Abstract
In China, fosfomycin alone or in combination with other antibiotics is commonly used in the treatment of infections caused by Gram-positive bacteria, including methicillin-resistant Staphylococcus aureus (MRSA). Although fosfomycin-resistant S. aureus strains have continued to emerge and increase, the research on them is rare. In order to determine the prevalence and mechanisms of fosfomycin resistance in MRSA clinical isolates, a total of 96 non-duplicate MRSA isolates were collected from blood and cerebrospinal fluid samples at Huashan Hospital in Shanghai, China between 2004 and 2014. Antimicrobial susceptibility testing was performed by agar dilution. Meanwhile, the fosfomycin-resistance-related genes, fosB, murA, glpT, and uhpT, were amplified by PCR and subjected to sequencing analysis. Multilocus sequence typing (MLST) was conducted to assess strain types. The minimum inhibitory concentration (MIC) of fosfomycin for the 96 MRSA strains ranged from 1.0 to >1,024 mg/L, and approximately 70% (67/96) of the isolates were resistant to fosfomycin (MIC ≥ 64.0 mg/L). Nine isolates with MICs ≥ 128 mg/L carried fosB gene. Twenty-five distinct mutations were detected in the murA (7), glpT (10), and uhpT (8) genes. While five of the murA mutations and five of the glpT mutations were observed only in fosfomycin-sensitive isolates and one of the murA mutation was found both in fosfomycin-resistant and fosfomycin-sensitive isolates, the remaining 14 mutations (1 murA, 5 glpT, and all uhpT mutations) were present only in fosfomycin-resistant isolates. MLST analysis demonstrated that the majority (46/67) of the glpT and/or uhpT mutants belong to ST5, the predominant sequence type among the fosfomycin-resistant MRSA isolates. In conclusion, there is a high rate of fosfomycin resistance in MRSA strains. The mutations in the murA, glpT, and uhpT genes are common in fosfomycin-resistant MRSA strains, and may play a greater role in the development of fosfomycin resistance than the presence of the fosB gene in these organisms.
Keywords: fosfomycin, resistance, mutations, Staphylococcus aureus, MRSA
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is one of the most common nosocomial pathogens, and resistant to most β-lactam antibiotics and even new antimicrobials, which has compelled clinicians to resort to some “old” antimicrobial agents (DeLeo and Chambers, 2009). Fosfomycin, which was first discovered in Streptomyces sp. in 1969, possesses broad-spectrum activity against both Gram-positive and Gram-negative bacteria by inhibiting bacterial cell wall synthesis (Michalopoulos et al., 2011). Specifically, fosfomycin deactivates the enzyme UDP-N-acetylglucosamine-3-enolpyruvyltransferase, also known as MurA (encoded by the murA gene), thereby irreversibly interfering with the first committed step of peptidoglycan biosynthesis (Michalopoulos et al., 2011). In recent years, the administration of fosfomycin alone or in combination with other antibiotics has been prescribed for treatment of MRSA (Falagas et al., 2009; Del Río et al., 2014; Sultan et al., 2015). However, fosfomycin-resistant S. aureus strains have continued to emerge and increase (Etienne et al., 1991). In China, the fosfomycin resistance rate in MRSA was as high as 29.5% in 2010 (Guo et al., 2013).
Bacterial fosfomycin resistance is attributed both to the acquisition of chromosomal mutations and to the expression of plasmid-encoded fosfomycin-modifying enzymes. Mutations in murA have been shown to reduce the affinity of fosfomycin for MurA (Jiang et al., 2011). Additionally, fosfomycin intake can be reduced in the presence of mutations in glpT and/or uhpT, which encode fosfomycin transport systems of bacteria (Takahata et al., 2010; Michalopoulos et al., 2011). Lastly, fosfomycin activity can be inhibited via the catalytic activity of FosA, FosB, FosC, and FosX, respectively (García et al., 1995; Fillgrove et al., 2007; Lee et al., 2012). Meanwhile, of the known plasmid-mediated fosfomycin resistance genes, only fosB has been detected in Staphylococcus sp. To date, however, most researches have focused on fosfomycin-resistant Gram-negative bacteria, and relatively few studies have been conducted in fosfomycin-resistant Gram-positive bacteria (Michalopoulos et al., 2011). The purpose of this study was to investigate the prevalence fosfomycin resistance and mutations in the murA, glpT, and uhpT genes in fosfomycin-resistant MRSA strains isolated in China.
