Abstract
PhoPQ-activated gene P (PagP) is an integral membrane enzyme that transfers the sn-1 palmitate chain from phospholipid to lipopolysaccharide in Gram-negative bacteria. A recent x-ray crystallographic study established that the sn-1 palmitate binds within a long cavity at the center of the PagP β barrel. The high mobility required to permit substrate entry into the central core of the barrel contrasts with the need to assemble a well defined structure in the peripheral loops, where many key catalytic residues are located. To gain insight into how dynamics relate to the function of PagP, the enzyme was reconstituted into CYFOS-7, a detergent that supports enzymatic activity. Under these conditions, PagP exists in equilibrium between two states, relaxed (R) and tense (T). The kinetics and thermodynamics of the interchange have been investigated by 1H-15N NMR spectroscopy, with ΔH = -10.7 kcal/mol and ΔS = -37.5 cal/mol·K for the R→ T transition. A comparison of chemical shifts between the two states indicates that major structural changes occur in the large extracellular L1 loop and adjacent regions of the β barrel. In addition to the R,T interconversion, other conformational exchange processes are observed in the R state, showing it to be quite flexible. Thus a picture emerges in which substrate entry is facilitated by the mobility of the R state, whereas the relatively rigid T state adopts a radically different conformation in a region of the protein known to be essential for catalysis. The ability to switch between dynamically distinct states may be a key feature of the catalytic cycle of PagP.
The structural plasticity of integral membrane proteins is critical to their function. Large-scale rearrangements drive a number of biological processes, including the opening and closing of channels (1, 2), active transport (3), and signal transduction across the bilayer (4). Although these changes may be triggered by an external stimulus such as ligand binding or membrane depolarization, they depend on a high intrinsic flexibility at key points within the protein. This intrinsic protein flexibility is underscored by the fact that x-ray studies of lactose permease (5), the mitochondrial ADP/ATP carrier (6), and a voltage-gated potassium channel (7), for example, were made possible only by freezing the protein into a single conformation, using either mutations (8) or the addition of ligands. Although such static representations are invaluable, it is important to recognize that crystal packing or the manipulations that are involved to obtain crystallization in the first place can lock flexible regions into a particular orientation (7, 9–11), and that this “locked orientation” may well be nonnative. It would clearly be beneficial to study the structure of a membrane protein “in motion,” not only to appreciate the full extent of mobility and how it contributes to function but also to develop a general understanding of how proteins behave in a membrane milieu. Solution NMR has been useful in characterizing structural dynamics in soluble proteins (12), and significant advances are now making it possible to study intact membrane proteins as well (13–15). Recently, backbone 15N T1, T2, and heteronuclear nuclear Overhauser enhancement experiments have been used to provide a detailed map of nanosecond-picosecond time scale motions in the bacterial outer membrane proteins OmpA (16) and PhoPQ-activated gene P (PagP) (17).
PagP is an outer membrane enzyme that catalyzes the transfer of a palmitate chain from phospholipid to the lipid A moiety of lipopolysaccharide (LPS) (18). This modification is important for virulence in a number of Gram-negative pathogens (19, 20) and has been shown to attenuate the host immune response by providing resistance to certain antimicrobial peptides (21) and reducing LPS-mediated signaling through toll-like receptor 4 (22). PagP is a remarkably small membrane enzyme, comprised of only an eight-stranded β barrel and an amphipathic α helix. The structure of the enzyme has recently been solved by solution NMR in dodecylphosphocholine (DPC) and n-octyl-β-d-glucoside (OG)/sodium dodecylsulfate (SDS) detergents (17) and by x-ray crystallography in lauryldimethylamine N-oxide (LDAO) (39), showing that the global folds of the protein, not surprisingly, are very similar in solution and crystal environments and essentially invariant to the three detergents that were used in the studies. Interestingly, x-ray studies identified a single detergent molecule buried in the core of the β barrel, and subsequent mutagenesis studies have revealed that this location corresponds to the binding site for the sn-1 palmitate of phospholipid, the acyl chain that is transferred to LPS (18). This creates an interesting dilemma in terms of the structural dynamics of PagP. The center of the β barrel in PagP is relatively rigid, with an increasing mobility gradient as one moves toward the flexible outer loops, a situation common to other β barrel membrane proteins (23, 24). Presumably, the dynamics gradient is necessary to facilitate substrate binding in the core of the enzyme. Remarkably, many of the active site residues in PagP are localized to the dynamic extracellular loops. In fact, the large extracellular L1 loop, which contains the greatest number of conserved polar (putative active site) residues, is the most mobile part of the entire molecule. NMR studies indicate the presence of processes with time scales ranging from picoseconds to milliseconds in this region of the protein, and chain density could not be found in the x-ray analysis. How can such a high level of dynamics and structural disorder involving the key L1 loop region of the protein be compatible with catalysis? In the current study, we have used NMR methods for examining microsecond–millisecond–second time-scale motions (25, 26) to address the potential relationships between dynamics and function in PagP. We show that in a detergent that maintains the activity of the enzyme, CYFOS-7, the protein exists in exchange between two states that differ significantly in mobility and in the structure of the L1 loop and neighboring sites. This duality of structure and dynamics may be important for function, with the more mobile state allowing facile substrate access, and the second conformation related to the active structure of the enzyme.
