Abstract
Cyclic nucleotide phosphodiesterases (PDEs) are enzymes controlling cellular concentrations of the second messengers cAMP and cGMP. The crystal structure of the catalytic domain of PDE9A2, a member of a PDE family specifically hydrolyzing cGMP, has been determined at 2.23-Å resolution. The PDE9A2 catalytic domain closely resembles the cAMP-specific PDE4D2 but is significantly different from the cGMP-specific PDE5A1, implying that each individual PDE family has its own characteristic substrate recognition mechanism. The different conformations of the H and M loops between PDE9A2 and PDE5A1 imply their less critical roles in nucleotide recognition. The nonselective inhibitor 3-isobutyl-1-methylxanthine (IBMX) binds to a similar subpocket in the active sites of PDE4, PDE5, and PDE9 and has a common pattern of the binding. However, significantly different orientations and interactions of IBMXs are observed among the three PDE families and also between two monomers of the PDE9A2 dimer. The kinetic properties of the PDE9A2 catalytic domain similar to those of full-length PDE9A imply that the N-terminal regulatory domain does not significantly alter the catalytic activity and the IBMX inhibition.
Twenty-one genes of human phosphodiesterase (PDE) are categorized into 11 PDE families and share a conserved catalytic domain with ≈300 aa (1–8). However, each PDE family has varying substrate preferences and selective inhibitors. The PDE4, PDE7, and PDE8 family members prefer to hydrolyze cAMP, whereas PDE5, PDE6, and PDE9 are cGMP-specific. PDE1, PDE2, PDE3, PDE10, and PDE11 show activities toward both cAMP and cGMP (7). Selective inhibitors of PDEs have been widely studied as therapeutics such as cardiotonics, vasodilators, smooth-muscle relaxants, antidepressants, antithrombotics, antiasthmatics, and agents for improving cognitive functions such as learning and memory (9–15). Many PDE inhibitors have been in clinical trials or have already entered the marketplace. For example, the PDE3 selective inhibitor cilostazol (Pletal) is a drug for the reduction of the symptoms of intermittent claudication and the PDE5 inhibitors sildenafil (Viagra), vardenafil (Levitra), and tadalafil (Cialis) are used for the treatment of male erectile dysfunction.
Structural studies have shown that the catalytic domains of PDE4 and PDE5 have similar folding, but the conformations of H and M loops that are involved in binding of sildenafil and rolipram are dramatically different (16–21). Thus, understanding whether the conformations of PDE4 and PDE5 are two representatives for the catalytic domains of other cAMP- and cGMP-specific PDE families and whether the H and M loops play a pivotal role in the substrate specificity and inhibitor selectivity is essential for the illustration of the PDE functions. Here, we report a crystal structure of the catalytic domain of human PDE9A2 that is a cGMP-specific PDE with the highest affinity for cGMP (Km = 70 nM) and a low affinity for cAMP (Km = 230 μM) (22, 23). The structural comparison surprisingly reveals that the catalytic domain of PDE9A2 is closer to PDE4D2 than PDE5A1.
Materials and Methods
Protein Expression and Purification. The catalytic domain of human PDE9A2 (GenBank accession no. BC009047) was purchased from American Type Culture Collection. A pair of oligonucleotide primers of gacgcgatcatatgacttaccccaagtacctg and tcaactcgagttacttcttctgtaactctttc were synthesized for amplification of the PDE9A2 coding region of amino acids 181–506 by PCR. The amplified PDE9A2 DNA and the expression vector pET15b were digested separately by the restriction enzymes NdeI and XhoI, purified from agarose gel, and then ligated by T4 DNA ligase. The resultant plasmid pET-PDE9A2 was transformed into Escherichia coli strain BL21 (Codonplus) for overexpression. The E. coli cell carrying pET-PDE9A2 was grown in LB medium at 37°C to absorption A600 = 0.7, and then 0.1 mM isopropyl β-d-thiogalactopyranoside was added for further growth at 15°C overnight. Recombinant PDE9A2 was purified by the chromatographic columns of Ni-NTA affinity (Qiagen, Valencia, CA), Q-Sepharose (Pharmacia), and Sephacryl S300 (Pharmacia). The Y424F mutant of the PDE9A2 catalytic domain was subcloned by the site mutagenesis and purified by using the same method as for the wild type. The PDE9A2 proteins had purity >95% as shown by SDS/PAGE. A typical batch of purification yielded >100 mg of PDE9A2 from a 2-liter cell culture.
