated from one library and assigned to same species Exiguobacterium indicum on the basis of 16S rRNA gene sequences. While ANI value amongst these two strains is 94.33%, very close to the cut-off for species delineation, suggesting that these two strains may belong to two different species.
Genus: Staphylococcus
Staphylococcus is also a gram-positive Firmicutes and 15 different strains were selected for sequencing from this genus belonging to four different libraries and three different year rice production lots. 16S rRNA sequences have assigned them to four different species Staphylococcus epidermidis (SA6, SA8, SB7b, SB7c), Staphylococcus warneri (SA9, SA10, NS346), Staphylococcus xylosus (NS341), and Staphylococcus sciuri (NS1, NS36, NS44, NS53, NS112, NS202, RSA37). Estimated ANI values also support the species distinction between the four groups as they are above the cut-off for species delineation.
Genus: Paenibacillus
Paenibacillus is the bacterium belonging to gram-variable Firmicutes. One strain NS115 belonging to Paenibacillus jamilae was isolated from rice seed environment.
Genus: Aureimonas
Aureimonas is a gram-negative bacterium belonging to α-Proteobacteria. Two strains belonging to species Aureimonas ureilytica (NS226, NS365) were isolated from the rice seeds in two different preparations and they have ANI value of 96.91%.
Genus: Methylobacterium
Another gram-negative bacterium belonging to α-Proteobacteria, Methylobacterium was also isolated from rice seed environment. Five isolates from two different year rice lots belonging to two different species were extracted, Methylobacterium radiotolerans (SB2, SB3), and Methylobacterium aquaticum (NS228, NS229, NS230). ANI values also confirmed the delineation of two species.
Genus: Novosphingobium
One bacterial isolate belonging to Novosphingobium barchaimii NS277 was isolated that is also a gram negative α-Proteobacteria.
Genus: Sphingomonas
Sphingomonas is another gram-negative α-Proteobacteria. Five isolates belonging to three different species Sphingomonas sanguinis (SB4, NS258, and NS319), Sphingomonas endophytica (NS334) and Sphingomonas yabuuchiae (NS355) were identified using 16S rRNA gene sequences. ANI values further confirmed the species delineation.
Genus: Pseudacidovorax
Pseudacidovorax is a gram-negative β-Proteobacteria. One isolate belonging to Pseudacidovorax intermedius NS331 was isolated from rice seeds.
Genus: Enterobacter
Enterobacter is a gram-negative bacteria belonging to family Enterobacteriaceae of γ-Proteobacteria. Bacterial isolates belonging to this genus were isolated from three different libraries and two rice lots. Eighteen isolates were sequenced belonging to three different species on the basis of 16S rRNA sequence i.e. Enterobacter asburia (NS7, NS23, and NS34), Enterobacter xiangfangensis (NS19, NS24, NS28, NS29, NS49, NS57, NS64, NS75, NS80, NS371, RSA8), and Enterobacter cancerogenus (NS31, NS104, NS111, NS188). ANI values amongst all Enterobacter asburia and E. xiangfangensis strains is more than 99.9%, suggesting that isolates actually belong to one species only while their ANI value with E. cancerogenus strains is around 86%.
Genus: Pantoea
Pantoea is also a gram-negative bacteria belonging to family Enterobacteriaceae of γ-Proteobacteria. Pantoea strains were isolated from four different libraries from all rice lots. Seventeen different bacterial isolates were sequenced belonging to this genus with three different species Pantoea dispersa (SA2, SA3, SA4, SA5, NS215, NS375, NS380, NS389, RSA31), Pantoea stewartii (NS381, RSA13, RSA30, RSA36), and Pantoea ananatis (NS296, NS303, NS311, and RSA47). These isolates grouping to different species were also supported by genome based taxonomy method ANI.
Genus: Pseudomonas
The gram-negative γ-Proteobacteria Pseudomonas was isolated from each year rice lot and each library preparation. Fifteen isolates were sequenced that were assigned to two different species on the basis of 16S rRNA gene sequences, Pseudomonas psychrotolerans (SB5, SB8, SB9, SB11, SB14, SB18, NS2, NS201, NS274, NS337, NS376, NS383, RSA46), and Pseudomonas parafulva (NS96, NS212). While ANI analysis suggests the presence of three different species as NS337 has ANI values less than 88.5% with all other isolates.
Author contributions
SM, KB, and VC carried out the bacterial isolations from rice seeds and their identification using 16S rRNA gene sequences. DNA isolation, QC and sequence assembly was performed by KB and SS. SM, KB, and SS did the library preparation for high throughput sequencing, run analysis and analyzed strains for genome based taxonomy. NK and PPP carried out the revival of strains for sequencing and helped in sequin file preparation. PBP conceived the study and participated in its design and coordination.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
SM, VC, and NK are supported by the fellowship from Council of Scientific and Industrial Research (CSIR). KB, SS, and PP are supported by the fellowship from University Grant Commission (UGC). We acknowledge the funding from CSIR Network projects (BSC-402H and BSC-117/PMSI). We thank Girish Sahni, director, IMTECH, for encouragement and support.
References
- Cottyn B., Regalado E., Lanoot B., De Cleene M., Mew T., Swings J. (2001). Bacterial populations associated with rice seed in the tropical environment. Phytopathology 91, 282–292. 10.1094/PHYTO.2001.91.3.282 [DOI] [PubMed] [Google Scholar]
- Jain R., Rivera M. C., Lake J. A. (1999). Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Natl. Acad. Sci. U.S.A. 96, 3801–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim O.-S., Cho Y.-J., Lee K., Yoon S.-H., Kim M., Na H., et al. (2012). Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721. 10.1099/ijs.0.038075-0 [DOI] [PubMed] [Google Scholar]
- Konstantinidis K. T., Tiedje J. M. (2005). Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. U.S.A. 102, 2567–2572. 10.1073/pnas.0409727102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M., Rosselló-Móra R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106, 19126–19131. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Eisen J. A. (2008). A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 9:R151. 10.1186/gb-2008-9-10-r151 [DOI] [PMC free article] [PubMed] [Google Scholar]