Materials and Methods
CYFOS-7 detergent was purchased from Anatrace (Maumee, OH), and perfluorooctanoic acid produced by Fluorochem (Old Glossop, Derbyshire, U.K.) was obtained from Oakwood Products (West Columbia, SC).
2H,15N,13C-labeled and 2H,15N-labeled PagP were expressed in Escherichia coli and purified as described previously (17). Denatured PagP was solubilized with 10% perfluorooctanoic acid (PFO), pH 6.0, and then 50 mg of CYFOS-7 was added. The sample was dialyzed (molecular mass cutoff, 3,500 Da) for 3 days against 50 mM sodium phosphate buffer, pH 6.0, to remove PFO. After concentration and addition of D2O, the resulting sample was 1.0 mM in PagP, 10% CYFOS-7, and 90%H2O/10%D2O.
Chemical shift assignments for PagP-CYFOS-7 at 25°C and 45°C were obtained by using a suite of transverse relaxation-optimized spectroscopy (TROSY)-based 3D NMR experiments, as described (17). A 3D 15N-edited nuclear Overhauser enhancement (NOE) experiment was used to confirm the assignments. 1H-15N TROSY-based heteronuclear single quantum correlation (HSQC) spectra were recorded on PagP at 15°C, 20°C, 25°C, 30°C, and 35°C on a Varian Inova 800 MHz spectrometer to quantify the temperature dependence of the relative populations of conformers in the sample. To obtain accurate peak ratios, relaxation losses during the magnetization transfer elements in the pulse sequence were taken into account, in a manner similar to that described previously (27).
Conformational exchange in PagP-CYFOS-7 at 25°C was quantified by using nitrogen Z-magnetization (NZZ) exchange spectroscopy. The published pulse sequence (28) was modified to use IPAP (29) for the 15N TROSY component selection and will be described in detail elsewhere (the pulse sequence code is available upon request). The decay of diagonal peaks and buildup of crosspeaks were fit to a model that accounts for T1 relaxation and a single exchange process, as described in Toll-inger et al. (27).
Conformational exchange on a millisecond time scale was probed by using 15N-CPMG, (CPMG, Carr–Purcell–Meiboom–Gill) relaxation dispersion experiments at 25°C and 45°C. Conventional TROSY methods were used, allowing for transverse relaxation optimization during the 40-ms constant time CPMG element, TCP (27), 15N evolution, and 1H detection periods, as described (30). Effective fields, νCPMG, ranging from 50 to 1,000 Hz, were used and R2,eff calculated as–1/TCPln(IνCPMG/I0), where IνCPMG and I0 are the intensities of correlations recorded in spectra with and without the TCP period, respectively. These experiments were performed at field strengths of 600 and 800 MHz. Data analysis followed Mulder et al. (31).
Results and Discussion
Chemical Shift Assignment. A crystal structure of PagP in lauryldimethylamine N-oxide (LDAO) establishes the presence of a single detergent molecule in the barrel cavity (39), and it is very likely that the detergents DPC and OG that were used in previous NMR studies (17) also penetrate the barrel. The bound detergent provides a likely explanation for the lack of enzyme activity in the three detergents used in structural studies to this point, because it may well compete with natural substrate for the binding site. To study PagP in an active form, it was reconstituted into CYFOS-7 detergent (Fig. 1a). CYFOS-7 was chosen because it is an analog of DPC with a bulky cyclohexane-containing aliphatic group that prohibits penetration into the enzyme cavity; indeed, the detergent was found to support PagP activity under the conditions used for NMR studies (see supporting information, which is published on the PNAS web site).
