Abstract
Heat shock factors (Hsfs) are important regulators of stress-response in plants. However, our understanding of Hsf genes and their responses to temperature stresses in two Pooideae cool-season grasses, Festuca arundinacea, and Lolium perenne, is limited. Here we conducted comparative transcriptome analyses of plant leaves exposed to heat or cold stress for 10 h. Approximately, 30% and 25% of the genes expressed in the two species showed significant changes under heat and cold stress, respectively, including subsets of Hsfs and their target genes. We uncovered 74 Hsfs in F. arundinacea and 52 Hsfs in L. perenne, and categorized these genes into three subfamilies, HsfA, HsfB, and HsfC based on protein sequence homology to known Hsf members in model organisms. The Hsfs showed a strong response to heat and/or cold stress. The expression of HsfAs was elevated under heat stress, especially in class HsfA2, which exhibited the most dramatic responses. HsfBs were upregulated by the both temperature conditions, and HsfCs mainly showed an increase in expression under cold stress. The target genes of Hsfs, such as heat shock protein (HSP), ascorbate peroxidase (APX), inositol-3-phosphate synthase (IPS), and galactinol synthase (GOLS1), showed strong and unique responses to different stressors. We comprehensively detected Hsfs and their target genes in F. arundinacea and L. perenne, providing a foundation for future gene function studies and genetic engineering to improve stress tolerance in grasses and other crops.
Keywords: heat shock factors (Hsfs), cold stress, heat stress, RNA-Seq, expression analysis, phylogenetic analysis, tall fescue, perennial ryegrass
Introduction
Climate change has resulted in harmful effects on plant growth (Hasanuzzaman et al., 2013; Miura and Furumoto, 2013). The environment in which a plant thrives is species-dependent, and as a result, each species has common and unique stress-response mechanisms (Yeh et al., 2012). Tall fescue (Festuca arundinacea) and perennial ryegrass (Lolium perenne) are two major species of forage and turf grasses, which are extensively planted in warm temperate to subtropical regions around the world because they are easy to plant, have better tolerance to abiotic stresses, and require low maintenance (Raeside et al., 2012; Huang et al., 2014). However, the regions that use tall fescue and perennial ryegrass experience high temperature over 38°C in the summer, and low temperature below −10°C in the winter almost every year, which greatly exceeds the growth temperature of these cool-season grasses, and reduces the forage yield and lawn quality. Accordingly, improving tall fescue and perennial ryegrass adaptability to high and/or low temperature can increase forage yield and lawn quality in regions where it is heavily planted.
Plants cope with adverse environmental changes by altering expression of stress-related genes (Krasensky and Jonak, 2012). Heat shock proteins (HSPs), which include HSP70 (DnaK), the chaperonins (GroEL and HSP60), HSP90, HSP100 (Clp), and the small HSP (sHSP) families (Wang et al., 2004), are important molecular chaperones, and their expression is induced by environmental stressors such as heat, waterlog, salinity, osmosis, cold, and oxidation (Timperio et al., 2008; Scharf et al., 2012; Hu et al., 2015). HSPs prevent aggregation of denatured proteins, and assist in folding of nascent polypeptides and refolding denatured proteins, as well as to re-solubilize aggregated denatured proteins (Wang et al., 2004; Timperio et al., 2008; Waters, 2013).
Transcription of HSPs is regulated by Heat shock factors (Hsfs) (Miller and Mittler, 2006; Scharf et al., 2012; Xue et al., 2014), which are transcriptional activators of heat shock genes. More recently, it has been shown that Hsfs not only regulate the expression of HSP genes, but also other Hsf genes, and genes encoding stress-induced metabolic enzymes, such as ascorbate peroxidase 2 (APX2), inositol-3-phosphate synthase 2 (IPS2), and galactinol synthase 1 (GOLS1) (Miller and Mittler, 2006; Scharf et al., 2012). Hsfs form homotrimers that binds to highly conserved heat shock elements (HSE; a palindromic motif of nGAAn) in the promoter region of target genes (Peteranderl et al., 1999). The plant Hsf gene family is divided into three subfamilies: HsfA, HsfB, and HsfC. Each subfamily is further divided into to different classes. All Hsf genes contain a DNA-binding domain (DBD), an oligomerization domain (HR-A and HR-B), and a nuclear localization sequence (NLS) (Scharf et al., 2012). Hsf A proteins contain an activator peptide motif (AHA motif) in the C-terminal domains (Döring et al., 2000; Kotak et al., 2004), and Hsf B proteins, except for Hsf B5, contain the tetrapeptide LFGV, which is suggested to be a repressor motif (Czarnecka-Verner et al., 2004; Ikeda and Ohme-Takagi, 2009).
Hsf family genes show high copy number across different model organisms. The smallest Hsf family that has been observed to date comprises of 18 or 19 Hsfs found in ricinus, vitis, citrus, and carica (Scharf et al., 2012). The largest number of Hsfs have been observed in wheat, which has 56 Hsf genes (Triticum aestivum) (Xue et al., 2014). Most Hsfs are present in both eudicots and monocots. However, Hsf B3 and Hsf B5 are confined to eudicots. The Hsf C group shows a substantially higher level of complexity in monocots, containing monocot-specific types C1b, C2a, and C2b.
Hsf A subfamily genes act as activators, and have been extensively characterized in various model plant species. All genes under the Hsf A1, A2, A3, and A4 category are involved in thermotolerance in plants (Doerks et al., 2002; Baniwal et al., 2007; Zhang et al., 2009; Scharf et al., 2012). Of the Hsf A1 members, HsfA1a functions as the main and irreplaceable regulator for acquired thermotolerance in tomato (Nishizawa-Yokoi et al., 2011); however, this is not the case in Arabidopsis thaliana (Lohmann et al., 2004). Transgenic A. thaliana plants overexpressing AtHsfA2 have increased higher thermotolerance, as well as an increased resistance to salt, osmosis, and oxidative stress (Ogawa et al., 2007; Zhang et al., 2009). Hsf A3 also participates in drought stress signaling that is dependent on DREB2A (dehydration-responsive element binding protein 2A) in Arabidopsis (Yoshida et al., 2008; Chen et al., 2010). Moreover, Hsf A9 regulates HSP expression during embryogenesis and seed maturation in sunflower (Helianthus annuus, Ha) and A. thaliana (Almoguera et al., 2002; Kotak et al., 2007), and is also involved in the hormonal control networks regulated by abscisic acid (ABA) and auxins (Carranco et al., 2010).
In contrast to the subfamily Hsf A, the subfamilies Hsf B and C have been less characterized. Most Hsf B members do not show activator function (Ikeda and Ohme-Takagi, 2009). However, Hsf B1 and Hsf A1a form a synergistic coactivator that regulates the different stages of tomato heat stress (HS) response (Bharti et al., 2004). Moreover, Hsf B1 cooperates with other transcriptional activators that control and/or restore expression of housekeeping genes during HS (Scharf et al., 2012). To date, the role of subfamily C Hsfs remains elusive.
Given the role Hsf family genes play in temperature-response in various plant species, our research group has been highly interested in determining whether Hsf family genes play a role in regulating heat and cold tolerance in grasses. However, there is no clear classification of Hsf genes and whether their expression changes under stress-response for tall fescue and perennial ryegrass. A comprehensive analysis of gene expression in the two species exposed to high or low temperature stress may allow for an improved understanding of their adaptability to temperature stress, and identify novel stress tolerance genes. Transcriptome analyses of tall fescue revealed that the heat-tolerant cultivar activated more genes compared to the heat-sensitive cultivar under heat stress, and the genes involving in sHSP, cell division and cell cycle genes showed increased expression in both cultivars (Hu et al., 2014). Previous studies have generated a single nucleotide polymorphism (SNP) library for perennial rygrass (Studer et al., 2012), and developed a reference transcriptome for multiple tissues from a highly inbred perennial ryegrass line (Farrell et al., 2014).
Tall fescue and perennial ryegrass belong to the subfamily Pooideae. The two species are likely closely related, as interspecific crosses occur naturally in the wild, and are employed in genetic breeding (Humphreys and Thomas, 1993; Yamada et al., 2005). To build a better reference for grass plants, we performed our experiments on the two species (population sample) instead of biological replicates of the same species.
In the present study, we generated transcriptomes of tall fescue and perennial ryegrass in response to high and low temperatures, and identified the genes closely related to temperature stress. We focused on Hsf genes in the two species, and classified their family members, predicted function, and analyzed the expression profile of Hsfs and their target genes under heat or cold stress. Our results could help elucidate the mechanism underlying the adaptability of tall fescue and perennial grass to temperature stress.
Materials and methods
Plant materials and temperature stress treatment
Tall fescue (Festuca arundinacea cv. Barlexas) and perennial ryegrass (Lolium perenne cv. Yatsyn) seeds were sowed in plastic pots (14 cm diameter, 25 cm length) filled with organic loam in March 2014, and seedlings were grown in a greenhouse at 18–25°C for 1 month in Chengdu (30.67°N, 104.06°E), Sichuan Province, China. The plants were clipped to a height of 8 cm, and watered by hand every 2 days. Prior to the experiment, the plants were transferred to a growth chamber set to a temperature and light cycle of 26/18°C (14 h day/10 h night) (Cool Daylight, 3070 Lm 85 Lm/W, Philips, Thailand), at relative humidity of 70%, and an irradiance of 200 mol·m−2s−1 (LI-6400/XT photometer, Li-Cor Inc., Lincoln, NE, USA). After 2 weeks, the samples from each species were divided into three parts and subjected to heat stress (HS) at 40°C, cold stress (CS) at −10°C, or maintained at the control temperature (22°C) for 10 h, with the same humidity and light conditions as above. After then, randomly selected leaves from at least 100 plants in each group were pooled, snap-frozen, and stored in liquid nitrogen. These samples were labeled FaCK (tall fescue in control temperature), FaHeat (tall fescue under HS), FaCold (tall fescue under CS), LpCK (perennial ryegrass in control temperature), LpHeat (perennial ryegrass under HS), and LpCold (perennial ryegrass under CS). Three independent sample pools from each treatment seedlings were collected by the same sampling method, snap-frozen immediately, and preserved in liquid nitrogen until use.
