Abstract
Class V myosin (myosin-V) was first found as a processive motor that moves along an actin filament with large (≈36-nm) successive steps and plays an important role in cargo transport in cells. Subsequently, several other myosins have also been found to move processively. Because myosin-V has two heads with ATP- and actin-binding sites, the mechanism of successive movement has been generally explained based on the two-headed structure. However, the fundamental problem of whether the two-headed structure is essential for the successive movement has not been solved. Here, we measure motility of engineered myosin-V having only one head by optical trapping nanometry. The results show that a single one-headed myosin-V undergoes multiple successive large (≈32-nm) steps, suggesting that a novel mechanism is operating for successive myosin movement.
Class V myosin (myosin-V) is a linear motor protein that moves unidirectionally along an actin filament with successive large (≈36-nm) steps (1–3). Myosin-V has two heads, each of which consists of a motor domain and a long neck domain. Based on this structural feature, a “hand-over-hand” mechanism has been proposed to explain a unidirectional and successive large stepping of myosin-V (4). The proposed mechanism is that the trailing head detaches upon ATP binding, and rapidly moves forward by ≈72 nm (5). Upon reattachment with actin, it becomes the new leading head. The partner and former leading head remains attached with the same actin monomer. Based on a “lever-arm tilting model” (6), it has been proposed that the tilting of the long neck domain of the attached leading head biases Brownian motion of the trailing head forward (7). Recently, it was demonstrated by using a single-molecule fluorescence polarization technique that the orientation of fluorophores on calmodulin light chain attached to the neck domain of myosin-V indeed alters before and after mechanical steps (8). This finding is consistent with the lever-arm tilting model. However, it is still unclear whether the observed tilting of the neck domain may not be the cause of steps but rather the effect of steps generated by other mechanisms. This uncertainty is because the time resolution of the fluorescence polarization was not sufficient to detect the change directly coupled to these steps.
On the other hand, we have recently reported that a short (≈4-nm) neck mutant of myosin-V can also generate successive large (≈35-nm) steps (9). That observation poses a challenge to the lever-arm tilting model because the neck domain is too short to lead the trailing head to the next target site ≈72 nm distant from the original site, especially at high loads. In support of this finding, class-VI myosin, which has a naturally very short (≈4-nm) neck domain, has been found to move processively with large steps as well as myosin-V (10, 11).
A central issue for the multiple successive large steps according to the hand-over-hand mechanism based on the lever-arm tilting model is that the two-headed structure is indispensable. Previous studies (7, 12) that measured the mechanical properties of myosin-V subfragment 1 in vitro failed to observe successive steps. In the present study, we used myosin cofilament technique, in which one-headed myosin-V smooth muscle myosin rod chimeras (M5SH) were incorporated into myosin rod filaments, allowing us to determine the orientation. As a result, we succeeded in observing successive multiple large steps of one-headed myosin-V.
Materials and Methods
Proteins. Actin and myosin rod were obtained from rabbit skeletal muscle and purified (13). To visualize under optical microscope, actin filaments were labeled with Alexa 647 phalloidin (Molecular Probes), and myosin rods with Cy5 (Amersham Biosciences, Piscataway, NJ). Recombinant calmodulin from Xenopus oocytes was expressed in Escherichia coli as described (14). α-Actinin was obtained from chicken gizzard and purified (15).
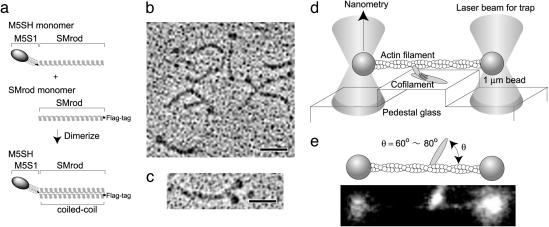
Recombinant M5SH and Two-Headed Myosin-V (M5DH). The myosin smooth muscle rod (SMrod) and myosin-V subfragment 1 (M5S1) SMrod chimera constructs (Fig. 1a) were produced as follows. A cDNA fragment of chicken gizzard SMrod encoding a part of the rod, Thr-1109/Arg-1936, was linked to the cDNA encoding the entire motor domain plus the IQ domain of myosin-V (Met-1/Gly-924) and inserted into a pFastBac baculovirus transfer vector. This expression vector was used to express M5DH and M5SH. A FLAG tag sequence was introduced at the 3′ side of (M5S1 SMrod) SMrod cDNA fragment encoding Thr-1109/Arg-1936 and then introduced into a pFastBac baculovirus transfer vector. M5SH was produced by coinfecting Sf9 cells with the viruses expressing M5S1 SMrod, FLAG SMrod, and calmodulin, respectively (Fig. 1a). The FLAG affinity chromatography completely eliminated M5DH having no FLAG tag. The purified sample was composed of M5SH monomer/SMrod (M5SH) heterodimers and SMrod homodimers. M5DH was produced by coinfecting Sf9 cells with M5S1 SMrod and calmodulin expressing viruses, respectively, and were purified as described (9).