Materials and Methods
Bacterial Strains
A total of 96 MRSA clinical strains were isolated from blood or cerebrospinal fluid specimens at Huashan hospital, a teaching hospital in Shanghai, China, between 2004 and 2014. S. aureus ATCC 29213 (American Type Tissue Culture Collection, Manassas, VA, USA) was used as a quality control strain in antimicrobial susceptibility testing experiments.
Antimicrobial Susceptibility Testing
The minimum inhibitory concentration (MIC) of fosfomycin for each clinical strain was determined by agar dilution supplemented with Glucose-6-phosphate (25 mg/L), according to the recommendations of the Clinical and Laboratory Standards Institute (Clinical and Laboratory Standards Institute [CLSI], 2014), and results were interpreted according to European committee on antimicrobial susceptibility testing criteria (European Committee on Antimicrobial Susceptibility Testing [EUCAST], 2012) (susceptible, ≤32 mg/L; resistant, ≥64 mg/L).
PCR Amplification
DNA of 96 MRSA strains was harvested using TIANamp bacteria DNA Kit (Tiangen, Beijing, China). The presence of the fosA, fosB, and fosC, was detected by PCR as described previously (Chen et al., 2014). murA, glpT and uhpT genes are amplified by PCR using primers listed in Table 1. The PCR products were sequenced to screen for mutations.
Table 1.
PCR primers of fosA, fosB, fosC, murA, glpT and uhpT gene.
| Primers | Gene | Primer sequences (5′ > 3′) | Product size | Reference |
|---|---|---|---|---|
| fosA-F fosA-R |
fosA | GCTGCACGCCCGCTGGAATA CGACGCCCCCTCGCTTTTGT |
217 bp | Chen et al., 2014 |
| fosB-F fosB-R |
fosB | CAGAGATATTTTAGGGGCTGACA CTCAATCTATCTTCTAAACTTCCTG |
312 bp | Chen et al., 2014 |
| fosC-F fosC-R |
fosC | GGGTTACATGCCCTTGCTCA AACCCGCACAACGACAGATG |
354 bp | Chen et al., 2014 |
| murA-F murA-R |
murA | GCCCTTGAAAGAATGGTTCGT GTTACAATACTCGACGCAGGT |
1600 bp∗ | NC_002745.2∗∗ |
| glpT-F glpT-R |
glpT | TGAATAAAACAGCAGGGCAA CACAGCTAGTATGTATAACGAC |
1699bp∗ | NC_002745.2∗∗ |
| uhpT-F uhpT-R |
uhpT | TGTGTTTATGTTCAGTATTTTGGA TCTTTCATCTCTTCACGCAC |
1571 bp∗ | NC_002745.2∗∗ |
∗PCR product including surrounding sequences adjacent to target gene.
∗∗GenBank-EMBL-DDBL accession number.
Multilocus Sequence Typing (MLST)
Staphylococcus aureus sequence types (STs) were determined by comparing the sequences of the housekeeping genes arcC, aroE, glp, gmk, pta, tpi, and yqiL, as described previously (Enright and Spratt, 1999), with those in the MLST database available at http://saureus.mlst.net/ (Aanensen and Spratt, 2005).
Nucleotide Sequence Accession Numbers
The sequences for the TypeAmurA, TypeImurA, TypeIImurA, TypeIII–VImurA, TypeA–EglpT, TypeIglpT, TypeIIglpT, TypeIIIglpT, TypeVIglpT, TypeVglpT, and TypeA–HuhpT strains have been deposited in GenBank with the following accession numbers: KT372186, KT372187, KT372185, KT372188–KT372191, KT372192–KT372196, KT372197, KT372198, KT372201, KT372199, KT372200, and KT372202–KT372209, respectively.