Illumina deep sequencing
Total RNA samples were prepared using Trizol™ reagent (Invitrogen, Carlsbad, CA, USA), and the mRNA library was constructed using the Truseq™ RNA Sample Prep Kit (Illumina, San Diego, CA, USA) following the manufacturer's instructions. Samples were then sequenced on Illumina HiSeq™ 2000 (Illumina) in BGI (Shenzhen, China), and each sample yielded more than 4 Gb of data.
De novo assembly and functional annotation
The raw reads from RNA-Seq were filtered to increase the quality of the data using the internal filter_fq software of BGI (Shenzhen, China). De novo assembly of the transcriptome of tall fescue and perennial ryegrass were conducted using Trinity (release-20130225) (http://trinityrnaseq.sourceforge.net/) under default parameters (Grabherr et al., 2011). The quality of the assembly was determined using length distribution, N50 number, average length, max length, and contig number at different length intervals. The assembled unigene sequences were aligned to protein databases including NR (release-20130408), Swiss-Prot (release-2013_03), the Kyoto Encyclopedia of Genes and Genomes (KEGG, Release 63.0), Cluster of Orthologous Groups of proteins (COG) (release-20090331) by blastx (e < 0.00001), nucleotide database NT (release-20130408) by blastn (e < 0.00001) (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and the Gene Ontology (GO) (http://www.blast2go.com/b2ghome) (Conesa et al., 2005). The resulting unigenes were functionally annotated, and assigned the proteins with the highest sequence similarity. GO enrichment analysis was performed by using the GO-TermFinder (v0.86) software (http://search.cpan.org/dist/GO-TermFinder/). All RNA-Seq reads were deposited to the Sequence Read Archive database (http://www.ncbi.nlm.nih.gov/Traces/sra/) under the accession numbers SRP059410 (tall fescue) and SRP059405 (perennial ryegrass).
Differential expression analysis
Unigene expression was calculated using the fragments per kilobase of exon model per million (FPKM) by the SOAP software (http://soap.genomics.org.cn/soapaligner.html) (Mortazavi et al., 2008). Differential expression levels were calculated according to the FPKM of each transcript between libraries (Audic and Claverie, 1997). Significant differences in transcript expression levels were determined by setting the thresholds for false discovery rate as “(FDR) < 0.001” and “|log2 Ratio| ≥ 1,” where ratio indicates the fold-change of FPKM values for the treatment and control samples. Venn diagrams for expression differences were generated online (http://bioinfogp.cnb.csic.es/tools/venny/index.html), and heatmaps were created by using the HemI 1.0 software (Deng et al., 2014).
Classification of Hsf proteins
Hsf members were determined using the Hsf proteins from Brachypodium distachyon and A. thaliana from the HSF database (www.cibiv.at/services/hsf) as references (Scharf et al., 2012). The phylogenetic tree of Hsf proteins with DNA-binding domain was generated using ClustalW, as well as using MEGA 6.0's un-rooted neighbor-joining method with pairwise deletion and poisson correction (Tamura et al., 2013). The level of statistical support for each tree node was determined using bootstrap analysis with 1000 replicates. Partial Hsf proteins lacking or having incomplete DNA-binding domain that were recovered in the transcriptome analysis were classified based on the highest sequence homology to full-length Hsf proteins in the NCBI database, but were excluded from phylogenetic analysis.
Quantitative real time-polymerase chain reaction (qRT-PCR) verification
cDNA was prepared from the purified RNA samples using the PrimeScript™ RT reagent kit with gDNA Eraser (Takara, Dalian, China). Gene-specific qRT-PCR primers (18–20 bp) were designed using the Premier 5.0 software (Premier Biosoft International, Palo Alto, CA). qRT-PCR was performed using AceQ SYBR Green Master Mix (Vazyme Biotech Co., Ltd, Nanjing, China) with a FTC3000 Real-Time PCR System (Funglyn, Canada). The qPCR conditions were as follows: 5 min at 95°C, 45 cycles of denaturation at 95°C for 10 s, annealing at 50–58°C (according to the Tm of primers) for 30 s, and extension at 68°C for 40 s. And all real-time reactions were performed in three biological replicates and three technical replicates, and a melting curve was used to check amplification specificity. Each gene's relative expression level was calculated based on the comparative threshold cycle Ct(2−ΔΔCt) method (Livak and Schmittgen, 2001) using tall fescue and perennial ryegrass β-actin 7 gene as the internal reference. The software SPSS 11.5 was used for statistical analysis.
Results
Global description of transcriptomes of tall fescue and perennial ryegrass
To identify genes that are differentially expressed in tall fescue and perennial ryegrass under temperature-stress, we subjected plants to heat (40°C), cold (−10°C), and control (22°C) temperatures, and collected RNA for de novo transcriptome assembly. After de novo assembly, we identified 97,565 unigenes representing a total length of 68,116,614 nt in the transcriptome of pooled tall fescue leaves in response to heat, cold, and control temperatures. The average length of the unigenes was 698 nt, with a N50 number of 1064 nt. Of these, we annotated 57,156, 55,389, 35,792, 34,063, 21,157, and 38,454 of the unigenes to the NR, NT, Swiss-Prot, KEGG, COG, and GO databases, respectively, thereby generating a total of 63,607 functionally annotated unigenes (Supplementary Table 1-S1). We obtained 62,399 coding region sequences (CDS) by translating all sequences in the transcriptome into amino sequences using the standard codon table. The CDS sequences were then aligned to protein databases in the priority order of NR, Swiss-Prot, KEGG, and COG; this resulted in 56,141 unigenes, with 6258 sequences that did not align to any database.
We also generated a de novo transcriptome of the perennial ryegrass leaf in response to heat, cold, and control temperatures, which resulted in a total of 73,125 unigenes. The total length of the unigenes was 52,879,856 nt, with an average length of 723 nt, and a N50 of 1133 nt. We generated 51,062 annotated unigenes by annotating 46,858, 45,050, 31,051, 29,644, 19,616, and 32,561 unigenes using the NR, NT, Swiss-Prot, KEGG, COG, and GO databases, respectively (Supplementary Table 2-S1). We then conducted a protein coding region prediction analysis, and identified a total of 51,200 CDSs, which were then mapped on to protein databases. We assigned 46,363 CDSs to a protein identity, whereas 4837 were unmapped.
The unigenes in the tall fescue or perennial ryegrass were classified into 55 GO functional groups, including 22 biological process groups, 17 cellular component groups, and 16 molecular function groups (Supplementary Figures 1A,B). KEGG analysis of the unigenes resulted in 128 pathways (Supplementary Tables 1-S2, 2-S2), and COG annotation analysis of the unigenes resulted in 25 functional categories (Supplementary Figures 2A,B). These results were consistent for both species.
Differentially expressed genes under temperature stress
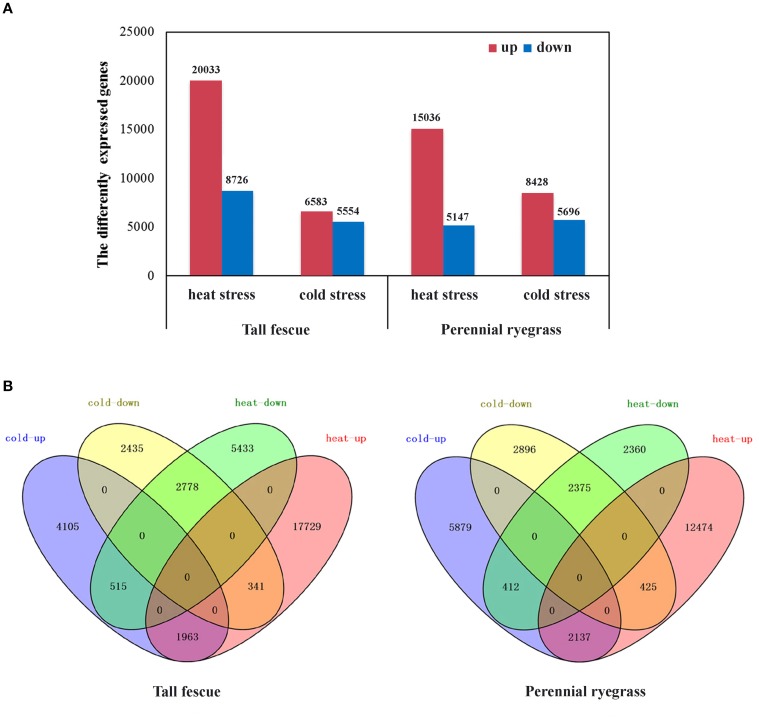
We next compared expression level of genes in the transcriptome to identify differentially expressed genes (DEGs) in tall fescue and perennial rye grass under temperature stress. DEGs were judged by the thresholds of “FDR < 0.001 and |log2Ratio| ≥ 1. Tall fescue under heat stress (HS) had 28,759 (29.48% of all the unigenes) DEGs, where 20,033 of the DEGs were upregulated and 8726 DEGs were downregulated. Tall fescue under cold-stress (CS) showed a total of 12 137 DEGs (12.44% of all unigenes), which consisted of 6583 upregulated DEGs and 5554 downregulated DEGs (Figure 1A). In both stress conditions, a total of 5597 DEGs showed overlapping differences in expression, including the following: (1) 1963 co-upregulated DEGs, (2) 2778 co-downregulated DEGs, (3) 341 DEGs upregulated in HS but downregulated in CS, and (4) 515 DEGs upregulated in CS but downregulated in HS (Figure 1B). The DEGs common between HS and CS accounted for 19.5% of the heat-DEGs and 46.1% of the cold-DEGs.