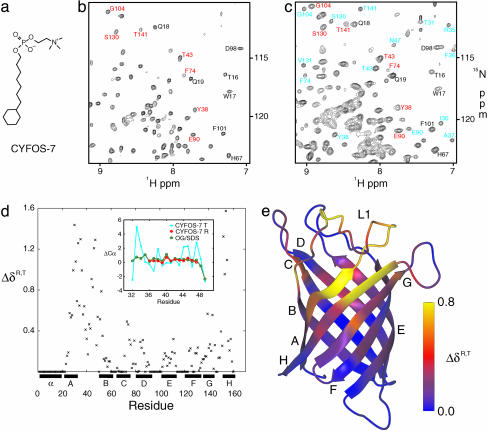
Fig. 1.
Comparison of the R and T forms of PagP in CYFOS-7 detergent. (a) Chemical structure of CYFOS-7 detergent. (b and c) 1H-15N HSQC spectrum of PagP-CYFOS-7 recorded at 45°C and 800 MHz (b) and at 25°C and 800 MHz (c). Some resonances from residues in the R state are labeled in red, and those in the T state are shown in blue. Residues that have only one set of chemical shifts in both R and T states are indicated in black. (d) Combined chemical shift difference between the R and T states, ΔδR,T = [(ωNΔδN)2 + (ωCaΔδCα)2 + (ωCOΔδCO)2]1/2, ωN = 0.154, ωCα = 0.276, ωCO = 0.341 (37), plotted as a function of sequence. For the R state, an average between the available chemical shifts in DPC, OG/SDS, and the CYFOS-7 R state was used. If a residue could not be observed in the T state or if the R and T states were identical (see text), ΔδR,T = 0. (Inset) Deviation from random coil Cα shifts in the L1 loop, residues 32–49, for the CYFOS-7 T state, CYFOS-7 R state, and PagP in OG/SDS (17). Some residues in the CYFOS-7 R state were broadened beyond detection. (e) Ribbon diagram of PagP colored according to ΔδR,T.
Favorable HSQC spectra of PagP in CYFOS-7 were obtained at 45°C (Fig. 1b), and this temperature was therefore chosen for initial studies. However, 25% of the protein could not be assigned because of exchange broadening in two extracellular regions, one comprised of residues 149–155, 28–35, 49–52, and 75 (strands H, A, B, and C), and the other made up of residues 82, 114–126, and 144–148 (strands D, E/F, and G). This pattern is similar to but more extensive than what was observed in spectra of PagP solubilized in DPC (17). The backbone chemical shifts that were obtained for the remainder of the protein suggest a structure very similar to that of PagP in DPC or OG/SDS at 45°C.
When the temperature is lowered to 25°C, new peaks appear throughout the 1H-15N correlation map, indicating the presence of a second minor conformer (Fig. 1c). To establish whether there are potential structural changes in the two forms of the protein and to characterize the system further, we have attempted to obtain backbone chemical shift assignments of both conformers. This is far from trivial. The spectral crowding caused by an approximate doubling of peaks, a low effective concentration of the minor conformer (≈300 μM, see below), the slow tumbling of the ≈50-kDa system at 25°C and most of all, the presence of multiple conformational exchange processes, all complicated the problem. Nevertheless, a surprisingly complete set of chemical shifts was obtained for the minor conformer, with the exception of residues 81–82, 114–123, and 146–148 from strands D, E/F, and G. Notably, the area comprised of residues in strands H, A, B, and C becomes entirely visible for the minor state at 25°C, although it remains invisible in the major conformer. Correlations from both major and minor forms of PagP are observed from most other regions of the structure with similar (but not identical) chemical shifts noted. Of interest, only a single set of cross peaks was obtained for a periplasmic region (residues 1–22, 60–70, and 88–102) consisting of the N-terminal amphipathic helix and turns T1 and T2. This part of the protein is furthest from the epicenter of the major structural changes (see below).