Fig. 1.
Structure of M5SH and measurements of mechanical steps. (a) Schematic diagram of M5SH. M5SH was obtained as a heterodimer of chicken SMrod monomers with and without the myosin-V head connected to the N terminus. FLAG was tagged at the C terminus of SMrod. M5S1, One-headed M5S1. The M5SH and SMrod mixture was obtained by FLAG affinity chromatography. (b) A rotary-shadowed electron micrograph of M5SH and SMrod mixture. The micrograph of M5DH is shown as a control in c. (Scale bars, 40 nm.) (d) Schematic drawing of optical trapping nanometry. Two ends of an actin filament were attached to optically trapped beads and the suspended actin filament was brought into contact with a sparse cofilament fixed on the pedestal. Steps were determined by measuring the deflection of a bead with a quadrant photodiode. (e) Angle between an actin filament and a cofilament. We measured the angles between the vectors along a cofilament toward its center and along an actin filament toward its pointed end. The polarity of actin filaments was determined according to the direction of steps developed by myosin. The angle was set to be 70 ± 10°. (Lower) A photograph of fluorescence images of the actin filament and the cofilament.
Cofilaments. M5SHs were copolymerized into long (6.6 μm on average) filaments with rabbit skeletal muscle myosin rods without heads. The total concentration of the proteins was set to be 0.15 μM, and the molar ratio of the M5SH to myosin rod in the mixture was adjusted to be 1:2,000 so that only a small number of M5SHs were incorporated into a cofilament (9, 16). The presence of SMrods in the preparation was not a problem in this assay, because M5SH/SMrod cofilaments contained a large excess of skeletal muscle myosin rods. M5DH/SMrod cofilaments were prepared the same way. To visualize cofilaments, a small amount of Cy5-labeled myosin rods were involved.
Single-Molecule Mechanical Assay. To attach beads to the two ends of an actin filament, the surface of polystyrene latex beads (0.945 μm in diameter) was coated with α-actinin by means of (1-(3-dimethylaminopropyl)-3-ethyl-carbodiimide hydrochloride; see Supporting Text, which is published as supporting information on the PNAS web site). Cofilaments applied to a flow chamber with pedestals on a glass slide surface were adsorbed onto the pedestal surface (9, 16). The pedestal surface was coated with casein to prevent α-actinin-coated beads from nonspecifically binding to the glass surface. An assay buffer (120 mM KCl buffer containing 1–1,000 μM ATP, 0.2 mg/ml calmodulin) containing Alexa 647 phalloidin-labeled actin filaments and α-actinin-coated beads was introduced into the chamber. Fig. 1d presents a schematic drawing of the experimental setup for optical trapping nanometry. Actin filament and myosin cofilament were visualized under an epifluorescence microscope. Two ends of an actin filament were attached to optically trapped beads through α-actinin, and the suspended actin filament was brought into contact with a cofilament on the pedestal. Angles between the actin filament and cofilament were chosen to be 70 ± 10° (Fig. 1e). This choice was critical for observing successive steps (Fig. 5, which is published as supporting information on the PNAS web site). A bright image of a bead, which was captured by optical tweezers and illuminated by a halogen lamp, was projected onto a quadrant photodiode detector, and displacement of the bead was determined by nanometer accuracy (16–18). The assay was carried out at 15°C. To reduce photobleaching, an oxygen scavenger system was added to the assay buffer (13).
Results
Construction of M5SH and Preparation of M5SH Cofilament. M5SH was produced by coexpressing a construct having the head domain of myosin-V heavy chain plus SMrod, along with a headless SMrod (Fig. 1a). Fig. 1b shows a rotary-shadowed electron micrograph of M5SH molecules. M5SHs and headless SMrods are seen. M5DH (Fig. 1c) was not found against the 200 molecules observed.
By using the purified M5SH, we examined whether an M5SH can produce multiple successive steps. To address this question, we used optical trapping nanometry and performed a single-molecule mechanical assay (Fig. 1d and refs. 16–18). A key issue is to ensure that the mechanical events produced by the preparation are actually exerted by a single M5SH. To achieve this end, we used a cofilament assay by which we directly determined the number of myosin molecules in the cofilament and the number of nucleotide-binding sites (thus the number of myosin heads).