Figure 1.
Analysis of differentially expressed genes (DEGs) in tall fescue and perennial ryegrass transcriptomes in response to heat and cold stress. (A) Up- and down-regulated DEGs under heat or cold stress in the two grass species. Up (red) indicates upregulated DEGs, and down (blue) represents downregulated DEGs. (B) Shared and divergent DEGs in the tall fescue or perennial ryegrass under heat and cold stress. Heat-up (red) indicates the upregulated DEGs under heat stress; heat-down (green) represents the downregulated DEGs under heat stress; cold-up (blue) denotes the upregulated DEGs under cold stress; and cold-down (yellow) represents the downregulated DEGs under cold stress.
In perennial ryegrass under HS, we detected a total of 20,183 DEGs (27.60% of all unigenes), including 15,036 upregulated and 5147 downregulated DEGs. Under CS, we detected a total of 14,124 DEGs (19.31% of all unigenes), with 8428 upregulated DEGs and 5696 downregulated DEGs (Figure 1A). Similar to our results in tall fescue, we detected 5349 overlapping DEGs under both stresses, which were expressed as follows: (1) 2137 co-upregulated, (2) 2375 co-downregulated, (3) 425 upregulated in HS but downregulated in CS, and (4) 412 upregulated in CS but downregulated in HS (Figure 1B). In perennial rye grass, common DEGs accounted for 26.5% of the heat-DEGs and 37.9% of the cold-DEGs.
In both species, GO-term analysis of the DEG-response to HS was enriched (P < 0.05) in various biological process categories, such as “response to heat,” “photosynthesis,” “response to oxidative stress,” and “response to inorganic substance,” whereas the DEG-response to CS was enriched (P < 0.05) in “oxidation-reduction process,” “response to inorganic substance,” and “response to cold” (Supplementary Tables 1-S3, 2-S3). In addition, the common DEGs under both stresses were transcription factors, starch synthases, protein kinases, heat shock proteins, cytochrome P450s, and several unidentified proteins. Interestingly, we observed a very strinking difference in expression of Hsf genes in both stresses. We focused our further analyses on the Hsf genes to better understand how both species respond to temperature stress.
Classification of Hsf proteins of the two grass species
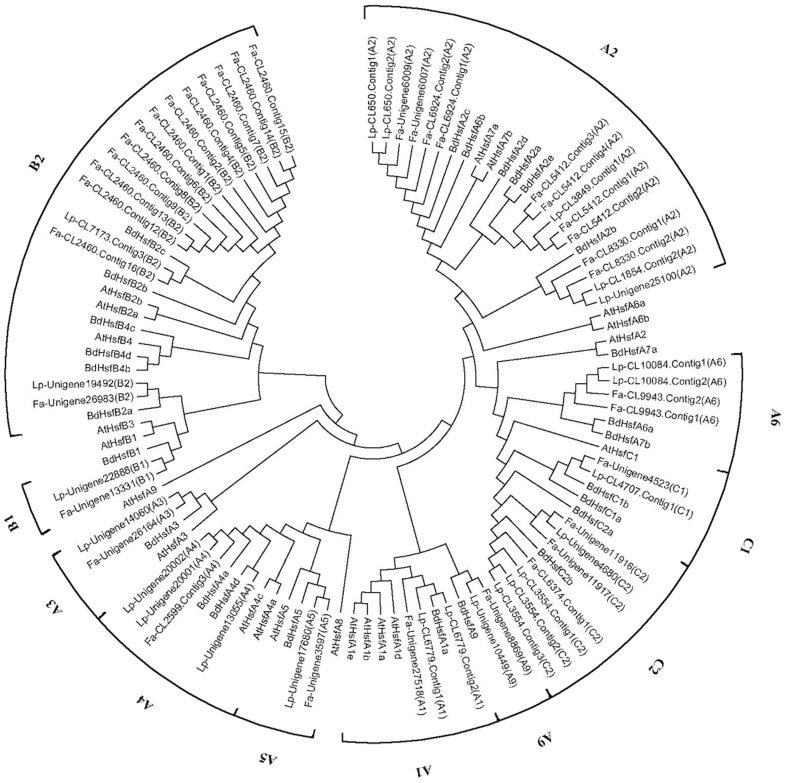
Hsf proteins have been categorized into different subfamilies and classes in model plant species such as A. thaliana, tomato, and B. distachyon (Scharf et al., 2012). Based on the characteristic sequences of Hsf genes identified in these model plant species, we identified 74 and 52 unigenes as Hsfs in the tall fescue and perennial ryegrass transcriptome, respectively. To better understand the potential function and homology of Hsf genes in both grass species, we conducted a phylogenetic analysis of Hsf proteins from the tall fescue, perennial ryegrass, B. distachyon, and A. thaliana (Figure 2). One of the main characteristics of Hsf proteins is the DNA-binding domain. In our de novo transcriptome assembly, we recovered some potential Hsf mRNA sequences that did not contain the characteristic Hsf DNA-bidning domain. These sequences were excluded from the phylogenetic analysis.
Figure 2.
Neighbor–joining phylogenetic tree of Hsf proteins from the tall fescue, perennial ryegrass, Brachypodium distachyon, and Arabidopsis thaliana. Hsfs in Brachypodium distachyon are prefixed by Bd, and Hsfs in Arabidopsis thaliana are prefixed by At. Hsfs in tall fescue are marked by a “Fa” prefix and sequence-ID in the transcriptome. Hsfs in perennial ryegrass are marked by a “Lp” prefix and a sequence-ID in the transcriptome.
Hsf proteins are categorized into subfamilies HsfA, HsfB, and HsfC. Our phylogenetic analysis uncovered seven smaller clusters within subfamily HsfA, whereas subfamilies HsfB and HsfC formed distinct groups. The same Hsf subfamily members for tall fescue and perennial ryegrass showed high sequence similarity. Hsfs from the two grass species were more closely related to B.distachyon Hsfs than A. thaliana Hsfs. In tall fescue and perennial ryegrass, we recovered seven HsfA classes (A1, A2, A3, A4, A5, A6, and A9), two HsfB classes (B1 and B2), and two HsfC classes (C1 and C2). Both grass species contained a large number of A2 and B2 class members, and we also detected C2 members that are unique in monocot species. However, we did not detect sequences belonging to subclasses A7, A8, B3, and B4 in the two grass species (Supplementary Tables 1-S4, 2-S4).
Expression profile of Hsf family genes
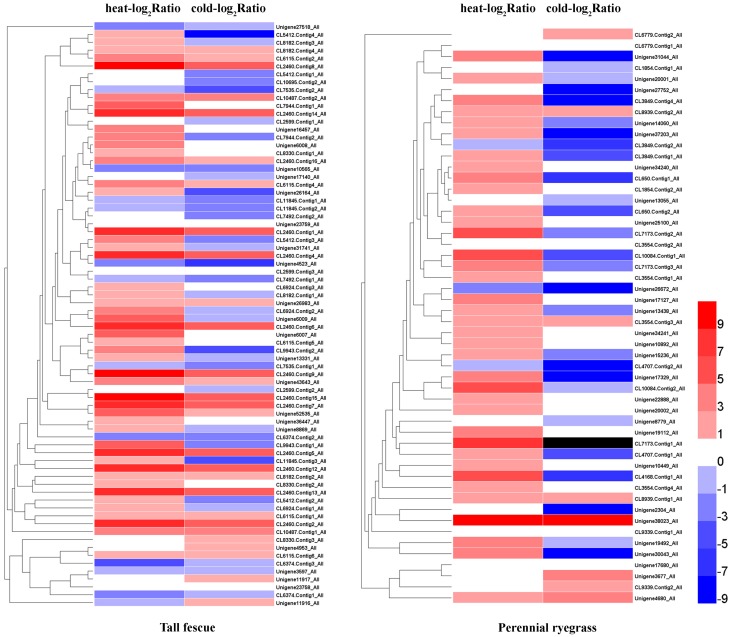
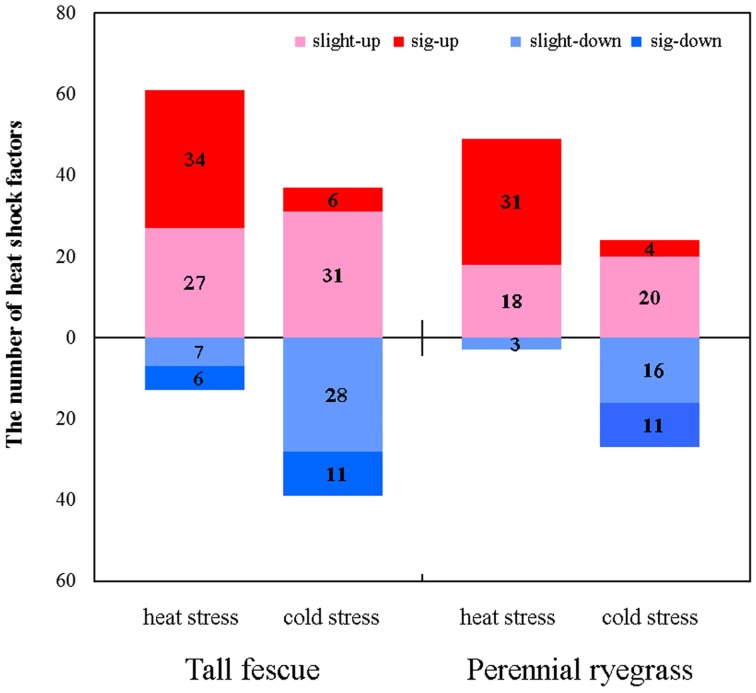
In both grass species, HS resulted in higher number of up-regulated Hsf genes compared to CS (Figure 3). In tall fescue, HS resulted in upregulation of 61 Hsfs including 34 significantly differentially expressed Hsfs (DE-Hsfs) and downregulation of 13 Hsfs (6 DE-Hsfs), whereas CS resulted in 37 upregulated Hsfs (6 DE-Hsfs) and 39 downregulated Hsfs (11 DE-Hsfs) (Figure 3). The expression differences of Hsf genes were more pronounced in perennial ryegrass, where HS resulted in 49 upregulated Hsfs (31 DE-Hsfs) and 3 downregulated Hsfs (no DE-Hsfs), and CS resulted in 24 upregulated Hsfs (4 DE-Hsfs) and 27 downregulated Hsfs (11 DE-Hsfs) (Figure 4). In addition, one Hsf (CL7173.Contig1_All) in perennial ryegrass was not expressed under CS, but was slightly upregulated under HS (Figure 4).