Results
Fosfomycin Susceptibility
The MICs of fosfomycin for the MRSA strains ranged from 1.0 mg/L to >1,024 mg/L. Furthermore, MIC50 and MIC90 values of these strains were 1,024 mg/L and >1,024 mg/L, respectively. According to the EUCAST criteria, 67 out of the 96 examined MRSA isolates were characterized as fosfomycin-resistant (MIC ≥ 64 mg/L).
Prevalence of Fosfomycin Resistance Genes
Of the 96 MRSA isolates, 9 fosfomycin-resistant strains with MIC ≥ 128 mg/L contained fosB (Table 2 and Supplementary Table S1), and no isolates were positive for fosA and fosC.
Table 2.
Characteristics of 96 methicillin-resistant Staphylococcus aureus isolates.
| MLST types | Strain No. | fosB gene | Mutation in glpT alone |
Mutation in uhpT alone |
Mutation in both glpT and uhpT |
No mutation in glpT and uhpT |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. | Fosfomycin MIC range(mg/L) | No. | Fosfomycin MIC range(mg/L) | No. | Fosfomycin MIC range(mg/L) | No. | Fosfomycin MIC range(mg/L) | |||
| ST5 | 40 | Negative | 4 | 64 ~ > 1024 | 6 | 128 ~ >1024 | 30 | 1024 ~ >1024 | 0 | – |
| ST239 | 34 | Negative | 2 | 128 | 2 | 128 ~ >1024 | 5 | 1024 ~>1024 | 25 | 1 ~ 256 |
| ST6 | 1 | Negative | 0 | - | 0 | - | 0 | - | 1 | 1 |
| ST30 | 1 | Negative | 1 | 2 | 0 | - | 0 | - | 0 | – |
| ST59 | 4 | Negative | 4 | 1 | 0 | - | 0 | - | 0 | – |
| ST88 | 1 | Negative | 0 | - | 0 | - | 0 | - | 1 | 1 |
| ST121 | 1 | Negative | 1 | 2 | 0 | – | 0 | - | 0 | – |
| ST398 | 1 | Negative | 1 | 1 | 0 | - | 0 | - | 0 | – |
| ST764 | 3 | Negative | 0 | - | 3 | 256 ~ 512 | 0 | - | 0 | – |
| ST863 | 1 | Negative | 0 | - | 0 | - | 0 | - | 1 | 32 |
| ST5 | 6 | Positive | 1 | 512 | 2 | >1024 | 3 | >1024 | 0 | – |
| ST239 | 1 | Positive | 0 | - | 0 | - | 0 | - | 1 | 128 |
| ST764 | 1 | Positive | 0 | - | 0 | - | 1 | >1024 | 0 | – |
| ST2590 | 1 | Positive | 1 | >1024 | 0 | - | 0 | - | 0 | – |
Mutations in the murA Gene
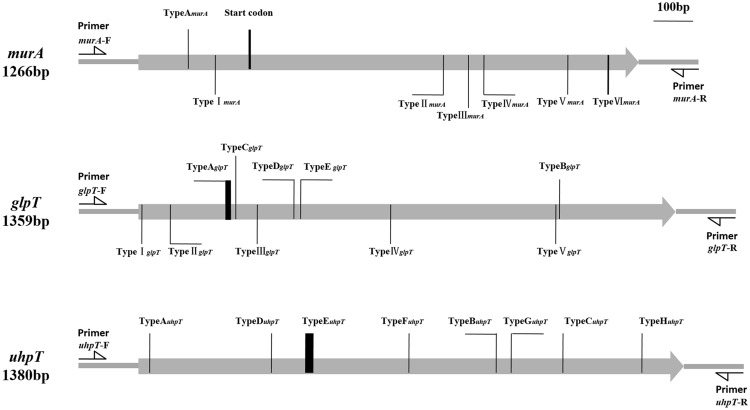
Seven distinct mutations were detected in the murA gene of the 96 MRSA isolates. Mutation TypeAmurA, which resulted in the creation of a stop codon (TA125A) at position 42 and a possible new start codon was found at position 94 (Figure 1), was contained by one of the fosfomycin-resistant MRSA isolates. In contrast, the other six mutations (TypeI–VImurA), which resulted in distinct amino acid substitutions within the MurA protein, could be found in fosfomycin-sensitive MRSA isolates, though one mutation (TypeIImurA) also could be found in fosfomycin-resistant MRSA isolates. (Table 3).