A striking difference in chemical shifts between conformers is observed in the large extracellular L1 loop connecting strands A and B. Most of this loop is invisible in the x-ray crystal structure, and 15N NMR relaxation measurements indicate that it is highly mobile (17). In the major conformer, the chemical shifts in this region resemble those from PagP-DPC and PagP-OG/SDS (see Fig. 1d Inset) and are typical of flexible unstructured segments. In what follows, therefore, we refer to this form of the protein as the relaxed or “R” state. The new conformation is the tense or “T” state, because most of its L1 loop residues display well dispersed chemical shifts suggestive of a more rigid structure. The structural transition in the L1 loop associated with the R,T interconversion is accompanied by a marked rearrangement in the extracellular half of strand A. Although this region is not observed in the R state of PagP-CYFOS-7, in the “R-like” structure of PagP in OG/SDS the regular hydrogen bonding pattern between strands B, A, and H is interrupted by a β bulge in strand A (P28, A29). Beyond the β bulge, additional hydrogen bonds are observed between strands A and H (I30-A155 and W32-Y153). However, in the T state, drastic chemical shift changes are seen for A29 (Cα, -3 ppm; Cβ, -5 ppm) and I30 (Cα, +3 ppm; Cβ, -4 ppm; N, +7 ppm), suggesting that the “β bulge” is no longer formed, and a close examination of 1HN-1HN nuclear Overhauser enhancements reveals the absence of the I30-A155 and W32-Y153 hydrogen bonds. The major structural changes between the R and T states are thus localized to the L1 loop, the β bulge of strand A, and the adjacent region on strand H, as highlighted by a detailed comparison of chemical shifts (see Fig. 1 d and e).
Kinetic and Thermodynamic Characterization of the R-T Interconversion. NMR measurements at 25°C and 45°C indicate that the population of the T state is very temperature-dependent. The relative populations of the R and T states were calculated from 1H-15N HSQC spectra recorded at 15, 20, 25, 30, and 35°C from which equilibrium constants, K, were obtained.
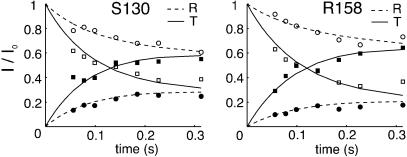
The R→T and T→R conversion rates were quantified at 25°C by recording the transfer of nitrogen Z-magnetization between the two states via NZZ exchange spectroscopy (28). The magnetization buildup/decay curves for S130 and R158 are shown in Fig. 2 and analyzed as described by Tollinger et al. (27). Similar kinetics were obtained for all residues that were sufficiently well resolved for quantification, as expected, with average values of the R→T(kRT) and T→R(kTR) conversion rates of 2.8 ± 0.5 s-1 and 6.5 ± 0.9 s-1, respectively, so that the fractional population of the T state is ≈0.30 at 25°C.
Fig. 2.
Decay [correlations at (ωNi,ωHi); open symbols] and buildup [correlations at (ωNi,ωHj); filled symbols] curves for S130 and R158 in the NZZ exchange experiment (27). The curves are normalized so that the starting intensities of the diagonal peaks, (ωNi,ωHi), are 1.0 in both R and T states.
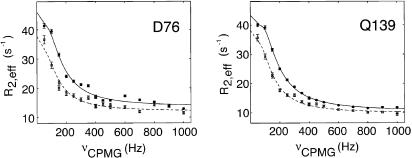
To study the R,T interconversion at 45°C, where only a single set of correlations is observed, we have used 15N-CPMG relaxation dispersion experiments, in which the presence of the “invisible” T state affects the line width of cross peaks derived from the R conformation, R2,eff/π. This exchange broadening can be refocused by a train of 180° pulses (... τ - 180° - τ...), leading to a decrease in R2,eff as νCPMG = 1/(4τ) is increased (Fig. 3). It is therefore possible to extract the exchange parameters, including the rate of interconversion and the relative populations of the exchanging states, as well as the differences in 15N chemical shifts between them by measuring R2,eff as a function of τ at multiple static magnetic fields (25). 15N-CPMG relaxation dispersion experiments were performed on PagP-CYFOS-7 at 45°C (Fig. 3). When these profiles were fit individually, similar values of rates and populations were obtained for many residues, suggesting that they share a single conformational exchange process. A global fit, performed according to Mulder et al. (32), yields kex of 331 ± 40 s-1 (kex = kTR + kRT, kTR = 298 s-1, kRT = 33 s-1) and a fractional population of the minor conformation of 0.10 ± 0.01. The extracted exchange parameters explain why a single set of crosspeaks is observed in correlation spectra of PagP recorded at 45°C. First, the population of the minor T conformation decreases 3-fold from 25°C. Second, peaks for the T state are severely broadened at elevated temperature due to the short lifetime of this conformer (1/kTR).
Fig. 3.
15N-CPMG relaxation dispersion curves at 45°C for D76 and Q139. The 800 MHz data are represented by circles and the 600 MHz data by squares (lower traces), with the solid and dashed lines indicating the fits to the experimental 800 and 600 MHz data, respectively, assuming a two-site exchange model (38). The value of νCPMG is 1/(4τCP), where 2τCP is the spacing between successive π pulses, with R2,eff defined in Materials and Methods. Experimental errors were estimated as described by Mulder et al. (31) and are indicated by vertical lines through the experimental data points.