Figure 3.
Heatmaps of the expression profiles of Hsf genes in the two grass species subjected to heat and cold stress. Heat-log2 Ratio indicates the fold-change of Hsf FPKM values under HS, and cold-log2 Ratio represents the fold-change of Hsf FPKM values under CS. The black grid denotes genes where we did not detect expression.
Figure 4.
The expression patterns of heat shock factors in the tall fescue and perennial ryegrass under heat and cold stress. Slight-up (pink) indicates insignificantly upregulated Hsf genes, sig-up (red) represents significantly upregulated Hsf genes, slight-down (light blue) denotes insignificantly downregulated Hsf genes, and sig-down (blue) denotes significantly downregulated Hsf genes. Significant differences in transcript expression levels were determined by setting the thresholds for false discovery rate as “(FDR) < 0.001” and “|log2 Ratio| ≥ 1,” where ratio indicates the fold-change of FPKM values for the treatment and control samples.
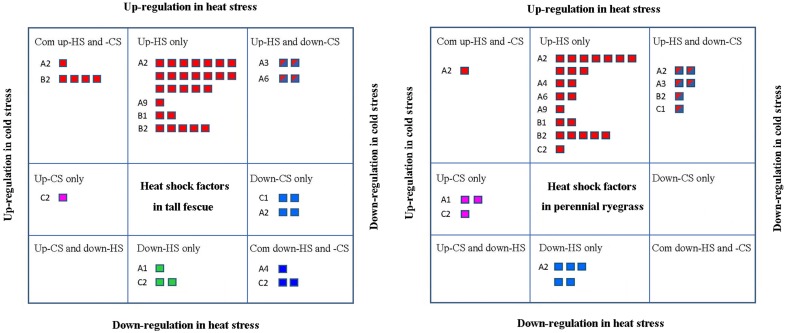
Next, we assessed the responses of different class Hsf genes to heat and cold stresses in the two grass species. Hsfs that showed significant differential expression under at least one stress conditions were classified and listed in Figure 5. In tall fescue, there were 25 Hsfs that showed significantly increases only in HS-treatment, including classes A2, A9, B1, and B2, and in perennial ryegrass, 24 Hsfs showed HS-specific upregulation, including Hsf A2, A4, A6, A9, B1, B2, and C2 classes. Of these, the classes Hsf A2 and B2 in the tall fescue and Hsf A2 in perennial ryegrass were significantly upregulated under HS and CS stresses. Members of classes Hsf A2 and A4 were significantly downregulated in CS in both grass species. Class Hsf A3 in both species and Hsf A6 in tall fescue were significantly upregulated in HS but downregulated in CS. In tall fescue, some Hsf C2 class genes showed significant upregulation only under CS, whereas other Hsf C2 class genes showed downregulation under both temperature stresses. In contrast, perennial ryegrass Hsf C2 class genes were upregulated by both temperature stresses, and downregulated only under CS (Figure 5).
Figure 5.
Stress-response analyses of different Hsf classes showing significant expression differences at least one stress condition. “Up-HS only” indicates upregulated heat shock factors (Hsfs) only under heat stress (HS); “Down-HS only” represents downregulated Hsfs only under HS; “Up-CS only” represents upregulated Hsfs only under cold stress (CS); and “Down-CS only” denotes downregulated Hsfs only under CS. “Com up-HS and CS” indicates upregulated Hsfs under HS and CS, and “Com down-HS and CS” represents downregulated Hsfs under HS and CS. “Up-HS and Down-CS” shows Hsfs upregulated under HS and downregulated under CS, and “Up-CS and Down-HS” denotes Hsfs downregualted under HS and upregulated under CS.
Analysis of Hsf target genes in the transcriptomes of the two grass species
HSP genes are one of the main targets of Hsf proteins under stress conditions. In the transcriptomes, we detected 371 transcripts encoding HSP genes in tall fescue, and 347 transcripts in perennial ryegrass (Table 1). These HSPs represented all five families. In both species and stress conditions, the strongest response was observed in the Hsp20 and Hsp70 families, then by the Hsp90 and Hsp60 families. The weakest response was detected in the Hsp100 family (Supplementary Tables 1-S5, 2-S5). HS resulted in significant upregulation of 199 and 192 HSPs in tall fescue and perennial ryegrass. HS also showed significant downregulation of 34 and 23 HSPs in tall fescue and perennial ryegrass, respectively. CS resulted in significant upregulation of 69 and 45 HSPs, and downregulation of 24 and 45 HSPs in tall fescue and perennial ryegrass, respectively (Table 1).
Table 1.
Genes encoding Heat shock proteins (HSPs), ascorbate peroxidases (APXs), inositol-3-phosphate synthases (IPSes), and four galactinol synthases (GOLSes) that show expression level changes responding to heat and cold stress in tall fescue and perennial ryegrass.
| Gene sum | Heat stress | Cold stress | ||||
|---|---|---|---|---|---|---|
| Upregulated | Downregulated | Upregulated | Downregulated | |||
| Tall fescue | HSP | 371 | 199 | 34 | 69 | 24 |
| APX | 36 | 7 | 0 | 0 | 1 | |
| IPS | 0 | 0 | 0 | 0 | 0 | |
| GOLS | 16 | 2 | 9 | 6 | 0 | |
| Perennial ryegrass | HSP | 347 | 192 | 23 | 45 | 45 |
| APX | 25 | 5 | 4 | 1 | 5 | |
| IPS | 5 | 3 | 0 | 0 | 0 | |
| GOLS | 5 | 0 | 0 | 3 | 0 | |
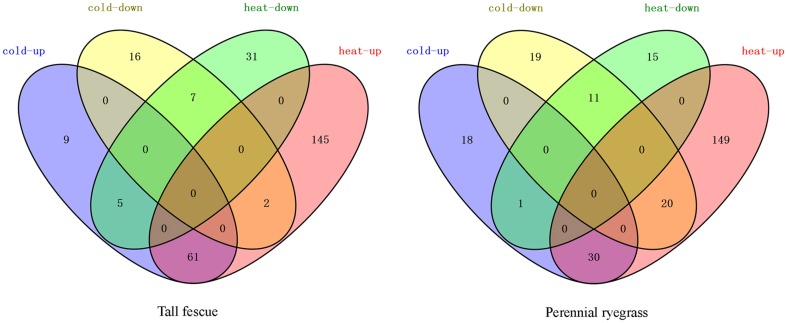
We next analyzed other known Hsf targets in the transcriptome. In tall fescue, we detected 36 APXs, 16 GOLS genes, but did not detect any IPS genes. In perennial ryegrass, we detected 25 APXs, five IPSes, and five GOLSes (Table 1, Supplementary Tables 1-S6, 2-S6). HS resulted in significant increase in the expression of APXs in both grass species, and IPSes in perennial ryegrass. In contrast, CS resulted in a significant increase in the expression of GOLSes in two species (Table 1). The Venn diagram in Figure 6 shows the distribution of shared and divergent target genes under two temperature stresses in each grass species.
Figure 6.
Venn diagram of differentially expressed target genes of Hsfs in the tall fescue or perennial ryegrass under heat and cold temperature stress. Red (heat-up) represents the upregulated target transcripts under heat stress; green (heat-down) represents the downregulated target transcripts under heat stress; blue (cold-up) indicates the upregulated target transcripts under cold stress; and yellow (cold-down) represents the down-regulated target transcripts under cold stress.