Enzymatic Properties. The enzymatic activities of the PDE9A2 domains were assayed by using 3H-cAMP and 3H-cGMP as substrates. The PDE9A2 catalytic domain was incubated with the reaction mixture of 50 mM Tris·HCl (pH 7.8), 10 mM MgCl2, and 3H-cAMP or 3H-cGMP (40,000 cpm per assay) at 24°C for 30 min. The reactions were terminated by addition of 0.2 M ZnSO4 and 0.2 M Ba(OH)2. The reaction product 3H-AMP or 3H-GMP was precipitated by BaSO4, whereas unreacted 3H-cAMP or 3H-cGMP remained in the supernatant. Radioactivity in the supernatant was measured by liquid scintillation in a Beckman–Coulter LS 6500 counter. The activity was measured at eight concentrations of cAMP and cGMP, and each measurement was repeated three times. Vmax and Km values were calculated by linear plot of Lineweaver–Burk and nonlinear regression of velocity versus substrate concentration. Eight concentrations of 3-isobutyl-1-methylxanthine (IBMX) were used in the determination of IC50.
Crystallization and Structure Determination. Crystals of PDE9A2–IBMX were grown by vapor diffusion. The catalytic domain of 10–15 mg/ml PDE9A2 (amino acids 181–506) in a storage buffer of 50 mM NaCl, 20 mM Tris·HCl (pH 7.5), 1 mM 2-mercaptoethanol, and 1 mM EDTA was mixed with 2 mM IBMX. The PDE9A2–IBMX complex was crystallized by hanging drops at 4°C. The protein drops contain 3 μl of PDE9A2–IBMX complex and 1 μl of well buffer of 0.1 M Hepes (pH 7.5), 2.2 M sodium formate, and 5% xylitol. The well buffer plus 20% glycerol was used as the cryosolvent for freezing the crystals in liquid nitrogen. Diffraction data were collected on beamline X12C at Brookhaven National Laboratory (Table 1) and processed by program hkl (24). The PDE9A2–IBMX crystal has the space group P41212 with cell dimensions of a = 103.5 and c = 269.7 Å. The structure of PDE9A2–IBMX was solved by the molecular replacement program amore (25), with the catalytic domain of PDE4D2 as the initial model. The atomic model was rebuilt by program o (26) against the electron density map improved by the density modification package of ccp4. The structure was refined by cns (27) (Table 1).
Table 1. Statistics on diffraction data and structure refinement.
| Data collection | PDE9A2-IBMX |
| Space group | P41212 |
| Unit cell (a, b, c), Å | 103.5, 269.7 |
| Resolution, Å | 2.23 |
| Total measurements | 476,768 |
| Unique reflections | 70,925 |
| Completeness, % | 98.2 (84.5) |
| Average I/σ | 27.8 (3.0) |
| Rmerge | 0.072 (0.489) |
| Structure refinement | |
| R factor | 0.215 |
| Rfree | 0.231 |
| Resolution, Å | 30-2.23 |
| Reflections | 67,397 |
| rms deviation for | |
| Bond, Å | 0.0063 |
| Angle, ° | 1.20 |
| Average B factor, Å2 (atoms) | |
| All atoms | 35.8 (5625) |
| Protein | 35.8 (5390) |
| IBMX | 38.2 (32) |
| Waters | 32.8 (195) |
| Metals | 37.3 (4) |
The numbers in parentheses are for the highest-resolution shell.