A major challenge in the interpretation of relaxation dispersion data is to understand the underlying molecular process responsible for the dispersions in the first place. In the case of PagP, insight is provided by comparing the 15N chemical shift differences between states, Δω, extracted from the dispersion data at 45°C with values of Δω obtained directly from correlations at 25°C (where both R and T states are observed). Fig. 4 Inset shows a good correlation between ΔωCPMG (45°C) and ΔωHSQC (25°C) values, providing strong evidence that the dispersion profiles for the residues mentioned above derive from the R,T interconversion.
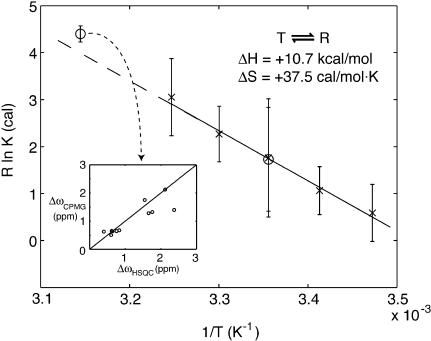
Fig. 4.
Plot of R ln K vs. 1/T,R = 1.9858 cal/mol·K. X represents values derived directly from 1H-15N HSQC spectra recorded at 15, 20, 25, 30, and 35°C. Error bars show twice the standard deviation of values obtained for different residues. Circles represent equilibrium constants derived from NZZ exchange and relaxation dispersion experiments at 25°C and 45°C, respectively. The error bars for the value at 45°C were estimated from 100 simulations in which 20% of the residues were randomly excluded from the global fit (see Results and Discussion). (Inset) Comparison of 15N Δω values (in ppm) for individual residues, calculated from the global fit of relaxation dispersion profiles at 45°C vs. Δω values obtained directly from measurements of 15N chemical shift differences at 25°C, where chemical shifts for both the R and T states are available.
Fig. 4 plots ln K vs. 1/T, obtained using the different methodology described above. The equilibrium constant derived from the NZZ exchange experiment at 25°C agrees well with the value obtained directly from cross peak intensities in the 1H-15N HSQC spectrum (corrected to account for differential relaxation losses during the transfer steps; see Materials and Methods). As well, the equilibrium constant derived from the relaxation dispersion experiments at 45°C is consistent with the value obtained by extrapolating the ln K vs. 1/T values measured in cases where correlations from both R and T states are observed in spectra. Assuming a linear relation between ln K and 1/T (i.e., ΔCp∼0), it is also possible to extract ΔH and ΔS (ΔG = -RT ln K = ΔH - TΔS), with ΔH = +10.7 ± 4.2 kcal/mol, ΔS = +37.5 ± 14.2 cal/mol·K for the T-to-R transition (note that the errors represent two standard deviations). The assumption that ΔCp∼0 is supported by the linearity in the ln K vs. 1/T profile and by the fact that the R,T interconversion involves only significant structural changes for a relatively small region of the protein (i.e., the β barrel conformation remains unchanged). It is important to emphasize that the thermodynamic parameters extracted correspond to those of the ternary system, which includes protein, detergent, and water. The simplest interpretation is that the minor T conformation is enthalpically more stable, with the R conformation preferred at temperatures above 10°C because of the entropic cost of ordering the protein. We show below that it is possible to establish that indeed the R state is more flexible than the T conformation.