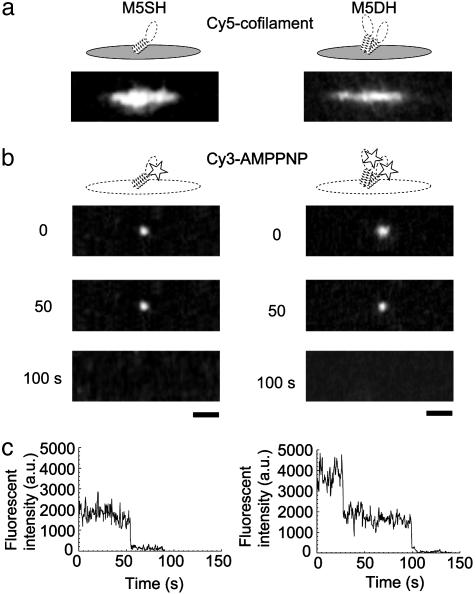
Estimation of the Number of M5SH Molecules in a Cofilament. We determined the number of M5SH molecules in a cofilament by observing the fluorescent spots resulting from the binding of f luorescent nucleotide analogue Cy3-adenosine 5′-[β,γ-imido]triphosphate (AMP-PNP) to myosin-V heads, which was hardly hydrolyzed by myosin and thus stably binds to myosin heads (Fig. 2). The experiments were performed at the conditions that the concentration of Cy3-AMP-PNP loaded on the cofilaments was 1 μM and the fluorescence intensity from Cy3-AMP-PNP bound to cofilaments was measured within 10 min after washing out free Cy3-AMP-PNP. The dissociation rate and constant (1/122 min-1 and 144 nM, respectively; Fig. 6, which is published as supporting information on the PNAS web site) give the probability that myosin heads bind Cy3-AMP-PNP at the experimental conditions as 0.92 × 0.87 = 0.80. Because the illumination time for measuring the fluorescence (<5 sec) was much shorter than the photobleaching time (54 sec, n = 89), the effect of photobleaching was negligibly small.
Fig. 2.
Measurements of the number of myosin heads in cofilaments. (a) Fluorescence images of cofilaments labeled with Cy5, including M5SH (Left) and M5DH (Right). (b) Fluorescence images of Cy3-AMP-PNP bound to M5SH (Left) and M5DH (Right) in cofilaments, observed after continuous illumination for 0, 50, and 100 sec. (Bars, 2 μm.) (c) The time courses of fluorescence intensity for M5SH (Left) and M5DH (Right).
Fig. 2b shows fluorescence of Cy3-AMP-PNP bound to M5SH (Left) and M5DH (Right) cofilaments. For M5SH cofilaments, 89 fluorescent spots due to bound Cy3-AMP-PNP were observed in 63 of 151 cofilaments, and the average number of spots per cofilament was 0.59 ± 0.85 (mean ± SD). Considering the dissociation rate and constant, the average number of myosin heads per cofilament is 0.74 (= 0.59/0.80). The number of myosin molecules per cofilament, determined by monitoring the number of fragments of fluorescently labeled actin filament bound to myosin heads in cofilaments in the absence of nucleotide, was 0.63 ± 0.77 (n = 52; Table 1). Thus, the number of myosin molecules in cofilaments determined by the two distinct methods agrees well.
Table 1. Summary of experiments.
| M5SH | M5DH | |
|---|---|---|
| No. of myosin molecules/cofilament | 0.74 ± 1.1 (n = 151)* | 0.96 ± 1.2 (n = 89)* |
| 0.63 ± 0.77 (n = 52)† | ||
| No. of cofilaments tested | 220 | 144 |
| No. of cofilaments having myosin molecules | 115 (115)‡ | 92 (89)‡ |
| No. of cofilaments interacting with actin filaments | 30 | 18 |
| No. of cofilaments showing successive steps | 22 | 13 |
| Favorable angles for successive steps, ° | ≈70 | ≈45 |
| Step size of forward successive steps, nm | 32 ± 16 (n = 182) | 34 ± 15 (n = 217) |
| Maximum force,§ pN | 0.95 ± 0.24 (n = 89) | 2.4 ± 1.4 (n = 75) |
All ranges are SD.
Obtained from fluorescence of Cy3-AMP-PNP bound to myosin heads in coflaments
Obtained from the no. of short actin fragments bound to myosin molecules in cofilaments
Obtained from fluorescence of Cy3-AMP-PNP bound to myosin heads in cofilaments. Values in parentheses are obtained from the Poisson distribution based on the average no. of myosin molecules in cofilaments as 220 × [1 - [exp(-0.74)]
Maximum forces when myosin heads produced multiple steps successively
Next, we investigated photobleaching characteristics of the fluorescent spots in M5SH cofilament, verifying that the observed fluorescent spots were due to single Cy3-AMP-PNP molecules (Fig. 2 b Left and c Left). All observed spots (n = 89) photobleached in a single-step fashion, indicating that the spots were due to single Cy3-AMP-PNP molecules bound to myosin heads. On the other hand, a majority of the spots (22 of 30) in the cofilaments containing M5DH photobleached in a two-step fashion (Fig. 2 b Right and c Right). From the fraction of M5DH photobleaching in a two-step fashion, the probability (p) that myosin heads bind to fluorescent Cy3-AMP-PNP was calculated to be 0.85 by using an equation, p2: [p2 + 2p(1 - p)] = 22:30. This value is consistent with that (0.80) determined above based on the dissociation rate and constant. The average number of M5SH per cofilament is smaller than one but some of cofilaments should contain more than one M5SH. However, they distribute over long (5–8 μm) cofilaments, so that the probability that more than one M5SH simultaneously interact with the actin filament is negligibly small (see Discussion for details).