Results and Discussion
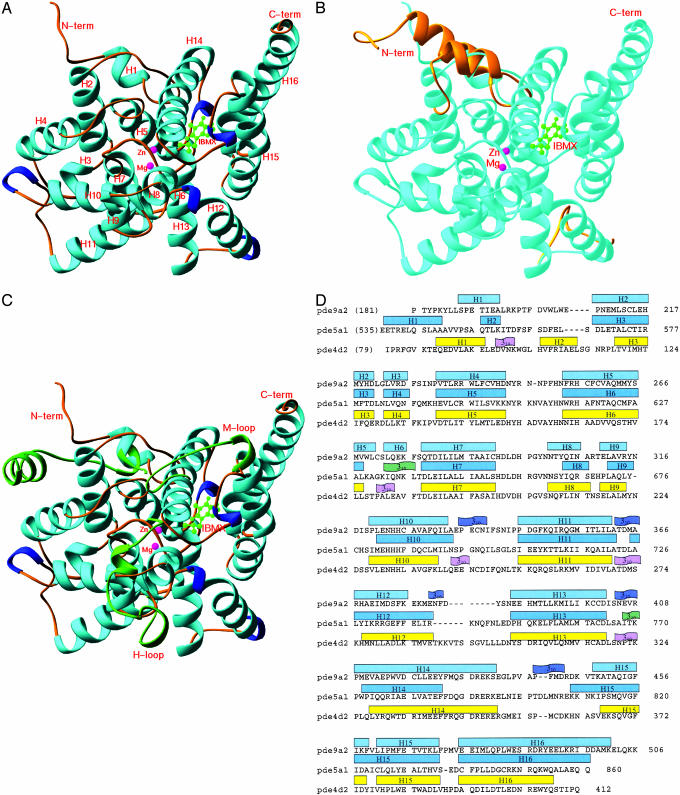
Overall Structure. The catalytic domain of PDE9A2 (residues 181–506) consists of 16 helices (Fig. 1A). The superposition of the PDE9A2 catalytic domain over PDE4D2 in PDE4D2–IBMX (21) yielded an rms deviation of 1.5 Å for the Cα atoms of residues 207–495 in PDE9A2 or residues 115–411 in PDE4D2, showing that the two structures are very similar. The structure variation is observed mainly in the N-terminal region that has a totally different 3D arrangement. Residues 79–114 of PDE4D2 consist of two α-helices and a 310-helix, but residues 181–206 of PDE9A2 contain a single α-helix. The residues after 207 of PDE9A2 are superimposed very well over those of PDE4D2 and possess similar elements of secondary structures, except for two new 310 helices around residues 380 and 440 of PDE9A2 and an extension of 10 residues in C-terminal helix H16 of PDE9A2 (Fig. 1 B and D). In addition, α-helix H6 (residues 272–275) in PDE9A2 corresponds to a 310-helix in PDE4D2.
Fig. 1.
Structures of the PDE–IBMX complexes. (A) Ribbon diagram of the catalytic domain of PDE9A2–IBMX. All the ribbon pictures in Figs. 1, 2, 3 are drawn by ribbons (34). The α-helices are cyan, the 310-helices are blue, the metal ions are pink, and IBMX is green. (B) Superposition of the N-terminal residues 79–119 and loop 287–300 of PDE4D2 (golden) over PDE9A2 (light cyan). The remaining structures of PDE4D2 are very similar to PDE9A2 and are not shown. (C) Superposition of three regions of PDE5A1 (green) over PDE9A2 (light cyan). These regions include the N-terminal residues 535–565 and H and M loops (residues 661–676 and 787–812). The remaining regions of PDE5A1 are similar to PDE9A2 and are not shown. (D) Sequence and secondary structures of the PDE catalytic domains.
On the other hand, the structure comparison of PDE9A2 with PDE5A1 (21) shows significant differences in the conformations of the two catalytic domains (Fig. 1C), as shown with an rms deviation of 2.8 Å for the Cα atoms of 272 residues. Three regions are not superimposable. The N-terminal residues 535–567 of PDE5A1 contain two helices and have different 3D arrangements from residues 181–206 of PDE9A2. The H loop residues 301–316 in PDE9A2 or residues 661–676 in PDE5A1 have positional differences of 6–7 Å for the Cα atoms of two short helices H8 and H9. The M loop residues 425–448 in PDE9A2 are well ordered and contain a 310-helix; in contrast, the majority of residues 787–812 in PDE5A1 are not traceable and presumably exist in random conformation.