FIGURE 1.
Types and positions of mutation in murA, glpT, and uhpT genes. TypeAmurA (KT372186): T125A (Result in a possible start codon in position 94th aa); TypeImurA (KT372187): G193C; TypeIImurA (KT372185): G770A; TypeIIImurA (KT372188): C834A; TypeIVmurA (KT372189): A873T; TypeVmurA (KT372190): A1085G; TypeVImurA (KT372191): CG1187-1188AT. TypeAglpT (KT372192): Deletion of 8bp from 225T to 232A; TypeBglpT (KT372193): G1064A; TypeC glpT (KT372194): Deletion of 248G; TypeD glpT (KT372195): Insertion of 392T; TypeE glpT (KT372196): T409C; TypeIglpT (KT372197): T7A; TypeIIglpT (KT372198): C79T; TypeIIIglpT (KT372201): C299T; TypeIVglpT (KT372199): G637A; TypeVglpT (KT372200): G1055A. TypeAuhpT (KT372202): Deletion of 27T; TypeBuhpT (KT372203): Insertion of 904T; TypeCuhpT (KT372204): G1073T; TypeDuhpT (KT372205): G335A; TypeEuhpT (KT372206): Deletion of 12 bp from 431A to 442T; TypeFuhpT (KT372207): G683A; TypeGuhpT (KT372208): C942A; TypeHuhpT (KT372209): T1273C.
Table 3.
Characteristics of MRSA isolates contained murA mutations.
| Types of mutation | Strain No. | Mutations in murA∗ | Fosfomycin MIC range (mg/L) | fosB gene |
|---|---|---|---|---|
| TypeAmurA | 1 | T125A (Truncated to 41 aa) | 1024 | Negative |
| TypeImurA | 1 | G193C (Val 65 Leu) | 2 | Negative |
| TypeIImurA | 29 | G770A (Gly 257 Asp) | 2 ~ >1024 | Negative |
| TypeIIImurA | 2 | C834A (Asp 278 Glu) | 1 ~ 2 | Negative |
| TypeIVmurA | 7 | A873T (Glu 291 Asp) | 1 ~ 2 | Negative |
| TypeVmurA | 1 | A1085G (Gln 362 Arg) | 2 | Negative |
| TypeVImurA | 5 | CG1187-1188AT (Thr 396 Asn) | 1 ~ 2 | Negative |
∗Deduced amino acid substitutions or sequence variations are shown in brackets.
Mutations in glpT and uhpT Genes
Of the 67 fosfomycin-resistant MRSA isolates, 46 contained one of the five different mutations (TypeA–EglpT) observed in the glpT gene. While TypeEglpT resulted in an amino acid substitution at position 137 (Trp-137→Arg) of GlpT, the other four mutations produced premature stop codons within the glpT coding sequence, thereby resulting in the generation of truncated proteins (Figure 1). In addition, we detected five other mutations within the glpT gene that were present only in the fosfomycin-sensitive MRSA isolates. Each of these mutations (TypeI–VglpT) resulted in amino acid substitutions within the GlpT protein (Supplementary Table S2 and Table 4).
Table 4.