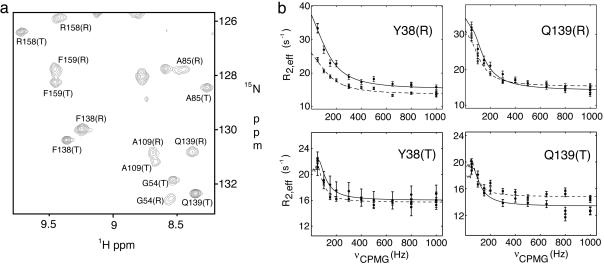
Differential Dynamics of the R and T States. HSQC spectra recorded at 25°C on the PagP system establish that most peaks from the R state of the protein are more broad than their T state counterparts (see Fig. 5a). It is possible to obtain a more quantitative description of these differences by recording 15N-CPMG relaxation dispersion profiles of both conformers at this temperature. Although the R:T interconversion at 25°C has been characterized via NZZ exchange spectroscopy, the CPMG experiments may be able to detect additional conformational exchange processes. The slow exchange between R and T states would be expected to produce a dispersion profile with characteristic oscillations in R2,eff at low νCPMG fields (27). Optimal extraction of exchange parameters would require very high quality data at these low fields, which cannot be obtained in the present application. Therefore, in fits of dispersion curves of the T state at this temperature, values of kTR were constrained to <15 s-1, consistent with results from the NZZ data. Fig. 5b shows that the dispersion profiles of Y38(T) and Q139(T) can be completely accounted for by the T→R transition, suggesting the absence of other major exchange processes on the microsecond to millisecond time scale. Notably, 15N Δω values extracted from the fits (2.1 ± 0.3 and 2.0 ± 0.4 ppm for Y38 and Q139) are consistent with what is observed in spectra (2.4 and 1.8 ppm), and the fitted kTR values (5.3 ± 1.0 and 5.0 ± 2.0 s-1 for Y38 and Q139) agree with the value measured by NZZ exchange spectroscopy (kTR = 6.5 ± 0.9 s-1). (Note that the dispersion curves are not sensitive to kRT because the exchange is in the slow regime.)
Fig. 5.
Differential dynamics of the R and T states. (a) Selected region of the 1H-15N TROSY-HSQC spectrum of PagP at 25°C. (b) 15N-CPMG relaxation dispersion curves for Y38 and Q139 in both R and T states at 25°C, recorded at 800 (solid lines) and 600 MHz (dashed lines).
If the R:T interconversion were the only process giving rise to the dispersion profiles at 25°C, one would expect Rex,R = kRT and Rex,T = kTR for slowly exchanging sites (kex << Δω), where Rex is the exchange contribution to the transverse relaxation rate, R2, and is approximated by R2,eff(νCPMG = 0) - R2,eff(νCPMG = 1,000 Hz). This is certainly the case for Y38(T) and Q139(T), for which Rex,T is ≈6 s-1 (the height of the dispersion curves in Fig. 5b, essentially the same as kTR = 6.5 ± 0.9 s-1). In contrast, very different behavior is noted for Y38 and Q139 in the R state. The R→T transition alone would give rise to small values of Rex, on the order of kRT = 2.8 ± 0.5 s-1. Instead, Rex values in excess of 20 s-1 are observed at 800 MHz, indicating the presence of additional conformational exchange processes in the R state. Values of kex on the order of 103 s-1 are obtained from fits of the dispersion curves of Y38(R) or Q139(R) assuming a single exchange process, much faster than the kinetics describing the R:T equilibrium (kex = 9.3 s-1). This faster time scale process is limited to the R state and is the main contributor to Rex at 25°C. It is not detected at 45°C, where the dispersion profiles can be completely explained by the R:T transition, presumably because the exchange rate of the faster process becomes too fast to give rise to any exchange broadening that can be refocused by applied νCPMG fields. Residues that are most strongly affected by this fast exchange process are located in the vicinity of the region that is broadened beyond detection in the R form (residues from strands H, A, B, and C) and thought to undergo a significant structural rearrangement between the two states. Thus, the most flexible portion of the β barrel in the R form on the millisecond time scale matches the region which changes most drastically in the R-to-T transition. In this regard, it seems best to view the L1 loop and portions of strands H, A, B, and C in the R state in terms of an ensemble of conformers, with the T state representing a more defined configuration.
One can imagine a simple enzyme with a fixed conformation, in which substrate is able to freely diffuse into the active site and react. This is unlikely in the case of PagP, because of the difficulty in adopting a stable “open” conformation with a large cleft in the β barrel. Instead, the barrel must possess enough flexibility to allow transient openings wide enough to accept substrate. Such extreme mobility would be detrimental to the formation of a precisely structured active site, but interestingly, the L1 loop and adjacent regions in PagP are able to alternate between a flexible form (R) that may facilitate substrate entry and one that is more structured (T) that may be important for catalysis. Although it is unknown to what extent the T state in the current study resembles the active conformation of PagP, it is nevertheless remarkable that the major structural reorganization observed in the T conformer takes place in a region known to be essential to catalysis. The ability of PagP to switch between R and T in the absence of substrate may be an intrinsic property of the polypeptide chain, analogous to the conformational equilibrium in the signaling proteins NtrC and Spo0F that occurs in the absence of phosphorylation (33, 34).