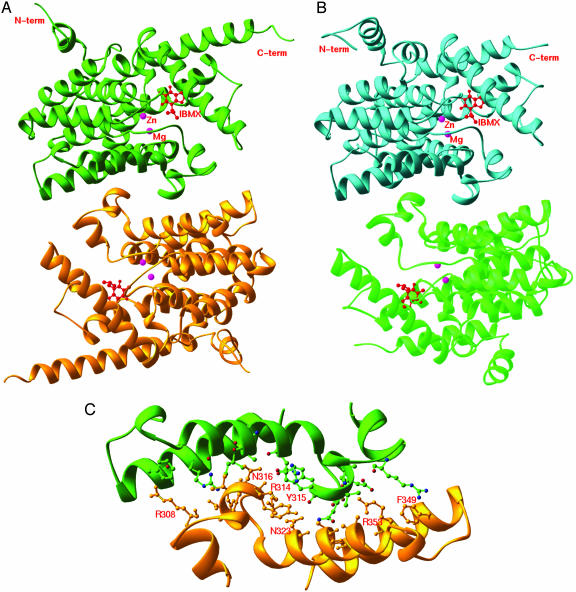
The crystallographic asymmetric unit of PDE9A2–IBMX contains two molecules of the catalytic domains. The solvent content is estimated to be ≈84% in the PDE9A2 crystal and is unusually high in comparison with the solvent contents of 50% and 47% in the crystals of PDE4D2 and PDE5A1. Despite high solvent content, the surface loops involved in crystallographic lattice contacts in PDE9A2 do not show significant conformation differences from those of PDE4D2. Two molecules of PDE9A2 form a dimer in the crystal state (Fig. 2A). The dimerization scheme of PDE9A2 is the same as that of PDE4D2, and the key residues for the dimer interface are identical between the two structures (Fig. 2 B and C). The interfacial residues Tyr-315, Asn-316, Asp-317, Asn-323, and Arg-353 in PDE9A2 form hydrogen bonds with one another and contribute to the major force for formation of the dimer. These residues correspond, respectively, to Tyr-223, Asn-224, Asp-225, Asn-231, and Arg-261 in PDE4D2. However, the dimers of PDE9A2 do not further associate into a tetramer, but PDE4D2 does. The Sephacryl S300 gel filtration shows a peak position equivalent to a molecular weight for a dimer of the PDE9A2 catalytic domain, but further study is needed to identify the biologically active oligomeric form of PDE9. Nevertheless, the identification of the dimerization interface of PDE9 in the crystal state probably rules out the potential role of the C-terminal residues in dimerization.
Fig. 2.
Ribbon presentation of dimers of PDE9A2 (A) and PDE4D2 (B) are shown, and the interface of the PDE9A2 dimer is shown in C.
Metal Binding. At the bottom of the active site, the two strongest peaks ≈10 and ≈6 times the background in the omitted (2Fo - Fc) map were assigned, respectively, as zinc and magnesium for purposes of structural refinement. Although the metal assignment is supported by the comparable B factors of the two metal ions with protein residues in the refinement (Table 1), the nature of the physiological ions is unknown. Although the first metal is probably a zinc ion as identified in PDE4 (19), it remains uncertain what is the identity of the second metal ion and whether different PDE families have different catalytic metal ions. Biochemical studies showed that manganese activated PDE9A twice as much as magnesium or calcium did (23, 28), and zinc was proposed to be the catalytic metal in the PDE5 family (29). Our structural study showed that the two metal ions in PDE9A2 occupy the same location as in the PDE4 and PDE5 structures (16, 19, 21). The first metal ion, presumably zinc, forms six coordinations with His-256, His-292, Asp-293, Asp-402, and two waters in an octahedral configuration. The second metal ion also forms an octahedron with Asp-293 and five water molecules.
Enzyme Activity. The catalytic domain of PDE9A2 has Km of 139 ± 18 nM and Vmax of 1.53 ± 0.24 μmol·min-1·mg-1 for cGMP, which are comparable with Km of 70–390 nM and Vmax of 0.96–4.9 μmol·min-1·mg-1 reported for the various isoforms of full-length PDE9 (22, 23, 28). The IC50 value for nonselective inhibitor 3-isobutyl-1-methylxanthine (IBMX) is ≈500 μM for the PDE9A2 catalytic domain, also consistent with IC50 = 230 μM for the IBMX inhibition of the full-length PDE9A1 (23). Thus, our data suggest that the N-terminal regulatory domain of PDE9 may not have a significant impact on the enzymatic activities nor potency of IBMX inhibition. The PDE9A2 catalytic domain has Km of 181 ± 17 μM and Vmax of 0.08 ± 0.015 μmol·min-1·mg-1 for cAMP, ≈1,000 and 20 times worse than cGMP, thus confirming that PDE9 is cGMP-specific.