Changes in the expression levels of Hsfs and target genes
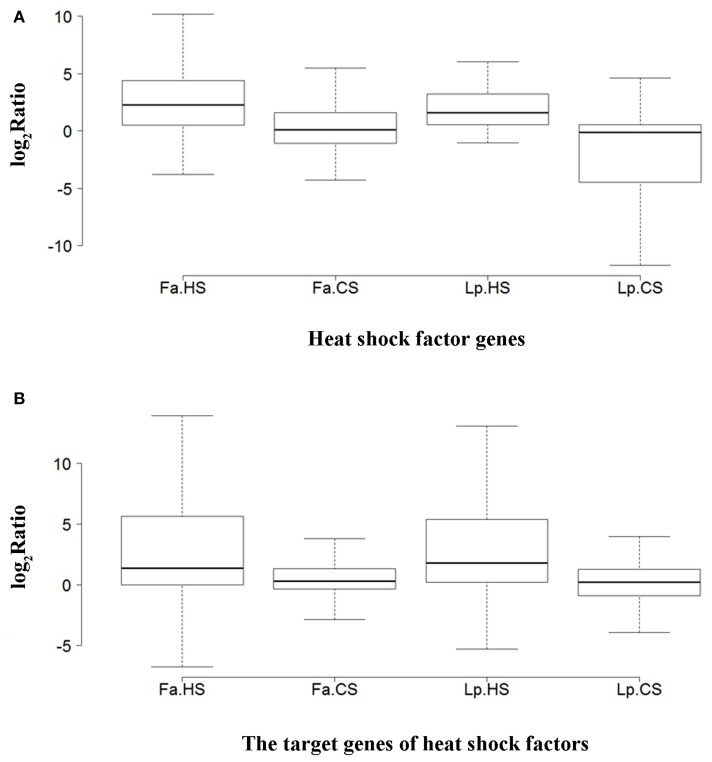
To understand the difference of expression profiles between species and stress responses, we analyzed the expression of Hsfs and their target genes under different temperatures in tall fescue and perennial ryegrass. Hsfs were expressed at low levels in the control, where only three Hsfs were detected to be >10 FPKM in tall fescue, with a peak FPKM value of 40.80, and two Hsfs in perennial ryegrass, with highest FPKM level of 12.76. Under HS, more than 30% of the Hsfs in the two grass species were expressed at >10 FPKM, and the highest FPKM value was 102.75 in the tall fescue and 104.25 in perennial ryegrass. In contrast, under CS, seven Hsfs showed >10 FPKM in tall fescue, with a peak FPKM value of 30.72, and one Hsf in perennial ryegrass with a 10.62 FPKM value. Comparison changes in Hsf expression levels in HS and CS, we observed that HS had a greater effect on Hsfs in the tall fescue relative to perennial ryegrass. The median log2Ratio in the tall fescue Hsfs was slightly higher than that observed in perennial ryegrass under the two temperature stresses (Figure 7A).
Figure 7.
Overall expression differences (log2 Ratio) of heat shock factors and their target genes between species and stresses. (A) Heat shock factor genes. (B) The target genes of heat shock factors. Abbreviations are as follows: Fa, tall fescue (Festuca arundinacea); Lp, perennial ryegrass (Lolium perenne); HS, heat stress; and CS, cold stress. The log2 Ratio represents the fold-change of Hsf FPKM values under heat or cold treatment relative to that of the control samples.
We next analyzed the expression changes of Hsf target genes, including HSP, APX, IPS, and GOLS genes. Under the control temperature, the highest level of HSP expression was 431.15 FPKM in the tall fescue and 453.01 FPKM in perennial ryegrass. Under HS, the highest FPKM of HSPs in the tall fescue and perennial ryegrass was 4436.84 and 3586.88, respectively. Under CS, the highest FPKM value was 458.05 and 210.59, respectively. For APXs, the peak value of FPKM under control, HS and CS was 383.27, 827.92, and 472.44 in tall fescue, and 277.42, 267.16, and 314.56 in perennial ryegrass, respectively. For IPSes, these peak values were 10.53, 27.03, and 156.44 in tall fescue, respectively, and 8.96, 9.83, and 17.39 in perennial ryegrass. For GOLSes, they were 33.63, 9.12, and 38.51 in tall fescue, and 10.12, 8.96, and 22.90 in perennial ryegrass, respectively. The expression levels of Hsf target genes did not differ between the two grass species, whereas differences were observed between the two temperature stresses conditions (Figure 7B).
qRT-PCR validation of differentially expressed transcripts from RNA-seq
To confirm the accuracy and reproducibility of the transcriptome results, 64 unigenes (32 from each grass transcriptome) were randomly chosen for quantitative real-time -PCR (qRT-PCR). These included several Hsfs and other genes. The primer sequences, FPKM, and qRT-PCR values are listed in Supplementary Table 3. The qPCR results showed a general agreement (75–78%) with the transcript abundance as determined by RNA-Seq. Specifically, we observed an agreement of 75% in the tall fescue and 78% in perennial ryegrass, which suggest that the RNA-Seq data was a reliable read-out for differential expression (Supplementary Table 3).
Discussion
Comparative analysis of transcriptomes of the tall fescue and perennial ryegrass under different temperature stresses
In subtropical regions, the growth of grass is limited by high temperatures in the summer and low temperatures during winter. However, heat and cold stress are not generally lethal because plants have a variety of protective mechanisms to tolerate temperature changes by altering expression of stress-response genes (Krasensky and Jonak, 2012). The leaves of plants, being exposed to air, generally receive the stress signals first, and rapidly respond to these environmental changes. We therefore used leaf samples of the tall fescue and perennial ryegrass under heat, cold, and normal temperatures for deep RNA-sequencing on an Illumina HiSeq 2000 platform. After de novo assembly, 97,565 and 73,125 unigenes were generated in the tall fescue and perennial ryegrass transcriptomes. Of these, about two-thirds were functionally annotated using various reference databases. In the transcriptome of heat-tolerant and heat-sensitive cultivars of tall fescue responding to high-temperature stress, 31,803 unigenes were assembled (Hu et al., 2014). In a transcriptome of perennial ryegrass generated by 454 sequencing, 9399 non-redundant contigs were acquired from 25,744 high-quality EST reads after de novo assembly (Studer et al., 2012). By using the RNA-seq strategy, de novo assembly of the perennial ryegrass transcriptome resulted in 185,833 transcripts, in which over 50,000 transcripts were functionally annotated (Farrell et al., 2014). Comparing our transcriptome to previously published ones suggest that the data obtained in our whole-genome-wide transcriptional sequencing were larger and sufficient in size for analyzing differentially expressed genes and mining for temperature-resistance genes in the two species.
In our study, approximately 30% and 25% of all genes in the transcriptome were identified as DEGs (differentially expressed genes) during heat or cold stress, with a greater number of DEGs showing upregulation than downregulation. HS exhibited a higher number of DEGs than CS, which may be attributed to the fact that the two grass species being more heat-tolerance. In addition, being exposed to below-freezing temperatures can result in to lethal mechanical damages to the plant, such as the formation of ice crystals within cells (intracellular freezing) and between cell walls (extracellular freezing) (Xin and Browse, 2000). We may have recovered a low number of gene expression changes in the below-freezing temperature conditions due to the mechanical damage that occurred before the plant could induce the cold acclimation gene-expression response. This suggests that sudden freezing stress can result in more severe injury to the plants compared to that observed during HS.
Heat shock factors in response to heat or cold stress
Transcription factors are one of the master regulators that lead to changes in the level of expression of downstream genes, thereby influencing plant metabolism (Ramirez and Basu, 2009). In the present study, the expression the transcription factor gene family Hsf were remarkably impacted by heat and cold stresses. A high proportion of Hsfs showed significant upregulation upon HS, whereas some Hsfs were upregulated by CS. Recent studies involving various plant models show that Hsfs respond not only to HS, but also to other abiotic stimuli such as waterlogging, salt, osmosis, and oxidative stresses (Ogawa et al., 2007; Yoshida et al., 2008; Shim et al., 2009; Liu et al., 2011; Xue et al., 2014). We conducted a transcriptome analysis of two non-model species in response to temperature stresses, and mainly focused on Hsfs and their target genes.
We identified 74 Hsfs in the tall fescue and 52 Hsfs in perennial ryegrass transcriptomes. The numbers of Hsf genes in model organisms are 21 in A. thaliana, 24 in B. distachyon, 25 in rice, 30 in poplar (Populus trichocarpa), 40 in Gossypium raimondii, and 56 in wheat (Scharf et al., 2012; Xue et al., 2014); therefore the Hsf gene family in the tall fescue was larger than those in most known species. The three plant Hsf subfamilies, Hsf A, Hsf B, and Hsf C were detected in the tall fescue and perennial ryegrass, and we assigned each gene to A1 to A6 and A9, B1 and B2, C1 and C2 classes. All subfamilies/classes are represented in monocots and eudicots, except for C2, which is exclusive to monocots (Scharf et al., 2012). We uncovered most members in the subfamily Hsf A, which was in agreement with known plant Hsf families. However, classes A7, A8, and B4, which are widely distributed in most plant species, were not detected in the two grass species. At present, Hsf A9 genes are considered to be confined to eudicots and play a special role in seed development (Almoguera et al., 2002; Kotak et al., 2007; Carranco et al., 2010). Nevertheless, we retrieved one Hsf A9 gene in tall fescue and two in perennial ryegrass. Hsf A9 genes were induced by HS, suggesting that Hsf A9 may also be related to heat tolerance, but potentially not involved in cold tolerance. Overall, general patterns were similar to model species, but we recovered some distinct differences in Hsf members between grasses and other monocots or eudicot species.
Analysis of Hsfs showing significant differences in gene expression in both grass species exposed to HS or CS suggested that Hsfs responded more strongly to heat. Among all of the Hsf classes, the members of class Hsf A2 were highest in quantity, and most of the Hsf A2 genes were upregulated under HS. A related study in tomato, A. thaliana, and rice also showed that Hsf A2 genes were most strongly induced upon exposure to long-term HS or repeated cycles of HS and recovery (Charng et al., 2007; von Koskull-Doring et al., 2007; Chan-Schaminet et al., 2009; Nishizawa-Yokoi et al., 2011; Liu and Charng, 2013). These studies suggested that HsfA2 improves thermotolerance based not only on expression abundance during heat-stress, but also based on the formation of the superactivator complex HsfA1/A2, which has a much higher level of activation for HSP-encoding genes than single Hsf proteins. However, in our HS analysis, Hsf A1 in perennial ryegrass was weakly upregulated, and was downregulated in the tall fescue. These results may suggest that Hsf A2 tall fescue and perennial ryegrass independently function to enhance thermotolerance. With a few exceptions, the Hsf A2d gene, which is a member of class Hsf A2, was upregulated during CS. To date, direct evidence on how Hsf A2d improves cold tolerance remains elusive. A. thaliana Hsf A2 knockout lines and Hsf A2-overexpression lines indicate that Hsf A2 is one of the key regulators in protecting organelles against oxidative damage (Miller and Mittler, 2006; Zhang et al., 2009). Plant stress often results in the overproduction of reactive oxygen species; therefore Hsf A2 may be involved in improving cold tolerance by alleviating oxidative damage.