Characteristics of fosfomycin-resistant MRSA isolates contained glpT and uhpT mutations.
| Mutation and its Combination | Strain No. | ST5 |
ST239 |
Other ST types |
|||
|---|---|---|---|---|---|---|---|
| No. | Fosfomycin MIC range (mg/L) | No. | Fosfomycin MIC range (mg/L) | No. | Fosfomycin MIC range (mg/L) | ||
| TypeAglpT | 5 | 2 | 1024 ~ >1024 | 2 | 128 | 1 | >1024 |
| TypeAglpT and TypeAuhpT | 1 | 0 | – | 0 | – | 1 | >1024 |
| TypeAglpT and TypeBuhpT | 26 | 25 | >1024 | 1 | 1024 | 0 | – |
| TypeAglpT and TypeEuhpT | 1 | 0 | – | 1 | 1024 | 0 | – |
| TypeBglpT | 2 | 2 | 64 | 0 | – | 0 | – |
| TypeBglpT and TypeBuhpT | 1 | 1 | >1024 | 0 | – | 0 | – |
| TypeBglpT and TypeCuhpT | 5 | 5 | 1024 | 0 | – | 0 | – |
| TypeBglpT and TypeFuhpT | 1 | 1 | >1024 | 0 | – | 0 | – |
| TypeCglpT | 1 | 1 | 512 | 0 | – | 0 | – |
| TypeDglpT and TypeHuhpT | 1 | 0 | – | 1 | >1024 | 0 | – |
| TypeEglpT and TypeCuhpT | 1 | 0 | – | 1 | >1024 | 0 | – |
| TypeEglpT and TypeGuhpT | 1 | 0 | – | 1 | 1024 | 0 | – |
| TypeAuhpT | 7 | 3 | >1024 | 1 | 128 | 3 | 256 ~ 512 |
| TypeBuhpT | 5 | 5 | 128 ~ >1024 | 0 | – | 0 | – |
| TypeCuhpT | 1 | 1 | 1024 | 0 | – | 0 | – |
| TypeDuhpT | 1 | 0 | – | 1 | >1024 | 0 | – |
Eight distinct mutations were detected in the uhpT gene (TypeA–HuhpT) of the fosfomycin-resistant MRSA isolates. Conversely, no uhpT mutations were detected in the fosfomycin-sensitive isolates (Supplementary Table S3 and Table 4). The TypeCuhpT, TypeDuhpT, and TypeHuhpT mutations led to amino acid substitutions within the UhpT protein, while each of the remaining mutations created premature stop codons within the uhpT gene, resulting in the production of truncated protein (Figure 1).
Molecular Typing
The 96 MRSA isolates were categorized into 11 ST types, including ST5, ST6, ST30, ST59, ST88, ST121, ST239, ST398, ST764, ST863, and ST2590 (Table 2). Notably, all ST5 strains and 45.7% (16/35) of the ST239 isolates were resistant to fosfomycin (Supplementary Table S1 and Table 2).
Discussion
The fosfomycin resistance rate of the MRSA isolates examined in this study was approximately 70% (67/96), which was higher than previously detected (Falagas et al., 2010). In consideration of the average fosfomycin use was as high as 300,977 DDDs (defined daily doses) per year in Shanghai hospitals between 2009 and 2014 (Unpublished data), this elevated resistance rate may have been due to the common usage of this antimicrobial in the region. However, while Etienne et al. (1991) previously detected the fosB gene in 46% (18/39) of fosfomycin-resistant S. aureus isolates, only 13.4% (9/67) of the strains examined in this study harbored the fosB gene. Although this gene was detected only in fosfomycin-resistant isolates, the majority of such isolates were fosB-negative, indicating that other mechanisms contribute to fosfomycin resistance in S. aureus.
Sequencing analyses detected 25 distinct mutations in the murA, glpT, and uhpT genes of the MRSA isolates tested (Figure 1 and Supplementary Table S1). Of the seven mutations present in murA, 6 (TypeI–VImurA) were contained by fosfomycin-sensitive isolates, indicating that these modifications did not confer fosfomycin resistance. Also, while the TypeAmurA mutation was only detected in a single fosfomycin-resistant isolate, this strain also harbored mutations within glpT and uhpT. Thus, it is unclear what role, if any, the TypeAmurA mutation plays in fosfomycin resistance in MRSA.