The sequence alignment shows that PDE9 and PDE8 have a tyrosine (Tyr-424 in PDE9A2) involved in interactions with IBMX. Because PDE9 and PDE8 are insensitive to the inhibition of IBMX (6) and this tyrosine corresponds to a phenylalanine in the other PDEs that are inhibited by IBMX, it was interesting to know whether the tyrosine to phenylalanine mutation sacrifices the IBMX inhibition. We subcloned and purified the Y424F mutant of the PDE9A2 catalytic domain. This mutant has a Km of 135 ± 21 nM and Vmax of 1.59 ± 0.14 μmol·min-1·mg-1 for cGMP, and an IC50 of 450 μM for IBMX. These enzymatic data are comparable with those of the wild type of the PDE9A2 catalytic domain, thus suggesting that Tyr-424 does not play a major role in IBMX binding. In the crystal of PDE9A2, the hydroxide oxygen Tyr-424 is in a hydrogen bond distance (3.2 Å) to N3 of xanthine of IBMX in molecule B, but interacts with the isobutyl group in molecule A. Modeling of the Y424F mutation shows that Phe-424 will be at a position perpendicular to the xanthine ring and thus forms hydrophobic interactions with IBMX. Because both the wild type and the Y424F mutant interact with IBMX, the mutation may not have a significant impact on the inhibitor binding. Thus, the nonsensitivity of IBMX inhibition must be determined by other elements, which are yet to be further explored.
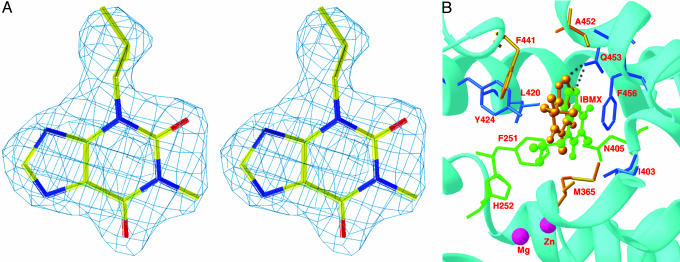
Common and Distinct Interactions of IBMX in the PDE9A2 Dimer. The electron density maps that were calculated from the PDE9A2 structure when IBMX was omitted reveal the specific binding of IBMX at the active site with interactions similar to those in the PDE4 and PDE5 structures (Fig. 3). The dimer of PDE9A2 contains two isolated sites for the binding of each IBMX. The xanthine ring of IBMX stacks against Phe-456 of PDE9A2 on one side and contacts residues Leu-420 and Tyr-424 on another side. The N7 atom of xanthine forms a hydrogen bond with Ne2 of Gln-453. These interactions of IBMX within each monomer are conserved in the PDE9A2 dimer. On the other hand different orientations and interactions of two IBMXs in the PDE9A2 dimer are observed. The xanthine rings of the two respective IBMX molecules have a rotation difference of ≈50° about an axis perpendicular to the xanthine ring, whereas the stacking against Phe-456 is conserved in the dimer. As a result, the atoms of IBMX have translational shifts of 0.6–4.7 Å, and IBMX interacts uniquely with residues of Phe-251, His-252, and Asn-405 in one monomer and Met-365 and Phe-441 in another. In addition, isobutyl groups of the IBMX molecules have different orientations in the two monomers and interact with different residues: His-252 and Tyr-424 in one monomer and Ile-403 and Phe-441 in the other. In contrast to the orientation differences of IBMX in the dimer, two monomers of PDE9A2 protein have similar conformations, as shown by an rms deviation of 0.36 Å from the superposition of Cα atoms of the entire catalytic domain. In addition, the graphic display of the super-position shows very similar conformations for the side chains of the IBMX-binding residues.
Fig. 3.