Steps of Single M5SHs. Steps of single M5SHs were measured by optical trapping nanometry (see Materials and Methods and Fig. 1d). Cofilaments were adsorbed on the surface of a pedestal made on a quartz glass surface. Long cofilaments allowed us to easily find the location of M5SHs. Displacements due to actomyosin interactions were determined by monitoring deflection of a bead by a quadrant photo detector with nanometer accuracy (16–18). The interaction was detected only when the myosin heads were incorporated into the cofilaments, and we never saw the interaction of actin with cofilaments without the myosin head nor with the glass surface.
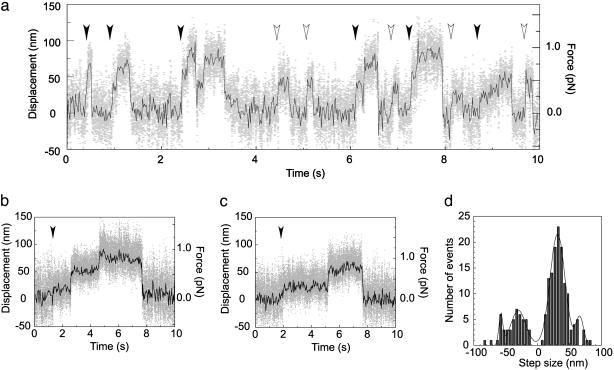
Typical traces of the time courses of displacements caused by single M5SHs at 100, 10, and 2 μM ATP concentrations are shown in Fig. 3 a–c, respectively. The displacements developed in a successive multistep fashion (successive steps, Fig. 3 a–c, filled arrowheads), although the displacements sometimes developed in a single-step fashion (nonsuccessive steps, Fig. 3a, open arrowheads). The number of successive steps per displacement was two to four. M5DHs often showed larger displacements, including five to six steps (Fig. 7, which is published as supporting information on the PNAS web site), and the mean maximum force was approximately twice as large as that of M5SH (Table 1). Fig. 3d depicts a histogram of successive steps of M5SHs except the first steps. The step size was independent of the concentration of ATP. The major direction was denoted as “forward.” The forward steps fit to Gaussian distribution with major and minor values of 32 (SD, 16 nm) and 66 nm (SD, 8 nm), respectively. The small Gaussian is probably a result of two successive steps rapidly taking place within the temporal resolution of the experimental system (1.5 ms), because the mean size was twice as large. The step developed in the opposite direction as well. The step size was similar to that observed in the forward direction. The ratio of the number of steps in the forward and backward directions was 4:1. The size of the first steps of successive and nonsuccessive steps, respectively, cannot be determined directly for individual steps, because the actual start positions of steps are random due to Brownian motion of the beads (18). Therefore, the mean size for these steps was determined from a histogram of the step size measured based on the plateau positions after steps to be 25 nm (SD, 17 nm, n = 61) for the first steps and 25 nm (SD, 21 nm, n = 205) for the nonsuccessive steps. These values are similar to those of the first steps of M5DH (7, 9) and nonsuccessive steps of M5S1 (7, 12).
Fig. 3.
Mechanical steps. Typical traces of the time courses of steps of M5SH at 100 (a), 10 (b), and 2 (c) μM ATP. Gray dots, raw data; black lines, same data passed through a low-pass filter of 25-Hz bandwidth. Filled and open arrowheads show start positions of successive and nonsuccessive steps, respectively. (d) Histogram of step size of an M5SH. Histogram shows step size of successive steps except for first steps. Data were fit with four Gaussians (solid line, see text). Medium: 120 mM KCl/5 mM MgCl2/1 mM EGTA/0.2 mg/ml calmodulin/2–100 μM ATP/20 mM Hepes (pH 7.8). The medium contained 5 μM phalloidin to stabilize actin filaments and an oxygen scavenger system to reduce photobleaching (13). Trap stiffness, 0.02–0.025 pN/nm. Temperature, 15°C.
Mean dwell times between adjacent two steps of successive steps were 0.34 (n = 85), 1.2 (n = 109), and 1.8 sec (n = 66) at 2, 10, and 100 μM ATP, respectively. Mean dwell times for nonsuccessive steps were 0.36 (n = 140), 1.3 (n = 222), and 2.3 sec (n = 128) at the above ATP concentrations, respectively. The mean dwell times of successive and nonsuccessive steps both fit well with a monophasic Michaelis–Menten kinetics, suggesting that each step corresponds to a single ATP hydrolysis event. The Vmax and Km for successive steps were 3.7 s-1 and 30 μM, respectively. These kinetic parameters were obtained from experiments at 15°C because Alexa 647 phalloidin-labeled actin filaments could be more clearly observed at a lower temperature. Successive large steps of single M5SH molecules were observed at 25°C as well (data not shown), and the Vmax (10 s-1) was similar to that of M5DH previously obtained at 25°C (3, 9, 19). The Vmax (10 s-1) and Km (30 μM) obtained by this technique were respectively similar to those biochemically obtained in solution (20, 21).