Likewise, HS also resulted in a significant increase in the expressions of Hsf A3, A4, and A6 in the two grasses, which was consistent to results observed in wheat under HS (Xue et al., 2014). It has been suggested that these Hsf genes participate in the process of acquiring thermotolerance. In Arabidopsis undergoing heat and drough stress, Hsf A3 is involved in the transcriptional cascade that is downstream of the DREB2A stress-regulatory system (Chen et al., 2010). Interestingly, we observed a significant upregulation of DREB transcripts in the two grass species. It is therefore possible that Hsf A3 and DREB may interact in these two species, although this requires further testing. HsfA4a in A. thaliana was regulated by the mitogen-activated protein kinases MPK3 and MPK6 to confer enhanced tolerance to salt stress (Pérez-Salamó et al., 2014). HsfA4a in wheat and rice confers Cd tolerance by upregulating the gene expression of metallothionein (Shim et al., 2009). A. thaliana HsfA6a responds strongly to exogenous ABA, NaCl, and drought, and acts as a transcriptional activator for stress-responsive genes via the ABA-dependent signaling pathway (Hwang et al., 2014). In our transcriptome analysis, we observe a HS-induced response in most members of subfamily HsfA in the two grasses. Although HsfA genes are currently the most extensively studied in the Hsf family, the function and mechanism of all of its members still remain unclear.
Hsf subfamily B classes B1 and B2 in the tall fescue and perennial ryegrass were upregulated under HS, whereas Hsf B2 genes in tall fescue were upregulated under CS. Based on the protein domains present in Hsf subfamily B genes, it is generally suggested that subfamily B does not have transcription activator function (von Koskull-Doring et al., 2007). However, tomato HsfB1 shows strong synergistic activation by cooperating with Hsf subfamily A proteins or other transcriptional activators (Bharti et al., 2004). A. thaliana HsfB1 and HsfB2b also appear to be necessary for the expression of HS-inducible HSP genes (Ikeda et al., 2011; Pick et al., 2012). HsfB2b in A. thaliana represses the expression of PRR7 and sustains circadian rhythm during heat and salt stress, which are important functions for plant growth and fitness (Kolmos et al., 2014). Arabidopsis HsfB2a is involved in gametophyte development, and a heat-inducible long non-coding antisense RNA controls its expression (Wunderlich et al., 2014). Our results supports the hypothesis that Hsf B subfamily genes are involved in temperature stress-response.
Hsf subfamily C genes were particularly enhanced in tall fescue and perennial ryegrass undergoing CS. To date, the function of Hsf subfamily C genes remain elusive. Our results suggested that Hsf C genes may be involved in cold tolerance. Interestingly, similar results have been reported in wheat and rice. An activation motif in the C-terminal domain of the wheat TaHsfC2a contains an activation motif in the C-terminal domain, and regulates the expression of HSP genes (Xue et al., 2014). Of the 26 rice Hsfs, 10 showed a response to low temperature stress (Mittal et al., 2009).
Targets genes of Hsfs in response to heat and cold stresses
The expression of Hsfs triggers changes in expression of downstream target genes. One of the major targets of Hsfs is the HSP genes, and HSPs play a role in enhancing thermotolerance in various plants (Timperio et al., 2008; Tripp et al., 2009). Several studies have also shown that HSPs can improve cold tolerance. For example, Hsp70 serves as a molecular chaperone at low temperature in vitro (Zhang and Guy, 2006). DcHsp17.7 from carrot can prevent cold-induced protein degradation, and confer cold tolerance (Song and Ahn, 2010). HSPs expression levels change under various stressors, which indicate similarities in plant adaption mechanisms in response to abiotic stresses (Wang et al., 2004; Krasensky and Jonak, 2012). Here, we found that HSPs show strong response to HS and CS in the tall fescue and perennial ryegrass, indicating that they may be involved in protecting cells against temperature stresses.
Other target genes of Hsfs include APX, IPS, and GOLS. It has been suggested that Hsfs act as molecular sensors that directly sense ROS signals, which in turn activate the expression of HSPs and oxidative stress response genes. Hsf-binding motifs have been detected in the promoter region of genes involved in the ROS gene network, including defense genes, H2O2-scavenging enzyme APX, and related transcription factors (Miller and Mittler, 2006). Three stress-related genes in A. thaliana, (APX2, IPS2, and GOLS1) are regulated by HsfA2 (Miller and Mittler, 2006; Scharf et al., 2012). Here, we specifically investigated the expression levels of these three genes under HS and CS. The expression of APXs and IPSes significantly increased upon HS, and GOLSes decreased under HS, but increased under CS. These results suggest that there may be a crosstalk between Hsfs and stress genes during ROS signaling.
Difference between two stresses and similarity of two species
Most DEGs were different between HS and CS in the tall fescue or perennial ryegrass, and a small portion of DEGs were commonly expressed under the two stress conditions. A previous report involving A. thaliana indicated the existence of a greater crosstalk between drought and high-salinity stress signaling processes compared to those involved in cold and high-salinity (Matsui et al., 2008). Transcriptome response of Zea mays to various adverse environments show a greater gene overlap between heat and salt stress compared to those induced by heat and cold (Fernandes et al., 2008). The results of the present study are in agreement to these findings, and show that genes responding to HS and CS show differences, irrespective of species. However, the DEGs were classified under the same GO category. Each GO class or gene family included many genes with functional divergence. Consequently, it is possible that different members in the same gene family or functional cluster respond to stressors differently. Some genes can also be induced by several different stresses. The difference in how Hsf family and their targets genes respond to different temperature treatments in the present study support this hypothesis.
Overall, the gene expression profiles of tall fescue and perennial ryegrass showed a similar response to the temperature stress. In terms of Hsfs and their target genes in the two grasses, their responses were almost identical in the same stress conditions. Previous reports have considered that the tall fescue has a greater heat tolerance compared to the perennial ryegrass (Raeside et al., 2012). However, in a previous study, we did not detect any physiological differences in how these plants responded to thermotolerance (Wang et al., 2014). In the present study, the two grasses showed similar changes in gene expression when subjected to HS or CS. The response mechanism of the two grasses to temperature may therefore be similar. Therefore, our results suggest that there are less differences between the two grass species, and more differences between the two temperature stresses.
Conclusions
By using RNA-seq analysis, we compared the transcriptomes of tall fescue and perennial ryegrass under HS and CS. We generated a total of 97,565 unigenes in the tall fescue and 73,125 unigenes in the perennial ryegrass through de novo transcriptome assembly. In addition, 28,759 and 12,137 significant DEGs under HS and CS were obtained in the tall fescue, and 20,183 and 14,124 in the perennial ryegrass, respectively. In each species, HS resulted in more DEGs than that observed under CS, and less overlap was detected under the two temperature conditions. However, DEGs in the two grasses exposed to the same stress showed similar GO-term functions. Using sequence similarity and phylogenetic analysis, we identified 74 Hsf members in the tall fescue and 52 Hsfs in the perennial ryegrass, and assigned these to three subfamilies, HsfA, HsfB, and HsfC. This included seven HsfA classes, two HsfB classes, and two HsfCs classes. Hsf expression was significantly affected by HS and CS, with a larger enhancement observed under HS. We recovered the highest number of members in subfamily Hsf A, and classes A2, A3, A4, A6, and A9 genes were upregulated by HS. Class HsfA2 was quite expansive in the two species, which showed a greater change of expression level under HS. Accordingly, HsfA2 members may be key regulators of HS response in these species. Subfamily HsfB was upregulated by both HS and CS, and the expression of HsfC genes were mainly upregulated under CS. These results suggested that subfamily HsfA are likely involved in acclimating the two grasses to heat tolerance, and HsfB plays a role in adaptation to the both stresses, whereas subfamily HsfC is related to the acquirement of cold tolerance. We also analyzed the expression profiles of the target genes of Hsfs. HSPs, the major target genes of Hsfs, were significantly upregulated under the two stresses, suggesting that HSPs may play important roles in protecting plant cells against the high and low temperature damage. For other target genes of Hsfs, the expression of APX and IPS in both grasses were significantly enhanced by HS, whereas that of the GOLS genes increased under CS. Similar to the Hsfs, different members of these target genes showed specific response to HS and/or CS. The results from the two species serve as useful references to investigate how other grass species respond to temperature-stress. Further, studies will clarify the function of potential candidate Hsf members, and identify the gene regulation network involving Hsfs under HS or CS conditions.
Author contributions
YW and XM designed the experimental plan and drafted and revised the manuscript. YD, JW, and HY participated in preparing, treating, and collecting samples. XT, HC, and YW analyzed and interpreted the sequence data. All authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by National Science and Technology Support Program (2014BAC05B04), National Natural Science Foundation of China (31502003), and the West Light Foundation of the Chinese Academy of Sciences (Y3C4011100 and Y5C4011100).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.01226
Gene Ontology (GO) annotation of transcripts in (A) tall fescue and (B) perennial ryegrass.