IBMX binding. (A) Stereoview of the electron density for IBMX bound to PDE9A2. The (2Fo - Fc) map was calculated from the structure omitting IBMX and contoured at 2.0σ. (B) IBMX binding to the active site of PDE9A2. The xanthine group stacks against Phe-456 and forms a hydrogen bond with Gln-453. The green balls-sticks represent IBMX in molecule A. The golden balls-sticks represent IBMX from molecule B, which is superimposed over molecule A. The dotted line represents a hydrogen bond. The PDE9A2 residues that interact with IBMX in both monomers are shown in blue sticks, whereas the unique residues interacting with IBMX in one monomer are drawn in green sticks (monomer A) and golden sticks (monomer B).
The IBMX binding to the catalytic domain of PDE9A2 in the crystal state is apparently inconsistent with the enzymatic data that IBMX does not effectively inhibit the PDE9A2 catalytic domain and full-length PDE9s (22, 23). A possible interpretation may be that 2 mM IBMX, ≈10 times the protein concentration of PDE9A2 in the crystallization drop, may be sufficient for high occupancy of the binding pocket. A similar case has been observed in the crystal structures of cyclophilin in complex with the peptide substrates, where the peptides have dissociation constants at the millimolar level (30, 31). Another explanation may be that the tight binding of cGMP to PDE9A2 (Km = 70 nM) requires a higher concentration of IBMX for an effective inhibition.
Variation of IBMX Binding in the Structures of PDE4, PDE5, and PDE9. IBMX binds to the similar subpocket of the active sites of PDE4, PDE5, and PDE9, which was proposed as a common site for binding of nonselective inhibitors of PDEs (21). Two key factors for binding of IBMX are absolutely conserved in the three structures: stacking of xanthine against a phenylalanine (Phe-456 in PDE9A2) and a hydrogen bond between N7 of xanthine and the glutamine side chain (Gln-453 in PDE9A2). The hydrophobic interactions of IBMX with Leu-420 and Tyr-424 of PDE9A2 are relatively conserved, as seen from the mutations of leucine to valine/isoleucine and tyrosine to phenylalanine across PDE families.
On the other hand, the orientation of IBMX varies significantly in the three structures. The xanthine ring of IBMX in monomer A of the PDE9A2 dimer shows ≈15° rotation variation and 1–1.5 Å translational shift from PDE4D2 and PDE5A1, whereas IBMX in monomer B shows ≈2 Å of translation shift from the structures of PDE4D2 and PDE5A1 (21). The explanation of the orientation variation of IBMX binding is not clear. A plausible interpretation may be that the much larger volume of the active sites (≈450 Å3) than IBMX (≈180 Å3) would allow variable orientations of IBMX under conservation of the hydrogen bond and stacking, because hydrophobic interaction is nonspecific and one hydrogen bond is not sufficient to lock the orientation of IBMX. Another possibility is that the subtly different active sites of the PDE families require adjustment of the IBMX orientation for the best fit.
The binding strength of IBMX in various PDEs may be impacted by the conformation variation of the glutamine side chain (Gln-369 in PDE4D2, Gln-817 in PDE5A1, and Gln-453 in PDE9A2) that forms the hydrogen bond with N7 of xanthine. In PDE5A, the side-chain conformation of Gln-817 is fixed by the hydrogen bond to Gln-775, which in turn hydrogen-bonds to the carbonyl oxygen of the backbone Ala-767 and Ne1 of Trp-853. In PDE4D2, the configuration of the Gln-369 side chain is also fixed by its hydrogen bonding to Oh1 of Tyr-329, although its side chain amide is ≈180° different from that of Gln-817 in PDE5A1 (21). However, Gln-453 in PDE9A2, which has a similar conformation to Gln-369 in PDE4D2, is not hydrogen-bonded with Tyr-329-equivalent residue Ala-413 but a nearby surrogate Glu-406. Although Glu-406 forms a hydrogen bond with Ser-486, the pocket appears to have more room in PDE9A2 for fluctuation of the Gln-453 conformation. As a result, the rigidity of the Gln-453 conformation is weakened, and its capacity for binding of IBMX is reduced. In addition, the carbonyl oxygen O6 of xanthine forms a hydrogen bond with Ne2 of Gln-817 in PDE5A or a water molecule bound to Asn-321 in PDE4D2 but no obvious interactions with protein residues or solvents in PDE9A2. Therefore, the absence of the interactions may be a factor to account for the weak binding affinity of IBMX for PDE9A2.