Each of the uhpT mutations (TypeA–HuhpT) and five of the glpT (TypeA–EglpT) mutations were present only in fosfomycin-resistant isolates. Indeed, approximately 90% (60/67) of the fosfomycin-resistant strains harbored a mutation in glpT and/or uhpT. Moreover, strains that contained mutations in both glpT and uhpT exhibited high levels of fosfomycin resistance (MIC > 1,024 mg/L; Table 2), indicating that each of these 13 mutations might contribute to fosfomycin resistance. In contrast, the remaining five glpT mutations (TypeI–VglpT), which were harbored only by fosfomycin-sensitive strains, likely do not contribute to fosfomycin resistance. Notably, several fosfomycin-resistant isolates were fosB-negative, and did not harbor mutations in murA, glpT, or uhpT. As such, the mechanism that governs fosfomycin resistance in these isolates requires further study.
Multilocus sequence typing analyses indicated that all of the ST5 and 45.7% of the ST239 isolates were resistant to fosfomycin. The majority of the glpT and/or uhpT mutants were typed as ST5 (Table 2), and several strains harbored identical mutations (Table 4), indicating that clonal dissemination may exist. The first ST5 strain emerged in 2004 (Supplementary Table S1). The spread of this clone may be associated with the increased occurrence of fosfomycin resistant MRSA. However, fosfomycin resistance-associated mutations were also present in isolates within other STs, suggesting that the emergence of fosfomycin resistance is due to not only clonal dissemination, but also fosfomycin selective pressure on isolates from different clones, and the emergence of spontaneous mutations.
Conclusion
We observed a high rate of fosfomycin resistance in MRSA strains isolated from Shanghai, China. Our findings indicate that mutations within the glpT and/or uhpT genes play a critical role in conferring this resistance. Thus, the continuous monitoring of fosfomycin susceptibility in S. aureus, particularly in MRSA strains is important. Furthermore, our results indicate that caution should be used when employing fosfomycin for the empiric treatment of MRSA infections.
Author Contributions
Designed and conceived the experiments: YL, XX, and MW. Performed the experiments: ZF, YM, YG, and CC. Analyzed the data: FH, XX, and ZF. Wrote and reviewed the manuscript: ZF, YM, CC, YL, and XX.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (81171613 to XX and 81120108024 to MW) and the National Major Scientific and Technological Special Project (2014ZX09507-009).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01544
References
- Aanensen D. M., Spratt B. G. (2005). The multilocus sequence typing network, mlst.net. Nucleic Acids Res. 33 W728–W733. 10.1093/nar/gki415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Xu X., Qu T., Yu Y., Ying C., Liu Q., et al. (2014). Prevalence of the fosfomycin-resistance determinant, fosB3, in Enterococcus faecium clinical isolates from China. J. Med. Microbiol. 63 1484–1489. 10.1099/jmm.0.077701-0 [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute [CLSI]. (2014). Performance Standards for Antimicrobial Susceptibility Testing: Twenty Fourth Informational Supplement, M100-S24 Vol. 34 Wayne, PA: Clinical and Laboratory Standards Institute [Google Scholar]
- DeLeo F. R., Chambers H. F. (2009). Reemergence of antibiotic-resistant Staphylococcus aureus in the genomics era. J. Clin. Invest. 119 2464–2474. 10.1172/JCI38226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Río A., Gasch O., Moreno A., Peña C., Cuquet J., Soy D., et al. (2014). Efficacy and safety of fosfomycin plus imipenem as rescue therapy for complicated bacteremia and endocarditis due to methicillin-resistant Staphylococcus aureus, A multicenter clinical trial. Clin. Infect. Dis. 59 1105–1112. 10.1093/cid/ciu580 [DOI] [PubMed] [Google Scholar]