Discussion
It is apparent that the basal mechanical characteristics such as step size, successive steps, and stepping kinetics of M5SH were essentially the same as those of M5DH. A critical argument is whether the observed successive stepping is due to the M5SHs but not the M5DHs. We observed >200 molecules under electron microscope, and no M5DH were observed. The result indicates that our preparation contained M5SH exclusively, and it is unlikely that the observed mechanical events are due to M5DH. However, we further evaluated our results quantitatively as follows and concluded that the observed steps were indeed due to single M5SHs.
For M5SH cofilaments: (i) 89 fluorescent spots derived from Cy3-AMP-PNP were observed for 63 of 151 cofilaments tested, and of the 63, 18 cofilaments showed multiple fluorescent spots; and (ii) all observed 89 spots photobleached in a single-step fashion. For cofilaments prepared in the same manner by using M5DH instead of M5SH: (i) 30 fluorescent spots were observed for 20 of 39 cofilaments; and (ii) 22 of 30 spots photobleached in a two-step fashion with the others in a one-step fashion. The probability (p) of the heads with bound florescent Cy3-AMP-PNP was obtained to be 0.80, independently from the kinetic analysis of Cy3-AMP-PNP binding to the heads (0.80), and from the photobleaching characteristics of M5DH cofilaments (0.85; see Results). Based on this value (p = 0.80), the fraction of M5DH having two, one and no bound nucleotide can be estimated to be 0.64 (= p2), 0.32 [= 2p(1 - p)] and 0.04 [= (1 - p)2], respectively. Because all 89 fluorescent spots we observed showed single-phase photobleaching, it is impossible that a significant number of M5DHs contributed to these fluorescent spots. Therefore, assuming that the “90th” spot shows a two-step photobleaching, the number of contaminant M5DHs can be estimated as follows.
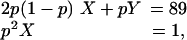 |
where X and Y are the numbers of M5DHs and M5SHs, respectively. Because p = 0.80, the equation determines the number for X and Y to be 1.6 and 111, respectively. In other words, the total number of contaminant M5DHs involved in 220 cofilaments used for the experiments would be, at the most, 2. Because successive large steps were observed for 22 of 220 cofilaments (Table 1), those observed in at least 20 cofilaments would be due to single M5SHs. It should be emphasized that this calculation is based on the assumption that the 90th spot photobleaches in a two-step fashion, but that possibility would be small. Therefore, it is highly unlikely that the observed mechanical activity is due to M5DH.
It is also unlikely that an actin filament interacted with more than one M5SH molecule at the same time. Because 18 of 151 cofilaments showed more than one fluorescent spot, the probability that more than one M5SH was involved in one cofilament is calculated as (18/151)/p = 0.15 after correction for the probability of the heads with bound Cy3-AMP-PNP. This finding agrees with the value calculated as 1 - (1 + 0.74) × exp(-0.74) = 0.17 based on the Poisson distribution by using the mean number of M5SH per cofilament (0.74). Based on this value (0.15), the mean distance between adjacent two M5SHs is calculated to be 3.0 μm if the M5SHs are randomly distributed on cofilaments of 6.6 μm in length. The probability that two M5SH are located within 120 nm in cofilaments, which is approximately twice as the distance from the tip of myosin head to the far end of S-2 of the M5SH molecule, is calculated as 0.15 × [1 - exp(-120/3000)] = 0.006. That is, the number of cofilaments of 220 cofilaments tested in which more than one M5SH is located within 120 nm is 0.006 × 220 = 1.3. We did not observe two fluorescent spots within the spatial resolution (≈500 nm) of the optical microscope for 151 cofilaments tested. The actin filament could not simultaneously interact with two myosin heads separated by 120 nm from each other when the actin filament was almost perpendicularly interacted with a cofilament. This estimation indicates that the actin filaments interacted with a single M5SH for at least 21 of 22 cofilaments, in which successive steps were observed.
This estimation was performed on the assumption that all myosin heads in cofilaments could interact with the actin filament. However, some of the myosin heads that faced the glass surface would be unavailable for interaction with actin filaments. Therefore, the possibility that more than one M5SH simultaneously interacted with the actin filament should be smaller.
Actin filaments interacted with 30 of 220 M5SH cofilaments tested (Table 1). Because 115 of 220 cofilaments are expected to include M5SH molecules, ≈25% (30 of 115 cofilaments) of M5SH bound cofilaments, including those that interacted with actin filaments and ≈70% (22 of 30 cofilaments) of the cofilaments interacting with actin filaments showed successive steps. These percentages are both similar to those for M5DHs. It is reasonable that only 25% of M5SH bound cofilaments interacted with actin filaments, because myosin heads are expected to radially project from the backbone of the cofilament, and hence the number of heads facing an actin filament would be limited.