Implication for Substrate Specificity. Because IBMX and other inhibitors do not significantly change the overall conformations of PDE4 and PDE5 (16–21), we believe that the PDE9A2–IBMX structure likely represents the native conformation of PDE9A2 and thus is useful for discussions of substrate specificity, a central issue of enzymology. The classic lock–key and induced-fit models both emphasize that a substrate fits the exact conformation of the enzyme whether or not the active site conformation is native or induced by substrate binding (32, 33). On the basis of this conformation determination for substrate specificity, one would predict that the active site of PDE9 resembles the cGMP-specific PDE5 more than it does the cAMP-specific PDE4. However, the structural work in this article shows an opposite result. The core catalytic domain of PDE9A2 (residues 207–495) has identity of 86 and 85 residues to PDE4D2 and PDE5A1, respectively. The structural superposition of the PDE9A2 core catalytic domain over those of PDE4 and PDE5 yields rms deviations of 1.5 and 2.8 Å, respectively, for 288 PDE4D2 and 272 PDE5A1 residues. This comparison shows that the core catalytic domain of PDE9A2 has a conformation closer to PDE4D2 than PDE5A1 and thus does not apparently obey the prediction from the lock–key and induced-fit theories. However, the superposition between the PDE9A2 and PDE5A1 catalytic domains, after exclusion of the H and M loops, yields an rms deviation of 1.7 Å for 243 residues of PDE5A1. This finding implies that the H and M loops do not play critical roles in the specificity of nucleotide binding, although they are involved in the binding of sildenafil and rolipram, respectively (18, 19). Thus, the substrate specificity in the PDE families must be determined by subtle conformation differences and amino acid variations of the remaining residues in the active sites, including the environment around the absolutely conserved glutamine (Gln-369 in PDE4D2, Gln-817 in PDE5A1, and Gln-453 in PDE9A2). We postulate that each PDE family has a characteristic recognition mechanism for substrate specificity, yet further structural and biochemical studies are needed.
Acknowledgments
We thank beamline X12C at National Synchrotron Light Source for the collection of diffraction data. This work was supported in part by National Institutes of Health Grants GM59791 (to H.K.) and P01 HL64943 (to R.W.C.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: IBMX, 3-isobutyl-1-methylxanthine; PDE, phosphodiesterase.
Data deposition: The coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1TBM).
References
- 1.Manganiello, V. C., Taira, M., Degerman, F. & Belfrage, P. (1995) Cell. Signalling 7, 445-455. [DOI] [PubMed] [Google Scholar]
- 2.Müller, T., Engels, P. & Fozard, J. R. (1996) Trends Pharmacol. Sci. 17, 294-298. [DOI] [PubMed] [Google Scholar]
- 3.Houslay, M. D., Sullivan, M. & Bolger, G. B. (1998) Adv. Pharmocol. 44, 225-343. [DOI] [PubMed] [Google Scholar]
- 4.Torphy, T. J. (1998) Am. J. Respir. Crit. Care Med. 157, 351-370. [DOI] [PubMed] [Google Scholar]
- 5.Corbin, J. D. & Francis, S. H. (1999) J. Biol. Chem. 274, 13729-13732. [DOI] [PubMed] [Google Scholar]
- 6.Soderling, S. H. & Beavo, J. A. (2000) Curr. Opin. Cell Biol. 12, 174-179. [DOI] [PubMed] [Google Scholar]