Functional classification using Clusters of Orthologous Groups of proteins (COG) in tall fescue (A) and perennial ryegrass (B).
Annotation of tall fescue transcriptome in response to heat and cold stress.
Supplementary Table 1-S1: Expression levels and functional annotation of transcripts of the tall fescue transcriptome.
Supplementary Table 1-S2: KEGG pathway mapping of the tall fescue transcriptome.
Supplementary Table 1-S3: GO-Biological Process enrichment (p < 0.05) of the differentially-expressed genes (DEGs) showing significant expression differences in tall fescue under heat and cold stress.
Supplementary Table 1-S4: Genes encoding heat shock factor (Hsf) genes in the tall fescue transcriptome.
Supplementary Table 1-S5: Genes encoding heat shock protein (HSP) genes in the tall fescue transcriptome.
Supplementary Table 1-S6: Genes encoding Ascorbate peroxidase (APX), Inositol-3-phosphate synthase (IPS) and Galactinol synthase (GOLS1) in the tall fescue transcriptome.
The annotation of perennial ryegrass transcriptome in response to heat and cold stress.
Supplementary Table 2-S1: Expression levels and functional annotation of transcripts in the perennial ryegrass transcriptome.
Supplementary Table 2-S2: KEGG pathway mapping of the perennial ryegrass transcriptome.
Supplementary Table 2-S3: GO-Biological Process enrichment (p < 0.05) of differentially-expressed genes (DEGs) showing significant expression differences in perennial ryegrass under heat and cold stress.
Supplementary Table 2-S4: Genes encoding heat shock factor (Hsf) genes in the perennial ryegrass transcriptome.
Supplementary Table 2-S5: Genes encoding heat shock protein (HSP) genes in the perennial ryegrass transcriptome.
Supplementary Table 2-S6: Genes encoding Ascorbate peroxidase (APX), Inositol-3-phosphate synthase (IPS) and Galactinol synthase (GOLS1) in the perennial ryegrass transcriptome.
Comparison of gene expression patterns between RNA-seq and quantitative real-time PCR results.
References
- Almoguera C., Rojas A., Díaz-Martín J., Prieto-Dapena P., Carranco R., Jordano J. (2002). A seed-specific heat-shock transcription factor involved in developmental regulation during embryogenesis in sunflower. J. Biol. Chem. 277, 43866–43872. 10.1074/jbc.M207330200 [DOI] [PubMed] [Google Scholar]
- Audic S., Claverie J. M. (1997). The significance of digital gene expression profiles. Genome Res. 7, 986–995. 10.1101/gr.7.10.986 [DOI] [PubMed] [Google Scholar]
- Baniwal S. K., Chan K. Y., Scharf K. D., Nover L. (2007). Role of heat stress transcription factor HsfA5 as specific repressor of HsfA4. J. Biol. Chem. 282, 3605–3613. 10.1074/jbc.M609545200 [DOI] [PubMed] [Google Scholar]
- Bharti K., von Koskull-Döring P., Bharti S., Kumar P., Tintschl-Körbitzer A., Treuter E., et al. (2004). Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein ortholog HAC1. Plant Cell 16, 1521–1535. 10.1105/tpc.019927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carranco R., Espinosa J. M., Prieto-Dapena P., Almoguera C., Jordano J. (2010). Repression by an auxin/indole acetic acid protein connects auxin signaling with heat shock factor-mediated seed longevity. Proc. Natl. Acad. Sci. U.S.A. 107, 21908–21913. 10.1073/pnas.1014856107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan-Schaminet K. Y., Baniwal S. K., Bublak D., Nover L., Scharf K. D. (2009). Specific interaction between tomato HsfA1 and HsfA2 creates hetero-oligomeric superactivator complexes for synergistic activation of heat stress gene expression. J. Biol. Chem. 284, 20848–20857. 10.1074/jbc.M109.007336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charng Y. Y., Liu H. C., Liu N. Y., Chi W. T., Wang C. N., Chang S. H., et al. (2007). A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 143, 251–262. 10.1104/pp.106.091322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Hwang J. E., Lim C. J., Kim D. Y., Lee S. Y., Lim C. O. (2010). Arabidopsis DREB2C functions as a transcriptional activator of HsfA3 during the heat stress response. Biochem. Biophys. Res. Commun. 401, 238–244. 10.1016/j.bbrc.2010.09.038 [DOI] [PubMed] [Google Scholar]
- Conesa A., Götz S., García-Gómez J. M., Terol J., Talón M., Robles M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. 10.1093/bioinformatics/bti610 [DOI] [PubMed] [Google Scholar]
- Czarnecka-Verner E., Pan S. Q., Salem T., Gurley W. B. (2004). Plant class BHSFs inhibit transcription and exhibit affinity for TFIIB and TBP. Plant Mol. Biol. 56, 57–75. 10.1007/s11103-004-2307-3 [DOI] [PubMed] [Google Scholar]
- Deng W., Wang Y., Liu Z., Cheng H., Xue Y. (2014). HemI: a toolkit for illustrating heatmaps. PLoS ONE 9:e111988. 10.1371/journal.pone.0111988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerks T., Copley R. R., Schultz J., Ponting C. P., Bork P. (2002). Systematic identification of novel protein domain families associated with nuclear functions. Genome Res. 12, 47–56. 10.1101/gr.203201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring P., Treuter E., Kistner C., Lyck R., Chen A., Nover L. (2000). The role of AHA motifs in the activator function of tomato heat stress transcription factors HsfA1 and HsfA2. Plant Cell 12, 265–278. 10.1105/tpc.12.2.265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell J. D., Byrne S., Paina C., Asp T. (2014). De novo assembly of the perennial ryegrass transcriptome using an RNA-Seq strategy. PLoS ONE 9:e103567. 10.1371/journal.pone.0103567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes J., Morrow D. J., Casati P., Walbot V. (2008). Distinctive transcriptome responses to adverse environmental conditions in Zea mays L. Plant Biotechnol. J. 6, 782–798. 10.1111/j.1467-7652.2008.00360.x [DOI] [PubMed] [Google Scholar]
- Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasanuzzaman M., Nahar K., Alam M. M., Roychowdhury R., Fujita M. (2013). Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 14, 9643–9684. 10.3390/ijms14059643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T., Sun X., Zhang X., Nevo E., Fu J. (2014). An RNA sequencing transcriptome analysis of the high-temperature stressed tall fescue reveals novel insights into plant thermotolerance. BMC Genomics 15:13. 10.1186/1471-2164-15-1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Wu L., Zhao F., Zhang D., Li N., Zhu G., et al. (2015). Phosphoproteomic analysis of the response of maize leaves to drought, heat and their combination stress. Front. Plant Sci. 6:298. 10.3389/fpls.2015.00298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., DaCosta M., Jiang Y. (2014). Research advances in mechanisms of turfgrass tolerance to abiotic stresses: from physiology to molecular biology. Crit. Rev. Plant Sci. 33, 141–189. 10.1080/07352689.2014.870411 [DOI] [Google Scholar]
- Humphreys M. W., Thomas H. (1993). Improved drought resistance in introgression lines derived from lolium-multiflorum x festuca-arundinacea hybrids. Plant Breeding 111, 155–161. 10.1111/j.1439-0523.1993.tb00622.x [DOI] [Google Scholar]
- Hwang S. M., Kim D. W., Woo M. S., Jeong H. S., Son Y. S., Akhter S., et al. (2014). Functional characterization of Arabidopsis HsfA6a as a heat-shock transcription factor under high salinity and dehydration conditions. Plant Cell Environ. 37, 1202–1222. 10.1111/pce.12228 [DOI] [PubMed] [Google Scholar]
- Ikeda M., Mitsuda N., Ohme-Takagi M. (2011). Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol. 157, 1243–1254. 10.1104/pp.111.179036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M., Ohme-Takagi M. (2009). A novel group of transcriptional repressors in arabidopsis. Plant Cell Physiol. 50, 970–975. 10.1093/pcp/pcp048 [DOI] [PubMed] [Google Scholar]
- Kolmos E., Chow B. Y., Pruneda-Paz J. L., Kay S. A. (2014). HsfB2b-mediated repression of PRR7 directs abiotic stress responses of the circadian clock. Proc. Natl. Acad. Sci. U.S.A. 111, 16172–16177. 10.1073/pnas.1418483111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S., Port M., Ganguli A., Bicker F., von Koskull-Döring P. (2004). Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J. 39, 98–112. 10.1111/j.1365-313X.2004.02111.x [DOI] [PubMed] [Google Scholar]
- Kotak S., Vierling E., Baumlein H., von Koskull-Döring P. (2007). A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. Plant Cell 19, 182–195. 10.1105/tpc.106.048165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasensky J., Jonak C. (2012). Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 63, 1593–1608. 10.1093/jxb/err460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. C., Charng Y. Y. (2013). Common and distinct functions of Arabidopsis class A1 and A2 heat shock factors in diverse abiotic stress responses and development. Plant Physiol. 163, 276–290. 10.1104/pp.113.221168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. C., Liao H. T., Charng Y. Y. (2011). The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 34, 738–751. 10.1111/j.1365-3040.2011.02278.x [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using realtime quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lohmann C., Eggers-Schumacher G., Wunderlich M., Schöffl F. (2004). Two different heat shock transcription factors regulate immediate early expression of stress genes in Arabidopsis. Mol. Genet. Genomics 271, 11–21. 10.1007/s00438-003-0954-8 [DOI] [PubMed] [Google Scholar]