The probability of generating successive steps depended on the angle between the actin filament and cofilament (Table 1, and Fig. 5, which is published as supporting information on the PNAS web site). The present constructs of the M5SH and the M5DH were myosin-V head SMrod chimeras and the head and backbone of cofilament was linked through a coiled coil. Short actin filaments attached to myosin heads on cofilaments did not rotate smoothly but remained stably at an angle, around which the probability of successive steps was high. Therefore, the orientation dependence of successive steps may be due to the interaction of the S-2 with the backbone of cofilament. S-2 protrudes from the filament backbone with a certain angle and therefore, only when actin filaments approach the heads with a favorable angle both M5SH and M5DH moved with successive steps.
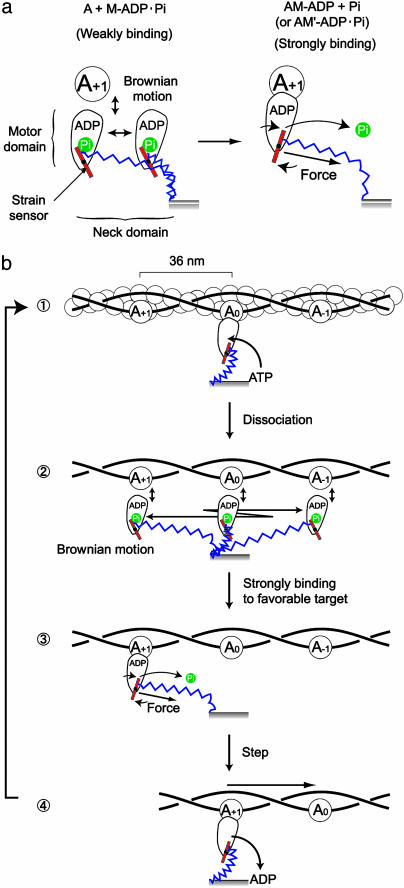
The results show that single M5SHs develop multiple successive large steps. How do they develop these successive large steps? The movement of M5DH is explained by the hand-over-hand mechanism based on the lever-arm tilting model, in which one head bound to the actin pivots the long neck domain connected with the head domain and biases Brownian motion of the detached partner head to lead it to the next helical pitch of an actin filament 36 nm apart (4). The step of 36 nm is expected to consist of the working stroke of ≈20 nm of the attached head and the Brownian motion of ≈16 nm of the partner head (7). However, it is apparent that this model could not be applied to the movement of one-head myosin-V because the net working stroke of one head should be 32 nm for producing successive 32-nm steps by one head. We propose a possible model based on a strain sensor mechanism (22) in Fig. 4. An actin hot spot that is evoked by an interaction with myosin heads and attracts myosin heads (11) or a toe-up-down mechanism that facilitates the landing of myosin heads on the forward target zone (23), which we have proposed, could be alternatives or cooperators of the strain sensor for favorable binding to the forward target zone. The present model does not require the rigid long lever arm for large processive steps and so could be applied to the movements of myosin-VI (10, 11) and truncation myosin-V mutants (9) with very short neck domains, if the coiled coil linking the two heads unzips and acts as a spring. Myosin-VI moves in the reverse direction to myosin-V (24). This reverse motion could be explained if the strain sensor is set to respond to force in the reverse direction; e.g., Pi binds to the left side of sensor (described by a red bar) and releases when the sensor is pulled in reverse and rotated counter clockwise (Fig. 4b). If the position of bound Pi relative to the sensor is responsible for the direction of movement, this model could explain our previous report (14) that replacement of the neck and converter (sensor) domains of myosin-V by those of myosin-VI does not change the direction of movement. This model is also consistent with a recent report (25) that when the neck domain is modified to be rotated in reverse, the direction of movement is reversed, because if the elastic domain is connected to other side of the sensor so that the sensor is rotated in reverse, the direction of movement would be reversed. Furthermore, our model is consistent with previous reports (12, 26, 27) that the step size is proportional to the neck domain length because the reach of the myosin head with the neck domain determines the step size.
Fig. 4.