- 7.Mehats, C., Andersen, C. B., Filopanti, M., Jin, S. L. & Conti, M. (2002) Trends Endocrinol. Metab. 13, 29-35. [DOI] [PubMed] [Google Scholar]
- 8.Houslay, M. D. & Adams D. R. (2003) Biochem J. 370, 1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Movsesian, M. A. (2000) Expert Opin. Invest. Drugs 9, 963-973. [DOI] [PubMed] [Google Scholar]
- 10.Truss, M. C., Stief, C. G., Uckert, S., Becker, A. J., Wafer, J., Schultheiss, D. & Jonas, U. (2001) World J. Urol. 19, 344-350. [DOI] [PubMed] [Google Scholar]
- 11.Liu, Y., Shakur, Y., Yoshitake, M. & Kambayashi, J. J. (2001) Cardiovasc. Drug Rev. 19, 369-386. [DOI] [PubMed] [Google Scholar]
- 12.Huang, Z., Ducharme, Y., MacDonald, D. & Robinchaud, A. (2001) Curr. Opin. Chem. Biol. 5, 432-438. [DOI] [PubMed] [Google Scholar]
- 13.Rotella, D. P. (2002) Nat. Rev. Drug Discovery 1, 674-682. [DOI] [PubMed] [Google Scholar]
- 14.Corbin, J. D. & Francis, S. H. (2002) Int. J. Clin. Pract. 56, 453-459. [PubMed] [Google Scholar]
- 15.Crowe, S. M. & Streetman, D. S. (2004) Ann. Pharmacother. 38, 77-85. [DOI] [PubMed] [Google Scholar]
- 16.Xu, R. X., Hassell, A. M., Vanderwall, D., Lambert, M. H., Holmes, W. D., Luther, M. A., Rocque, W. J., Milburn, M. V., Zhao, Y., Ke, H. & Nolte, R. T. (2000) Science 288, 1822-1825. [DOI] [PubMed] [Google Scholar]
- 17.Lee, M. E., Markowitz, J., Lee, J. O. & Lee, H. (2002) FEBS Lett. 530, 53-58. [DOI] [PubMed] [Google Scholar]
- 18.Sung, B. J., Yeon Hwang, K., Ho Jeon, Y., Lee, J. I., Heo, Y. S., Hwan Kim, J., Moon, J., Min Yoon, J., Hyun, Y. L., Kim, E., et al. (2003) Nature 425, 98-102. [DOI] [PubMed] [Google Scholar]
- 19.Huai, Q., Wang H., Sun, Y., Kim, H. Y., Liu, Y. & Ke, H. (2003) Structure (London) 11, 865-873. [DOI] [PubMed] [Google Scholar]
- 20.Huai, Q., Colicelli, J. & Ke, H. (2003) Biochemistry 42, 13220-13226. [DOI] [PubMed] [Google Scholar]
- 21.Huai, Q., Liu, Y., Francis, S. H., Corbin, J. D. & Ke, H. (2004) J. Biol. Chem. 279, 13085-13101. [DOI] [PubMed] [Google Scholar]
- 22.Soderling, S. H., Bayuga, S. J. & Beavo, J. A. (1998) J. Biol. Chem. 273, 15553-15558. [DOI] [PubMed] [Google Scholar]
- 23.Fisher, D. A., Smith, J. F., Pillar, J. S., St. Denis, S. H. & Cheng, J. B. (1998) J. Biol. Chem. 273, 15559-15564. [DOI] [PubMed] [Google Scholar]
- 24.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 25.Navaza, J. & Saludjian, P. (1997) Methods Enzymol. 276, 581-594. [DOI] [PubMed] [Google Scholar]
- 26.Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 27.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 28.Wang, P., Wu, P., Egan, R. W. & Billah, M. M. (2003) Gene 314, 15-27. [DOI] [PubMed] [Google Scholar]
- 29.Francis, S. H., Colbran, J. L., McAllister-Lucas, L. M. & Corbin, J. D. (1994) J. Biol. Chem. 269, 22477-22480. [PubMed] [Google Scholar]
- 30.Zhao, Y. & Ke, H. (1996) Biochemistry 35, 7356-7361. [DOI] [PubMed] [Google Scholar]
- 31.Zhao, Y. & Ke, H. (1996) Biochemistry 35, 7362-7368. [DOI] [PubMed] [Google Scholar]
- 32.Jorgensen, W. L. (1991) Science 254, 954-955. [DOI] [PubMed] [Google Scholar]
- 33.Monod, J., Wyman, J. & Changeux, J. P. (1965) J. Mol. Biol. 12, 88-118. [DOI] [PubMed] [Google Scholar]
- 34.Carson, M. (1991) J. Appl. Crystallogr. 24, 958-961. [Google Scholar]