- Matsui A., Ishida J., Morosawa T., Mochizuki Y., Kaminuma E., Endo T. A., et al. (2008). Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 49, 1135–1149. 10.1093/pcp/pcn101 [DOI] [PubMed] [Google Scholar]
- Miller G., Mittler R. (2006). Could heat shock transcription factors function as hydrogen peroxide sensors in plants? Ann. Bot. 98, 279–288. 10.1093/aob/mcl107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal D., Chakrabarti S., Sarkar A., Singh A., Grover A. (2009). Heat shock factor gene family in rice: genomic organization and transcript expression profiling in response to high temperature, low temperature and oxidative stresses. Plant Physiol. Biochem. 47, 785–795. 10.1016/j.plaphy.2009.05.003 [DOI] [PubMed] [Google Scholar]
- Miura K., Furumoto T. (2013). Cold signaling and cold response in plants. Int. J. Mol. Sci. 14, 5312–5337. 10.3390/ijms14035312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- Nishizawa-Yokoi A., Nosaka R., Hayashi H., Tainaka H., Maruta T., Tamoi M., et al. (2011). HsfA1d and HsfA1e involved in the transcriptional regulation of HsfA2 function as key regulators for the Hsf signaling network in response to environmental stress. Plant Cell Physiol. 52, 933–945. 10.1093/pcp/pcr045 [DOI] [PubMed] [Google Scholar]
- Ogawa D., Yamaguchi K., Nishiuchi T. (2007). High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased themotolerance but also salt/osmotic stress tolerance and enhanced callus growth. J. Exp. Bot. 58, 3373–3383. 10.1093/jxb/erm184 [DOI] [PubMed] [Google Scholar]
- Pérez-Salamó I., Papdi C., Rigó G., Zsigmond L., Vilela B., Lumbreras V., et al. (2014). The heat shock factor A4A confers salt tolerance and is regulated by oxidative stress and the mitogen-activated protein kinases MPK3 and MPK6. Plant Physiol. 165, 319–334. 10.1104/pp.114.237891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peteranderl R., Rabenstein M., Shin Y. K., Liu C. W., Wemmer D. E., King D. S., et al. (1999). Biochemical and biophysical characterization of the trimerization domain from the heat shock transcription factor. Biochemistry 38, 3559–3569. 10.1021/bi981774j [DOI] [PubMed] [Google Scholar]
- Pick T., Jaskiewicz M., Peterhänsel C., Conrath U. (2012). Heat shock factor HsfB1 primes gene transcription and systemic acquired resistance in Arabidopsis. Plant Physiol. 159, 52–55. 10.1104/pp.111.191841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raeside M. C., Friend M. A., Behrendt R., Lawson A. R., Clark S. G. (2012). A review of summer-active tall fescue use and management in Australia's high-rainfall zone. N.Z. J. Agric. Res. 55, 393–411. 10.1080/00288233.2012.708663 [DOI] [Google Scholar]
- Ramirez S. R., Basu C. (2009). Comparative analyses of plant transcription factor databases. Curr. Genomics 10, 10–17. 10.2174/138920209787581253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf K. D., Berberich T., Ebersberger I., Nover L. (2012). The plant heat stress transcription factor (Hsf) family: structure, function and evolution. BBA Gene Regul. Mech. 1819, 104–119. 10.1016/j.bbagrm.2011.10.002 [DOI] [PubMed] [Google Scholar]
- Shim D., Hwang J. U., Lee J., Lee S., Choi Y., An G., et al. (2009). Orthologs of the class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice. Plant Cell 21, 4031–4043. 10.1105/tpc.109.066902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song N. H., Ahn Y. J. (2010). DcHsp17.7, a small heat shock protein from carrot, is upregulated under cold stress and enhances cold tolerance by functioning as a molecular chaperone. Hortscience 45, 469–474. [Google Scholar]
- Studer B., Byrne S., Nielsen R. O., Panitz F., Bendixen C., Islam M. S., et al. (2012). A transcriptome map of perennial ryegrass (Lolium perenne L.). BMC Genomics 13:140. 10.1186/1471-2164-13-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timperio A. M., Egidi M. G., Zolla L. (2008). Proteomics applied on plant abiotic stresses: role of heat shock proteins (HSP). J. Proteomics 71, 391–411. 10.1016/j.jprot.2008.07.005 [DOI] [PubMed] [Google Scholar]
- Tripp J., Mishra S. K., Scharf K. D. (2009). Functional dissection of the cytosolic chaperone network in tomato mesophyll protoplasts. Plant Cell Environ. 32, 123–133. 10.1111/j.1365-3040.2008.01902.x [DOI] [PubMed] [Google Scholar]
- von Koskull-Döring P., Scharf K. D., Nover L. (2007). The diversity of plant heat stress transcription factors. Trends Plant Sci. 12, 452–457. 10.1016/j.tplants.2007.08.014 [DOI] [PubMed] [Google Scholar]
- Wang W., Vinocur B., Shoseyov O., Altman A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9, 244–252. 10.1016/j.tplants.2004.03.006 [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang J., Li J.-L., Ma X.-R. (2014). Exogenous hydrogen peroxide enhanced the thermotolerance of Festuca arundinacea and Lolium perenne by increasing the antioxidative capacity. Acta Physiol. Plant. 36, 2915–2924. 10.1007/s11738-014-1661-2 [DOI] [Google Scholar]
- Waters E. R. (2013). The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 64, 391–403. 10.1093/jxb/ers355 [DOI] [PubMed] [Google Scholar]
- Wunderlich M., Gross-Hardt R., Schöffl F. (2014). Heat shock factor HSFB2a involved in gametophyte development of Arabidopsis thaliana and its expression is controlled by a heat-inducible long non-coding antisense RNA. Plant Mol. Biol. 85, 541–550. 10.1007/s11103-014-0202-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Z., Browse J. (2000). Cold comfort farm: the acclimation of plants to freezing temperatures. Plant Cell Environ. 23, 893–902. 10.1046/j.1365-3040.2000.00611.x [DOI] [Google Scholar]
- Xue G. P., Sadat S., Drenth J., McIntyre C. L. (2014). The heat shock factor family from Triticum aestivum in response to heat and other major abiotic stresses and their role in regulation of heat shock protein genes. J. Exp. Bot. 65, 539–557. 10.1093/jxb/ert399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Forster J. W., Humphreys M. W., Takamizo T. (2005). Genetics and molecular breeding in Lolium/Festuca grass species complex. Grassl. Sci. 51, 89–106. 10.1111/j.1744-697X.2005.00024.x [DOI] [Google Scholar]
- Yeh C. H., Kaplinsky N. J., Hu C., Charng Y. Y. (2012). Some like it hot, some like it warm: phenotyping to explore thermotolerance diversity. Plant Sci. 195, 10–23. 10.1016/j.plantsci.2012.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Sakuma Y., Todaka D., Maruyama K., Qin F., Mizoi J., et al. (2008). Functional analysis of an Arabidopsis heat-shock transcription factor HsfA3 in the transcriptional cascade downstream of the DREB2A stress-regulatory system. Biochem. Biophys. Res. Commun. 368, 515–521. 10.1016/j.bbrc.2008.01.134 [DOI] [PubMed] [Google Scholar]
- Zhang C., Guy C. L. (2006). In vitro evidence of Hsc70 functioning as a molecular chaperone during cold stress. Plant Physiol. Biochem. 44, 844–850. 10.1016/j.plaphy.2006.09.012 [DOI] [PubMed] [Google Scholar]
- Zhang L., Li Y., Xing D., Gao C. (2009). Characterization of mitochondrial dynamics and subcellular localization of ROS reveal that HsfA2 alleviates oxidative damage caused by heat stress in Arabidopsis. J. Exp. Bot. 60, 2073–2091. 10.1093/jxb/erp078 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene Ontology (GO) annotation of transcripts in (A) tall fescue and (B) perennial ryegrass.
Functional classification using Clusters of Orthologous Groups of proteins (COG) in tall fescue (A) and perennial ryegrass (B).
Annotation of tall fescue transcriptome in response to heat and cold stress.
Supplementary Table 1-S1: Expression levels and functional annotation of transcripts of the tall fescue transcriptome.
Supplementary Table 1-S2: KEGG pathway mapping of the tall fescue transcriptome.
Supplementary Table 1-S3: GO-Biological Process enrichment (p < 0.05) of the differentially-expressed genes (DEGs) showing significant expression differences in tall fescue under heat and cold stress.
Supplementary Table 1-S4: Genes encoding heat shock factor (Hsf) genes in the tall fescue transcriptome.
Supplementary Table 1-S5: Genes encoding heat shock protein (HSP) genes in the tall fescue transcriptome.
Supplementary Table 1-S6: Genes encoding Ascorbate peroxidase (APX), Inositol-3-phosphate synthase (IPS) and Galactinol synthase (GOLS1) in the tall fescue transcriptome.
The annotation of perennial ryegrass transcriptome in response to heat and cold stress.
Supplementary Table 2-S1: Expression levels and functional annotation of transcripts in the perennial ryegrass transcriptome.
Supplementary Table 2-S2: KEGG pathway mapping of the perennial ryegrass transcriptome.
Supplementary Table 2-S3: GO-Biological Process enrichment (p < 0.05) of differentially-expressed genes (DEGs) showing significant expression differences in perennial ryegrass under heat and cold stress.
Supplementary Table 2-S4: Genes encoding heat shock factor (Hsf) genes in the perennial ryegrass transcriptome.
Supplementary Table 2-S5: Genes encoding heat shock protein (HSP) genes in the perennial ryegrass transcriptome.
Supplementary Table 2-S6: Genes encoding Ascorbate peroxidase (APX), Inositol-3-phosphate synthase (IPS) and Galactinol synthase (GOLS1) in the perennial ryegrass transcriptome.
Comparison of gene expression patterns between RNA-seq and quantitative real-time PCR results.