Clearly, a single static conformation of PagP cannot adequately describe its function. Moreover, the static structures that have been determined of the enzyme to date are those of an inactive form. The present study identifies at least two major states of the enzyme, R and T, and characterizes the differences in the dynamics accessible to each. These differential dynamics might well account for competing requirements of binding and catalysis. NMR studies of large numbers of membrane proteins have provided strong evidence of slow time scale dynamics (17, 24, 35, 36). Analysis of these dynamics and a characterization of the states that these dynamical processes connect will ultimately provide a detailed understanding of the relation between structure, dynamics, and function in this important class of proteins.
Supplementary Material
Acknowledgments
We thank Dr. Ranjith Muhandiram for assistance in setting up some of the NMR experiments. We also thank Oscar Millet, Martin Tollinger, and Tony Mittermaier (University of Toronto, Toronto) for assistance in analyzing NZZ exchange and CPMG relaxation dispersion experiments. P.M.H. acknowledges the Canadian Institutes of Health Research (CIHR) for a predoctoral award. L.E.K. holds a Canada Research Chair in Biochemistry. This research was supported by operating grants from the CIHR (to R.E.B. and L.E.K.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TROSY, transverse relaxation-optimized spectroscopy; PagP, PhoPQ-activated gene P; DPC, dodecylphosphocholine; OG, n-octyl-β-d-glucoside; SDS, sodium dodecylsulfate; HSQC, heteronuclear single quantum correlation; CPMG, Carr–Purcell–Meiboom–Gill; R state, relaxed state; T state, tense state.
Data deposition: The NMR chemical shifts for the R state at 45°C have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession no. 6234).
References
- 1.Jiang, Y., Lee, A., Chen, J., Cadene, M., Chait, B. T. & MacKinnon, R. (2002) Nature 417, 523-526. [DOI] [PubMed] [Google Scholar]
- 2.Perozo, E. & Rees, D. C. (2003) Curr. Opin. Struct. Biol. 13, 432-442. [DOI] [PubMed] [Google Scholar]
- 3.Chang, G. (2003) FEBS Lett. 555, 102-105. [DOI] [PubMed] [Google Scholar]
- 4.Kim, M., Carman, C. V. & Springer, T. A. (2003) Science 301, 1720-1725. [DOI] [PubMed] [Google Scholar]
- 5.Abramson, J., Smirnova, I., Kasho, V., Verner, G., Kaback, H. R. & Iwata, S. (2003) Science 301, 610-615. [DOI] [PubMed] [Google Scholar]
- 6.Pebay-Peyroula, E., Dahout-Gonzalez, C., Kahn, R., Trezeguet, V., Lauquin, G. J. & Brandolin, G. (2003) Nature 426, 39-44. [DOI] [PubMed] [Google Scholar]
- 7.Jiang, Y., Lee, A., Chen, J., Ruta, V., Cadene, M., Chait, B. T. & MacKinnon, R. (2003) Nature 423, 33-41. [DOI] [PubMed] [Google Scholar]
- 8.Smirnova, I. N. & Kaback, H. R. (2003) Biochemistry 42, 3025-3031. [DOI] [PubMed] [Google Scholar]
- 9.Jiang, Y., Ruta, V., Chen, J., Lee, A. & MacKinnon, R. (2003) Nature 423, 42-48. [DOI] [PubMed] [Google Scholar]
- 10.Chang, G. (2003) J. Mol. Biol. 330, 419-430.12823979 [Google Scholar]
- 11.Campbell, J. D., Biggin, P. C., Baaden, M. & Sansom, M. S. (2003) Biochemistry 42, 3666-3673. [DOI] [PubMed] [Google Scholar]
- 12.Kay, L. E. (1998) Nat. Struct. Biol. 5 Suppl, 513-517. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez, C. & Wider, G. (2003) Curr. Opin. Struct. Biol. 13, 570-580. [DOI] [PubMed] [Google Scholar]