- Enright M. C., Spratt B. G. (1999). Multilocus sequence typing. Trends Microbiol. 7 482–487. 10.1016/S0966-842X(99)01609-1 [DOI] [PubMed] [Google Scholar]
- Etienne J., Gerbaud G., Fleurette J., Courvalin P. (1991). Characterization of staphylococcal plasmids hybridizing with the fosfomycin resistance gene fosB. FEMS Microbiol. Lett. 68 119–122. 10.1111/j.1574-6968.1991.tb04580.x [DOI] [PubMed] [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing [EUCAST] (2012). Available at: http://www.eucast.org [Google Scholar]
- Falagas M., Maraki S., Karageorgopoulos D. E., Kastoris A. C., Kapaskelis A., Samonis G. (2010). Antimicrobial susceptibility of Gram-positive non-urinary isolates to fosfomycin. Int. J. Antimicrob. Agents 35 497–499. 10.1016/j.ijantimicag.2010.01.010 [DOI] [PubMed] [Google Scholar]
- Falagas M., Roussos N., Gkegkes I., Rafailidis P. I., Karageorgopoulos D. E. (2009). Fosfomycin for the treatment of infections caused by Gram-positive cocci with advanced antimicrobial drug resistance, a review of microbiological, animal and clinical studies. Exp. Opin. Investig. Drugs 8 921–944. 10.1517/13543780902967624 [DOI] [PubMed] [Google Scholar]
- Fillgrove K., Pakhomova S., Schaab M. R., Newcomer M. E., Armstrong R. N. (2007). Structure and mechanism of the genomically encoded fosfomycin resistance protein, fosx, from Listeria monocytogenes. Biochemistry 46 8110–8120. [DOI] [PubMed] [Google Scholar]
- García P., Arca P., Evaristo S. J. (1995). Product of fosC, a gene from Pseudomonas syringae, mediates fosfomycin resistance by using ATP as cosubstrate. Antimicrob. Agents Chemother. 39 1569–1573. 10.1128/AAC.39.7.1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Zhu D., Hu F., Wang F., Yu Y., Yang Q., et al. (2013). CHINET 2010 surveillance and analysis of antibiotic resistance in Staphylococcus spp. in China. Chin. J. Infect. Chemother. 13 86–92. [Google Scholar]
- Jiang S., Gilpin M. E., Attia M., Ting Y. L., Berti P. J. (2011). Lyme disease enolpyruvyl-UDP-GlcNAc synthase: fosfomycin-resistant MurA from Borrelia burgdorferi, a fosfomycin-sensitive mutant, and the catalytic role of the active site Asp. Biochemistry 12 2205–2212. 10.1021/bi1017842 [DOI] [PubMed] [Google Scholar]
- Lee S. Y., Park Y. J., Yu J. K., Jung S., Kim Y., Jeong S. H., et al. (2012). Prevalence of acquired fosfomycin resistance among extended-spectrum b-lactamase-producing Escherichia coli and Klebsiella pneumoniae clinical isolates in Korea and IS26-composite transposon surrounding fosA3. J. Antimicrob. Chemother. 67 2843–2847. 10.1093/jac/dks319 [DOI] [PubMed] [Google Scholar]
- Michalopoulos A. S., Livaditis I. G., Gougoutas V. (2011). The revival of fosfomycin. Int. J. Infect. Dis. 15 e732–e739. 10.1016/j.ijid.2011.07.007 [DOI] [PubMed] [Google Scholar]
- Sultan A., Rizvi M., Khan F., Sami H., Shukla I., Khan H. M. (2015). Increasing antimicrobial resistance among uropathogens, Is fosfomycin the answer? Urol. Annal. 7 26–30. 10.4103/0974-7796.148585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata S., Ida T., Hiraishi T., Sakakibara S. (2010). Molecular mechanisms of fosfomycin resistance in clinical isolates of Escherichia coli. Int. J. Antimicrob. Agents 2010 333–337. 10.1016/j.ijantimicag.2009.11.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.