A possible model of multiple successive large steps of M5SH based on a strain sensor (23). (a) Operation of a strain sensor. We postulate that the neck domain (blue) is elastic for stretching and bending, and the junction (red) between the motor and neck domains acts as a strain sensor that governs the transition from the weak to strong binding between actin and myosin, depending on the strain exerted on the sensor. (b) Steps of M5SH. ATP binds to the motor domain and the strain sensor is reset ( ). The motor domain dissociates from actin and diffuses back and forth over the actin filament derived by thermal stretching and bending of the neck domain (
). The motor domain dissociates from actin and diffuses back and forth over the actin filament derived by thermal stretching and bending of the neck domain ( ). Because the Brownian motion of the motor domain would be much faster (microseconds; ref. 31) than that of the actin filament with the two ends attached to beads (hundred microseconds), the myosin head could repeat steps without diffusing away from the actin filament. When the motor reaches to the next actin helical pitch in the forward direction (A+1) and binds to it, the strain sensor is pulled by the neck domain and the motor domain strongly bind to actin (A+1) probably coupled to the release of phosphate (Pi) or the isomerization of myosin (AM-ADP Pi → AM′-ADP Pi; ref. 32) (
). Because the Brownian motion of the motor domain would be much faster (microseconds; ref. 31) than that of the actin filament with the two ends attached to beads (hundred microseconds), the myosin head could repeat steps without diffusing away from the actin filament. When the motor reaches to the next actin helical pitch in the forward direction (A+1) and binds to it, the strain sensor is pulled by the neck domain and the motor domain strongly bind to actin (A+1) probably coupled to the release of phosphate (Pi) or the isomerization of myosin (AM-ADP Pi → AM′-ADP Pi; ref. 32) ( ). The actin is then pulled while the stretched and bent neck domain is relaxed to be in an equilibrium state (
). The actin is then pulled while the stretched and bent neck domain is relaxed to be in an equilibrium state ( ). When the motor domain thermally moves in the opposite direction, the strain sensor does not operate so that the motor domain returns to the original position. As a result, the myosin head moves an actin in one direction. Chemical states are coupled to mechanical states to fit the biochemical data (20) as much as possible. The stretch of a spring in the neck domain is overdrawn to be more easily understood.
). When the motor domain thermally moves in the opposite direction, the strain sensor does not operate so that the motor domain returns to the original position. As a result, the myosin head moves an actin in one direction. Chemical states are coupled to mechanical states to fit the biochemical data (20) as much as possible. The stretch of a spring in the neck domain is overdrawn to be more easily understood.
What is the role of a two-headed structure? Our results indicate that the two-headed structure is not essential for the successive large step. However, the number of steps per displacement for M5SH (Fig. 3 a–c) was smaller than that of two-headed ones. We did not observe by fluorescence microscopy movement over a long (>200 nm) distance of GFP-fused M5SH along actin filaments fixed on a glass surface, although long distance movements of M5DH were observed (data not shown). Therefore, the coordination between two heads, e.g., the hand-over-hand mechanism (Fig. 8, which is published as supporting information on the PNAS web site), may help stabilizing the basal motility so that myosin-V travels a long distance in physiological circumstances. A similar argument has been advanced recently for conventional kinesin (28) and a one-headed kinesin superfamily (Unc104/KIF1A; refs. 29 and 30).
Supplementary Material
Acknowledgments
We thank our colleagues at the Single Molecule Process Project (International Cooperative Research Project), R. Cooke (University of California, San Francisco), and K. Iwasaki (Osaka University, Osaka) for valuable discussion.
Abbreviations: myosin-V, class V myosin; SMrod, myosin smooth muscle rod; M5S1, myosin-V subfragment 1 SMrod chimera; M5SH, one-headed myosin-V SMrod chimera; M5DH, two-headed myosin-V SMrod chimera; AMP-PNP, adenosine 5′-[β,γ-imido]-triphosphate.
References
- 1.Mehta, A. D., Rock, R. S., Rief, M., Spudich, J. A., Mooseker, M. S. & Cheney, R. E. (1999) Nature 400, 590-593. [DOI] [PubMed] [Google Scholar]