- 14.Oxenoid, K., Sonnichsen, F. D. & Sanders, C. R. (2001) Biochemistry 40, 5111-5118. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez, C. & Wuthrich, K. (2003) FEBS Lett. 555, 144-150. [DOI] [PubMed] [Google Scholar]
- 16.Tamm, L. K., Abildgaard, F., Arora, A., Blad, H. & Bushweller, J. H. (2003) FEBS Lett. 555, 139-143. [DOI] [PubMed] [Google Scholar]
- 17.Hwang, P. M., Choy, W. Y., Lo, E. I., Chen, L., Forman-Kay, J. D., Raetz, C. R., Prive, G. G., Bishop, R. E. & Kay, L. E. (2002) Proc. Natl. Acad. Sci. USA 99, 13560-13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bishop, R. E., Gibbons, H. S., Guina, T., Trent, M. S., Miller, S. I. & Raetz, C. R. (2000) EMBO J. 19, 5071-5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Preston, A., Maxim, E., Toland, E., Pishko, E. J., Harvill, E. T., Caroff, M. & Maskell, D. J. (2003) Mol. Microbiol. 48, 725-736. [DOI] [PubMed] [Google Scholar]
- 20.Robey, M., O'Connell, W. & Cianciotto, N. P. (2001) Infect. Immun. 69, 4276-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo, L., Lim, K. B., Poduje, C. M., Daniel, M., Gunn, J. S., Hackett, M. & Miller, S. I. (1998) Cell 95, 189-198. [DOI] [PubMed] [Google Scholar]
- 22.Kawasaki, K., Ernst, R. K. & Miller, S. I. (2004) J. Biol. Chem. 279, 20044-20048. [DOI] [PubMed] [Google Scholar]
- 23.Fernandez, C., Hilty, C., Bonjour, S., Adeishvili, K., Pervushin, K. & Wuthrich, K. (2001) FEBS Lett. 504, 173-178. [DOI] [PubMed] [Google Scholar]
- 24.Arora, A., Abildgaard, F., Bushweller, J. H. & Tamm, L. K. (2001) Nat. Struct. Biol. 8, 334-338. [DOI] [PubMed] [Google Scholar]
- 25.Palmer, A. G., 3rd, Kroenke, C. D. & Loria, J. P. (2001) Methods Enzymol. 339, 204-238. [DOI] [PubMed] [Google Scholar]
- 26.Akke, M. (2002) Curr. Opin. Struct. Biol. 12, 642-647. [DOI] [PubMed] [Google Scholar]
- 27.Tollinger, M., Skrynnikov, N. R., Mulder, F. A., Forman-Kay, J. D. & Kay, L. E. (2001) J. Am. Chem. Soc. 123, 11341-11352. [DOI] [PubMed] [Google Scholar]
- 28.Farrow, N. A., Zhang, O., Forman-Kay, J. D. & Kay, L. E. (1994) J. Biomol. NMR 4, 727-734. [DOI] [PubMed] [Google Scholar]
- 29.Ottiger, M., Delaglio, F. & Bax, A. (1998) J. Magn. Reson. 131, 373-378. [DOI] [PubMed] [Google Scholar]
- 30.Loria, J. P., Rance, M. & Palmer, A. G., 3rd. (1999) J. Biomol. NMR 15, 151-155. [DOI] [PubMed] [Google Scholar]
- 31.Mulder, F. A., Hon, B., Mittermaier, A., Dahlquist, F. W. & Kay, L. E. (2002) J. Am. Chem. Soc. 124, 1443-1451. [DOI] [PubMed] [Google Scholar]
- 32.Mulder, F. A., Mittermaier, A., Hon, B., Dahlquist, F. W. & Kay, L. E. (2001) Nat. Struct. Biol. 8, 932-935. [DOI] [PubMed] [Google Scholar]
- 33.Volkman, B. F., Lipson, D., Wemmer, D. E. & Kern, D. (2001) Science 291, 2429-2433. [DOI] [PubMed] [Google Scholar]
- 34.Feher, V. A. & Cavanagh, J. (1999) Nature 400, 289-293. [DOI] [PubMed] [Google Scholar]
- 35.Fernandez, C., Hilty, C., Wider, G., Guntert, P. & Wuthrich, K. (2004) J. Mol. Biol. 336, 1211-1221. [DOI] [PubMed] [Google Scholar]
- 36.Oxenoid, K., Kim, H. J., Jacob, J., Sonnichsen, F. D. & Sanders, C. R. (2004) J. Am. Chem. Soc. 126, 5048-5049. [DOI] [PubMed] [Google Scholar]
- 37.Evenas, J., Tugarinov, V., Skrynnikov, N. R., Goto, N. K., Muhandiram, R. & Kay, L. E. (2001) J. Mol. Biol. 309, 961-974. [DOI] [PubMed] [Google Scholar]
- 38.Carver, J. P. & Richards, R. E. (1972) J. Magn. Reson. 6, 89-105. [Google Scholar]
- 39.Aho, V., Lo, E. I., Engel, C. K., Chen, L., Hwang, P. M., Kay, L. E., Bishop, R. E. & Privé, G. G. (2004) EMBO J., in press. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.