- 2.Sakamoto, T., Amitani, I., Yokota, E. & Ando, T. (2000) Biochem. Biophys. Res. Commun. 272, 586-590. [DOI] [PubMed] [Google Scholar]
- 3.Rief, M., Rock, R. S., Mehta, A. D., Mooseker, M. S., Cheney, R. E. & Spudich, J. A. (2000) Proc. Natl. Acad. Sci. USA 97, 9482-9486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker, M. L., Burgess, S. A., Sellers, J. R., Wang, F., Hammer, J. A., III, Trinick, J. & Knight, P. J. (2000) Nature 405, 804-807. [DOI] [PubMed] [Google Scholar]
- 5.Yildiz, A., Forkey, J. N., McKinney, S. A., Ha, T., Goldman, Y. E. & Selvin, P. R. (2003) Science 300, 2061-2065. [DOI] [PubMed] [Google Scholar]
- 6.Spudich, J. A. (1994) Nature 372, 515-518. [DOI] [PubMed] [Google Scholar]
- 7.Veigel, C., Wang, F., Bartoo, M. L., Sellers, J. R. & Molloy, J. E. (2002) Nat. Cell Biol. 4, 59-65. [DOI] [PubMed] [Google Scholar]
- 8.Forkey, J. N., Quinlan, M. E., Alexander Shaw, M., Corrie, J. E. & Goldman, Y. E. (2003) Nature 422, 399-404. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka, H., Homma, K., Iwane, A. H., Katayama, E., Ikebe, R., Saito, J., Yanagida, T. & Ikebe, M. (2002) Nature 415, 192-195. [DOI] [PubMed] [Google Scholar]
- 10.Rock, R. S., Rice, S. E., Wells, A. L., Purcell, T. J., Spudich, J. A. & Sweeney, H. L. (2001) Proc. Natl. Acad. Sci. USA 98, 13655-13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishikawa, S., Homma, K., Komori, Y., Iwaki, M., Wazawa, T., Iwane, A. H., Saito, J., Ikebe, R., Katayama, E., Yanagida, T. & Ikebe, M. (2002) Biochem. Biophys. Res. Commun. 290, 311-317. [DOI] [PubMed] [Google Scholar]
- 12.Purcell, T. J., Morris C., Spudich, J. A. & Sweeney, H. L. (2002) Proc. Natl. Acad. Sci. USA 99, 14159-14164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harada, Y., Sakurada, K., Aoki, T., Thomas, D. D. & Yanagida, T. (1990) J. Mol. Biol. 216, 49-68. [DOI] [PubMed] [Google Scholar]
- 14.Homma, K., Yoshimura, M., Saito, J., Ikebe, R. & Ikebe, M. (2001) Nature 412, 831-834. [DOI] [PubMed] [Google Scholar]
- 15.Craig, S. W., Lancashire, C. L. & Cooper, J. A. (1982) Methods Enzymol. 85, 316-321. [DOI] [PubMed] [Google Scholar]
- 16.Ishijima, A., Kojima, H., Funatsu, T., Tokunaga, M., Higuchi, H., Tanaka, H. & Yanagida, T. (1998) Cell 92, 161-171. [DOI] [PubMed] [Google Scholar]
- 17.Finer, J. T., Simmons, R. M. & Spudich, J. A. (1994) Nature 368, 113-119. [DOI] [PubMed] [Google Scholar]
- 18.Molloy, J. E., Burns, J. E., Kendrick-Jones, J., Tregear, R. T. & White, D. C. (1995) Nature 378, 209-212. [DOI] [PubMed] [Google Scholar]
- 19.Moore, J. R., Krementsova, E. B., Trybus, K. M. & Warshaw, D. M. (2001) J. Cell Biol. 155, 625-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De La Cruz, E. M., Wells, A. L., Rosenfeld, S. S., Ostap, E. M. & Sweeney, H. L. (1999) Proc. Natl. Acad. Sci. USA. 96, 13726-13731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trybus, K. M., Krementsova, E. & Freyzon, Y. (1999) J. Biol. Chem. 274, 27448-27456. [DOI] [PubMed] [Google Scholar]
- 22.Yanagida, T., Esaki, S., Iwane, A. H., Inoue, Y., Ishijima, A., Kitamura, K., Tanaka, H. & Tokunaga, M. (2000) Philos. Trans. R. Soc. London B 355, 441-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ali, M. Y., Homma, K., Iwane, A. H., Adachi, K., Itoh, H., Kinosita, K., Jr., Yanagida, T. & Ikebe, M. (2004) Biophys. J. 86, 3804-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wells, A. L., Lin, A. W., Chen, L. Q., Safer, D., Cain, S. M., Hasson, T., Carragher, B. O., Milligan, R. A. & Sweeney, H. L. (1999) Nature 401, 505-508. [DOI] [PubMed] [Google Scholar]
- 25.Tsiavaliaris, G., Fujita-Becker, S. & Manstein, D. J. (2004) Nature 427, 558-561. [DOI] [PubMed] [Google Scholar]
- 26.Warshaw, D. M., Guilford, W. H., Freyzon, Y., Krementsova, E., Palmiter, K. A., Tyska, M. J., Baker, J. E. & Trybus, K. M. (2000) J. Biol. Chem. 275, 37167-37172. [DOI] [PubMed] [Google Scholar]
- 27.Sakamoto, T., Wang, F., Schmitz, S., Xu, Y., Xu, Q., Molloy, J. E., Veigel, C. & Sellers, J. R. (2003) J. Biol. Chem. 278, 29201-29207. [DOI] [PubMed] [Google Scholar]
- 28.Inoue, Y., Iwane, A. H., Miyai, T., Muto, E. & Yanagida, T. (2001) Biophys. J. 81, 2838-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okada, Y. & Hirokawa, N. (1999) Science 283, 1152-1157. [DOI] [PubMed] [Google Scholar]
- 30.Tomishige, M., Klopfenstein, D. R. & Vale, R. D. (2002) Science 297, 2263-2267. [DOI] [PubMed] [Google Scholar]
- 31.Kinosita, K., Jr., Ishiwata, S., Yoshimura, H., Asai, H. & Ikegami, A. (1984) Biochemistry 23, 5963-5975. [Google Scholar]
- 32.Goldman, Y. E. (1987) Annu. Rev. Physiol. 49, 637-654. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.